Abstract
The tomato pathotype of Alternaria alternata produces host-specific AAL toxin and causes Alternaria stem canker on tomato. A polyketide synthetase (PKS) gene, ALT1, which is involved in AAL toxin biosynthesis, resides on a 1.0-Mb conditionally dispensable chromosome (CDC) found only in the pathogenic and AAL toxin-producing strains. Genomic sequences of ALT1 and another PKS gene, both of which reside on the CDC in the tomato pathotype strains, were compared to those of tomato pathotype strains collected worldwide. This revealed that the sequences of both CDC genes were identical among five A. alternata tomato pathotype strains having different geographical origins. On the other hand, the sequences of other genes located on chromosomes other than the CDC are not identical in each strain, indicating that the origin of the CDC might be different from that of other chromosomes in the tomato pathotype. Telomere fingerprinting and restriction fragment length polymorphism analyses of the A. alternata strains also indicated that the CDCs in the tomato pathotype strains were identical, although the genetic backgrounds of the strains differed. A hybrid strain between two different pathotypes was shown to harbor the CDCs derived from both parental strains with an expanded range of pathogenicity, indicating that CDCs can be transmitted from one strain to another and stably maintained in the new genome. We propose a hypothesis whereby the ability to produce AAL toxin and to infect a plant could potentially be distributed among A. alternata strains by horizontal transfer of an entire pathogenicity chromosome. This could provide a possible mechanism by which new pathogens arise in nature.
Fungi produce a huge variety of secondary metabolites. Some plant-pathogenic fungi, especially necrotrophic pathogens that kill plant cells during invasion, produce phytotoxic metabolites to impair host tissue functions (20, 30, 42, 47). Phytotoxins produced by fungal plant pathogens are generally low-molecular-weight secondary metabolites that exert toxic effects on host plants. Among these phytotoxins, host-specific toxins (HSTs) are critical determinants of pathogenicity or virulence in several plant-pathogen interactions (13, 30, 33, 40, 42, 47, 49).
Recent advances in molecular biological techniques for fungi have led to the identification of fungal genes involved in pathogenesis, as exemplified by those used in the biosynthesis of toxic secondary metabolites, such as HSTs. Genes involved in the biosynthesis of secondary metabolites are typically clustered in filamentous fungi, including plant pathogens (20, 24, 44). The origins and evolutionary processes of these gene clusters, however, are largely unknown. Analysis of the arrangement and sequences of genes in the clusters would shed light on how the clusters themselves and their ability to produce toxic secondary metabolites evolved (20, 24, 44).
The involvement of horizontal gene transfer (HGT) in the evolution of fungal secondary-metabolite gene clusters has been discussed (34, 44). HGT events are well known in prokaryotes (21, 29), and the genomic regions that have undergone HGT are referred to as pathogenicity or genomic islands (7). In prokaryotes, the mechanisms of HGT are also associated with conjugation, transformation, and transduction (21, 29). Although these transfer mechanisms are generally unknown in eukaryotes such as fungi, interspecific transfer of a virulence gene encoding the production of a critical toxin has been reported in Pyrenophora tritici-repentis (14). There is also clear evidence of recent lateral gene transfer of the ToxA gene from Stagonospora nodorum to P. tritici-repentis (14, 30).
In Alternaria alternata plant pathogens (37), we have shown that all strains of the A. alternata pathotypes harbor small extra chromosomes of less than 1.7 Mb, whereas nonpathogenic isolates do not have these small chromosomes (5). A cyclic peptide synthetase gene, AMT, which is involved in host-specific AM toxin biosynthesis of the apple pathotype of A. alternata, was located on a small chromosome of 1.1 to 1.7 Mb, depending on the strain (22, 23). The AF toxin biosynthesis gene cluster was also present on a single small chromosome of 1.05 Mb in the strawberry pathotype of A. alternata (18). Based on biological and pathological observations, those small chromosomes were regarded as supernumerary chromosomes, or conditionally dispensable chromosomes (CDCs) (10, 18, 22). Fungal supernumerary chromosomes, which are not important for normal growth but confer advantages for colonizing an ecological niche, such as infecting host plants, are regarded as CDCs (21). The functions and pathological roles of CDCs have been studied in the pea pathogen Nectria haematococca (11, 17, 25, 32, 43, 46).
The origin and evolution of CDCs have been intriguing issues in the study of plant-microbe interactions. The supernumerary chromosomes of certain strains of N. haematococca have been suggested to have a different evolutionary history than essential chromosomes (ECs) in the same genome, and they might have been introduced into the genome by horizontal transfer from another strain (10, 12, 36). In Colletotrichum gloeosporioides, the 2-Mb supernumerary chromosome was transferred from a biotype A strain to a vegetative incompatible biotype B strain (19, 31). Transfer of the chromosome, however, did not affect the pathogenicity of the recipient fungus, perhaps because it did not harbor pathogenicity genes (19, 31). These results suggest that supernumerary chromosomes of fungi might have the capacity for horizontal transfer across an incompatibility barrier between two distinct strains.
AAL toxins are HSTs produced by the tomato pathotype of A. alternata (synonym A. alternata f. sp. lycopersici, synonym Alternaria arborescens), the causal agent of Alternaria stem canker disease in tomatoes, which causes severe necrosis of susceptible tomato cultivars (15, 26, 35). AAL toxins and fumonisins of the maize pathogen Gibberella moniliformis are structurally related to sphinganine and termed sphinganine-analogue mycotoxins. AAL toxins and fumonisins are sphinganine-analogue mycotoxins, which are toxic to some plant species and mammalian cells (16, 48). They cause apoptosis in susceptible tomato cells and mammalian cells by inhibiting ceramide biosynthesis (9, 41, 45). In the tomato pathotype of A. alternata-tomato interactions, a major factor in pathogenicity is the production of host-specific AAL toxins capable of inducing cell death only in susceptible cultivars (3, 9, 48).
