Abstract
When the genome organizations of 30 native isolates belonging to a wine spoilage yeast, Dekkera (Brettanomyces) bruxellensis, a distant relative of Saccharomyces cerevisiae, were examined, the numbers of chromosomes varied drastically, from 4 to at least 9. When single gene probes were used in Southern analysis, the corresponding genes usually mapped to at least two chromosomal bands, excluding a simple haploid organization of the genome. When different loci were sequenced, in most cases, several different haplotypes were obtained for each single isolate, and they belonged to two subtypes. Phylogenetic reconstruction using haplotypes revealed that the sequences from different isolates belonging to one subtype were more similar to each other than to the sequences belonging to the other subtype within the isolate. Reanalysis of the genome sequence also confirmed that partially sequenced strain Y879 is not a simple haploid and that its genome contains approximately 1% polymorphic sites. The present situation could be explained by (i) a hybridization event where two similar but different genomes have recently fused together or (ii) an event where the diploid progenitor of all analyzed strains lost a regular sexual cycle, and the genome started to accumulate mutations.
Recent achievements in genome sequencing have revealed that gene contents vary among distantly related organisms but are relatively constant among closely related species. For example, among hemiascomycete yeasts, which originated more than 250 million years ago and include well-studied yeasts such as Saccharomyces cerevisiae and Candida albicans (3, 4), an average genome contains approximately 5,000 genes. Approximately one-half of the protein-coding gene families are preserved in all of the yeasts sequenced to date. However, there is a large variation in the gene order and configuration of chromosomes among different species.
Chromosome configuration is usually well preserved among populations belonging to the same species. Only rarely do geographically separated populations, for example, Mus musculus (8, 32), differ in the number and form of chromosomes. The mutability of the genome enhances the adaptability of the species, but it also decreases the viability of the new variant. In addition, these changes can preclude successful reproduction and can be a decisive factor in the emergence of new species (2; for a review, see references 6 and 7).
Among closely related yeasts belonging to the Saccharomyces sensu stricto clade (including S. cerevisiae), which originated approximately 20 million years ago, the gene contents are relatively similar (13). Their genomes are almost colinear and consist of 16 chromosomes. Some inter- and intraspecific variations are observed predominantly at the chromosome ends (18, 19). Sensu stricto species are semifertile, meaning that they can successfully mate and produce F1 offspring but that the hybrids are largely sterile. It appears that this clade has still not completed the speciation process (7). The relatively low chromosome variability among Saccharomyces sensu stricto yeasts is probably promoted by regular sexual cycles. These yeasts are diploid, but heterozygosity is almost absent because of the homothallic life-style, which enables haploid spores from the same yeast cell to mate. Only for “sterile” hybrids, such as the lager brewing yeast Saccharomyces pastorianus (Saccharomyces carlsbergensis), originating upon the mating of two different species, has a pronounced heterozygosity been observed (14). The parental genomes came from S. cerevisiae and a close relative, Saccharomyces bayanus. A study of allotetraploid hybrids between a diploid S. cerevisiae strain and a diploid S. bayanus strain demonstrated that these hybrids behave essentially as diploids regarding meiosis and sporulation and had 77% spore viability (1, 22). The extent of intra- and interspecific genome variability is not well known for other yeasts, especially among distant relatives of S. cerevisiae. The only well-studied exception is a pathogen, Candida albicans, that is believed to be predominantly asexual. This yeast diverged from the S. cerevisiae lineage prior to the origin of the efficient homothallic life-style (reviewed in reference 25). The genome is diploid and shows a low level of heterozygosity (12), and large variations in the configurations of the chromosomes among different isolates have been reported (reviewed in reference 29).
Dekkera bruxellensis is often isolated in wineries and is well known as a major microbial cause of wine spoilage. The lineages of D. bruxellensis and S. cerevisiae separated at approximately the same time as the lineages of S. cerevisiae and C. albicans separated, approximately 200 million years ago (40). However, D. bruxellensis and S. cerevisiae share several characteristics, such as the production of ethanol, the ability to propagate in the absence of oxygen (anaerobic growth), and petite positivity (the ability to produce offspring without mitochondrial DNA [mtDNA]), that are rarely found among other yeasts (16, 20). So far, a sexual cycle in D. bruxellensis has not been found.
In this paper, we analyzed the genome structures of 30 isolates of D. bruxellensis originating from different geographical localities around the world. We show that these isolates have different numbers and sizes of chromosomes and also that the numbers of copies of several analyzed genes and their sequences vary. In addition, we could detect heterozygosity in the partial genome sequence of strain Y879.
MATERIALS AND METHODS
Yeast strains and culture conditions.
We analyzed 30 strains of D. bruxellensis originating from the Centraalbureau Vor Schimmelcultures collection, isolated primarily from alcohol production environments from all over the world (Table 1). The yeast collection was preserved in 25% glycerol at −80°C. Under sterile conditions, the 30 strains were propagated on yeast extract-peptone-dextrose (1% yeast extract [Formedium], 2% Bacto peptone [Merck], 2% glucose [Merck]) agar plates at 25°C. Single colonies were obtained after transferring selected colonies onto fresh solid medium.
TABLE 1.
Strains and their molecular markersa
| Strain designation | Laboratory designation | No. of PFGE bands | GenBank accession no. |
|
|---|---|---|---|---|
| Nuclear 26S rDNA | Mitochondrial 15S rDNA | |||
| CBS 1940 | Y869 | 8 | FJ805762 | FJ769558 |
| CBS 1941 | Y870 | ND | FJ805763 | FJ769559 |
| CBS 1942 | Y871 | 7 | FJ805764 | FJ769560 |
| CBS 1943 | Y872 | 5 | FJ805765 | FJ769561 |
| CBS 2336 | Y878 | 6 | FJ805766 | FJ769562 |
| CBS 2499 | Y879 | 7 | FJ805767 | FJ769563 |
| CBS 2547 | Y880 | 5 | FJ805768 | FJ769564 |
| CBS 2796 | Y881 | 7 | FJ805769 | FJ769565 |
| CBS 2797 | Y882 | 4 | FJ805770, FJ805771 | FJ769566 |
| CBS 3025 | Y883 | 8 | FJ805772 | FJ769567 |
| CBS 4459 | Y888 | 6 | FJ805773 | FJ769568 |
| CBS 4480 | Y899 | 5 | FJ805778 | FJ769572 |
| CBS 4481 | Y900 | 5 | FJ805779 | FJ769573 |
| CBS 4482 | Y901 | 6 | FJ805780 | FJ769574 |
| CBS 4601 | Y902 | 5 | FJ805781 | FJ769575 |
| CBS 4602 | Y891 | 4 | FJ805774 | FJ769569 |
| CBS 4914 | Y906 | 6 | FJ805782 | FJ769576 |
| CBS 5206 | Y908 | 7 | FJ805783 | FJ769577 |
| CBS 5512 | Y911 | 7 | FJ805784 | FJ769578 |
| CBS 5513 | Y912 | 8 | FJ805785 | FJ769579 |
| CBS 6055 | Y895 | 7 | FJ805775, FJ805776 | FJ769570 |
| CBS 72 | Y858 | 5 | FJ805754 | FJ769550 |
| CBS 73 | Y859 | 7 | FJ805755 | FJ769551 |
| CBS 74 | Y860 | 6 | FJ805756 | FJ769552 |
| CBS 75 | Y861 | 6 | FJ805757 | FJ769553 |
| CBS 78 | Y864 | 6 | FJ805758 | FJ769554 |
| CBS 8027 | Y897 | 5 | FJ805777 | FJ769571 |
| CBS 96 | Y865 | 7 | FJ805759 | FJ769555 |
| CBS 97 | Y866 | 3 | FJ805760 | FJ769556 |
| CBS 98 | Y867 | 7 | FJ805761 | FJ769557 |
| CBS 8139 | D. anomala | ND | FJ805786 | FJ769580 |
The Centraalbureau Vor Schimmelcultures (CBS) collection of Dekkera bruxellensis strains was used in this study. Shown are GenBank accession numbers for nuclear 26S rDNA (D1/D2 fragment) and mitochondrial 15S rDNA. The PFGE bands were deduced from data shown in Fig. 2. ND, not determined.
