Abstract
The Oxa1 protein plays a central role in facilitating the cotranslational insertion of the nascent polypeptide chains into the mitochondrial inner membrane. Mitochondrially encoded proteins are synthesized on matrix-localized ribosomes which are tethered to the inner membrane and in physical association with the Oxa1 protein. In the present study we used a chemical cross-linking approach to map the Saccharomyces cerevisiae Oxa1-ribosome interface, and we demonstrate here a close association of Oxa1 and the large ribosomal subunit protein, MrpL40. Evidence to indicate that a close physical and functional relationship exists between MrpL40 and another large ribosomal protein, the Mrp20/L23 protein, is also provided. MrpL40 shares sequence features with the bacterial ribosomal protein L24, which like Mrp20/L23 is known to be located adjacent to the ribosomal polypeptide exit site. We propose therefore that MrpL40 represents the Saccharomyces cerevisiae L24 homolog. MrpL40, like many mitochondrial ribosomal proteins, contains a C-terminal extension region that bears no similarity to the bacterial counterpart. We show that this C-terminal mitochondria-specific region is important for MrpL40's ability to support the synthesis of the correct complement of mitochondrially encoded proteins and their subsequent assembly into oxidative phosphorylation complexes.
The mitochondrial genome encodes a small, but important, number of proteins (8). These proteins are predominantly essential components of the mitochondrial oxidative phosphorylation (OXPHOS) machinery. In the yeast Saccharomyces cerevisiae the proteins encoded by the mitochondrial DNA (mtDNA) include cytochrome c oxidase subunits Cox1, Cox2, and Cox3, cytochrome b of the cytochrome bc1 complex, F1Fo-ATP synthase subunits Atp6, Atp8, and Atp9, and the small ribosomal subunit component Var1. With the exception of Var1, these mitochondrially encoded proteins are integral membrane proteins which become inserted into the inner membrane during their synthesis on mitochondrial ribosomes tethered to the inner membrane (11, 19, 29, 32, 34). The cotranslational membrane insertion of these proteins is achieved by maintaining a close physical association of the ribosomes to the inner membrane at sites where the insertion machinery exists (19, 31, 32).
Oxa1 is an inner membrane protein that forms a central component of the insertion machinery, whose presence is required for the cotranslational membrane insertion of the mitochondrially encoded proteins (4-6, 15-17). The Oxa1 protein has been shown to physically associate with the ribosomes and more specifically with the large ribosomal subunit. Matrix-exposed elements of the Oxa1 protein, such as its hydrophilic C-terminal tail, support this Oxa1-ribosome interaction (19, 32). Furthermore, in intact mitochondria we have previously demonstrated that Oxa1 can be chemically cross-linked to Mrp20, a component of the large ribosomal subunit (19). Mrp20 is homologous to the bacterial ribosomal protein L23, a component known from the structural analysis of the ribosomes to be located next to the polypeptide exit site of the large ribosomal subunit (3, 10, 23, 27, 30). Thus, it was concluded that Oxa1, the site of membrane insertion into the inner membrane, exists in close physical proximity to the large ribosomal subunit and specifically to that region of the ribosomes where the nascent chain emerges. This close physical relationship between ribosomal components and the Oxa1 insertion site has been proposed to support a tight coordination between the protein translation and membrane insertion events (19, 31, 32). Given the strong hydrophobicity of the OXPHOS complex subunits which are encoded by the mitochondrial DNA and synthesized by these ribosomes, a close coupling of the translation and insertion events is proposed to ensure that the hydrophobic nascent chains are directly inserted into the membrane during their synthesis. The exposure of hydrophobic nascent chains to the hydrophilic matrix space may promote their aggregation and thus incompetency for subsequence membrane insertion.
In bacteria, the L23 protein has been implicated to play a direct role in the cotranslational insertion of proteins into the membrane (7, 13, 24, 33). Thus, it is possible that proteins adjacent to the polypeptide exit site of mitochondrial ribosomes may be directly involved in targeting ribosomes to specific regions of the inner membrane where the membrane insertion and subsequent assembly events occur. The mitochondrial ribosomes resemble their prokaryotic ancestors in some respects, e.g., antibiotic sensitivity, but they differ in a number of important ways (1, 12, 22, 30). In general, the protein content of the mitochondrial ribosomes is greater than their bacterial counterparts. This increase in protein content is largely attributed to the fact that the mitochondrial ribosomal proteins are larger in size than their bacterial homologs. Over the course of evolution, many of the mitochondrial ribosomal proteins have acquired novel extensions, new domains, in addition to their bacterial homology domains. These acquired extensions not only include N-terminal (often cleavable) signals to target these proteins (nuclear encoded) to the mitochondria but also in many instances large C-terminal extensions, which are unique to the mitochondrial ribosomal proteins and have thus been termed “mitospecific domains” (12, 30). Largely uncharacterized, the functional relevance of these various mitospecific domains of the ribosomal proteins remains unknown. It is speculated that some (or all) of these mitospecific domains serve to ensure that the ribosome becomes assembled and is translationally active while bound to the inner membrane surface.
In the present study we sought to further characterize the interaction of the mitochondrial ribosome with the Oxa1 protein. We show here that MrpL40, a large ribosomal subunit component, is physically close to both the Mrp20 and Oxa1 proteins, demonstrating the proximity of MrpL40 to both the ribosomal polypeptide exit site and the Oxa1 membrane insertion site. MrpL40 contains a large C-terminal mitospecific domain, which includes a predicted α-helical region at its extreme C-terminal end. The results presented here highlight that the integrity of this domain of MrpL40 is crucial to ensure ribosome translational fidelity and subsequent OXPHOS complex assembly.
MATERIALS AND METHODS
Yeast strains.
Unless otherwise stated, all strains used in this study were rho+. Yeast strains used in this study were wild-type W303-1A (Mata, leu2 trp1 ura3 his3 ade2), rho0 W303-1A (Mata, leu2 trp1 ura3 his3 ade2) (16), and the oxa1 null mutant, Δoxa1 overexpressing the Oxa1His or nontagged Oxa1 protein (W303-1A leu2 trp1 ura3 ade2 OXA1::HIS3 Yip351-GAL10-OXA1[+/−]His12-LEU2) (19). The galactose-induced overexpression of Oxa1His in this manner does not perturb a wild-type cell's ability to grow aerobically (results not shown). Construction of the strain expressing the C-terminal hemagglutinin (HA)-tagged MrpL40 protein was performed by homologous recombination at the MRPL40 gene locus of wild-type cells, resulting in the introduction of a DNA sequence encoding one HA epitope prior to the translational stop codon of the MRPL40 open reading frame (ORF), followed by a new stop codon and the HIS3 auxotrophic gene. Correct tagging of the MRPL40 gene in this manner and the insertion of the HIS3 gene at the 3′end of the MRPL40 ORF were verified by PCR analysis of the MRPL40 genomic region. Expression of the HA-tagged MrpL40 protein in the resulting strain (MrpL40HA) was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis of the mitochondria isolated from the MrpL40HA strain and using an HA-specific antiserum. The Oxa1His protein was expressed in this strain by transforming the MrpL40HA cells with the Yip351GAL10-OXA1His plasmid (19) and selecting for LEU+ transformants. Galactose-dependent Oxa1His expression was verified in the resulting strain (MrpL40HA+Oxa1His) by using Oxa1- and His-specific antisera. The mrpL40ΔC strain was generated through homologous recombination in the W303-1A wild-type yeast strain. A premature translational stop codon followed by the HIS3 auxotrophic marker gene, resulting in a partial deletion (the final 28 codons) on the 3′ end of the MRPL40 open reading frame, was performed essentially as previously described (35). Correct homologous recombination of the HIS3 gene at the MRPL40 gene locus was verified by PCR analysis (results not shown).
Cross-linking assays.
Mitochondria (200 μg total protein) were suspended in 600 μl SH buffer (0.6 M sorbitol, 20 mM HEPES-KOH, pH 7.2), and cross-linking was performed with disuccinimidyl glutarate (DSG; 0.3 mM) or maleimidobenzoyl-N-hydroxysuccinimide (MBS; 0.5 mM), as indicated for 30 min on ice. MBS cross-linking was performed in the presence of 3 mM NADH. Both cross-linking reagents were dissolved in dimethyl sulfoxide, so the mock-treated samples received dimethyl sulfoxide alone. Excess cross-linker was quenched by adding glycine (80 mM, pH 8.0). Mitochondria were reisolated by centrifugation and washed with SH buffer prior to further analysis. For direct analysis of the proteins and cross-linked adducts the mitochondria were solubilized in the SDS-sample buffer and analyzed by SDS-PAGE and Western blotting.