In this study, we describe evidence showing that the ability to produce the host-specific AAL toxin and to infect host tomato plants could potentially be distributed among a population of strains of the A. alternata tomato pathotype by horizontal transfer of an entire pathogenicity chromosome of the pathogen.
MATERIALS AND METHODS
Fungal strains.
The isolates used in this study are described in Table 1. All isolates were maintained on potato dextrose agar (Difco, Detroit, MI) slants or as 20% glycerol mycelial fragments at −80°C.
TABLE 1.
Strains of A. alternata used in this study
| Strain | Host | Origin |
|---|---|---|
| As-27 | Solanum lycopersicum | United States |
| VU2001 | S. lycopersicum | The Netherlands |
| H6 | S. lycopersicum | Greece |
| AL4 | S. lycopersicum | Mie, Japan |
| O-227 | S. lycopersicum | Mie, Japan |
| O-94 | Unknown | Tottori, Japan |
| O-276 | Pyrus pyrifolia var. culta | Tottori, Japan |
| AC320 | Citrus jambhiri | United States |
| AC325 | Citrus reticulata | United States |
| O-267 | Nicotiana tabacum | Unknown |
| M-30 | Fragaria ananassa | Tottori, Japan |
| M-71 | Malus pumila var. domestica | Nagano, Japan |
| IFO8984 | Malus pumila var. domestica | Hyogo, Japan |
Preparation of fungal chromosomes and PFGE.
Chromosomal DNA was prepared as described by Akamatsu et al. (5). Contour-clamped homogeneous electric field (CHEF) pulsed-field gel electrophoresis (PFGE) was used to separate intact chromosomes with a CHEF DR-III apparatus (Bio-Rad). The running conditions are described in the relevant figure legends and Table S1 in the supplemental material.
Chromosomal probes.
Chromosomal DNA of As-27 prepared as described above was separated by CHEF gel electrophoresis as follows. A 0.8% agarose gel was run at 6 V/cm with a switching interval of 120 to 180 s for 20 h. After ethidium bromide staining, the 1.0-Mb chromosome was excised from the gel and purified from the agarose using a GeneClean II Kit (Bio 101). Labeling was performed with a digoxigenin random-primed labeling reaction (Roche Diagnostics) to obtain a chromosome-specific probe.
DNA manipulation.
All PCR primers used in this study are shown in Table S2 in the supplemental material. The nucleotide sequences of representative clones were determined using a DNA-sequencing kit (Applied Biosystems) and an automated sequencing system (ABI Prism 310; Applied Biosystems). DNA was sequenced in both directions, and sequence construction was performed with Genetyx-Mac (Genetyx Corporation, Tokyo, Japan). Alignment phylogenetic analyses of nucleotide sequences were conducted using Genetyx-Mac. For DNA-fingerprinting analysis, probes were prepared from repetitive sequences (AAR1, AAR9, AAR27, and pAR274) derived from an A. alternata Japanese pear pathotype (T. Tsuge, personal communication), rDNA of A. alternata (Alt1), and a telomere sequence (TTAGGG)n. To digest chromosomal DNA with the rare-cutting enzyme NotI, agarose blocks containing chromosomal DNA were rinsed 10 times in wash buffer (20 mM Tris-HCl, 50 mM EDTA, pH 8.0), transferred to a new tube containing 1 ml of wash buffer, and incubated on ice for 30 min. The blocks were then incubated in NotI digestion buffer for 1 h at room temperature, followed by digestion of the chromosomal DNA with NotI overnight at 37°C.
Protoplast fusion.
Protoplasts of the geneticin-resistant tomato pathotype (27G-1) and the hygromycin B-resistant strawberry pathotype (TP1) of A. alternata were fused by electrofusion as described previously (2, 38, 39).
Nucleotide sequence accession numbers.
The nucleotide sequences used for phylogenetic analysis in this study have been deposited at DDBJ/EMBL/GenBank under accession numbers AB465630 to AB465687 and AB468132 to AB468151.
RESULTS AND DISCUSSION
A CDC in the tomato pathotype strains.
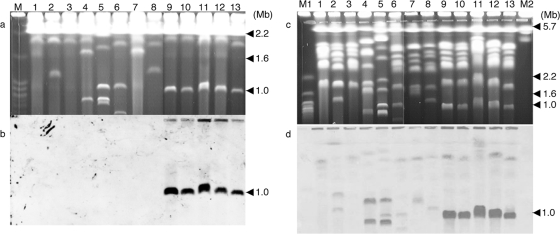
The chromosomal location of the polyketide synthetase (PKS) gene ALT1, which is involved in AAL toxin biosynthesis, virulence, and pathogenicity of the tomato pathotype (4, 48), was examined by PFGE for five tomato pathotype isolates (AAL toxin producers) and eight other strains of A. alternata (non-AAL toxin producers) collected from distant geographic areas (Japan, the United States, The Netherlands, Greece, etc. [Table 1]). The non-AAL toxin producers included nonpathogenic A. alternata and several pathotypes of A. alternata, such as the Japanese pear apple and the tomato pathotypes, producing AK, AM, and AAL toxins, respectively (26). A. alternata has about 10 chromosomes and a total genome size of approximately 30 Mb (5). All isolates of the tomato pathotype had a 1.0-Mb chromosome, and the ALT1 gene was located on the 1.0-Mb chromosome specifically detected in the tomato pathotype (Fig. 1a and b). The DNA of the 1.0-Mb chromosome present in the tomato pathotype As-27 was recovered and used as a probe (CDC probe) for hybridization to the gel blot. The CDC probe hybridized strongly to 1.0-Mb chromosomes of all tomato pathotype isolates, but not to ECs of the tomato pathotype (Fig. 1c and d). The weak signal obtained for ECs in almost all isolates was probably due to hybridization of the probe to rDNA, given that rDNA is a highly multicopy gene and it is difficult to remove the trace amounts of contaminating rDNA from the probe DNA. The chromosomes demonstrating weak signal corresponded to those harboring rDNA sequences (5). The nature of the 1.0-Mb chromosome is in agreement with the criteria determined for supernumerary chromosomes (10).