Genomic DNA extraction from single colonies.
Cells originating from a colony grown from a single cell of each of the D. bruxellensis strains were inoculated in 5 ml of yeast extract-peptone-dextrose medium. Under constant shaking, the samples were grown overnight at 25°C. The culture, grown overnight, was transferred into a 2-ml Eppendorf tube and centrifuged at 8,000 rpm for 3 min. The resulting pellet was washed with 1 ml distilled sterile water and centrifuged again. The supernatant was removed, and the cleaned pellet was dissolved in a 500-μl solution (0.9 M sorbitol, 0.1 M EDTA [pH 7.4], 0.05 M Tris [pH 7.4], 1% β-mercaptoethanol, 20 mg/ml 20T Zymolase [20 μl per sample]). The samples were incubated for 60 min at 37°C. After incubation the samples were centrifuged for 5 min at 12,000 rpm. The resultant pellet was washed with a 500-μl solution (50 mM Tris [pH 7.4] and 20 mM EDTA) and was shaken vigorously for 50 s. Sodium dodecyl sulfate was added (1/5 volume of 10%, which is 100 μl per sample), and the mixture was then incubated for 10 min at 65°C. After the addition of 5 M potassium acetate (100 μl per sample), tubes were incubated on ice for 1 h. The samples were then centrifuged for 15 min at 12,000 rpm. Four hundred microliters of the resulting supernatant was transferred into a new 1.5-ml Eppendorf tube. The DNA was precipitated with 2 volumes of 96% ethanol and washed with 70% ethanol. The clean and dry pellet of DNA was diluted in 250 μl of distilled sterile water. The isolated DNA was checked on a 1% agarose gel, and the concentration was measured with a NanoDrop ND-1000 spectrophotometer. The DNA was stored at −20°C.
Phylogenetic analysis.
To ensure the correct species assignment, a conserved region of the nuclear 26S ribosomal DNA (rDNA) locus and a segment of the mitochondrial 15S locus were amplified by PCR and sequenced for all isolates presumed to be D. bruxellensis isolates and one control (outgroup) isolate of its closest relative, Dekkera anomala. The sequences were manually aligned, and the relationships between them were inferred using the neighbor-joining method (30). Phylogenetic analyses were conducted using MEGA4 software (36), with bootstrap values (500 replicates) shown as percentages next to the branches (5). Evolutionary distances were computed as the number of base substitutions per site using the maximum composite likelihood method (37). All positions containing gaps and missing data were eliminated from the data set.
Chromosome variability.
Yeast chromosomal DNA was prepared in plugs as previously described (24). Pulsed-field gel electrophoresis (PFGE) was performed using a Chef Mapper apparatus (Bio-Rad). The gels were run in a multistate program for 110 h with four blocks (block 1, 25 h at 1.5 V/cm with a 2,700-s pulse time and an angle of 53°; block 2, 25 h at 1.5V/cm with a 2,200-s pulse time and an angle of 60°; block 3, 30 h at 2 V/cm with a 1,500-s pulse time and an angle of 60°; block 4, 30 h at 2.5 V/cm with a 500-s pulse time and an angle of 60°) at a constant temperature (12°C). As a control for chromosome length, two standards, Schizosaccharomyces pombe and Hansenula wingei (Bio-Rad), were used, with a span from 1.05 to 5.7 Mbp. To visualize the chromosome pattern, the gel was soaked in 1 μg/ml ethidium bromide for 1 h with constant shaking and visualized in UV light.
Southern blot analysis.
Five different loci in this study, derived from the sequenced strain of D. bruxellensis, were multiplied by PCR and used as probes. To map these loci to different chromosomes, we transferred the separated chromosomes from the PFGE gel with vacuum transfer onto a Hybond-XL membrane (GE Healthcare). The gel was depurinated for 30 min, denatured for 30 min, and finally neutralized according to the manufacturer's protocol, with Southern blotting as a neutral transfer gel treatment. The different loci were made into probes by amplifying them in a PCR, followed by purification with Qiagen purification for PCR products, and then labeling them with radioactive [32P]dCTP from GE Healthcare by use of the Amersham Rediprime II Random Prime labeling system. To remove unincorporated nucleotides from the DNA labeling reaction, Illustra ProbeQuant G-50 microcolumns (GE Healthcare) were used according to the manufacturer's instructions. The labeled probes were hybridized to the membrane in hybridization solution (0.5 M Na2HPO4) overnight at 60°C before being washed three times in washing solution, also at 60°C, to remove unincorporated and unspecifically bound probes. The washed membrane was then sealed in plastic and exposed to an imaging screen (Bio-Rad) in a closed cassette for 2 to 24 h before scanning using a Molecular Imager FX apparatus (Bio-Rad). The membranes could be stripped (the radioactive probe was removed from the membrane in hot 0.1% sodium dodecyl sulfate) and reused several times.
Primer design and PCRs.
To analyze species-specific sequence variation, five regions from the D. bruxellensis sequence were randomly chosen (40): three of them are located upstream from an open reading frame (ORF) (DbYER090 [870 bp], DbYDL040 [660 bp], and DbYDR513 [750 bp]), one is an intron (DbYLR084 [620 bp]), and one coding gene originated from horizontal transfer from a bacterium (HAD1 [620 bp]). With the exception of HAD1, the genes in this species are named after their S. cerevisiae orthologues, with the prefix Db to indicate that they are D. bruxellensis genes. The primer sites for the noncoding fragments were chosen in the nearby coding region to ensure that the sites were as conserved as possible. The primer sequences used (from 5′ to 3′) were as follows: TAACCAATGGCTCCACCACT and CAAACTCGGTCGTTGTTCAA for DbYER090, GAACACAGCCGTTTATGGAT and CTCTTGTAATGCTTGTTTTCGTA for DbYDL040, GGGCAGTAAGTCTTTGAGG and CTGATATGTTGATTAGCCAGAA for DbYDR513, GGCTTGTTGTGTACGATTAC and ATAGTGAAGCGGATACCACTT for DbYLR084, and AAGGTTGAACTTCACTGTCA and CCGTGATCAACTCGATC for HAD1.
For PCRs, 100 ng of genomic DNA was added to a 28-μl reaction mix (5 U/μl Phusion high-fidelity DNA polymerase [Finnzymes], 50 μM of each primer [forward and reverse], 5× Phusion buffer [Finnzymes], 20 μM deoxynucleoside triphosphate). Reactions were run on a Robocycler Gradient 40 temperature cycler (AH Diagnostics, Stratagene) programmed as follows: an initial denaturing temperature of 98°C for 1 min, followed by 7 cycles of denaturation at 98°C for 30 s, annealing at 65°C for 45 s, and polymerization at 72°C for 1 min. This was followed by 30 cycles of denaturation at 98°C for 30 s, annealing at 55°C for 45 s, and polymerization at 72°C for 1 min. The polymerization was completed in 10 min of incubation at 72°C.