Ni-NTA purification of Oxa1His and cross-linked adducts.
For the Ni-nitrilotriacetic acid (NTA) purification of Oxa1His and cross-linked adducts, mitochondria (200 μg total protein) were solubilized in 200 μl TNT buffer (1% Triton X-100, 300 mM NaCl, 60 mM Tris-HCl, pH 7.4) for 30 min on ice. Where indicated, mitochondria were initially solubilized in 0.1% SDS prior to the addition of the TNT buffer. After a clarifying spin (20,860 × g; 15 min at 4°C), the supernatants were incubated for 1 h at 4°C with the Ni-NTA beads (equilibrated in the TNT buffer containing 30 mM imidazole). The beads were washed three times with TNT-imidazole buffer, and bound proteins were eluted with SDS-sample buffer containing 5% (vol/vol) β-mercaptoethanol and 0.5 M imidazole.
Mrp20 and cross-linked adduct immunoprecipitation.
For the immunoprecipitation of Mrp20 and its cross-linked adducts, mitochondria (200 μg total protein) following cross-linking were solubilized in SDS (1%) buffer and cooked (5 min at 95°C), followed by dilution into immunoprecipitation (IP) buffer (1% Triton X-100, 300 mM NaCl, 10 mM Tris-HCl, pH 7.4) for 30 min on ice. After a clarifying spin (20,860 × g; 15 min at 4°C), the supernatants were incubated overnight at 4°C with protein A-Sepharose beads and 30 μl of culture supernatant containing the Mrp20 monoclonal antibody. The beads were washed three times with IP buffer and twice with IP buffer without Triton X-100. Bound proteins were eluted with SDS-sample buffer containing 5% (vol/vol) β-mercaptoethanol. The immunoprecipitated Mrp20 and cross-linked adducts were analyzed by SDS-PAGE, Western blotting, and immunodecoration with MrpL40 antiserum.
MrpL40 antiserum generation.
The region of the MRPL40 ORF encompassing codons 1 to 284 was amplified by PCR and cloned in frame with an N-terminal His tag into the pET-28a(+) vector. The resulting His-tagged protein (35 kDa) was expressed in an isopropyl-β-d-thiogalactopyranoside-inducible manner in Escherichia coli. Following sonication to disrupt the bacterial cells, the soluble His-tagged MrpL40(1-284) was purified by Ni-NTA chromatography and subjected to thrombin cleavage to remove the His tag. The resulting MrpL40(1-284) 33-kDa fragment was injected into rabbits to generate a polyclonal antiserum.
Triton X-100 solubilization of mitochondria and sucrose gradient centrifugation.
Sucrose gradient analysis of detergent-solubilized mitochondrial ribosomes was performed essentially as previously published (36, 37). Mitochondria (300 μg protein) were solubilized with 300 μl of lysis buffer (0.5% Triton X-100, 10 mM Mg-acetate, 0.1 M NaCl, 20 mM HEPES-KOH, pH 7.4, 1 mM phenylmethylsulfonyl fluoride) for 30 min on ice. The lysate was clarified by centrifugation at 30,000 × g for 30 min at 4°C, and the supernatant was layered onto an 11-ml continuous sucrose gradient (15 to 30%) containing 500 mM NH4Cl, 10 mM Tris-HCl, pH 7.4, 10 mM Mg-acetate, 7 mM β-mercaptoethanol, and 0.5 mM phenylmethylsulfonyl fluoride. Gradients were centrifuged at 20,500 rpm for 17 h at 4°C in a Beckman SW41 Ti rotor. Fractions (750 μl) were collected, trichloroacetic acid precipitated, and subjected to SDS-PAGE and Western blot analysis.
Antisera used in this study.
Antisera against Cox2, Su e, MrpL40 were generated as described above and as published previously (2, 18). The following antisera were generously obtained from the following sources: MrpL36 (J. M. Herrmann, University of Kaiserlautern), MrpL32 (T. Langer, University of Cologne), Mrp47 (M. Boguta, Polish Academy of Sciences, Warsaw, Poland), Mrp10 (A. Tzagoloff, Columbia University, New York, NY), Mrp20 monoclonal (T. Mason, University of Massachusetts, Amherst), Tim17 (W. Neupert, University of Munich), Atp6 (J. Velours, Bordeaux, France), and the F1 sector (D. Mueller, Rosalind Franklin Medical School, Chicago, IL).
Miscellaneous.
Mitochondria were isolated from cultures grown at 30°C in yeast extract-peptone (YP)-0.5% lactate, 2% galactose medium. Isolation of mitochondria and in organello labeling with [35S]methionine were performed essentially as described earlier (17, 19). Standard procedures were used for SDS-PAGE, Blue native-PAGE, and Western blotting (20). The enzyme activity measurements were performed as described previously (9), with the exception that antimycin A-sensitive cytochrome c reductase activity was specifically determined.
RESULTS
Oxa1 can be cross-linked to a number of ribosomal proteins.
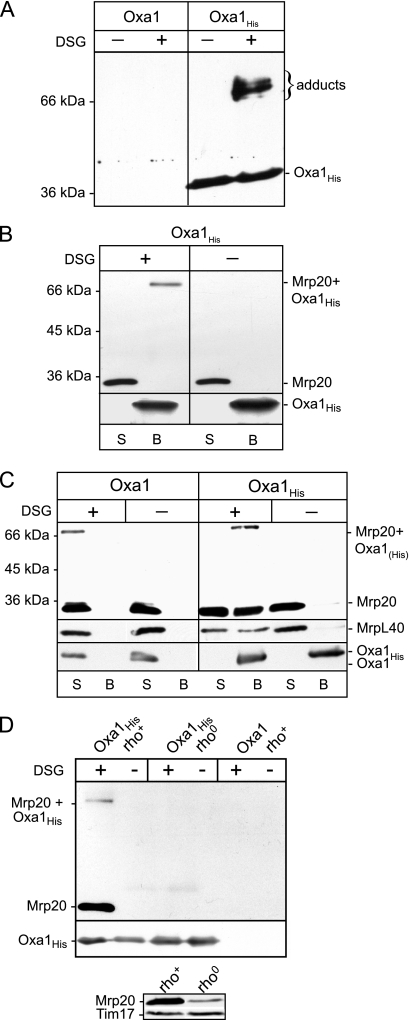
We embarked on a chemical cross-linking approach to identify other ribosomal proteins which, like Mrp20, exist in close proximity to Oxa1. Isolated mitochondria harboring a C-terminal histidine-tagged (12 His residues) Oxa1 derivative (Oxa1His) or control mitochondria containing nontagged Oxa1 (Oxa1) were subjected to cross-linking using DSG, an amino-specific, noncleavable cross-linking reagent (7.7-Å spacer arm). Following the cross-linking reaction, the mitochondria were solubilized under denaturing conditions (using the detergent SDS), and Oxa1His and covalently associated cross-linked adducts were purified by Ni-NTA chromatography. The Ni-NTA-purified material was analyzed by SDS-PAGE and Western blotting (Fig. 1A). Probing with an Oxa1-specific antibody indicated that Oxa1His (38 kDa) was recovered on the Ni-NTA beads together with at least three Oxa1His-containing adducts (size range, 66 to 75 kDa). These larger Oxa1His-containing species were recovered on the beads in a cross-linking-dependent manner, and from their sizes they indicate the ability of Oxa1His to become cross-linked to several proteins in the size range of 28 to 37 kDa. No specific bands were recognized by the Oxa1 antiserum in the Ni-NTA-purified material, in either the DSG- or mock-treated control samples, i.e., from mitochondria which harbored the non-His-tagged Oxa1 protein (Fig. 1A). We conclude from these data that Oxa1His can be cross-linked to at least three different proteins which can be recovered as cross-linked adducts together with Oxa1His in a specific manner.
FIG. 1.