FIG. 1.
Electrophoretic karyotypes of the tomato pathotype of A. alternata and other Alternaria strains. Chromosome-size DNA was separated by PFGE under conditions for <2.0 Mb DNA (a and b) and 1.0 to 6.0 Mb DNA (c and d). The blots were probed with ALT1 (b) and with the 1.0-Mb chromosomal DNA of As-27 (CDC probe). (a and b) Chromosome-size DNA was separated in a 0.8% agarose (pulsed-field certified agarose; Bio-Rad) gel run at 5.5 V/cm with pulse intervals of 120 s for 12 h and 180 s for 12 h. Lanes: M, Saccharomyces cerevisiae; 1, nonpathogenic A. alternata O-94; 2, A. alternata Japanese pear pathotype O-276; 3, A. alternata tangerine pathotype AC320; 4, A. alternata rough lemon pathotype AC325; 5, A. alternata tobacco pathotype O-267; 6, A. alternata strawberry pathotype M-30; 7, A. alternata apple pathotype M-71; 8, A. alternata apple pathotype IFO8984; 9, A. alternata tomato pathotype As-27; 10, A. alternata tomato pathotype VU2001; 11, A. alternata tomato pathotype H6; 12, A. alternata tomato pathotype AL4; 13, A. alternata tomato pathotype O-227. (c and d) Chromosome-size DNA was separated in a 0.8% agarose (pulsed-field certified agarose; Bio-Rad) gel run at 1.5 V/cm for 115 h with pulse intervals of 3,600 to 1,800 s; for 24 h with 1,800 to 1,300 s; at 1.8 V/cm for 28 h with 1,300 to 800 s; and 2.4 V/cm for 28 h with 800 to 600 s. Lanes: M1, S. cerevisiae; 1, O-94; 2, O-276; 3, AC320; 4, AC325; 5, O-267; 6, M-30; 7, M-71; 8, IFO8984; 9, As-27; 10, VU2001; 11, H6; 12, AL4; 13, O-227; M2, Schizosaccharomyces pombe.
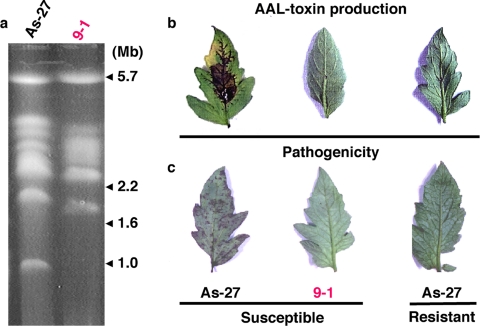
Transformation of the tomato pathotype of A. alternata As-27 by restriction enzyme-mediated integration generated mutants defective in AAL toxin production and pathogenicity (3). PFGE analysis of one of the mutants, 9-1, which showed the toxin- and pathogenicity-minus phenotype, revealed loss of the entire 1.0-Mb chromosome (Fig. 2). The colony growth and morphology of 9-1 and wild-type As-27 were compared and found to be indistinguishable (data not shown). Because this supernumerary chromosome could be entirely lost without affecting growth, we propose that it is a CDC.
FIG. 2.
Transformation-mediated loss of the 1.0-Mb CDC of the A. alternata tomato pathotype. (a) Electrophoretic karyotypes of the CDC-deficient mutant. As-27, the wild-type strain of the A. alternata tomato pathotype; 9-1, the mutant strain; (b) Leaf necrosis bioassay for AAL toxin production by the wild-type and mutant strains. Leaves of the susceptible cultivar Aichi-first and the resistant cultivar Ace were treated with culture filtrates of the strain at 25°C for 3 days. (c) Pathogenicity test of the wild-type and mutant strains. Leaves were inoculated with a spore suspension (105 spores/ml) and incubated in a moist chamber at 25°C for 3 days.
The CDCs in tomato pathotype strains from different geographical origins were identical, although the genetic backgrounds of the strains differed.
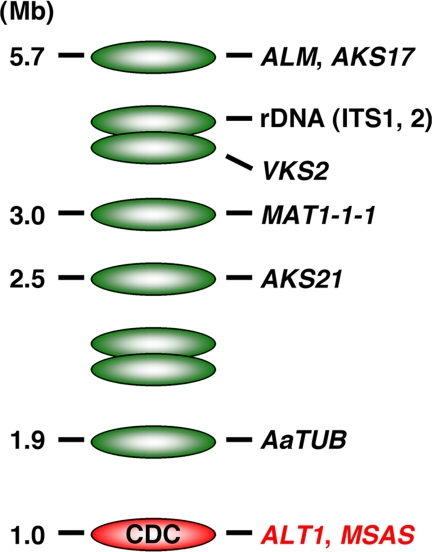
The nature of the CDC in the tomato pathotype strains collected worldwide was examined. As indicated in Fig. 1, all five of the tomato pathotype strains isolated in the United Sates, Europe, and Japan had the 1.0-Mb CDC including the ALT1 gene, whereas the banding pattern of the middle- to large-size ECs was different. With regard to the origin of the CDC of the tomato pathotype, we postulated either that those geographically diverse isolates of the tomato pathotype of A. alternata have common origins or that the isolates have different genetic backgrounds but harbor identical CDCs. To test these hypotheses, phylogenetic analysis of the genes on the CDC and the ECs was conducted for several strains. Two genes on the CDC, the Tox gene ALT1 and the MSAS-type PKS gene (8, 27) (another PKS gene located on the CDC not related to toxin production), were used as representative CDC genes. In addition, seven genes on the different ECs, including β-tubulin (AaTUB), a melanin biosynthesis PKS gene (ALM), mating-type genes (MAT1-1-1 and MAT1-2-1) (6), PKS genes with unknown functions (AKS17, AKS21, and VKS2), and rDNA internal transcribed spacers 1 and 2 (ITS1 and ITS2) (28) (Fig. 3), were sequenced for phylogenetic analysis.