Sequencing of PCR products.
Single-extension sequencing from both strands of the PCR products from the genomic DNA (isolated from single colonies) was performed by Macrogen Ltd. Sequencing System, Seoul, South Korea. Procedure details can be found at http://www.macrogen.com/eng/sequencing/extension.jsp. Sequencing was conducted under BigDye terminator cycling conditions. The products were purified using ethanol precipitation and run by using a 3730xl automatic sequencer. The sequences were edited and manually aligned by using BioEdit 7.0.5.3 (10).
Subcloning of PCR-multiplied loci.
The sequencing chromatograms from genomic DNA isolated from a yeast colony grown from a single cell showed evidence of two different sequences for one locus in almost all the strains and fragments. To obtain the haplotype sequences for the strains where the genomic sequences differed the most, we subcloned the PCR products into a pCR4-TOPO vector (TOPO TA cloning kit for sequencing; Invitrogen). Eight to 12 plasmids per fragment and a strain carrying the yeast locus were isolated, and each individual plasmid was separately amplified and sequenced as described above. In other words, if the original yeast cell contained a polymorphic gene template, this would result in a pool of plasmids carrying different haplotype inserts. As a control, subcloning and sequencing were reproduced twice for the DbYLR084 intron, with different starting single cells from each original yeast strain.
Sequence analysis.
Sequence alignment of the subcloned fragments (haplotypes) was done manually by using BioEdit 7.0.5.3 (10), and analyses of nucleotide polymorphism, haplotype differentiation, total divergence, gene conversion, and recombination events were performed using DnaSP 4.10 software (28). Phylogenetic trees of the haplotype sequences were constructed using two software packages, MEGA4 (36) and TREECON1.3b (39), to ensure the consistency of the trees.
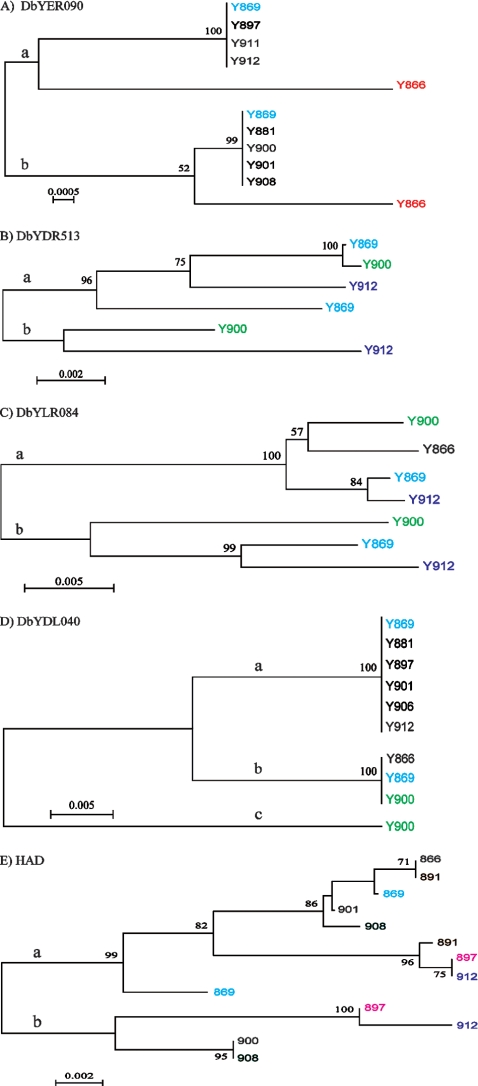
For the strains where more than two different haplotype sequences were obtained from a single fragment, the two most divergent sequences (which also corresponded to the sequences with the highest frequency) were chosen for genetic analyses and phylogenetic reconstruction. All different haplotype sequences were analyzed manually for additional variants, but the less representative sequences could all be generated from two “parental” sequences by recombination. The a, b, and c branches (see Table 3) correspond to the subcloned sequences that make up each branch shown in Fig. 5. Genetic analyses were performed by using sequences from four strains (Y866, Y869, Y900, and Y912).
TABLE 3.
Genetic analysis of the five loci multiplied by PCR, subcloned, and then sequenceda
| Locus | Sequence | No. of sequences used | No. of variable sites | No. of haplotypes | Pi | Avg no. of nucleotide differences | K | Ka | Ka/Ks ratio |
|---|---|---|---|---|---|---|---|---|---|
| DbYER090 | a branch | 3 | 11 | 2 | 0.0086 | 7.333 | 0.0132 | ||
| b branch | 3 | 5 | 2 | 0.0039 | 3.333 | ||||
| Y869 | 2 | 10 | 2 | 0.0117 | 10 | 0.0117 | |||
| Y866 | 2 | 9 | 2 | 0.0105 | 9 | 0.0105 | |||
| Y900 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| Y912 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| DbYDR513 | a branch | 4 | 29 | 4 | 0.0236 | 16 | 0.0399 | ||
| b branch | 2 | 20 | 2 | 0.0295 | 20 | ||||
| Y869 | 2 | 23 | 2 | 0.0336 | 23 | 0.0336 | |||
| Y900 | 2 | 23 | 2 | 0.0330 | 23 | 0.0330 | |||
| Y912 | 2 | 32 | 2 | 0.0456 | 32 | 0.0456 | |||
| DbYLR084 | a branch | 3 | 9 | 3 | 0.0098 | 6 | 0.0419 | ||
| b branch | 3 | 25 | 3 | 0.0272 | 16.667 | ||||
| Y869 | 2 | 25 | 2 | 0.0409 | 25 | 0.0409 | |||
| Y900 | 2 | 26 | 2 | 0.0425 | 26 | 0.0425 | |||
| Y912 | 2 | 27 | 2 | 0.0441 | 27 | 0.0441 | |||
| DbYDL040 | a branch | 2 | 0 | 1 | 0 | 0 | 0.02947 | ||
| b branch | 2 | 0 | 1 | 0 | 0 | 0.05741* | |||
| c branch | 1 | 0 | 1 | 0 | 0 | 0.05704** | |||
| Y869 | 2 | 17 | 2 | 0.0293 | 17 | 0.0293 | |||
| Y900 | 2 | 32 | 2 | 0.0570 | 32 | 0.0570 | |||
| Y912 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| HAD1 | a branch | 8 | 14 | 6 | 0.0122 | 6.4286 | 0.1095 | 0.0069 | 0.063 |
| b branch | 4 | 10 | 3 | 0.0120 | 6.3333 | ||||
| Y869 | 2 | 8 | 2 | 0.0152 | 8 | 0.0575 | 0.0025 | 0.042 | |
| Y900 | 2 | 3 | 2 | 0.0057 | 3 | 0.0248 | 0 | 0 | |
| Y912 | 2 | 19 | 2 | 0.0361 | 19 | 0.1242 | 0.0099 | 0.073 | |
| Y897 | 2 | 17 | 2 | 0.0323 | 17 | 0.1158 | 0.0074 | 0.059 | |
| Y908 | 2 | 12 | 2 | 0.0228 | 12 | 0.0822 | 0.0050 | 0.057 | |
| Y866 | 2 | 2 | 2 | 0.0038 | 2 | 0.0164 | 0 | 0 | |
| Y891 | 2 | 9 | 2 | 0.0170 | 9 | 0.0656 | 0.0025 | 0.036 |
Shown are data for divergence between the paralogous sequences within each strain and between sequences in different strains (a, b, and c branches), based on analysis of the subcloned sequences (haplotypes). The a, b, and c branches correspond to the subcloned sequences that make up each branch in Fig. 5. Pi, nucleotide diversity (per site); K, divergence between the a and b branches (based on the sequences within each of these) and the divergence between paralogues in each strain. In the coding sequence HAD1, K is substituted with Ks (synonymous divergence between sequences in the a and b branches), and for each strain, the paralogues are compared. For HAD1, the Ka values (nonsynonymous divergence) and the ratio of Ka/Ks is also calculated between the sequences in the a and b branches and for the paralogues in each strain. In DbYDL040, K for the a and c branches (*) and b and c branches (**) is also indicated.