Oxa1 can be cross-linked to ribosomal proteins. (A) Mitochondria harboring expressed wild-type or histidine-tagged Oxa1 (Oxa1 and Oxa1His, respectively) were incubated in the presence or absence of cross-linking reagent DSG (0.3 mM), as indicated. Following quenching, mitochondria were reisolated, lysed initially in SDS, and then diluted into Triton X-100 buffer and subjected to Ni-NTA affinity chromatography. The Ni-NTA-bound material was eluted with SDS-sample buffer and subjected to SDS-PAGE and Western blotting, followed by immunodecoration with an Oxa1-specific antiserum. (B) Oxa1His-containing mitochondria were subjected to mock or DSG cross-linking, as indicated. Samples were further processed as described for panel A, again following the initial lysis of the mitochondria in an SDS-containing buffer. The Ni-NTA-bound material (B lanes) and the free (nonbound) material remaining in the supernatant (S lanes) after isolation of the Ni-NTA beads were analyzed by SDS- PAGE and Western blotting. Immunodecoration was performed with Mrp20- and Oxa1-specific antibodies, as indicated. The position of the Mrp20-Oxa1His cross-linked adduct is indicated, and the presence of monomeric Mrp20 and Oxa1His in the samples is indicated. (C) Mitochondria harboring the Oxa1 or Oxa1His proteins were subjected to cross-linking analysis and Ni-NTA affinity chromatography, as described above for panels A and B, with the exception that the mitochondria were lysed in Triton X-100-containing buffer, i.e., the SDS lysis step was omitted. The presence of monomeric MrpL40, in addition to Mrp20, was also analyzed through immunodecoration with specific antibodies. (D, upper panel) Oxa1His mitochondria, rho+ and rho0, as indicated and control Oxa1 (rho+) mitochondria were subjected to cross-linking, Triton X-100 lysis, and subsequent analysis as described for panel C. Only the Ni-NTA-bound material is shown. (Lower panel) Mitochondria (50 μg protein) isolated from wild-type rho+ and rho0 cells were analyzed by SDS-PAGE, Western blotting, and immunodecoration with Mrp20 and Tim17 (loading control) antibodies.
One of these Oxa1His adducts is most likely the Mrp20-Oxa1His adduct. As previously mentioned, we have demonstrated that Oxa1 can be cross-linked with DSG to the large ribosomal subunit protein Mrp20 (33 kDa) (19). When Oxa1His-DSG cross-linked adducts were purified by Ni-NTA chromatography following lysis of the mitochondrial membranes with the denaturing detergent SDS, Oxa1His and the Mrp20-Oxa1His cross-linked adduct were purified on the Ni-NTA beads (Fig. 1B). Monomeric Mrp20 was not recovered on the Ni-NTA beads with the affinity-purified Oxa1His protein, in either the presence or absence of prior cross-linking. Thus, the purification of the Mrp20-Oxa1His adduct on the Ni-NTA beads is due to the His-tagged Oxa1 protein being covalently bound to Mrp20, rather than because of a possible nonspecific association of Mrp20 with the beads.
In order to investigate whether the other Oxa1His cross-linked adducts recovered on the Ni-NTA beads represent Oxa1-ribosomal protein adducts, Oxa1His mitochondria were solubilized after the cross-linking step with the detergent Triton X-100 rather than with SDS (Fig. 1C). These Triton X-100 solubilization conditions disrupt the interaction between Oxa1 and the ribosome, but in contrast to the SDS lysis procedure previously used (Fig. 1B), they do not perturb the integrity of the assembled large ribosomal subunit. Following cross-linking with DSG, the Triton X-100-solubilized Oxa1His and associated proteins were purified by Ni-NTA affinity chromatography and subsequently analyzed by SDS-PAGE and Western blotting. In addition to the Mrp20-Oxa1His adduct, monomeric Mrp20 was recovered with Oxa1His on the Ni-NTA beads. The recovery of monomeric Mrp20 on the Ni-NTA beads was specific for the presence of the His-tagged Oxa1 derivative. No monomeric Mrp20 or cross-linked Mrp20 adduct was recovered on the Ni-NTA beads from the control Oxa1-containing mitochondria (Fig. 1C). Furthermore, in the absence of prior cross-linking, monomeric Mrp20 was not recovered with Oxa1His on the Ni-NTA beads, because the detergent conditions used were sufficient to disrupt an Oxa1-ribosome interaction (19, 23, 32). Thus, the recovery of monomeric Mrp20 with Oxa1His was dependent on the prior cross-linking step, suggesting that other ribosomal proteins associated with Mrp20 are cross-linked to Oxa1His. To further support this conclusion, the monomeric form of another large ribosomal subunit protein, MrpL40, was also recovered with Oxa1His on the Ni-NTA beads, in a cross-linking-dependent fashion (Fig. 1C). Like Mrp20, MrpL40 was not recovered on the Ni-NTA beads when cross-linking was performed in the Oxa1 (i.e., not His-tagged Oxa1) mitochondria.
To demonstrate that the cross-linking-dependent recovery of the monomeric Mrp20 with Oxa1His was due to its association with the assembled ribosomes cross-linked to Oxa1His, DSG cross-linking was performed in rho0 mitochondria harboring Oxa1His, i.e., containing Mrp20 which was not assembled into a ribosome (Fig. 1D, upper panel). Following cross-linking, the rho0 mitochondria were solubilized by Triton X-100, and Ni-NTA purification of Oxa1His and associated proteins was performed. Although the overall levels of Mrp20 were reduced in the rho0 mitochondria (relative to rho+ mitochondria) (Fig. 1D, lower panel), no monomeric Mrp20 or Mrp20-Oxa1His adduct was detected with Oxa1His in the Ni-NTA-purified material. We conclude therefore that the recovery of Mrp20 with Oxa1His appears to depend on Mrp20 being present in a ribosome assembled state.
In summary, Oxa1 can be cross-linked to a number of partner proteins, including Mrp20 and at least one other large ribosomal subunit component. Monomeric ribosomal proteins, such as Mrp20 and MrpL40, can be copurified with Oxa1 in a cross-linking-dependent fashion and when solubilization of the mitochondrial membranes is performed under conditions which preserve the integrity of the large ribosomal subunit. The recovery of monomeric Mrp20 with Oxa1His in a cross-linking-dependent fashion supports the idea that other ribosomal components in a complex with Mrp20 become independently cross-linked to Oxa1. Finally, copurification of monomeric Mrp20 with Oxa1His was found to be dependent on the presence of an assembled ribosome, indicating that Mrp20 may not have the capacity to independently interact with Oxa1.
The ribosomal protein MrpL40 exists in close proximity to Oxa1.
We have demonstrated here that Oxa1 can form cross-linked adducts in the size range of 66 to 75 kDa, and given that the monomeric Mrp20 protein is recovered with Oxa1His in a cross-linking-dependent fashion, we can conclude that at least one of the observed Oxa1His cross-linked adducts must represent an Oxa1His-ribosomal protein adduct which is distinct from the observed Mrp20-Oxa1His adduct. To identify other large ribosomal subunit proteins which may become cross-linked and hence affinity purified with Oxa1His, we searched the database of known proteins of the mitochondrial large ribosomal subunit for possible candidates using the following criteria: (i) in the molecular mass range of 28 to 40 kDa, (ii) has been described to have a physical or functional relationship with Mrp20, and (iii) like Mrp20, may be located at the polypeptide nascent chain exit site of the ribosome. One of these candidate proteins was MrpL40, encoded by the YPL173w gene and with a predicted mature size of 34 kDa. MrpL40 is known to be a large ribosomal subunit component but has not yet been annotated as a homolog of a known bacterial ribosomal protein (1, 12).
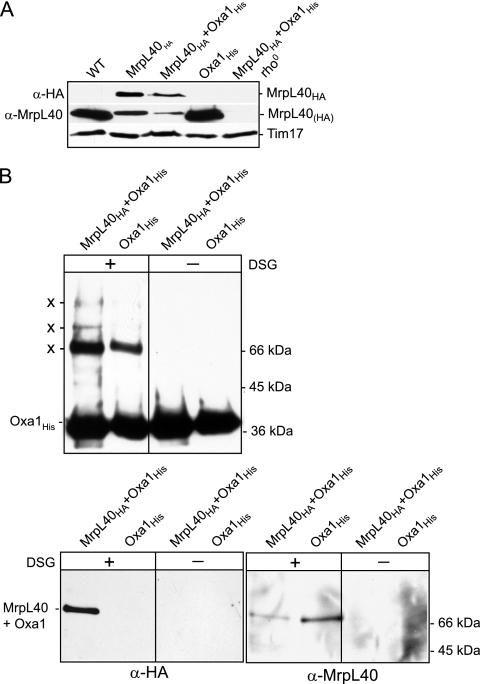
We therefore investigated whether one of the Oxa1 cross-linked adducts corresponds to an MrpL40-Oxa1 adduct. In order to facilitate the detection of MrpL40 initially, an HA epitope was added at the C-terminal end of MrpL40 through homologous recombination at the MrpL40 gene locus, thereby ensuring the HA-tagged MrpL40 protein (MrpL40HA) was expressed under the control of its endogenous promoter. The resulting tagged MrpL40HA protein was detected using an HA-specific antisera in mitochondria isolated from the MrpL40HA and MrpL40HA+Oxa1His strains (where, interestingly, in the latter it was present at slightly reduced levels), but not in the control wild-type or Oxa1His mitochondria, analyzed in parallel (Fig. 2A). Like many mitochondrial ribosomal proteins, the steady-state levels of MrpL40HA were found to be severely impacted in rho0 mitochondria, indicating that stability of MrpL40 may be dependent on its ability to assemble into ribosomes (Fig. 2A).