FIG. 3.
Diagram of electrophoretically separated A. alternata chromosomes and the distribution of marker genes on each chromosome. ECs and the 1.0-Mb CDC are indicated by green and red shading, respectively.
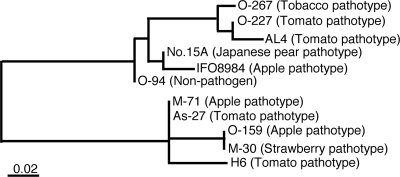
The sequences of both CDC genes were identical among five A. alternata tomato pathotype strains having different geographical origins. With regard to the ALT1 gene fragment, a sequenced 689-bp fragment including about 80 bp of intron was identical among all five strains. On the other hand, the EC genes, excluding ITS1, demonstrated polymorphisms among the five strains (Table 2). Among those genes, we found that the mating-type gene MAT1-1-1 showed high variation among A. alternata strains. The highly resolved phylogeny of the MAT gene is shown in Fig. 4. The tomato pathotype strains did not fall into a distinct group. Moreover, the VU2001 strain had another mating-type ideomorph, MAT 1-2-1, that differed almost completely from the MAT1-1-1 gene. Other genes on the ECs showed the same results, indicating that these tomato isolates are independent strains with different genetic backgrounds and origins.
TABLE 2.
Sequence comparison of genes on the CDC and other chromosomes of A. alternataa
| Strain | ALT1 | AaMSAS | ALM | AKS17 | AKS21 | VKS2 | MAT1-1-1 | ITS1c | ITSc | AaTUB |
|---|---|---|---|---|---|---|---|---|---|---|
| As-27 | A | A | A | A | A | A | A | 1-2 | 2-2 | A |
| VU2001 | A | A | A | A | A | B | 1-2b | 1-2 | 2-2 | A |
| H6 | A | A | A | B | B | C | B | 1-2 | 2-2 | A |
| AL4 | A | A | B | C | D | C | C | 1-2 | 2-1 | B |
| O-227 | A | A | B | C | D | D | D | 1-2 | 2-1 | B |
| O-94 | C | D | F | E | E | 1-2 | 2-1 | B | ||
| O-276 | D | E | E | F | 1-2 | 1-2 | 2-2 | A | ||
| M-71 | A | A | C | A | A | 1-2 | 2-2 | C |
The same letters for each gene in the table indicate that the sequences are identical. A portion of the following genes were sequenced (the lengths of the sequenced DNAs are indicated in parentheses): ALT1 (668 bp), PKS gene for AAL toxin biosynthesis; AaMSAS (474 bp), PKS gene for 6-methyl salicylic acid; ALM (432 bp), PKS gene for melanin biosynthetase; AKS17 (439 bp), PKS gene; AKS21 (474 bp), PKS gene; VKS2 (341 bp), PKS gene; MAT1-1-1 (576 bp), mating-type I gene; ITS1 (120 bp), ITS1; ITS2 (160 bp), ITS2; AaTUB (239 bp), β-tubulin gene.
1-2 indicates the strain has an alternative mating-type gene (MAT1-2-1), the sequence of which is completely different from that of MAT1-1-1.
The ITS1 and ITS2 sequences of A. alternata are divided into two groups.
FIG. 4.
Phylogenetic tree based on the MAT1-1-1 genes of the A. alternata strains. Sequence alignment and phylogenetic analysis were performed with Genetyx-Mac software. The name of each pathotype of A. alternata is indicated next to the strain names.
Telomere-fingerprinting analysis of A. alternata strains, including five tomato pathotype strains, revealed that the tomato pathotype strains had different genetic backgrounds, except for two Japanese strains (AL4 and O-227) (Fig. 5). The two Japanese strains seemed to be identical, indicating the common origin of these strains. DNA fingerprinting with repetitive sequences of A. alternata, Alt1, AAR1, AAR9, AAR27, and pAR274 (T. Tsuge, personal communication) also demonstrated the diverse genetic backgrounds of the tomato pathotype strains.
FIG. 5.
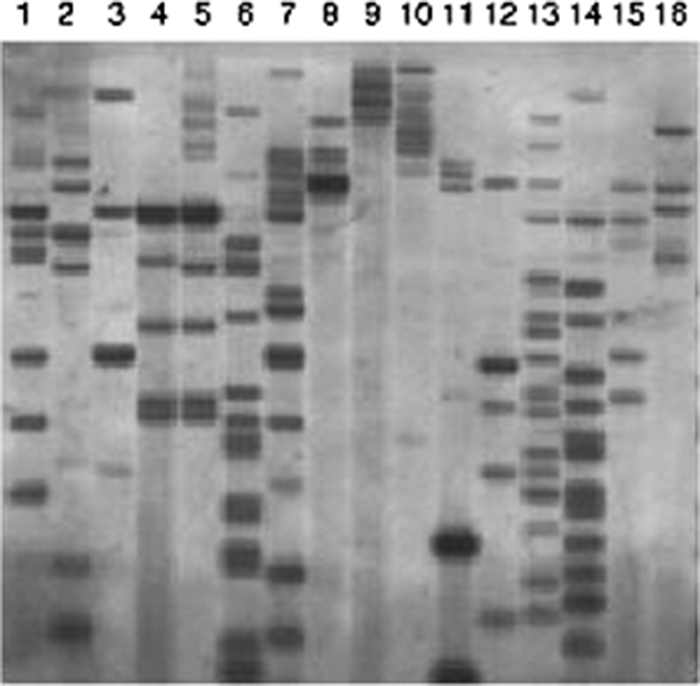
Telomere fingerprint of HindIII-digested DNA from the A. alternata tomato pathotype and other Alternaria strains. The blots were probed with a telomere repeat sequence (TTAGGG)n. Lanes: 1, As-27; 2, VU2001; 3, H6; 4, AL4; 5, O-227; 6, O-94; 7, O-276; 8, AC320; 9, AC325; 10, O-267; 11, M-30; 12, M-71; 13, IFO8984; 14, O-159; 15, FIV cont 45; 16, O-210.