FIG. 5.
Phylogenetic relationships of haplotype sequences. Phylogenetic relationships of haplotype sequences are based on subcloned fragments originating from various isolates. (A) DbYER090 (896 bp). (B) DbYDR513 (144 bp). (C) DbYDL040 (665 bp). (D) DbYLR084 (680 bp). (E) HAD1 gene (602 bp). The a and b branches inferred on every tree represent the two clades separating the two copies from each isolate. Phylogenetic analyses were conducted by using MEGA4 (36), with bootstrap values (500 replicates) shown as percentages next to the branches (5). The evolutionary distances were computed using the maximum composite likelihood method (37) as the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the data set. The black strains represent lineages that are monomorphic for the particular locus.
Reanalysis of the partially sequenced genome.
D. bruxellensis Y879 was previously partially sequenced and analyzed, assuming that it was as a haploid organism (40). Here we reanalyzed the sequence data and assumed that it did not have a haploid genome organization. The assembled contigs were blasted against the trimmed reads (GenBank accession numbers EF364424 to EF364429 and EI011584 to EI026443) using BLASTN. To ensure homologous sequences and to avoid very short alignments, a cutoff E value of 1−50 was chosen. From these different settings, only contigs with more than one trimmed read that were not identical to each other were selected.
To avoid flanking regions with large polymorphisms, further selection was made where the aligned length had to be as long as the trimmed read length or the contig length. The differences in the flanking regions may indicate paralogues rather than orthologues. Depending on the presence of a divergent flanking sequence and the E cutoff value, the trimmed read was either included in the “selected list” or discarded.
The selected reads were compared to the PHRAP output described previously (40) for heterozygote sites. Since Jones et al. (12) previously found PHRAP analyses to be too stringent for diploid organisms, we used both data sets to analyze the heterozygosity from the PHRAP output and the BLASTN output, the latter with a filter that removed reads with flanking regions that were too divergent. The most divergent sequences in the PHRAP analyses were not used either, since the PHRAP alignments were compared with the output of the different BLASTN outputs, and sequences in the PHRAP alignments that diverged too much were removed.
In order to obtain polymorphic sites, two or more different reads had to overlap each other; therefore, contigs with two or more overlapping trimmed reads were selected, and the lengths of the overlapping regions were calculated. The selected E cutoff value for the BLAST analyses of 1−50 rendered the largest overlapping region compared to E cutoff values of between 1−30 and 1−100. The unique sequence variation in the overlapping region was then determined by taking away duplicate hits and mismatches with a quality score of under 20.
The heterozygosity in the coding and noncoding regions was analyzed using the coding regions homologous to S. cerevisiae determined previously (40). For the coding regions, the distribution of the polymorphic sites was estimated for positions in a codon and the amino acid changes.
Since the genome is only partially sequenced, not all contigs have overlapping sequences, and even if they did, all alleles might not be sequenced, and the variation is therefore underestimated. The program and documentation can be found at http://www.cob.lu.se/yeastandenzymes/ruby.html.
Nucleotide sequence accession numbers.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. FJ805754 to FJ805834 and FJ769550 to FJ769580.
RESULTS
All strains belong to the same species.
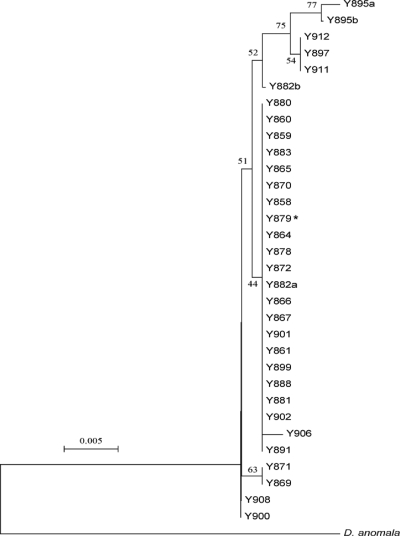
Thirty isolates (Table 1) that were previously determined to be D. bruxellensis isolates were analyzed for their nuclear rDNA locus, in particular the 26S D1/D2 domain (17), and an 832-bp fragment from the mitochondrial 15S rDNA locus. Both loci are members of the structural RNA components, which are highly repetitive and conserved through gene conversion. Their analysis is in general very useful for studies of genome origin and stability (11). The phylogenetic relationship presented in Fig. 1 (see also supplemental material file 1) clearly demonstrates that all of the strains indeed belonged to the same species, D. bruxellensis. The outgroup is represented by the closest D. bruxellensis relative, D. anomala. A majority of isolates had only one version of the 26S D1/D2 domain and the mitochondrial 15S rDNA locus. However, two isolates, Y882 and Y895, had apparently at least two different copies of the 26S rDNA locus, with one polymorphic site each corresponding to two different single nucleotide polymorphisms (SNPs) observed in the other isolates. All sequences were obtained from yeast colonies originating from a single cell.
FIG. 1.
Phylogenetic relationships among Dekkera bruxellensis isolates. Phylogenetic relationships are based on nuclear 26S rDNA (D1/D2 fragment) among 30 Dekkera bruxellensis isolates using its closest relative, Dekkera anomala, as an outgroup. The asterisk indicates the partially sequenced strain. The a and b sequences for Y882 and Y895 indicate the presence of two different sequences with one polymorphic site each, corresponding to the SNP seen in the other isolates. Phylogenetic analyses were conducted with MEGA4 (36), with bootstrap values (500 replicates) shown as percentages next to the branches (5). The evolutionary distances were computed using the maximum composite likelihood method (37) as the number of base substitutions per site. All positions containing gaps and missing data were eliminated from the data set.
Karyotypes vary drastically.
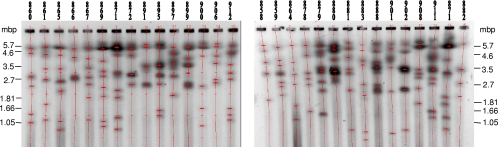
Karyotypes can be used to distinguish different yeast species (35). However, differences in karyotypes within the isolates belonging to the same yeast species are usually low (34). We used PFGE to separate chromosomes. Gel separation could be successfully reproduced, giving the same sizes and numbers of chromosome bands for each analyzed strain. Karyotypes showed that D. bruxellensis strains contained between 4 and 9 chromosomes. In some strains, some bands showed a higher intensity (Fig. 2; see supplemental material file 2), suggesting that some chromosomes overlap in size. The chromosome sizes ranged from less than 1 Mbp to over 6 Mbp. Such a high degree of karyotype variation suggests that the genome has rearranged very fast upon the separation of single lineages.