FIG. 2.
Oxa1 can be cross-linked to ribosomal protein MrpL40. (A) MrpL40HA and MrpL40 were detected in mitochondria isolated from yeast strains, as indicated, following SDS-PAGE, Western blotting, and immunodecoration with HA-specific (α-HA) or MrpL40-specific (α-MrpL40) antisera, as indicated. The levels of Tim17, a loading control, are also indicated. (B) MrpL40HA+Oxa1His and Oxa1His control mitochondria were subjected to chemical cross-linking with DSG, quenching, and lysis in an SDS-containing buffer. Oxa1His and cross-linked adducts (indicated by an X) were purified by Ni-NTA chromatography and further analyzed as described for Fig. 1B. Only the Ni-NTA-bound material is shown. Immunodecoration was performed with antiserum specific for Oxa1 (upper panels), HA (lower left panels), or MrpL40 (lower right panels).
In order to test whether MrpL40 exists in close proximity to Oxa1, mitochondria isolated from the MrpL40HA+Oxa1His strain and from the control Oxa1His strain (i.e., expressing authentic non-HA-tagged MrpL40) were subjected to DSG cross-linking followed by SDS solubilization and Ni-NTA purification (Fig. 2B). Analysis of the Ni-NTA-purified material using the Oxa1 antiserum confirmed the recovery of Oxa1His on the beads and the cross-linking of Oxa1His to other proteins, as was shown above in Fig. 1A (Fig. 2B, upper panel). The ability of Oxa1 to form cross-linked adducts did not appear to be perturbed in the mitochondria expressing the HA-tagged MrpL40 derivative (Fig. 2B, upper panel). When the Ni-NTA-purified material was probed with the HA antiserum, an Oxa1His cross-linked adduct of approximately 70 kDa was detected in the MrpL40HA+Oxa1His mitochondria, but not in the control (Oxa1His) mitochondria, harboring the nontagged MrpL40 protein (Fig. 2B, lower left panels). The recovery of this 70-kDa HA-containing Oxa1His adduct was dependent on the prior cross-linking reaction. Furthermore, the size of the adduct from the MrpL40HA+Oxa1His mitochondria was consistent with that expected from MrpL40HA (36 kDa) and Oxa1His (38 kDa) (Fig. 2B, lower left panels).
In light of the finding that the MrpL40HA protein can be cross-linked to Oxa1His, we generated an antiserum for MrpL40 so that we could directly assay whether the authentic MrpL40 protein could also be cross-linked to Oxa1His, and thus we could rule out that the observed MrpL40-Oxa1 adduct formation was not an artifact of the HA tagging of the MrpL40 protein at its C terminus. The MrpL40 protein (residues 1 to 284) was expressed in bacteria, purified, and used as an antigen to generate an MrpL40-specific polyclonal antiserum in rabbits. Both endogenous and HA-tagged forms of MrpL40 could be detected with the generated MrpL40 antiserum (Fig. 2A). The levels of MrpL40 were reduced in the mitochondria bearing the HA-tagged version of the protein, suggesting that the addition of the C-terminal HA tag may have compromised the stability of the MrpL40 protein (discussed further in Fig. 5B, below).
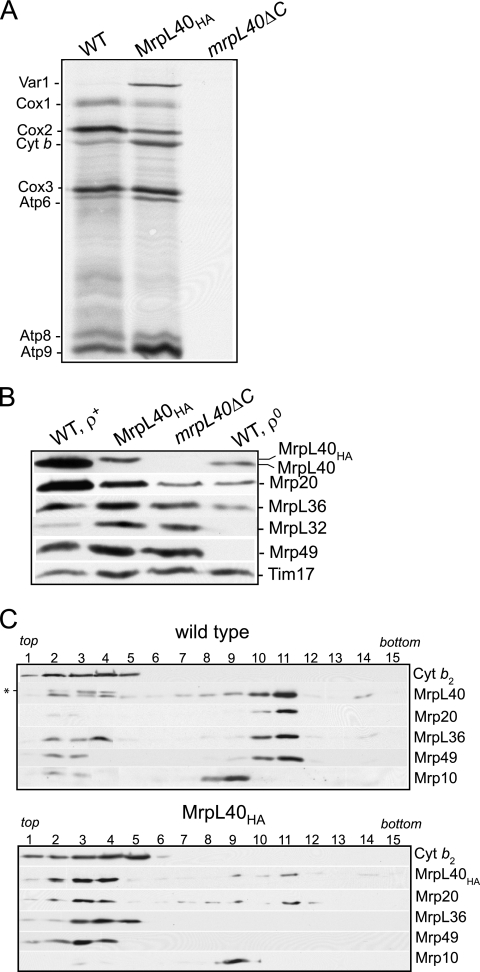
FIG. 5.
Modification of the C-terminal region of MrpL40 has adverse consequences for ribosome function and stability. (A) In organello translation was monitored in mitochondria isolated from wild-type, MrpL40HA, and mrpL40ΔC strains at 25°C for 15 min, following the addition of [35S]methionine. Following chase with excess cold methionine and puromycin for 10 min at 25°C, mitochondria were reisolated, lysed in SDS-sample buffer, and subjected to SDS-PAGE, Western blotting, and autoradiography. Abbreviations: Cox1, Cox2, and Cox3, cytochrome c oxidase subunits 1, 2, and 3, respectively; Cyt b, cytochrome b; Atp6, Atp8, and Atp9, subunits 6, 8, and 9 of the Fo sector, respectively. (B) Mitochondria (50 μg of protein) were subjected to SDS-PAGE and analyzed by Western blotting for the presence of various large ribosomal subunit proteins, as indicated. Tim17 was used as a loading control. (C) Wild-type and MrpL40HA mitochondria were lysed with a Triton X-100-containing buffer and subjected to a clarifying spin. The resulting solubilized material was fractionated with a linear sucrose gradient (see Materials and Methods for further details). Fractions were collected (1 to 15, top to bottom of the gradient) and were analyzed by SDS-PAGE and Western blotting. Immunodecoration was performed with antibodies directed against large ribosomal subunits, MrpL40, Mrp20, MrpL36, and Mrp49, and the small ribosomal subunit protein Mrp10. The * indicates a cross-reactivity signal from the MrpL40 antiserum. The fractionation behavior of a control soluble (non-ribosome-associated) protein, cytochrome b2 (Cyt b2), was also studied.
The MrpL40 antiserum was then used to probe the Oxa1His and cross-linked adduct material purified by the Ni-NTA beads (Fig. 2B, lower right panels). MrpL40 cross-linked adducts were recovered with the Oxa1-purified Oxa1His protein in a cross-linking-dependent manner. We conclude therefore that the authentic MrpL40 protein, like the MrpL40HA derivative, can be cross-linked to and affinity purified with Oxa1His.
In summary, we have demonstrated here that the large ribosomal subunit component MrpL40 exists in close proximity to the inner membrane protein Oxa1.
MrpL40 is located close to the Mrp20 protein of the large ribosomal subunit.