Restriction fragment length polymorphism (RFLP) analysis using the rare-cutting enzyme NotI also indicated polymorphisms in the tomato pathotype strains (Fig. 6). On the other hand, a Southern blot of the RFLP gel probed with the CDC ALT1 gene revealed the same band (130 kb) in all tomato strains, indicating that at least a 130-kb region in the CDC of the tomato pathotype might be identical in all strains (Fig. 6b). The CDC probe prepared from the As-27 strain also resulted in identical banding patterns in all of the tomato pathotype strains, whereas a repetitive DNA probe, pAR274, showed polymorphisms (Fig. 6c and d), indicating the common structure of the CDC in these tomato pathotype strains.
FIG. 6.
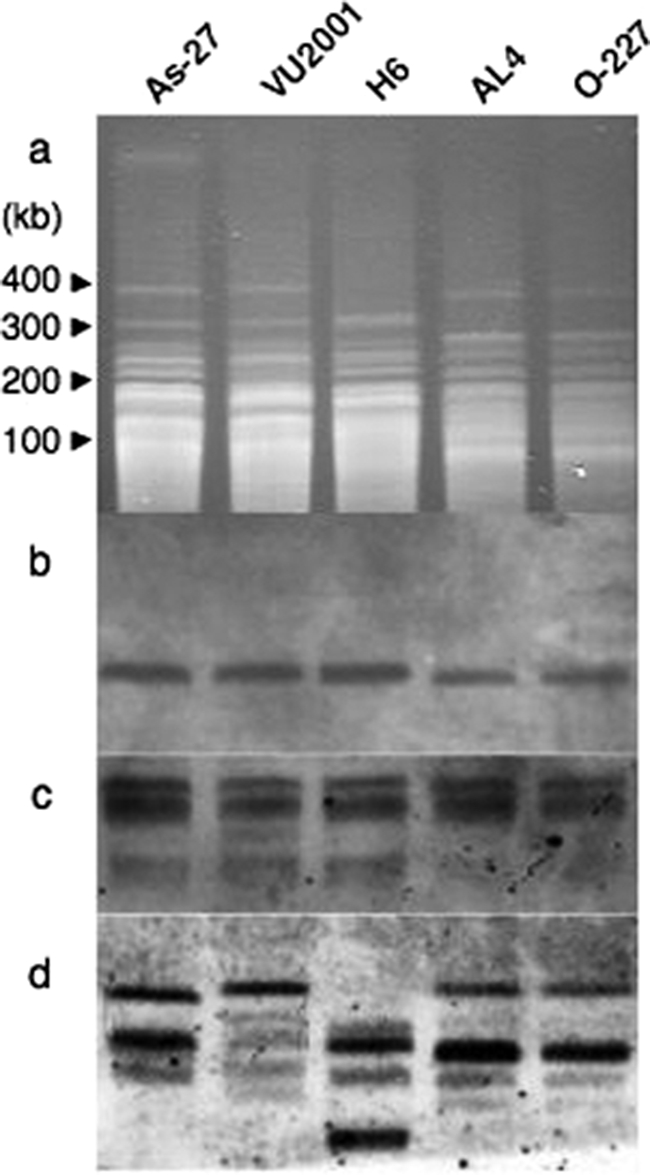
RFLPs of chromosomal DNA obtained from the A. alternata tomato pathotype. Chromosomal DNA samples were digested in agarose blocks with the rare-cutting enzyme NotI, electrophoresed (a), transferred to a membrane, and hybridized with the following probes: ALT1 (b), 1.0-Mb CDC DNA of As-27 (c), and repetitive DNA of A. alternata pAR274 (d).
In summary, the CDCs in tomato pathotype strains from different geographical origins were identical, although the genetic backgrounds of the strains differed. When these results are taken into account, the CDC appears to have an evolutionary history different from those of other ECs in the same genome, and horizontal transfer might be involved in the distribution of the CDC among tomato pathotype strains.
The CDC derived from the tomato pathotype strain could be maintained stably in a new genetic background with an expanded range of pathogenicity.
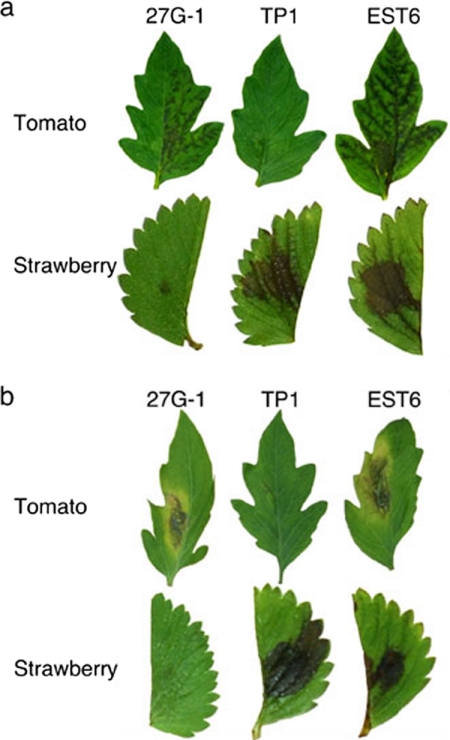
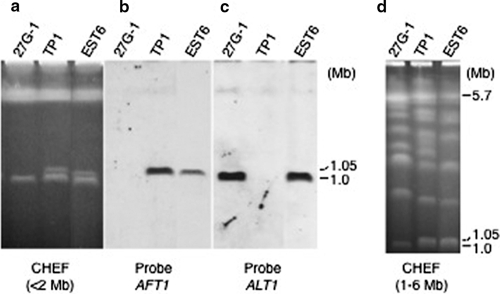
If this is the case, what are the mechanisms involved in CDC HGT? In Colletotricam, there is experimental evidence that a supernumerary chromosome can be transferred by coculturing two different strains (19, 31). In our work, although it is an artificial condition, a new pathogen with extended pathogenicity could be generated through protoplast fusion (2, 38, 39). A fusant (EST6) between the tomato (27G-1) and strawberry (TP1) pathotypes produced both AAL and AF toxins and was pathogenic to tomato and strawberry host plants (Fig. 7). By PFGE analysis, the parental tomato and strawberry strains were shown to harbor a 1.0- and a 1.05-Mb CDC, respectively. The hybrid strain, EST6, had CDCs derived from both parental strains (Fig. 8). Sequence analysis of the marker genes ALM, AKS17, VKS2, and MAT1-1-1 indicated that the genetic background of the fusant was the strawberry pathotype, in addition to the 1.0-Mb tomato CDC. The hybrid strain showed a stable phenotype after being subcultured on nonselective medium. Therefore, the CDC could be transmitted from one strain to another and was stably maintained in the new genome.