FIG. 2.
Chromosome separation in Dekkera bruxellensis isolates. PFGE of 29 strains of Dekkera bruxellensis shows a large variation in the numbers and sizes of chromosomes. Strains are indicated above each lane. The vertical lines indicate one lane, and the larger horizontal lines indicate a chromosome band (the smaller horizontal lines are artifacts derived from the program). The size ladder on each side is in megabase pairs (Mbp). For the raw figure without any vertical lines, see supplemental material file 2.
Duplications.
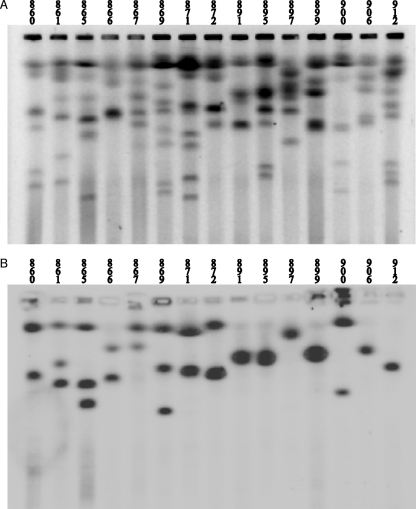
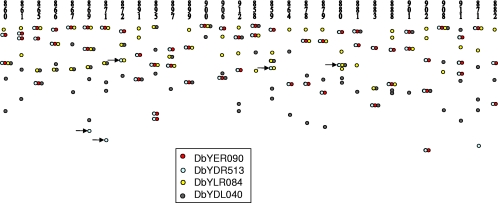
For Southern analysis of duplications, we used the same noncoding loci (DbYER090, DbYDL040, DbYDR513, and DbYLR084) as those used for species-specific sequencing. These loci mapped to different chromosomes in 29 D. bruxellensis strains (Fig. 3 and 4; see supplemental material file 6). In Fig. 3, which shows chromosome separation and Southern analysis with a functional gene encoding allantoinase as a probe, one can see that out of 15 strains, only 6 strains gave a signal with only one chromosome band, while 6 strains gave a signal with two bands, and 3 strains gave a signal with three bands. Interestingly, in a majority of cases, the intensities of both or all three bands belonging to the same strain were similar. However, in some cases, the intensities were clearly different (see supplemental material file 6). Surprisingly, each single probe hybridized to more than one chromosome band in a majority of strains. When the noncoding loci (mentioned above) were used as a single probe, 60% of all hybridizations mapped to two chromosomes, 20% mapped to one chromosome, and 19% mapped to three chromosomes. In one case (1%), a single probe mapped to four different chromosomes (Fig. 4 and Table 2).
FIG. 3.
PFGE gel of D. bruxellensis strains (A) and a corresponding Southern analysis (B) of the blot using 850 bp from the gene encoding allantoinase (homologous to S. cerevisiae YIL166) as a probe. Note that in a majority of the strains, at least two chromosome bands hybridized to the probe, and the intensities of such pairs are often similar.
FIG. 4.
Single-probe hybridization to different chromosomes in Dekkera bruxellensis isolates. Southern blots originating from PFGE gels hybridized with a single probe, DbYER090, DbYDR513, DbYLR084, and DbYDL040, are shown. The arrows point out chromosomes where the DbYDR513 locus but not DbYER090 is found.
TABLE 2.
Isolates with one, two, or three (or more) chromosome bands hybridizing to a single gene probe
| Gene probe | No. of hybridizing bands |
|||
|---|---|---|---|---|
| Single signal | 2 signals | 3 signalsa | Total signals | |
| DbYER090 | 8 | 17 | 4 | 29 |
| DbYDR513 | 4 | 20 | 5 | 29 |
| DbYLR084 | 7 | 18 | 4 | 29 |
| DbYDL040 | 4 | 15 | 9 (1) | 29 |
| Total | 23 | 70 | 22 (1) | 116 |
Numbers in parentheses indicate the number of additional signals.
Of the tested loci, two of them, DbYER090 and DbYDR513, gave almost the same signal pattern and were apparently linked (Fig. 4 and see supplemental material file 3). There was no obvious correlation between the other probes, which hybridized to different chromosome bands (Fig. 4). The tightly linked loci DbYER090 and DbYDR513 were always assigned to the same chromosome, but in five cases, the DbYDR513 signal was also found on a smaller chromosome, while DbYER090 was not detected on this chromosome (Fig. 4 and see supplemental material file 3). In three of these cases, DbYDR513 was linked to DbYLR084, and in one case, it was also linked to DbYDL040 (Fig. 4). This suggests that the loci can be randomly rearranged on the chromosomes.
Intrastrain variations.
The above-described locus-mapping analysis showed that each locus is frequently present on the genome in at least two copies. Furthermore, we amplified the four loci plus HAD1 from a majority of strains by PCR and carefully analyzed the directly obtained sequences (see supplemental material file 4). Almost all strains had polymorphic sites at each locus analyzed. The positions of the variable sites (SNPs) in each locus were very similar across all strains (see supplemental material file 4). This suggests that each locus has at least two original sequences that are preserved among the strains, suggesting a polyploidy status of the strains. For the HAD1 gene, 95% of the strains (21 out of 22) had variable sites corresponding to two different sequences. In the noncoding fragments, the proportions of strains with variable sites were 76% (22/29) for DbYER090, 100% (9/9) for DbYDR513, 40% (4/10) for DbYDL040, and 95% (21/22) for DbYLR084. The overall divergence between the two different sequences in all noncoding loci per strain was 0.03482 (calculated from all strains with more than one sequence per locus). This indicates that the two or more “genomes” present in the cell could be diverged by more than 3% within the noncoding DNA.
Haplotypes.
PCR products of the five loci analyzed as described above were subcloned from several strains, and approximately 50 resulting plasmids from each subcloning experiment were verified for the insert sequence. In general, the PCR products that showed the highest variability on directly obtained sequences were selected for the subcloning experiment. We subcloned strains where the genomic sequences differed the most according to variable sites. As a control we also subcloned some strains where the genomic sequences obtained upon the direct sequencing of the PCR products did not show any heterozygosity at all. In short, the sequences obtained from the subcloned-fragment experiments confirmed the previously deduced variable sites.
