Abstract
Saccharomyces cerevisiae produces extracellular glycerophosphoinositol through phospholipase-mediated turnover of phosphatidylinositol and transports glycerophosphoinositol into the cell upon nutrient limitation. A screening identified the RAS GTPase-activating proteins Ira1 and Ira2 as required for utilization of glycerophosphoinositol as the sole phosphate source, but the RAS/cyclic AMP pathway does not appear to be involved in the growth phenotype. Ira1 and Ira2 affect both the production and transport of glycerophosphoinositol.
Membrane phospholipids are continually synthesized and degraded as cells grow and respond to environmental conditions. A major pathway of phosphatidylinositol (PI) turnover in Saccharomyces cerevisiae is its deacylation to produce extracellular glycerophosphoinositol (GroPIns) (3). Plb3, an enzyme with phospholipase B (PLB)/lysophospholipase activity, is thought to be primarily responsible for the production of extracellular GroPIns, with Plb1 playing a lesser role (11, 12, 13). GroPIns is transported into the cell by the Git1 permease (17). GIT1 expression is upregulated by phosphate limitation and inositol limitation. In fact, GroPIns can act as the cell's sole source of both inositol (17) and phosphate (1).
A screening for gene products involved in the process by which GroPIns enters the cellular metabolism identified Ira1 and Ira2, yeast homologs of the mammalian protein neurofibromin. Alterations in NF1, the gene encoding neurofibromin, are associated with the pathogenesis of neurofibromatosis type 1, an autosomal dominant genetic disease (4, 5, 25). Ira1 and Ira2 and neurofibromin function as RAS GTPase-activating proteins (RAS GAPs). S. cerevisiae Ras1 and Ras2 activate adenylate cyclase to modulate cyclic AMP (cAMP) levels. The binding of cAMP to the regulatory subunits of protein kinase A (Bcy1) results in dissociation and activation of the catalytic subunits (Tpk1 to Tpk3). Ira1 and Ira2 inactivate RAS and thereby downregulate the pathway (18, 19). Hydrolysis of cAMP by the phosphodiesterases encoded by PDE1 and PDE2 also downregulate the pathway (7, 20, 23). The RAS/cAMP pathway responds to nutrient signals to modulate fundamental cellular processes, including stress resistance, metabolism, and cell proliferation (7, 20, 21).
Identification of IRA genes as affecting GroPIns metabolism.
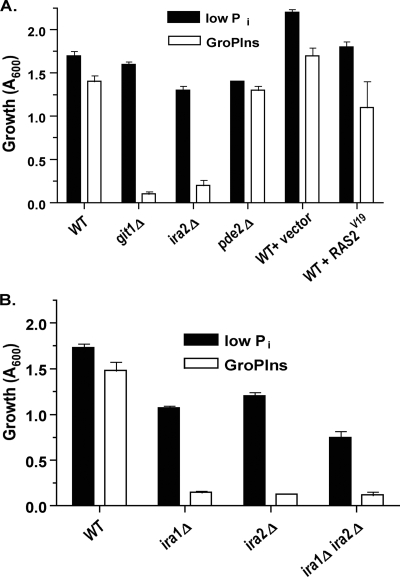
To identify genes involved in the metabolic process by which GroPIns is used as a phosphate source, we screened the MATα viable yeast knockout collection (Research Genetics) for strains displaying compromised growth when GroPIns rather than low Pi was supplied as the phosphate source. Synthetic complete medium, high-Pi (10 mM KH2PO4) medium, low-Pi (0.2 mM KH2PO4) medium, and GroPIns+ (70 μM) medium were made as described previously (2). Mutants were transferred by hand pinner from a master plate to a 96-well plate containing low-Pi medium and allowed to grow at 30°C for 3 days. From low-Pi medium, cell inocula were transferred to plates containing GroPIns+ medium. Growth was monitored at 595 nm after 4 days of incubation at 30°C. The ratio of absorbance in low-Pi medium to absorbance in GroPIns+ medium was determined. This screening was performed twice. Mutants with a value of 3 or greater for the ratio of absorbance in low-Pi medium to absorbance in GroPIns+ medium were subjected to a second screening by the monitoring of their growth in test tubes. As expected, the GIT1 gene was required for growth, and no other mutant displayed a complete growth abatement phenotype (Fig. 1). In particular, we did not identify a gene likely to encode a glycerophosphodiesterase responsible for GroPIns catabolism. A likely explanation for this result is that multiple gene products are involved in the mechanism(s) by which GroPIns enters cellular metabolism. However, a number of strains exhibited a slow-growth phenotype, one of which was the ira2Δ mutant (Fig. 1A). To confirm the validity of this result, we analyzed growth on GroPIns in strains of the unrelated Σ1278b background (gifts from M. Cardenas, Duke University), and found that not only the ira2Δ mutant but also the ira1Δ mutant and the ira1Δ ira2Δ double mutant (Table 1) exhibited greatly reduced growth on GroPIns (Fig. 1B). The ira1Δ mutant was not present in the deletion set screened initially.