FIG. 7.
Production of AAL and AF toxins and pathogenicity of the fusion strain EST6. (a) Leaves of tomato cultivar Aichi-first and strawberry cultivar Morioka-16 were wounded slightly, treated with culture filtrates of the parental and fusion strains, and incubated in a moist chamber at 25°C for 3 days. (b) Leaves were inoculated with mycelial pieces of the strains and incubated in a moist chamber at 25°C for 3 days. 27G-1, Geneticin-resistant parental strain derived from the A. alternata tomato pathotype (As-27) harboring an npt gene; TP1, hygromycin B-resistant parental strain derived from the A. alternata strawberry pathotype (NAF8) harboring an hph gene; EST6, the fusion strain between 27G-1 and TP1.
FIG. 8.
Electrophoretic karyotypes of parental and hybrid strains of A. alternata. Chromosome-size DNA was separated by PFGE under conditions for <2.0 Mb DNA (a, b, and c) and for 1.0 to 6.0 Mb DNA (d). The blots were probed with AFT1 (b) from the strawberry pathotype and ALT1 (c) from the tomato pathotype. Chromosome-size DNA was separated in a 0.8% agarose gel run at 5.5 V/cm with pulse intervals of 120 s for 12 h and 180 s for 12 h (a, b, and c) and at 1.5 V/cm for 48 h (d) with pulse intervals of 1,500 to 3,000 s, at 1.8 V/cm for 40 h with 900 to 1,500 s, and 2.4 V/cm for 57 h with 480 to 900 s. 27G-1, Geneticin-resistant parental strain derived from the A. alternata tomato pathotype (As-27) harboring an npt gene; TP1, hygromycin B-resistant parental strain derived from the A. alternata strawberry pathotype (NAF8) harboring an hph gene; EST6, the fusion strain between 27G-1 and TP1.
In naturally occurring lesions on host plants in fields, coinfection is very common during infection with A. alternata pathogens (i.e., about 20% of single lesions contain two or more different strains, pathogens and nonpathogens) (1). Thus, coinfected lesions could be important for genetic interaction between different strains, because coinfected lesions enable different isolates to coexist and interact in a very small space.
A hypothesis whereby the ability to produce AAL toxin determining specific pathogenicity of the pathogen could be potentially distributed among A. alternata strains by horizontal transfer of an entire pathogenicity chromosome.
Based on these results, we propose the horizontal chromosome transfer hypothesis to explain the evolution and differentiation of HST-producing Alternaria pathogens. The supporting data are as follows. (i) The ALT1 gene resides on a 1.0-Mb CDC found only in the tomato pathotype of A. alternata, and no ALT1-homologous gene exists in nonpathogenic strains of A. alternata. (ii) Tomato pathotype strains with diverse geographical origins have different genetic backgrounds but identical CDCs. (iii) The CDC could be transferred between two different A. alternata strains and stably maintained in a recipient strain having a different genetic background.
The advantage and driving force of retaining genes for toxin biosynthesis, pathogenicity, and virulence on the CDC are not clear; however, this phenomenon might be related to unique features of A. alternata pathogens. First, A. alternata is basically an asexual (mitosporic) species that lacks sexual reproduction; hence, highly variable karyotypes are stably maintained. Extra chromosomes (CDCs) can survive through parasexual recombination because no homologous pair chromosomes exist. Second, the saprophytic life cycle dominates over the parasitic life cycle (26, 37), and the pathogenicity chromosome (CDC) is costly in the absence of host plants. Finally, during the parasitic stage, a nonpathogenic strain could acquire parasitic capacity by gaining a CDC with toxin-biosynthetic and pathogenicity genes via horizontal transfer. In the saprophytic stage, on the other hand, the strain could become competitive by losing nonessential genes through loss of the CDC, along with the toxin genes.
The CDC in the tomato pathotype of A. alternata is regarded as a “pathogenicity chromosome.” The horizontal chromosome transfer hypothesis, in which the ability to produce a pathogenicity or virulence effector (HST) is potentially distributed among A. alternata strains by horizontal transfer of the CDC, could provide a possible mechanism whereby new pathogens arise in nature. To test this hypothesis, a sequencing project of the entire CDC in the A. alternata tomato pathotype is currently in progress in our laboratory.
Supplementary Material
Acknowledgments
We are grateful to S.P Briggs for valuable suggestions and critical reading of the manuscript. We thank R. D. Johnson and L. Johnson for helpful discussions. We also thank R. P. Oliver for providing pAN7-1; T. Tsuge for supplying repetitive DNA clones derived from A. alternata; and M. Yamamoto, J. Hille, B. F. Brandwagt, T. L. Peever, and D. G. Gilchrist for providing Alternaria strains.