When we constructed phylogenetic trees, two “parental” lineages became apparent for each locus (Fig. 5). Moreover, each strain with at least two different alleles for a locus was represented in the two different lineages, suggesting a duplication event before the radiation of the species. We could also see that haplotypes from different strains, orthologues (sequences within a single lineage) (Fig. 5), are more similar to each other than the two copies within the same strain, paralogues (sequences on the a and b branches) (Fig. 5 and Table 3). This is strong evidence that the ancestor of all strains had at least two copies, paralogues, of each locus. There was no correlation between the number of haplotypes found for each fragment and the number of chromosomes to which the fragment hybridized within a single strain (Table 4). A locus could in principle be present in several allelic forms or as several identical copies. For example, in two out of five fragments, DbYER090 and DbYDL040, we found some strains with only a single paralogue sequence (Fig. 5). Interestingly, for DbYER090, either paralogue appears to be “lost” with a very similar probability (3:4); however, in DbYDL040, one paralogue is much more likely to be lost (1:5). This nonrandom deletion of gene copies was also previously observed for other yeasts (31) and Arabidopsis thaliana (38). In addition, some of the observed results could be explained as gene conversion, for example, if only a single paralogue sequence was obtained but the locus actually hybridized to two or more chromosomes (Fig. 4 and Table 4). Evidence for a deletion can be seen in Fig. 4 (see also supplemental material file 3), where DbYER090 and DbYDR513 are both duplicated (and linked) in 17 strains and are present as a single linked group in only 8 strains (they are present in triplicate in 4 strains). In four out of five cases where a copy of DbYDR513 is present alone (without DbYER090), there is only a single linked group of DbYER090 and DbYDR513. In this case, a copy of DbYER090 was probably lost from a duplicated segment that still contains DbYDR513.
TABLE 4.
Haplotype and hybridization resultsa
| Fragment | Strain | Sequence type | No. of blot bands | Description |
|---|---|---|---|---|
| DbYER090 | Y866 | Polymorphic | 2 | |
| Y869 | Polymorphic | 2 | ||
| Y900 | Monomorphic | 1 | ||
| Y912 | Monomorphic | 2 | Gene conversion | |
| DbYDR513 | Y866 | Polymorphic | 2 | |
| Y869 | Polymorphic | 2 | ||
| Y900 | Polymorphic | 1 | ||
| Y912 | Polymorphic | 2 | ||
| DbYLR084 | Y866 | Monomorphic | 2 | Gene conversion |
| Y869 | Polymorphic | 3 | Multiple copies | |
| Y900 | Polymorphic | 1 | ||
| Y912 | Polymorphic | 2 | ||
| DbYDL040 | Y866 | Monomorphic | 2 | Gene conversion |
| Y869 | Polymorphic | 4 | Multiple copies | |
| Y900 | Polymorphic | 2 | ||
| Y912 | Monomorphic | 3 | Multiple copies/gene conversion | |
| HAD1 | Y866 | Polymorphic | 3 | Multiple copies |
| Y869 | Polymorphic | 1 | ||
| Y900 | Polymorphic | 2 | ||
| Y912 | Polymorphic | 2 |
Comparison between the sequence variation within a strain and the number of bands that hybridized to the respective probe by Southern blotting. The sequence analysis is based on the cloned PCR fragments. If only one sequence/haplotype was found in a strain, the locus was designated monomorphic. If there were more than two haplotypes, the locus was designated polymorphic. The blot band signals are derived from data shown in Fig. 4. The Southern blot was made from a PFGE gel, which separates chromosomes.
Reanalysis of the genomic sequence.
Previously, one strain, Y879, was partially sequenced (40). To gain further insights into the genome organization, we reanalyzed the deposited sequences. The total data set consisted of 14,860 trimmed reads, covering approximately 7.6 Mb. The total number of contigs was 5,407, including single contigs. For the BLASTN run, 3,528 contigs with two or more reads were found (E cutoff value of 10−50). The total length of the overlapping sequences was 1,811,317 bp. After taking away polymorphisms of loci sequenced more than once, the total number of unique SNPs with a quality score of 20 or more was 13,715. These SNPs were distributed on 2,012 contigs representing just over one-half of the total number of contigs. The level of heterozygosity, calculated as unique SNPs/total overlapping sequence (bp), was 0.7%. A similar calculation with the PHRAP output estimated the heterozygosity to be 0.4% (Table 5).
TABLE 5.
Genomic sequence analysesa
| Parameter | Value determined by: |
|
|---|---|---|
| BLASTN analysis | PHRAP analysis | |
| No. of contigs with 2 or more reads | 3,528 | 3,621 |
| No. of reads in selected fileb | 10,521 | |
| No. of ambiguous bases removedc | 708 | 613 |
| No. of contigs with polymorphic sites | 1,355 | 2,149 |
| No. of homozygote contigs | 2,210 | 1,416 |
| Ratio of heterozygote contigs | 0.380 | 0.602 |
| Total sequence size with overlapping reads (bp) | 2,071,659 | 2,434,896 |
| Size of coding region with overlap (bp) | 752,412 | 824,895 |
| Size of noncoding region with overlap (bp) | 1,319,247 | 1,610,001 |
| No. of polymorphic sitesc | 10,569 | 10,605 |
| No. of polymorphic sites in the coding region | 2,362 | 2,149 |
| No. of polymorphic sites in the noncoding region | 8,207 | 8,456 |
| Heterozygosity in the coding region (%) | 0.314 | 0.260 |
| Heterozygosity in the noncoding region (%) | 0.622 | 0.525 |
| Avg heterozygosity (%) | 0.510 | 0.436 |
Reanalyses of the partially sequenced genome of D. bruxellensis strain Y879. The contigs (assembled sequences) from the PHRAP analysis (40) are used for PHRAP analysis data shown, and they were also blasted against the original reads with an E cutoff value of 10−50 to obtain the alignments for the BLASTN analysis.
BLASTN only and removing sequences (reads) with a filter (see the text).
Base quality score of 20 or more.
The distributions of heterozygosity in the coding and noncoding regions from the BLASTN search were 0.4% and 1.0%, respectively. Among the 2,656 genes analyzed, 2,362 SNPs were found to be distributed on 581 genes (Table 6). In the coding regions the polymorphisms at the three different codon positions were 440:273:1,649 (or, in relative numbers, 1.61:1:6.04), meaning that the second codon position is the least variable one. This may suggest that both copies are still expressed and that neutral mutations (third positions) are more frequently fixed in the genome. The nonsynonymous changes in the ORFs, which result in amino acid polymorphisms between the two alleles, are not evenly distributed among all amino acids, and it seems that the exchanged amino acids have similar properties (Table 6 and see supplemental material file 5). The ratio between numbers of transitions and transversions in synonymous substitutions in polymorphic ORFs was almost twice as high as the ratio in nonsynonymous substitutions (Table 7).
TABLE 6.
Distribution of genes with SNPs among either ORFs or pseudogenes from the partially sequenced genomea
| Group analyzed | Total no. of ORFs and pseudogenes | No. of ORFs (% of total) | No. of pseudogenes (% of total) |
|---|---|---|---|
| Genes | 2,656 | 2,529 (95.2) | 127 (4.8) |
| Genes with SNPs | 581 | 545 (93.8) | 36 (6.2) |
| SNPs | 2,362 | 2,141 (90.7) | 221 (9.3) |
| Nonsynonymous SNPs | 707 | 587 (83.0) | 120 (17.0) |
The genes containing stop codons are considered to be pseudogenes.
TABLE 7.
Substitutions in ORFs divided into synonymous substitutions and nonsynonymous substitutionsa
| Type of substitution | No. of substitutions |
No. of transitions | No. of transversions | Transition/transversion ratio | |||||
|---|---|---|---|---|---|---|---|---|---|
| A-G | T-C | A-C | G-C | T-G | A-T | ||||
| Synonymous | 564 | 706 | 94 | 77 | 51 | 62 | 1,270 | 284 | 4.47 |
| Nonsynonymous | 276 | 133 | 41 | 40 | 51 | 46 | 409 | 178 | 2.3 |
| Total | 840 | 839 | 135 | 117 | 102 | 108 | 1,679 | 462 | 3.63 |
The number of transitions and transversions and their ratios are also shown. Note that the ratio between transitions and transversions for synonymous substitutions is almost twice as high as that for nonsynonymous substitutions (and the normal transition/transversion ratio found for other organisms).