FIG. 1.
Ira1 and Ira2 are required for optimal growth of cells using GroPIns as a phosphate source. Strains in the BY4742 (A) or Σ1278b (B) background were grown overnight in low-Pi medium. Cells were transferred to low-Pi or GroPIns+ medium (A600 = 0.005). Absorbance was recorded after 48 h of growth at 30°C. Wild-type cells transformed with empty vector pRS415 (vector) or plasmid pPHY453, containing the hyperactive RAS2 allele RAS2-V19 (RAS2V19), were also assayed (A). Readings are means ± standard errors of the means of results of at least two independent experiments.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Genotype | Source or reference |
|---|---|---|---|
| Σ1278b background | |||
| MLY41 | WTa | MATaura3-52 | M. Cardenas (26) |
| MLY187 | ras2Δ | MATaura3-52 ras2::kanMX | M. Cardenas (26) |
| MLY186 | ras1Δ | MATaura3-52 ras1::kanMX | M. Cardenas (26) |
| THY337 | ira1Δ | MATaura3-52 ira1::loxP-nat | M. Cardenas (26) |
| THY336 | ira2Δ | MATaura3-52 ira2::loxP-nat | M. Cardenas (26) |
| THY345 | ira1Δ ira2Δ | MATaura3-52 ira1::loxP-nat ira2::loxP-nat | M. Cardenas (26) |
| JPV469 | git1Δ | MATaura3-52 git1::kanMX | This study |
| BY4742 background | |||
| JPV203 | WT | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| JPV597 | pde2Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pde2::kanMX | Research Genetics |
| JPV212 | git1Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 git1::kanMX | Research Genetics |
| JPV574 | ira2Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ira2::kanMX | Research Genetics |
WT, wild type.
To probe the role of the RAS/cAMP in GroPIns metabolism, we analyzed other strains bearing alterations in the pathway. Interestingly, a pde2Δ mutant that, like the ira2Δ mutant, exhibits increased cAMP levels (15) was not defective for growth on GroPIns. Similarly, a strain bearing a plasmid-borne hyperactive allele of RAS2, RAS2-V19 (a gift from P. Herman, Ohio State University) (9, 10, 22), was not defective for growth on GroPIns (Fig. 1A). Thus, upregulation of the RAS/cAMP pathway may not be the primary reason for the growth defect seen in the absence of Ira1 or Ira2. Not surprisingly, ras1Δ and ras2Δ mutants, expected to downregulate the RAS/cAMP pathway, were not defective for growth on GroPIns (data not shown).
A trivial explanation for the inability of strains bearing mutations in the IRA genes to utilize GroPIns as a phosphate source is that the cells are less robust than wild-type cells and die before they are able to induce the metabolism required to utilize GroPIns. To test this, we compared the survival of the wild type and an ira1Δ ira2Δ mutant upon phosphate starvation and found no difference between the two strains (data not shown).