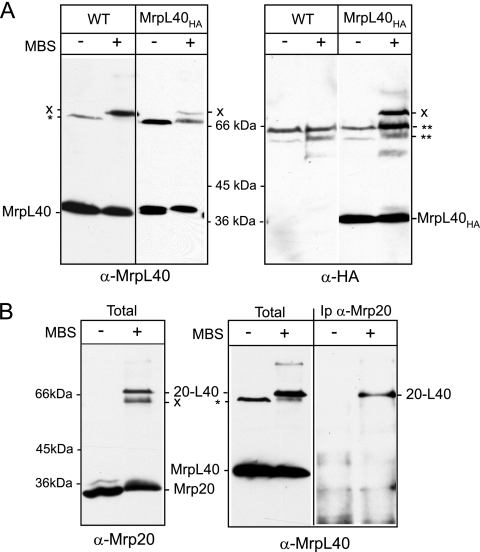
Given the finding that both MrpL40 and Mrp20 can be cross-linked to Oxa1, we decided to test if MrpL40 existed in close physical proximity to Mrp20, which is known to be located next to the polypeptide exist site of the large ribosomal subunit (3, 30). A chemical cross-linking approach to probe the molecular environment of the MrpL40 and MrpL40HA proteins in intact isolated mitochondria using the sulfhydryl-amino-specific heterobifunctional, noncleavable, cross-linking reagent MBS (9.9-Å spacer arm) (Fig. 3A) was undertaken. Following cross-linking and SDS-PAGE and Western blot analysis, both the MrpL40 and MrpL40HA proteins were found to form cross-linked adducts of approximately 70 kDa, as revealed by parallel decoration with MrpL40- and HA-specific antisera, respectively (Fig. 3A, left and right panels). To test whether this 70-kDa MrpL40 adduct may represent cross-linking of MrpL40 to Mrp20, we next addressed whether Mrp20 also formed an MBS-generated cross-linked adduct of similar size. Mrp20 was found to form two predominant cross-linked adducts of approximately 62 and 70 kDa in wild-type mitochondria (Fig. 3B, left panel), the larger of which comigrated with the MrpL40 cross-linked adduct (Fig. 3B, center panel). To demonstrate that this Mrp20 adduct represented a cross-linked product between Mrp20 and MrpL40, the Mrp20 protein and its cross-linked adducts were immunoprecipitated using an Mrp20 monoclonal antibody, analyzed by SDS-PAGE and Western blotting, and then probed with the MrpL40-specific antiserum (Fig. 3B, right panel). An MrpL40-reactive band of 70 kDa was observed in the Mrp20-immunoprecipitated material from the cross-linked samples. No MrpL40-containing material was detected in the Mrp20 immunoprecipitate in the absence of prior cross-linking. We conclude therefore that the 70-kDa MBS cross-linked adduct observed with both Mrp20- and MrpL40-specific antibodies represents an Mrp20-MrpL40 adduct.
FIG. 3.
MrpL40 and Mrp20 proteins exist in close proximity to each other. (A) Isolated wild-type (WT) or MrpL40HA mitochondria were subjected to cross-linking with MBS or mock treated, as indicated. Following quenching, mitochondria were reisolated and analyzed by SDS-PAGE, Western blotting, and immunodecoration with antiserum specific to MrpL40 (left panel) or HA (right panel). * and ** indicate cross-reactivity signals from the MrpL40 and HA antisera, respectively. X indicates an MrpL40(HA)-specific MBS adduct. (B) Wild-type mitochondria were subjected to MBS cross-linking or were mock treated, as indicated. Mitochondria were either lysed directly in SDS-sample buffer and subjected to SDS-PAGE and Western blotting analysis (“total” samples) or were lysed in SDS-buffer, diluted in Triton X-100 buffer, and Mrp20 and cross-linked adducts were immunoprecipitated using Mrp20 monoclonal antibodies (Ip α-Mrp20). Immunodecoration of the resulting Western blots was performed with either Mrp20 monoclonal antibodies (left panel) or with MrpL40 antisera (center and right panels). The position of the Mrp20-MrpL40 adduct is indicated by “20-L40”.
The abilities of Mrp20 and MrpL40 to be chemically cross-linked to each other demonstrate that a close physical relationship exists between these two proteins within the assembled ribosome, supporting that MrpL40, like Mrp20, the mitochondrial L23 homolog, is located close to the nascent chain exit site of the large ribosomal subunit.
The C-terminal region of MrpL40 is critical for function.
MrpL40 contains a KOW (Kyrpides, Ouzounis, Woese) motif (residues 62 to 89) (Fig. 4A), a motif found also in the ribosomal L24 protein family (Fig. 4A), where it is thought to be involved in rRNA binding (25). Like many mitochondrial ribosomal proteins, MrpL40 has a region (residues 90 to 289) which displays no conservation with prokaryotic ribosomal proteins and most likely constitutes a mitochondria-specific domain acquired through eukaryotic evolution of MrpL40 from its prokaryotic ancestors (Fig. 4A).
FIG. 4.
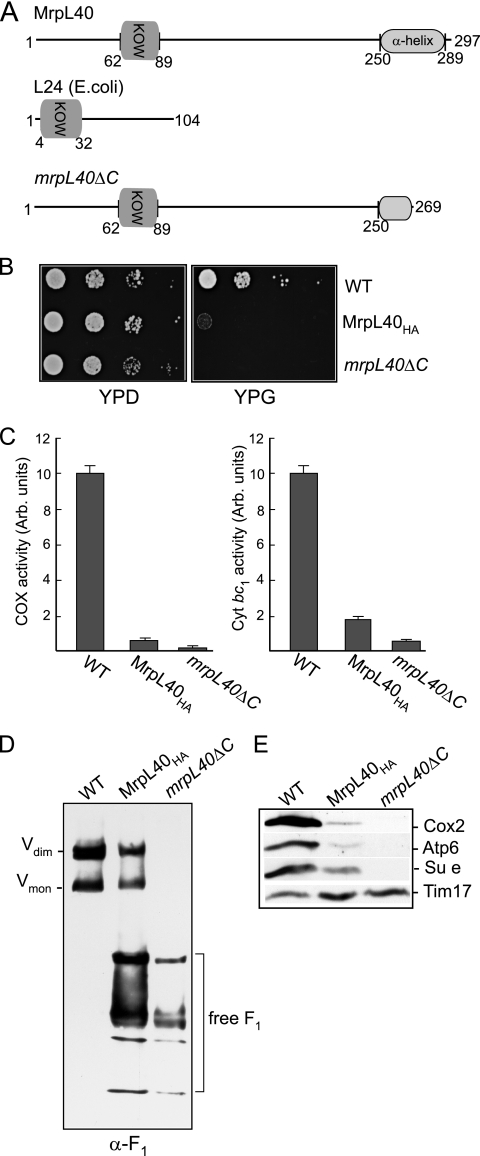
The C-terminal region of MrpL40 is crucial for its ability to support mitochondrial OXPHOS activity. (A) The S. cerevisiae MrpL40 and L24 (E. coli) proteins are depicted. The position of the conserved KOW motif is indicated for both proteins, together with the α-helical region in the C-terminal end of the yeast MrpL40. This region has been truncated in the MrpL40ΔC protein, from which the final 28 amino acid residues have been removed. (B) A dilution series of wild-type, MrpL40HA, and mrpL40ΔC strains was generated by serially diluting the cell suspensions 10-fold each time. A 2-μl aliquot of each of the resulting dilutions was spotted onto YPD (glucose) or YPG (glycerol) plates and incubated at 30°C. (C) The levels of cytochrome c oxidase (COX) (left panel) and antimycin-sensitive cytochrome c reductase (cyt bc1) enzyme activities were determined in mitochondria isolated from the wild-type, MrpL40HA, and mrpL40ΔC strains. Samples were measured in triplicate and averaged, and the relative specific activities in the MrpL40 mutant mitochondria were calculated and are expressed as a percentage of that measured in wild-type control mitochondria. Standard deviations are indicated. (D) Blue native-PAGE analysis of wild-type, MrpL40HA, and mrpL40ΔC mitochondria following digitonin (2%) solubilization, Western blotting, and immunodecoration with an antibody generated against the purified yeast F1 sector. The positions of free F1-containing complexes are indicated. (E) Steady-state levels of Cox2, Atp6, Su e, and Tim17 (loading control) were analyzed in mitochondria (50 μg of protein) isolated from the wild-type, MrpL40HA, and mrpL40ΔC strains.
Deletion of the MrpL40 gene causes an inability to grow on nonfermentable carbon sources (1). Cells expressing the C-terminally HA-tagged MrpL40 protein, MrpL40HA, were significantly compromised in their ability to grow on glycerol-based media (Fig. 4B). Thus, the ability of the mitochondria to support the assembly of functional oxidative phosphorylation complexes is compromised through the modification of the C-terminal region of MrpL40.
Essential proteins of the cytochrome bc1 (cytochrome b), cytochrome c oxidase (Cox1, Cox2, and Cox3), and ATP synthase complexes (Atp6, Atp8, and Atp9) are encoded by the mtDNA and are synthesized on the mitochondrial ribosomes. We therefore addressed whether the levels of these OXPHOS enzymes were adversely affected in the MrpL40HA mitochondria. Measurements of the enzyme levels of the cytochrome bc1 complex (cytochrome c reductase) and the cytochrome c oxidase complexes were both found to be strongly reduced in the isolated MrpL40HA mitochondria. The cytochrome bc1 complex enzyme levels in the MrpL40HA mitochondria were found to be only 15 to 20% those of wild-type mitochondria measured in the MrpL40HA mitochondria, whereas the cytochrome c oxidase enzyme levels were reduced to less than 10% of the wild-type control levels (Fig. 4C). Likewise, the levels of the assembled F1Fo-ATP synthase complex were found to be strongly reduced in the MrpL40HA mitochondria relative to the wild-type control (Fig. 4D). An abundance of free F1-containing species was observed in the mitochondria harboring the MrpL40HA protein, suggesting that the level of assembled Fo sector may have been limiting in these mitochondria. The strongly reduced levels of these OXPHOS complexes were also reflected in decreased steady-state levels of components of these complexes, such as Cox2 of the Cox complex and Atp6 and Su e of the F1Fo-ATP synthase (Fig. 4E). These proteins were chosen for this analysis because they are known to be susceptible to proteolytic turnover if the assembly of their respective complexes is hindered. Thus, given the overall decrease in the levels of these OXPHOS complexes, it appears that HA tagging of MrpL40 at its C terminus confers a pleiotrophic phenotype on the assembly of the mitochondrial OXPHOS system.