In 5% (127) of the analyzed genes, one variant of the gene contained one or several stop codons (Table 6). Therefore, in these pairs, one copy likely represents a pseudogene. Among these genes the number of additional SNPs (apart from the stop codon) was approximately two times higher than among other genes. In other words, the genes (5% of the total genes) that contained at least one stop codon contained 9% of the total SNPs and 16% of all nonsynonymous polymorphisms.
DISCUSSION
In nature, yeasts are one of the dominant groups of organisms involved in the breakdown of simple poly-, oligo-, and monosaccharides. Yeasts also include important industrial organisms, pathogens, and popular laboratory organisms that serve as general models to understand the eukaryotic cell. For decades, Saccharomyces cerevisiae, baker's yeast, has been one of the best-characterized organisms from a genetic, biochemistry, and physiology point of view and was the first eukaryote with a sequenced genome (9, 21). Also, the genomes of several S. cerevisiae relatives have recently been well characterized (for example, see references 24 and 25). However, a majority of wild yeasts are still poorly characterized, and the organization and dynamics of their genomes are largely unknown.
In Saccharomyces yeasts, new genetic combinations are generated through sex and diploidized through the switch of the mating type and mating of the mother and daughter cells (23, 41). However, meiosis and homothallism also help to decrease genetic variability and contribute to genome stability. Saccharomyces yeasts are diploids, and heterozygosity in their genomes has rarely been observed, likely because of their homothallic life-style. Also, their karyotypes show a relatively low degree of rearrangements within each species. On the other hand, Saccharomyces sensu stricto hybrids, which have lost a sexual cycle, show an increased level of rearrangements (27).
In this study we analyzed native isolates of D. bruxellensis for the structural properties of their genome. To confirm that these isolates belong to a closely related species complex, nuclear 26S rDNA domain D1/D2 and an 832-bp fragment from the mitochondrial 15S rDNA locus were analyzed. Indeed, all isolates belong to the same species. The two sequenced loci exhibited a very low degree of polymorphism among the 30 strains (Table 1 and Fig. 1). On the other hand, the numbers and sizes of the chromosomes varied enormously among the isolates (Fig. 2 and see supplemental material file 2). Pronounced karyotype variability in two pathogenic yeasts, Candida albicans (33) and Candida glabrata (26), was also previously observed and analyzed. Both these yeasts are predominantly asexual and apparently use a relaxed control over the chromosome structure to increase their genome variability and competitiveness (26). In D. bruxellensis, the observed karyotype variability also makes it difficult to believe that this species regularly undergoes standard meiosis. The mating of two strains with drastically different karyotypes would produce zygotes that would rarely be able to segregate chromosomes into viable combinations during meiosis. However, several putative meiosis genes, such as homologues of the FUS3, SGF29, and NAT1 genes, etc., are actually present on the partially revealed genome sequence. So far, there is no evidence for a sexual cycle in D. bruxellensis, and even though there have been some reports of spore formation in this species, these spores have never been successfully mated.
In our previous genome sequencing study we assumed that D. bruxellensis is a haploid yeast (40). This assumption has been based mainly on the observation that one could relatively easily isolate auxotrophic mutants in this yeast (our unpublished observations). However, our Southern analysis showed that all five gene probes hybridized to more than one chromosomal band in a majority of strains (Table 2 and Fig. 3 and 4), indicating that these genes were present in more than one copy. Although karyotype variability is extreme in D. bruxellensis, a small portion could be attributed to the nonindependence of the samples (some strains are very close relatives), which will result in a population structure and maybe an overestimation of the rate of rearrangement. When comparing the “similar” distributions of DbYER090 and DbYDR513 together (see supplemental material file 3) and the sequence tree based on mtDNA (see supplemental material file 1), the relationship between them is not perfect, but a population structure cannot be excluded. The mtDNA clade of Y895, Y897, Y911, and Y912 contains strains with both duplicate and triplicate copies of DbYER090 and DbYDR513. The other, less-well-supported groups in the mtDNA tree consisting of Y872, Y880, Y859, and Y870 (Y870 is not shown in Fig. 4) have a single copy of DbYER090 but a duplicate copy of DbYDR513. The other small group in the rDNA tree consisting of Y899, Y888, Y891, and Y902 has duplicate copies, except for Y902, which has three copies of DbYER090 and DbYDR513. These results point out that at least some parts of the genome are duplicated and that this yeast is not a simple haploid.
Further support for a polyploidy status comes from the direct sequencing of the above-described five genes (Table 3). In a haploid organism, only one sequence version would be expected. However, each of the analyzed genes was present in each strain in at least two haplotype versions (Fig. 5 and Table 3 and see supplemental material file 4). Subcloning of the five PCR-multiplied genes and sequence analysis of several plasmid inserts originating from each subcloning revealed that in several cases, even more than two haplotype copies were present in the culture, originating from a single cell. These data, summarized in Table 4, suggest that the tested isolates are at least diploids and that several regions may even be additionally duplicated. Similarly, recent studies of S. cerevisiae (15) and C. glabrata (26) showed that segmental duplications may be common in yeasts.
When different haplotypes of several loci were analyzed (Fig. 5), one could clearly see that orthologous and paralogous copies exist in the strains. Apparently, two sequence types exist (paralogous, represented by the a and b lineages in Fig. 5), and they could be found in almost every strain. The orthologous sequences originating from one of the paralogous types are in different isolates more similar to each other than to the sequences of the other paralogous type. This suggests that the present diploid-like status originated prior to the separation of the analyzed lineages and even before the divergence into two D. bruxellensis phylogenetic subgroups (Fig. 1).
In several cases only one haplotype subgroup was found for some yeast isolates (strains Y897, Y911, Y912, Y881, Y900, Y901, and Y908) (Fig. 5A). This could be explained as gene conversion or a loss of one of the subgroup copies. Mitotic gene conversion as a result of mitotic recombination can be seen in the HAD1 gene for strain Y869. The two most divergent sequences for this strain have the same sequence for the first 216 bases (see supplemental material file 4A), after which they start to diverge. In the tree (Fig. 5E), this can be seen where the two Y869 haplotypes appear on the same a branch, but one is basal and the other is nested at the top of this clade. Gene conversion is also the reason why the paralogous sequences in Y866 for DbYER090 look so basal in the phylogenetic tree (Fig. 5A). The apparent high level of polymorphism in each clade compared to the divergence between the paralogues is often attributed to the fact that gene conversion is homogenizing the two different parental sequences and thereby making new variants. The most obvious homogenization could be found in the 26S rDNA sequences. In 30 different strains of D. bruxellensis, we detected only 2 strains containing two different sequences for this region (strains Y882 and Y895) (Fig. 1). When Southern analysis was performed using the 26S D1/D2 probe, in several strains, more than one chromosome band gave a signal, and the band intensities could vary (data not shown).