An ira1Δ ira2Δ mutant exhibits altered GroPIns transport activity.
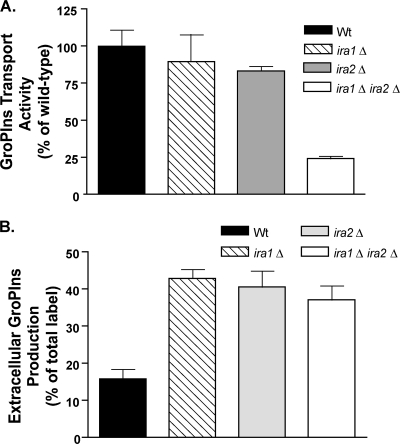
To determine the cause of the defect in GroPIns utilization that occurs upon deletion of IRA1 or IRA2, we measured GroPIns transport activity (1) under low-Pi conditions. In cells harboring single-deletion mutations, GroPIns transport was similar to that of the wild-type strain or only slightly reduced (Fig. 2A). However, the simultaneous deletion of IRA1 and IRA2 resulted in a clear reduction in activity. These results do not explain the growth defects seen in the single-deletion mutants, but they do indicate a role for Ira1 or Ira2 in regulating Git1. A complete understanding of the nature of the regulation of GroPIns transport by Ira1 or Ira2 will be the focus of future studies.
FIG. 2.
Ira1 and Ira2 affect GroPIns transport activity and GroPIns production. (A) Cells grown to log phase in low-Pi medium were assayed for GroPIns transport activity. Data represent means ± standard errors of the means of results of two independent experiments performed in duplicate. (B) Cells prelabeled with [3H]inositol were reinoculated into fresh high-Pi medium, and the amount of extracellular GroPIns was determined following 22 h of growth. Data are means ± standard errors of the means of results of at least three independent experiments and are reported as the percentage of total [3H]inositol label found as extracellular GroPIns normalized for culture density.
Deletion of IRA1 or IRA2 increases the extracellular accumulation of GroPIns.
We next monitored the effect of Ira1 or Ira2 upon extracellular production of GroPIns via PI deacylation. Cells were grown overnight in medium containing 3 μCi/ml of [2-3H]inositol. Cells were harvested, washed, and reinoculated to an A600 of 0.2 in high-Pi medium, the condition under which extracellular GroPIns accumulates (17). At various times, aliquots of the cultures were centrifuged, and the inositol compounds in the supernatant were separated (16). The ira1Δ, ira2Δ, and ira1Δ ira2Δ mutants accumulated roughly twice as much extracellular GroPIns as the wild-type strain (Fig. 2B) upon entrance to stationary phase. The lack of an additive effect upon GroPIns formation in the double mutant compared to formation in the single mutants may be explained by substrate availability. In other words, there may be a limited amount of plasma membrane-associated PI available for PLB-mediated turnover. Whereas an ira1Δ ira2Δ double mutant exhibits an increase in PLB1 and PLB3 transcript levels as measured by real-time reverse transcriptase PCR (data not shown), no such increase was detectable in the ira1Δ or ira2Δ mutant. Thus, Ira1 or Ira2 regulation of the PLB activity leading to GroPIns formation likely occurs at multiple levels and will be the focus of future studies. Another explanation is that yet another, uncharacterized, phospholipase exists for the formation of GroPIns in the absence of Ira1 or Ira2. Importantly, GroPIns transport does not occur under the growth conditions of the experiment (high Pi), so diminished transport cannot explain the increased accumulation of GroPIns in the absence of Ira1 or Ira2.