This work was supported by a Grant-in-Aid for Scientific Research (S) (19108001) from the Japanese Society for Promotion of Sciences and Global COE Program Advanced Utilization of Fungus/Mushroom Resources for Sustainable Society in Harmony with Nature), MEXT, Japan.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adachi, Y., and T. Tsuge. 1994. Coinfection by different isolates of Alternaria alternata in single black spot lesions of Japanese pear leaves. Phytopathology 84:447-451. [Google Scholar]
- 2.Akagi, Y., M. Taga, M. Yamamoto, T. Tsuge, Y. Fukumasa-Nakai, H. Otani, and M. Kodama. 2009. Chromosome constitution of hybrid strains constructed by protoplast fusion between the tomato and strawberry pathotypes of Alternaria alternata. J. Gen. Plant Pathol. 75:101-109. [Google Scholar]
- 3.Akamatsu, H., Y. Itoh, M. Kodama, H. Otani, and K. Kohmoto. 1997. AAL-toxin deficient mutants of Alternaria alternata tomato pathotype by restriction enzyme-mediated integration. Phytopathology 87:967-972. [DOI] [PubMed] [Google Scholar]
- 4.Akamatsu, H., H. Otani, and M. Kodama. 2003. Characterization of a gene cluster for host-specific AAL-toxin biosynthesis in the tomato pathotype of Alternaria alternata. Fungal Genet. Newsl. 50(Suppl.):355. [Google Scholar]
- 5.Akamatsu, H., M. Taga, M. Kodama, R. Johnson, H. Otani, and K. Kohmoto. 1999. Molecular karyotypes for Alternaria plant pathogens known to produce host-specific toxins. Curr. Genet. 35:647-656. [DOI] [PubMed] [Google Scholar]
- 6.Arie, T., I. Kaneko, T. Yoshida, M. Noguchi, Y. Nomura, and I. Yamaguchi. 2000. Mating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Mol. Plant-Microbe Interact. 13:1330-1339. [DOI] [PubMed] [Google Scholar]
- 7.Arnold, D. L., A. Pitman, and R. W. Jackson. 2003. Pathogenicity and other genomic islands in plant pathogenic bacteria. Mol. Plant Pathol. 4:407-420. [DOI] [PubMed] [Google Scholar]
- 8.Beck, J., S. Ripka, A. Siegner, E. Schiltz, and E. Schweizer. 1990. The multifunctional 6-methylsalicylic acid synthase gene of Penicillium patulum. Its gene structure relative to that of other polyketide synthases. Eur. J. Biochem. 192:487-498. [DOI] [PubMed] [Google Scholar]
- 9.Brandwagt, B. F., L. A. Mesbah, F. L. W. Takken, P. L. Laurent, T. J. A. Kneppers, J. Hille, and H. J. J. Nijkamp. 2000. A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc. Natl. Acad. Sci. USA 97:4961-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covert, S. F. 1998. Supernumerary chromosomes in filamentous fungi. Curr. Genet. 33:311-319. [DOI] [PubMed] [Google Scholar]
- 11.Covert, S. F., J. Enkerli, V. P. W. Miao, and H. D. VanEtten. 1996. A gene for maackiain detoxification from a dispensable chromosome of Nectria haematococca. Mol. Gen. Genet. 251:397-406. [DOI] [PubMed] [Google Scholar]
- 12.Enkerli, J., G. Bhatt, and S. F. Covert. 1997. Nht1, a transposable element cloned from a dispensable chromosome in Nectria haematococca. Mol. Plant-Microbe Interact. 10:742-749. [DOI] [PubMed] [Google Scholar]
- 13.Friesen, T. L., J. D. Faris, P. S. Solomon, and R. P. Oliver. 2008. Host-specific toxins: effectors of necrotrophic pathogenicity. Cell Microbiol. 10:1421-1428. [DOI] [PubMed] [Google Scholar]
- 14.Friesen, T. L., E. H. Stukenbrock, Z. Liu, S. Meinhardt, H. Ling, J. D. Faris, J. B. Rasmussen, P. S. Solomon, B. A. McDonald, and R. P. Oliver. 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38:953-956. [DOI] [PubMed] [Google Scholar]
- 15.Gilchrist, D. G., and R. G. Grogan. 1976. Production and nature of a host-specific toxin from Alternaria alternata f. sp. lycopersici. Phytopathology 66:165-171. [Google Scholar]
- 16.Gilchrist, D. G., H. Wang, and R. M. Bostock. 1995. Sphingosine-related mycotoxins in plant and animal disease. Can. J. Bot. 73:S459-S467. [Google Scholar]
- 17.Han, Y., X. Liu, U. Benny, H. C. Kistler, and H. D. Vanetten. 2001. Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J. 25:305-314. [DOI] [PubMed] [Google Scholar]
- 18.Hatta, R., K. Ito, Y. Hosaki, T. Tanaka, A. Tanaka, M. Yamamoto, K. Akimitsu, and T. Tsuge. 2002. A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 161:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, C., A. G. Rusu, A. M. Poplawski, J. A. G. Irwin, and J. M. Manners. 1998. Transfer of a supernumerary chromosome between vegetatively incompatible biotypes of the fungus Colletotrichum gloeosporioides. Genetics 150:1459-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howlett, B. J. 2006. Secondary metabolite toxins and nutrition of plant pathogenic fungi. Curr. Opin. Plant Biol. 9:371-375. [DOI] [PubMed] [Google Scholar]
- 21.Jain, R., M. C. Rivera, and J. A. Lake. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl. Acad. Sci. USA 96:3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, L. J., R. D. Johnson, H. Akamatsu, A. Salamiah, H. Otani, K. Kohmoto, and M. Kodama. 2001. Spontaneous loss of a conditionally dispensable chromosome from Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr. Genet. 40:65-72. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, R. D., L. Johnson, Y. Itoh, M. Kodama, H. Otani, and K. Kohmoto. 2000. Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM-toxin synthesis and pathogenicity. Mol. Plant-Microbe Interact. 13:742-753. [DOI] [PubMed] [Google Scholar]
- 24.Keller, N. P., G. Turner, and J. W. Bennett. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 25.Kistler, H. C., L. W. Meinhardt, and U. Benny. 1996. Mutants of Nectria haematococca created by a site-directed chromosome breakage are greatly reduced in virulence toward pea. Mol. Plant-Microbe Interact. 9:804-809. [Google Scholar]
- 26.Kohmoto, K., H. Otani, and T. Tsuge. 1995. Alternaria alternata pathogens, p. 3-22. In K. Kohmoto, U. S. Singh and R. P. Singh (ed.), Pathogenesis and host specificity in plant diseases: histopathological, biochemical, genetic and molecular bases, vol. II. Eukaryotes. Pergamon, Oxford, United Kingdom.