A detailed reanalysis of the genome sequences (40) confirms the diploid/polyploid nature of D. bruxellensis. The total observed heterozygosity of around 0.7% indicates that the two genome sets present are very similar and that most of the genes are still present in at least two copies (Fig. 5 and Tables 5 to 7). Are both copies still active? In approximately 5% of the analyzed ORFs, we could detect a stop codon, suggesting that the yeast can survive with only one active copy. On the other hand, the distribution of the heterozygous sites, where the second codon position is the most conserved one, suggests that in most cases, both genes are still operational. This is supported by a large bias among the heterozygous sites found in ORFs and resulting in synonymous changes versus nonsynonymous ones (Table 7). However, it seems that once a copy was inactivated by a stop codon, it started to accumulate mutations and was in the process of degeneration. The observation that nonsynonymous changes in the analyzed ORFs, which result in amino acid polymorphisms between different alleles, are not equally distributed suggests that selection pressure operates to keep the gene product functional. The ratio of the number of transitions to the number of transversions, which is significantly higher for synonymous than for nonsynonymous substitutions (Table 7), also suggests differences in selection pressure at different sites.
The observed heterozygosity could occur by mating between two sister species isolates, generating a hybrid that has kept both genomes. This scenario is reminiscent of S. pastorianus (S. carlsbergensis), which also generated a sterile outcome, but one should keep in mind that the putative D. bruxellensis parents were more related to each other. Another possible explanation is that the D. bruxellensis progenitor was a sexual diploid, which at some point turned into an asexual yeast that continued with clonal propagation. The presence of pseudogenes (Table 6) suggests that the hybrid/diploid status may now be in the process of degeneration. In addition, this yeast seems to now be very prone to generating further duplications and chromosome rearrangements (Fig. 3 and 5).
D. bruxellensis, known as a wine and beer yeast and as an active agent in bioethanol production, has now proven to be a very interesting organism to study evolution in action, especially genome rearrangements and the role of asexuality.
Supplementary Material
Acknowledgments
We thank Torbjörn Säll, Silvia Polakova, and Dag Ahren for discussion of some results; Meg Woolfit for English perusal; and Jouzas Sirkus and Dorte Jørck-Ramberg for help in the initial phylogenetic and karyotype analyses.
We thank the Crafoord, Futura, Fysiografen, Sörensen, and Trygger foundations for their financial support.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Banno, I., and Y. Kaneko. 1989. A genetic analysis of taxonomic relation between Saccharomyces cerevisiae and Saccharomyces bayanus. Yeast 5:S373-S377. [PubMed] [Google Scholar]
- 2.Delneri, D., I. Colson, S. Grammenoudi, I. N. Roberts, E. J. Louis, and S. G. Oliver. 2003. Engineering evolution to study speciation in yeasts. Nature 422:68-72. [DOI] [PubMed] [Google Scholar]
- 3.Dujon, B. 2006. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22:375-387. [DOI] [PubMed] [Google Scholar]
- 4.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44.15229592 [Google Scholar]
- 5.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson-Smith, M. A., and V. Trifonov. 2007. Mammalian karyotype evolution. Nat. Rev. Genet. 8:950-962. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, G., S. A. James, I. N. Roberts, S. G. Oliver, and E. J. Louis. 2000. Chromosomal evolution in Saccharomyces. Nature 405:451-454. [DOI] [PubMed] [Google Scholar]
- 8.Fraguedakis-Tsolis, S., H. C. Hauffe, and J. B. Searle. 1997. Genetic distinctiveness of a village population of house mice: relevance to speciation and chromosomal evolution. Proc. Biol. Sci. 264:355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed] [Google Scholar]
- 10.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 11.James, S. A., M. J. O'Kelly, D. M. Carter, R. P. Davey, A. van Oudenaarden, and I. N. Roberts. 2009. Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Res. 19:626-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 14.Kodama, Y., M. C. Kielland-Brandt, and J. Hansen. 2006. Lager brewing yeast, p. 145-164. In P. Sunnerhagen and J. Piskur (ed.), Comparative genomics using fungi as models. Springer-Verlag, Berlin, Germany.
- 15.Koszul, R., S. Caburet, B. Dujon, and G. Fischer. 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23:234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurtzman, C. P., and J. W. Fell. 1998. The yeasts, a taxonomic study. Elsevier Science BV, Amsterdam, The Netherlands.
- 17.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 18.Liti, G., D. B. Barton, and E. J. Louis. 2006. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics 174:839-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Louis, E. J., E. S. Naumova, A. Lee, G. Naumov, and J. E. Haber. 1994. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics 136:789-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merico, A., P. Sulo, J. Piskur, and C. Compagno. 2007. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 274:976-989. [DOI] [PubMed] [Google Scholar]
- 21.Mortimer, R. K. 2000. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 10:403-409. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson-Tillgren, T., C. Gjermansen, M. C. Kielland-Brandt, J. G. L. Petersen, and S. Holmberg. 1981. Genetic differences between Saccharomyces carlsbergensis and S. cerevisiae. Analysis of chromosome III by single chromosome transfer. Carlsberg Res. Commun. 46:65-76. [Google Scholar]
- 23.Oshima, Y., and I. Takano. 1971. Mating types in Saccharomyces: their convertibility and homothallism. Genetics 67:327-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen, R. F., T. Nilsson-Tillgren, and J. Piskur. 1999. Karyotypes of Saccharomyces sensu lato species. Int. J. Syst. Bacteriol. 49(Pt. 4):1925-1931. [DOI] [PubMed] [Google Scholar]
- 25.Piskur, J., and R. B. Langkjaer. 2004. Yeast genome sequencing: the power of comparative genomics. Mol. Microbiol. 53:381-389. [DOI] [PubMed] [Google Scholar]
- 26.Poláková, S., C. Blume, J. Zárate, M. Mentel, D. Jorck-Ramberg, and J. Piskur. 2009. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 106:2688-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rainieri, S., Y. Kodama, Y. Kaneko, K. Mikata, Y. Nakao, and T. Ashikari. 2006. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl. Environ. Microbiol. 72:3968-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 29.Rustchenko, E. 2007. Chromosome instability in Candida albicans. FEMS Yeast Res. 7:2-11. [DOI] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Scannell, D. R., A. C. Frank, G. C. Conant, K. P. Byrne, M. Woolfit, and K. H. Wolfe. 2007. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc. Natl. Acad. Sci. USA 104:8397-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Searle, J. B. 1998. Speciation, chromosomes, and genomes. Genome Res. 8:1-3. [DOI] [PubMed] [Google Scholar]
- 33.Selmecki, A., A. Forche, and J. Berman. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin, J. H., M. J. Chae, J. W. Song, S. I. Jung, D. Cho, S. J. Kee, S. H. Kim, M. G. Shin, S. P. Suh, and D. W. Ryang. 2007. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J. Clin. Microbiol. 45:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spirek, M., J. Yang, C. Groth, R. F. Petersen, R. B. Langkjaer, E. S. Naumova, P. Sulo, G. I. Naumov, and J. Piskur. 2003. High-rate evolution of Saccharomyces sensu lato chromosomes. FEMS Yeast Res. 3:363-373. [DOI] [PubMed] [Google Scholar]
- 36.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 37.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas, B. C., B. Pedersen, and M. Freeling. 2006. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16:934-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 40.Woolfit, M., E. Rozpedowska, J. Piskur, and K. H. Wolfe. 2007. Genome survey sequencing of the wine spoilage yeast Dekkera (Brettanomyces) bruxellensis. Eukaryot. Cell 6:721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yun, S. H., M. L. Berbee, O. C. Yoder, and B. G. Turgeon. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. USA 96:5592-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.