In summary, this study indicates a role for Ira1 and Ira2 in regulation of both the production of extracellular GroPIns and its transport into the cell. In addition, we show that Ira1 and Ira2 are required for the utilization of GroPIns as a phosphate source under conditions in which transport of GroPIns into the cell is normal. However, we were unable to show a linkage between hyperactivation of the RAS/cAMP pathway, which occurs upon deletion of IRA1 or IRA2, and the growth phenotype. Future studies will focus on determining the mechanism(s) for the growth disturbance, including whether it involves the RAS/cAMP pathway or other, less defined functions of Ira and Ira2. Growth is a multifactorial process, and a combination of disturbances, some involving heightened signaling through the RAS/cAMP pathway and some not, may contribute to alterations in the cell's ability to utilize GroPIns as a phosphate source. Ira1 and Ira2 are large proteins, each over 3,000 amino acids. The RAS GAP-related domain of each protein is contained within a few hundred amino acids, and C-terminal regions of approximately 200 amino acids bind to Gpb1 or Gpb2 and are important for protein stability (8). In addition, yeast Ira1 and Ira2 proteins and the human neurofibromin type 1 protein all contain bipartite phospholipid binding modules consisting of a Sec14 homologous segment and a pleckstrin homology-like domain (6). Both in vitro phospholipid binding and lipid exchange activity have been documented for neurofibromin (24), suggesting that Ira1 and Ira2 may have similar activities. Indeed, Ira1 appears to play a membrane-anchoring role for adenylate cyclase in addition to its role as a RAS GAP (14). It is tempting to speculate that Ira1 or Ira2 might also play a role in the membrane association or activation of an enzyme or enzymes responsible for the metabolism of internalized GroPIns. A test of that hypothesis awaits the identification of the gene products involved in the process by which GroPIns enters cellular metabolism.
Acknowledgments
This work was supported by National Institutes of Health grant GM59817 to J.P.-V.
We thank M. Cardenas for the gift of strains and P. Herman for the gift of plasmids. We thank Lindsay Chromiak and David Ekpin for technical assistance.
Footnotes
Published ahead of print on 28 August 2009.
REFERENCES
- 1.Almaguer, C., W. Cheng, C. Nolder, and J. Patton-Vogt. 2004. Glycerophosphoinositol, a novel phosphate source whose transport is regulated by multiple factors in Saccharomyces cerevisiae. J. Biol. Chem. 279:31937-31942. [DOI] [PubMed] [Google Scholar]
- 2.Almaguer, C., E. Fisher, and J. Patton-Vogt. 2006. Posttranscriptional regulation of Git1p, the glycerophosphoinositol/glycerophosphocholine transporter of Saccharomyces cerevisiae. Curr. Genet. 50:367-375. [DOI] [PubMed] [Google Scholar]
- 3.Angus, W. W., and R. L. Lester. 1975. The regulated catabolism of endogenous and exogenous phosphatidylinositol by Saccharomyces cerevisiae leading to extracellular glycerophosphorylinositol and inositol. J. Biol. Chem. 250:22-30. [PubMed] [Google Scholar]
- 4.Ballester, R., D. Marchuk, M. Boguski, A. Saulino, R. Letcher, M. Wigler, and F. Collins. 1990. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell 63:851-859. [DOI] [PubMed] [Google Scholar]
- 5.Buchberg, A. M., L. S. Cleveland, N. A. Jenkins, and N. G. Copeland. 1990. Sequence homology shared by neurofibromatosis type-1 gene and IRA-1 and IRA-2 negative regulators of the RAS cyclic AMP pathway. Nature 347:291-294. [DOI] [PubMed] [Google Scholar]
- 6.D'Angelo, I., S. Welti, F. Bonneau, and K. Scheffzek. 2006. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 7:174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dechant, R., and M. Peter. 2008. Nutrient signals driving cell growth. Curr. Opin. Cell Biol. 20:678-687. [DOI] [PubMed] [Google Scholar]