The MrpL40 protein contains a region (residues 250 to 289) with a predicted α-helical structure at its extreme C-terminal end (Fig. 4A), so it is possible that the addition of the HA tag to the end of this region may have adversely affected its structure or function. To assess the functional relevance of this C-terminal feature, we created a yeast strain expressing a truncated derivative of MrpL40, MrpL40ΔC, where the last 28 residues of the protein had been removed by a corresponding internal deletion in the chromosomal MRPL40 gene (Fig. 4A). This small C-terminal truncation was performed to test initially whether disruption rather than a complete deletion of the predicted α-helical segment would have a pronounced effect on MrpL40's ability to support OXPHOS assembly. Truncation of the MrpL40 protein in this manner was found to interfere completely with the ability of MrpL40 to support aerobic respiration, as the resulting strain, mrpL40ΔC, failed to grow on glycerol-containing medium (Fig. 4B). Analysis of the mrpL40ΔC mitochondria indicated an extreme decrease of cytochrome bc1 and cytochrome c oxidase enzyme levels (Fig. 4C). In addition no assembled F1Fo-ATP synthase complex was detected in the mrpL40ΔC mitochondria, but rather an accumulation of free F1 subcomplexes was detected (Fig. 4D).
We conclude that the extreme C-terminal region of MrpL40 is important for its function, and modification of this region, through either the addition of the HA tag or truncation of the last 28 residues, either partially or completely affects the ability of this protein to support the biogenesis of the mitochondrial OXPHOS system.
The function and assembly of mitochondrial ribosomes are adversely affected in mitochondria harboring MrpL40HA and truncated proteins.
A pleiotrophic effect on the OXPHOS complexes in the MrpL40HA and MrpL40ΔC strains may be indicative of a general decrease in the ribosomes' ability to synthesize the key OXPHOS subunits encoded by the mtDNA in these mutant mitochondria. We therefore performed an in organello translation in the presence of [35S]methionine using isolated mitochondria harboring either the HA-tagged, truncated, or authentic MrpL40 proteins (Fig. 5A). The MrpL40HA mitochondria displayed a capacity to synthesize the radiolabeled proteins; however, the pattern of protein synthesis in these mitochondria differed from that in the control wild-type mitochondria. Mitochondria harboring the MrpL40HA protein displayed a reduced capacity to synthesize Cox1 and Cox2 proteins, whereas the levels of Var1, cytochrome b, Atp6, and Atp9 appeared elevated compared to the control mitochondria containing the authentic MrpL40 protein. The levels of Atp8 and Cox3 synthesized in the MrpL40HA mitochondria did not differ significantly from the wild-type control. The ability of the mitochondrial ribosomes to synthesize all eight mtDNA-encoded proteins was severely compromised in the mitochondria harboring the MrpL40ΔC truncated protein (Fig. 5A). We conclude therefore that modulation of the C-terminal region of MrpL40 has consequences for the translational capacity of the mitochondrial ribosomes. Truncation of this region of MrpL40 hinders translation. While the addition of the HA tag allows the ribosomes to remain translationally active, they display an altered pattern of protein synthesis relative to the wild-type control.
When probing the isolated MrpL40HA mitochondria with the MrpL40 antisera, we observed that the levels of MrpL40HA were reduced compared to the authentic MrpL40 levels in the wild-type mitochondria (Fig. 5B and 2A). This observation indicates that the addition of the HA tag to the C terminus of MrpL40 may have partially compromised the stability of the MrpL40 protein. Interestingly, the levels of Mrp20 were found to be strongly reduced in the mitochondria harboring the HA-tagged MrpL40 protein compared to the wild-type control. The reduction in Mrp20 protein levels in these mitochondria did not simply reflect a general decrease in mitochondrial ribosomal protein levels, because the levels of other large ribosomal control proteins MrpL36, MrpL32, and Mrp49 (and Mrp10, a small subunit protein [reference 21; data not shown]) were not impacted in the MrpL40HA mitochondria (Fig. 5B). The levels of these proteins are strongly reduced in the rho0 mitochondria, where the complete ribosome assembly is defective due to the absence of mtDNA and hence the coding capacities for the rRNA and Var1 ribosomal protein (Fig. 5B). Truncation of the mitospecific region of MrpL40 also negatively impacted the steady-state levels of the MrpL40 and Mrp20 proteins (Fig. 5B). The mrpL40ΔC protein was undetectable, suggesting that possibly the levels of the truncated protein are so strongly reduced or a key epitope of the protein is compromised because of the truncation. Once again the reductions in Mrp20 and MrpL40 levels did not appear to be caused by a general loss of ribosome assembly, as the levels of MrpL36, MrpL32, and Mrp49 (and Mrp10 [data not shown]) were not reduced in the mrpL40ΔC mitochondria. On the contrary, the levels of the MrpL32 and Mrp49 proteins were observed to be somewhat elevated in the MrpL40HA and mrpL40ΔC mitochondria (Fig. 4B). The stability of ribosomal protein subunits known to be susceptible to proteolytic turnover when their assembly is defective due to the loss of rRNA is an indication that the MrpL40HA and mrpL40ΔC strains do not exhibit a high degree of rho−/rho0 formation. This conclusion has been supported through the level of 4′,6-diamidino2-phenylindole (DAPI)-stained mitochondrial DNA in the MrpL40HA and mrpL40ΔC cells (results not shown).
Taken together, we conclude that the reduced levels of Mrp20 in MrpL40HA- and mrpL40ΔC-containing mitochondria do not simply reflect a gross perturbation in overall ribosome assembly. Rather, we propose that these data argue for a dependency of the Mrp20 protein on MrpL40 for its full stability within the ribosome/mitochondria.
In addition to displaying an altered pattern of protein synthesis and protein composition, the mitochondrial ribosomes from the MrpL40HA mutant strain displayed an altered stability state compared to those isolated from the wild-type control mitochondria. Mitochondria isolated from both the MrpL40HA and wild-type control strains were solubilized with detergent and subjected to a clarifying spin, and the assembly state of the solubilized ribosomal proteins was analyzed by sucrose gradient centrifugation (Fig. 5C). The sedimentation behavior of the large and small ribosomal subunits was monitored by the immunological detection of proteins contained within these complexes, such as Mrp20, MrpL40, MrpL36, and Mrp49, components of the large ribosomal subunit, and Mrp10, a component of the small ribosomal subunit (21). Cofractionation of Mrp20, MrpL40, MrpL36, and Mrp49 toward the bottom of the gradient indicated the fractionation behavior of the large ribosomal subunit in the wild-type control sample. Mrp10 was recovered in the fractions just prior to the Mrp20/MrpL40/MrpL36/Mrp49 proteins, indicating that the large and small ribosomal subunits do not remain together under these fractionation conditions (Fig. 5C, upper panel). A fraction of the MrpL36 protein did not comigrate with the large ribosomal subunit under these conditions but rather was detected toward the top of the gradient, where smaller protein complexes such as cytochrome b2 (240 kDa) were recovered. The presence of MrpL36 toward the top of the gradient indicates that a significant fraction of this protein in the mitochondrial extract is assembled into the large ribosomal subunit, an observation consistent with a previous analysis of the MrpL36 protein (29, 36).
The stable assembly state of the mitochondrial ribosomes was significantly altered in the MrpL40HA mitochondria. In contrast to wild-type mitochondria, the majorities of the Mrp20, MrpL40HA, Mrp49, and MrpL36 proteins were recovered toward the top of the gradient. Only a minor fraction of these proteins was assembled into a complex, which from its migration behavior may correspond to the assembled large ribosomal subunit (Fig. 5C, lower panel). Note that MrpL40, which is not assembled into ribosomes (i.e., from rho0 mitochondria), is recovered exclusively at the top of the sucrose gradient under these conditions (results not shown). The assembly state of the small ribosomal subunit, as evidenced by the unaltered migration behavior of Mrp10, appeared unaffected in the MrpL40HA mitochondria, however.