- 27.Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100:15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusaba, M., and T. Tsuge. 1995. Phylogeny of Alternaria fungi known to produce host-specific toxins on the basis of variation in internal transcribed spacers of ribosomal DNA. Curr. Genet. 28:491-498. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence, J. 1999. Selfish operons: the evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 9:642-648. [DOI] [PubMed] [Google Scholar]
- 30.Markham, J. E., and J. Hille. 2001. Host-selective toxins as agents of cell death in plant-fungus interactions. Mol. Plant Pathol. 2:229-239. [DOI] [PubMed] [Google Scholar]
- 31.Masel, A. M., C. He, A. M. Poplawski, J. A. G. Irwin, and J. M. Manners. 1996. Molecular evidence for chromosome transfer between biotypes of Colletotrichum gloeosporioides. Mol. Plant-Microbe Interact. 9:339-348. [Google Scholar]
- 32.Miao, V. P. W., S. F. Covert, and H. D. VanEtten. 1991. A fungal gene for antibiotics resistance on a dispensable (B) chromosome. Science 254:1773-1776. [DOI] [PubMed] [Google Scholar]
- 33.Nishimura, S., and K. Kohmoto. 1983. Host-specific toxins and chemical structure from Alternaria species. Annu. Rev. Phytopathol. 20:87-116. [DOI] [PubMed] [Google Scholar]
- 34.Oliver, R. P., and P. S. Solomon. 2008. Recent fungal diseases of crop plants: is lateral gene transfer a common theme? Mol. Plant-Microbe Interact. 21:287-293. [DOI] [PubMed] [Google Scholar]
- 35.Peever, T. L., G. Su, L. Carpenter-Boggs, and L. W. Timmer. 2004. Molecular systematics of citrus-associated Alternaria species. Mycologia 96:119-134. [PubMed] [Google Scholar]
- 36.Rosewich, U. L., and H. C. Kistler. 2000. Role of horizontal gene transfer in the evolution of fungi. Annu. Rev. Phytopathol. 38:325-363. [DOI] [PubMed] [Google Scholar]
- 37.Rotem, J. 1994. The genus Alternaria: biology, epidemiology, and pathogenicity. American Phytopathological Society, St. Paul, MN.
- 38.Salamiah, H. Akamatsu, Y. Fukumasa-Nakai, H. Otani, and M. Kodama. 2001. Construction and genetic analysis of hybrid strains between apple and tomato pathotypes of Alternaria alternata by protoplast fusion. J. Gen. Plant Pathol. 67:97-105. [Google Scholar]
- 39.Salamiah, Y. Fukumasa-Nakai, H. Akamatsu, H. Otani, K. Kohmoto, and M. Kodama. 2001. Genetic analysis of pathogenicity and host-specific toxin production of Alternaria alternata tomato pathotype by protoplast fusion. J. Gen. Plant Pathol. 67:7-14. [Google Scholar]
- 40.Sheffer, R. P., and R. S. Livingston. 1984. Host-selective toxins and their role in plant diseases. Science 223:17-21. [DOI] [PubMed] [Google Scholar]
- 41.Spassieva, S. D., J. E. Markham, and J. Hille. 2002. The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 32:561-572. [DOI] [PubMed] [Google Scholar]
- 42.Thomma, B. P. H. J. 2003. Alternaria spp.: from general saprophyte to specific parasite. Mol. Plant Pathol. 4:225-236. [DOI] [PubMed] [Google Scholar]
- 43.VanEtten, H., S. Jorgensen, J. Enkerli, and S. F. Covert. 1998. Inducing the loss of conditionally dispensable chromosomes in Nectria haematococca during vegetative growth. Curr. Genet. 33:299-303. [DOI] [PubMed] [Google Scholar]
- 44.Walton, J. D. 2000. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet. Biol. 30:167-171. [DOI] [PubMed] [Google Scholar]
- 45.Wang, H., J. Li, R. M. Bostock, and D. G. Gilchrist. 1996. Apoptosis: a functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8:375-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wasmann, C. C., and H. D. VanEtten. 1996. Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol. Plant-Microbe Interact. 9:793-803. [Google Scholar]
- 47.Wolpert, T. J., L. D. Dunkle, and L. M. Ciuffetti. 2002. Host-selective toxins and avirulence determinants: what's in a name? Annu. Rev. Phytopathol. 40:251-285. [DOI] [PubMed] [Google Scholar]
- 48.Yamagishi, D., H. Akamatsu, H. Otani, and M. Kodama. 2006. Pathological evaluation of host-specific AAL-toxins and fumonisin mycotoxins produced by Alternaria and Fusarium species. J. Gen. Plant Pathol. 72:323-327. [Google Scholar]
- 49.Yoder, O. C. 1980. Toxins in pathogenesis. Annu. Rev. Phytopathol. 18:103-129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.