- 8.Harashima, T., S. Anderson, J. R. Yates III, and J. Heitman. 2006. The kelch proteins Gpb1 and Gpb2 inhibit Ras activity via association with the yeast RasGAP neurofibromin homologs Ira1 and Ira2. Mol. Cell 22:819-830. [DOI] [PubMed] [Google Scholar]
- 9.Howard, S. C., A. Hester, and P. K. Herman. 2003. The Ras/PKA signaling pathway may control RNA polymerase II elongation via the Spt4p/Spt5p complex in Saccharomyces cerevisiae. Genetics 165:1059-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kataoka, T., S. Powers, S. Cameron, O. Fasano, M. Goldfarb, J. Broach, and M. Wigler. 1985. Functional homology of mammalian and yeast RAS genes. Cell 40:19-26. [DOI] [PubMed] [Google Scholar]
- 11.Lee, K. S., J. L. Patton, M. Fido, L. K. Hines, S. D. Kohlwein, F. Paltauf, S. A. Henry, and D. E. Levin. 1994. The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269:19725-19730. [PubMed] [Google Scholar]
- 12.Merkel, O., M. Fido, J. A. Mayr, H. Pruger, F. Raab, G. Zandonella, S. D. Kohlwein, and F. Paltauf. 1999. Characterization and function in vivo of two novel phospholipases B/lysophospholipases from Saccharomyces cerevisiae. J. Biol. Chem. 274:28121-28127. [DOI] [PubMed] [Google Scholar]
- 13.Merkel, O., O. V. Oskolkova, F. Raab, R. El-Toukhy, and F. Paltauf. 2005. Regulation of activity in vitro and in vivo of three phospholipases B from Saccharomyces cerevisiae. Biochem. J. 387:489-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitts, M. R., J. Bradshaw-Rouse, and W. Heideman. 1991. Interactions between adenylate cyclase and the yeast GTPase-activating protein IRA1. Mol. Cell. Biol. 11:4591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park, J. I., C. M. Grant, and I. W. Dawes. 2005. The high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae is the major determinant of cAMP levels in stationary phase: involvement of different branches of the Ras-cyclic AMP pathway in stress responses. Biochem. Biophys. Res. Commun. 327:311-319. [DOI] [PubMed] [Google Scholar]
- 16.Patton, J. L., L. Pessoa-Brandao, and S. A. Henry. 1995. Production and reutilization of an extracellular phosphatidylinositol catabolite, glycerophosphoinositol, by Saccharomyces cerevisiae. J. Bacteriol. 177:3379-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patton-Vogt, J. L., and S. A. Henry. 1998. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka, K., B. K. Lin, D. R. Wood, and F. Tamanoi. 1991. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc. Natl. Acad. Sci. USA 88:468-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka, K., K. Matsumoto, and A. Toh-e. 1989. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:757-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 21.Thevelein, J. M., R. Gelade, I. Holsbeeks, O. Lagatie, Y. Popova, F. Rolland, F. Stolz, S. Van de Velde, P. Van Dijck, P. Vandormael, A. Van Nuland, K. Van Roey, G. Van Zeebroeck, and B. Yan. 2005. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem. Soc. Trans. 33:253-256. [DOI] [PubMed] [Google Scholar]
- 22.Toda, T., I. Uno, T. Ishikawa, S. Powers, T. Kataoka, D. Broek, S. Cameron, J. Broach, K. Matsumoto, and M. Wigler. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27-36. [DOI] [PubMed] [Google Scholar]
- 23.Weeks, G., and G. B. Spiegelman. 2003. Roles played by Ras subfamily proteins in the cell and developmental biology of microorganisms. Cell. Signal. 15:901-909. [DOI] [PubMed] [Google Scholar]
- 24.Welti, S., S. Fraterman, I. D'Angelo, M. Wilm, and K. Scheffzek. 2007. The sec14 homology module of neurofibromin binds cellular glycerophospholipids: mass spectrometry and structure of a lipid complex. J. Mol. Biol. 366:551-562. [DOI] [PubMed] [Google Scholar]
- 25.Xu, G. F., B. Lin, K. Tanaka, D. Dunn, D. Wood, R. Gesteland, R. White, R. Weiss, and F. Tamanoi. 1990. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell 63:835-841. [DOI] [PubMed] [Google Scholar]
- 26.Zurita-Martinez, S. A., and M. E. Cardenas. 2005. Tor and cyclic AMP-protein Kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 4:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]