Taken together, the presence of the HA tag at the C terminus of the MrpL40 protein appears to compromise the stability of the large ribosomal subunit. As functional, translationally active ribosomes can assemble in the MrpL40HA mitochondria, we conclude that the assembly state of the ribosome must be somewhat altered by this modification in the MrpL40 protein, such that the large ribosomal subunit does not remain as a single entity during the process of detergent/salt extraction and sucrose gradient centrifugation.
DISCUSSION
Oxa1 is a component of a protein insertion machinery which facilitates the insertion of nascent chains synthesized on the mitochondrial ribosomes tethered to the inner membrane. A physical interaction between the Oxa1 protein and the large ribosomal subunit has previously been demonstrated (19, 32). Furthermore, the large ribosomal subunit component, Mrp20, can be chemically cross-linked to Oxa1 (19). Mrp20 is the yeast mitochondrial homolog of the bacterial ribosomal protein L23, known to be located at the ribosomal polypeptide exit site (3, 10, 27, 30). Thus, the demonstrated proximity of Mrp20 and Oxa1 indicates that the region of the large ribosomal subunit where the nascent chain emerges is intimately related to the Oxa1 protein, a site where the nascent chains are integrated into the inner membrane. We have sought here to further map the ribosome-Oxa1 interface, with the goal of identifying further ribosomal components which are physically and possibly functionally close to the Oxa1 protein.
We have demonstrated here that Mrp20 does not represent the only large ribosomal protein which is in close proximity to Oxa1 in the inner membrane. Following chemical cross-linking, monomeric (i.e., non-cross-linked) ribosomal proteins can be affinity purified with Oxa1His in a cross-linking-dependent fashion and under detergent conditions which preserve the assembled large ribosomal subunit. Thus, within a given population of mitochondria different ribosomes may become cross-linked to Oxa1 by forming adducts with different ribosomal components. We demonstrated here that one of these large ribosomal subunit components close to Oxa1 is MrpL40. Initially, by using C-terminally HA-tagged MrpL40 and subsequently by analyzing the authentic MrpL40 protein with a specific polyclonal antibody, we demonstrated that MrpL40HA can be chemically cross-linked to, and affinity purified with, Oxa1His. Taking these results together with our previous analysis of Mrp20, we conclude that the ribosome-Oxa1 interface involves at least two large ribosomal subunit proteins in close proximity to Oxa1, Mrp20, the L23 homolog, and the MrpL40 protein.
We propose here that the MrpL40 protein represents the yeast mitochondrial homolog of the bacterial ribosomal protein L24. The bacterial L24 protein is located close to the L23 protein at the ribosomal polypeptide exit site (3, 23, 27, 30). In addition to containing a KOW motif, a feature which is conserved through L24 family members, BLASTp searches indicate homology of MrpL40 to bovine MrpL24 protein, the bovine mitochondrial L24 counterpart. Furthermore, our findings here support a close physical and functional relationship between the MrpL40 and Mrp20 proteins. First, MrpL40 can be chemically cross-linked to Mrp20 in intact wild-type mitochondria, indicating their close proximity to each other in the assembled ribosome. The close physical relationship of MrpL40 to Mrp20 places MrpL40, like the bacterial L24 protein, close to the polypeptide exit site of the large ribosomal subunit. Second, the truncation or alteration of the C-terminal region of MrpL40 was observed to negatively impact the steady-state levels of Mrp20 specifically, in contrast to other control ribosomal proteins, thus supporting their close physical relationship and possibly their dependence on each other for stability and hence stable accumulation within the mitochondria.
The bacterial L24 protein homologs are small proteins (usually in the range of 100 amino acid residues). The yeast MrpL40 protein is significantly larger (297 amino acid residues) and in addition to the conserved KOW motif has a predicted noncleavable N-terminal mitochondria targeting signal (C. Meisinger and N. Pfanner, personal communication). MrpL40 contains a large C-terminal extension, a mitospecific domain (residues 89 to 297). Conserved largely among fungal relatives, and only distantly related to the mitospecific domains of mammalian mitochondrial L24 proteins, we provide evidence here for the functional importance of this domain, in particular the extreme C-terminal region. HA tagging of the C terminus of MrpL40 caused an alteration in the translation pattern of the ribosome, with a noticeable decrease in Cox1 and Cox2 synthesis and an elevation in the levels of some of the newly synthesized proteins, in particular Var1, cytochrome b, and Atp9. Truncation of the C-terminal region of MrpL40 by the removal of the final 28 residues caused a severe disruption in the ability of the ribosomes to translate. Although the ribosomes from the MrpL40HA mitochondria remained translationally competent, sucrose gradient centrifugation analysis indicated that the stability of the large ribosomal subunit, but not of the small ribosomal subunit, was compromised as a result of tagging the MrpL40 in this manner. We conclude, therefore, that the extreme C-terminal region of MrpL40 is either directly or indirectly required (e.g., through affecting Mrp20 levels) to ensure stability of the assembled ribosomes to detergent extraction.
Modification of the C-terminal region of the mitospecific domain of MrpL40 severely compromised the mitochondria's ability to assemble functional OXPHOS machinery. The levels of the cytochrome bc1, cytochrome c oxidase enzymes, and the assembled F1Fo-ATP synthase complex were strongly impacted in the MrpL40HA and mrpL40ΔC mitochondria. The observed decrease in the OXPHOS complex assembly was to be anticipated from the mrpL40ΔC mitochondria due to their severe defect in mitochondrial translational capacity. The OXPHOS assembly defect was, however, unexpected for the MrpL40HA mitochondria, where mitochondrial protein translation, albeit at altered ratios, occurred at relatively normal levels. The observed detrimental effects of the MrpL40HA and MrpL40ΔC proteins on the OXPHOS complex may be directly due to the modification of the mitospecific domain of MrpL40 and/or indirectly due to the adverse impact that these modifications have on the levels of Mrp20 (and/or possibly on other as-yet-uncharacterized proteins located at the ribosomal polypeptide exit site). We propose that the mitospecific domain of MrpL40 (and/or Mrp20) may function to secure the assembly and/or activity of the mitochondrial ribosome to specific locations of the inner membrane, where the events of OXPHOS complex assembly could occur. It is conceivable that regions of the inner membrane exist which are enriched in various assembly chaperones and/or nonassembled nuclear-encoded proteins which coassemble with the mitochondrially encoded proteins following their synthesis and membrane insertion. Thus, according to this model the mitospecific regions of MrpL40, Mrp20, and possibly other ribosomal proteins may function to bind or recruit inner membrane-localized proteins with domains exposed to the matrix, which would ensure the targeting of/assembly of ribosomes to these specific assembly locations within the inner membrane.
Do the mitospecific domains of Mrp20 and MrpL40 directly interact with the C-terminal region of Oxa1? Known to support the ribosome-Oxa1 interaction, it is conceivable that the C-terminal region (approximately 90 residues) of Oxa1 directly binds to the mitospecific domain of MrpL40 and/or Mrp20. Using a bacterium-expressed Oxa1 C-terminal region and the mitospecific regions of MrpL40 and Mrp20, no evidence could be obtained to support a direct interaction between these regions of Oxa1, MrpL40, and Mrp20 (L. Jia and R. A. Stuart, unpublished data). Although these results are negative evidence, they may reflect that the mitospecific domains of MrpL40 and Mrp20 do not directly bind to the C-terminal region of Oxa1 in a stable manner but that they possibly bind to other inner membrane-anchored proteins in close association with Oxa1. Although the OXPHOS assembly is seriously perturbed in the MrpL40HA mitochondria, we propose that the initial ribosome-Oxa1 interaction can still productively occur despite the HA tagging of MrpL40. First, the MrpL40HA protein, like the authentic MrpL40, existed in close proximity to Oxa1, as evidenced by the abilities for both to cross-link with Oxa1His. Second, the in organello labeling experiment indicated that the newly synthesized Cox2 species was present in its mature form in the MrpL40HA mitochondria, and no evidence of the precursor form of Cox2 (pCox2) was obtained. In oxa1-defective mitochondria, pCox2 (1 to 2 kDa larger in size than the mature Cox2 species due to the presence of its uncleaved N-terminal signal sequence) is observed to accumulate. Maturation of pCox2 requires the prior Oxa1-dependent membrane insertion step, as the protease responsible for its maturation, Imp1, is exposed to the mitochondrial intermembrane space (14, 15, 28). Accumulation of newly synthesized Cox2 in its precursor form is observed in mitochondria harboring C-terminally truncated oxa1-encoded protein, i.e., under conditions where the Oxa1-ribosome interaction is known to be physically compromised (19, 32). The observed lack of pCox2 species in MrpL40HA mitochondria indicates that at least the initial Oxa1-dependent cotranslational events resulting in the N-terminal tail export of pCox2 species are not inhibited in the MrpL40HA mitochondria.
Finally, our work has furthered our understanding and mapping of the mitochondrial ribosome-Oxa1 interface, by identifying two large ribosomal subunit components, Mrp20 and MrpL40, homologs of bacterial ribosomal proteins L23 and L24, respectively, in close proximity to Oxa1. The findings reported here extend a recent study by Kohler et al. (23), who investigated the interactions of Oxa1 and YidC (the bacterial Oxa1 homolog) with the bacterial ribosomes. Using purified bacterial (E. coli) ribosomes and bacterially expressed and purified YidC and Oxa1 proteins, a mapping of the contact points between YidC/Oxa1 and the purified ribosome was performed, largely using cryo-electron microscopy analysis. This elegant study identified ribosomal proteins L23, L24, and L29 and the helix H59 of the 23S rRNA as physical contact points of YidC and the ribosome. Interestingly, these ribosomal contact points for YidC overlapped with those previously shown for the SecY translocon, although no direct sequence similarity exists between SecY and YidC (26). Overlapping contact points L23, L29, and H59 were observed with the Oxa1 protein and the E. coli ribosome; however, evidence for the L24 protein interacting with the Oxa1 protein was lacking. The L24 protein in E. coli is significantly shorter and is lacking the mitochondria-specific domain of the MrpL40 protein. Thus, it is possible that the mitospecific domain of MrpL40 may form a critical feature of the Oxa1-mitochondrial ribosome interface. Further characterization of the mitospecific domain of MrpL40 and the identification of possible interacting partners of this domain are currently being investigated in our laboratory.
Acknowledgments
We thank Mary K. Dienhart for valuable technical assistance with this project. We are very grateful for the generous gifts of many of the antisera used in this study; please see Materials and Methods for their sources.
This research was supported by NSF grants MCB 0347025 and MCB 0744067 to R.A.S.
Footnotes
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Accardi, R., E. Oxelmark, N. Jauniaux, V. de Pinto, A. Marchini, and M. Tommasino. 2004. High levels of the mitochondrial large ribosomal subunit protein 40 prevent loss of mitochondrial DNA in null mmf1 Saccharomyces cerevisiae cells. Yeast 21:539-548. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, I., K. Pfeiffer, W. Neupert, R. A. Stuart, and H. Schägger. 1998. Yeast mitochondrial F1Fo-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J. 17:7170-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, M., M. Behrens, K. Esser, G. Michaelis, and E. Pratje. 1994. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet. 245:272-278. [DOI] [PubMed] [Google Scholar]
- 5.Bonnefoy, N., F. Chalvet, P. Hamel, P. P. Slonimski, and G. Dujardin. 1994. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239:201-212. [DOI] [PubMed] [Google Scholar]
- 6.Bonnefoy, N., H. L. Fiumera, G. Dujardin, and T. D. Fox. 2009. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim. Biophys. Acta 1793:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornemann, T., J. Jöckel, M. V. Rodnina, and W. Wintermeyer. 2008. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15:494-499. [DOI] [PubMed] [Google Scholar]
- 8.Borst, P., and L. A. Grivell. 1978. The mitochondrial genome of yeast. Cell 15:705-723. [DOI] [PubMed] [Google Scholar]
- 9.Dienhart, M. K., and R. A. Stuart. 2008. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol. Biol. Cell 19:3934-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fearon, K., and T. L. Mason. 1992. Structure and function of Mrp20 and Mrp49, the nuclear genes for two ribosomal proteins of the 54S subunit of the yeast mitochondrial ribosome. J. Biol. Chem. 267:5162-5170. [PubMed] [Google Scholar]
- 11.Fiori, A., T. L. Mason, and T. D. Fox. 2003. Evidence that synthesis of the Saccharomyces cerevisiae mitochondrially encoded ribosomal protein Var1p may be membrane localized. Eukaryot. Cell 2:651-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graack, H. R., and B. Wittmann-Liebold. 1998. Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem. J. 329:433-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu, S. Q., F. Peske, H. J. Wieden, M. V. Rodnina, and W. Wintermeyer. 2003. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA 9:566-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, S., and T. D. Fox. 1997. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N- and C-termini and dependence on the conserved protein Oxa1p. Mol. Biol. Cell 8:1449-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hell, K., J. Herrmann, E. Pratje, W. Neupert, and R. A. Stuart. 1997. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 418:367-370. [DOI] [PubMed] [Google Scholar]
- 16.Hell, K., J. M. Herrmann, E. Pratje, W. Neupert, and R. A. Stuart. 1998. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl. Acad. Sci. USA 95:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hell, K., W. Neupert, and R. A. Stuart. 2001. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 20:1281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann, J. M., H. Koll, R. A. Cook, W. Neupert, and R. A. Stuart. 1995. Topogenesis of cytochrome oxidase subunit II: mechanisms of protein export from the mitochondrial matrix. J. Biol. Chem. 270:27079-27086. [DOI] [PubMed] [Google Scholar]
- 19.Jia, L., M. Dienhart, M. Schramp, M. McCauley, K. Hell, and R. A. Stuart. 2003. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22:6438-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia, L., M. K. Dienhart, and R. A. Stuart. 2007. Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex. Mol. Biol. Cell 18:1897-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, C., A. M. Myers, and A. Tzagoloff. 1997. Cloning and characterization of MRP10, a yeast gene coding for a mitochondrial ribosomal protein. Curr. Genet. 31:228-234. [DOI] [PubMed] [Google Scholar]
- 22.Koc, E. C., W. Burkhart, K. Blackburn, M. B. Moyer, D. M. Schlatzer, A. Moseley, and L. L. Spremulli. 2001. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 276:43958-43969. [DOI] [PubMed] [Google Scholar]
- 23.Kohler, R., D. Boehringer, B. Greber, R. Bingel-Erlenmeyer, I. Collinson, C. Schaffitzel, and N. Ban. 2009. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol. Cell 34:344-353. [DOI] [PubMed] [Google Scholar]
- 24.Kramer, G., T. Rauch, W. Rist, S. Vorderwulbecke, H. Patzelt, A. Schulze-Specking, N. Ban, E. Deuerling, and B. Bukau. 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature 419:171-174. [DOI] [PubMed] [Google Scholar]
- 25.Kyrpides, N. C., C. R. Woese, and C. A. Ouzounis. 1996. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 21:425-426. [DOI] [PubMed] [Google Scholar]
- 26.Ménétret, J. F., J. Schaletzky, W. M. Clemons, Jr., A. R. Osborne, S. S. Skånland, C. Denison, S. P. Gygi, D. S. Kirkpatrick, E. Park, S. J. Ludtke, T. A. Rapoport, and C. W. Akey. 2007. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol. Cell 28:1083-1092. [DOI] [PubMed] [Google Scholar]
- 27.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Nature 289:920-930. [DOI] [PubMed] [Google Scholar]
- 28.Nunnari, J., T. D. Fox, and P. Walter. 1993. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262:1997-2004. [DOI] [PubMed] [Google Scholar]
- 29.Prestele, M., F. Vogel, A. S. Reichert, J. M. Herrmann, and M. Ott. 2009. Mrpl36 is important for generation of assembly competent proteins during mitochondrial translation. Mol. Biol. Cell 20:2615-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma, M. R., E. C. Koc, P. P. Datta, T. M. Booth, L. L. Spremulli, and R. K. Agrawal. 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115:97-108. [DOI] [PubMed] [Google Scholar]
- 31.Stuart, R. A. 2002. Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochim. Biophys. Acta 1592:79-87. [DOI] [PubMed] [Google Scholar]
- 32.Szyrach, G., M. Ott, N. Bonnefoy, W. Neupert, and J. M. Herrmann. 2003. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22:6448-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ullers, R. S., E. N. Houben, A. Raine, C. M. Ten Hagen-Jongman, M. Ehrenberg, J. Brunner, B. Oudega, N. Harms, and J. Luirink. 2003. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 161:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel, F., C. Bornhövd, W. Neupert, and A. S. Reichert. 2006. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 36.Williams, E. H., X. Perez-Martinez, and T. D. Fox. 2004. MrpL36p, a highly diverged L31 ribosomal protein homolog with additional functional domains in Saccharomyces cerevisiae mitochondria. Genetics 167:65-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams, E. H., N. Bsat, N. Bonnefoy, C. A. Butler, and T. D. Fox. 2005. Alteration of a novel dispensable mitochondrial ribosomal small-subunit protein, Rsm28p, allows translation of defective COX2 mRNAs. Eukaryot. Cell 4:337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]