Abstract
Coordinated control of hyphal elongation and branching is essential for sustaining mycelial growth of filamentous fungi. In order to study the molecular machinery ensuring polarity control in the industrial fungus Aspergillus niger, we took advantage of the temperature-sensitive (ts) apical-branching ramosa-1 mutant. We show here that this strain serves as an excellent model system to study critical steps of polar growth control during mycelial development and report for the first time a transcriptomic fingerprint of apical branching for a filamentous fungus. This fingerprint indicates that several signal transduction pathways, including TORC2, phospholipid, calcium, and cell wall integrity signaling, concertedly act to control apical branching. We furthermore identified the genetic locus affected in the ramosa-1 mutant by complementation of the ts phenotype. Sequence analyses demonstrated that a single amino acid exchange in the RmsA protein is responsible for induced apical branching of the ramosa-1 mutant. Deletion experiments showed that the corresponding rmsA gene is essential for the growth of A. niger, and complementation analyses with Saccharomyces cerevisiae evidenced that RmsA serves as a functional equivalent of the TORC2 component Avo1p. TORC2 signaling is required for actin polarization and cell wall integrity in S. cerevisiae. Congruently, our microscopic investigations showed that polarized actin organization and chitin deposition are disturbed in the ramosa-1 mutant. The integration of the transcriptomic, genetic, and phenotypic data obtained in this study allowed us to reconstruct a model for cellular events involved in apical branching.
The formation of complex structures of multicellular organisms is a pivotal question in biology. Breaking cell symmetry has been recognized as the first step in patterning an organism. This step from apolar to polar growth involves the perception and transmission of environmental and/or internal signals and results in the formation of a cellular axis. Establishing and maintaining cell polarity are thus fundamental prerequisites for the morphogenesis of organisms. Examples for a polarized mode of cell growth can be found in yeast (budding), filamentous fungi (hyphal tip growth), algae (rhizoids), plants (root hairs and pollen tubes), and animals (neurons). As tip-growing hyphal cells provide examples of highly polarized growth, filamentous fungi are attractive eukaryotic model systems to study the mechanisms underlying this process.
In addition, filamentous fungi such as Aspergillus niger are also used as cell factories for the production of chemicals, pharmaceuticals, and proteins. During the last years, however, it has become clear that the morphology of filamentous fungi seriously limits the product yields obtained (37, 64, 72). Previous studies suggested a link between protein production and the abundance of actively growing hyphal tips (36, 107); however, only contradictory results have been reported so far. An increase in the number of hyphal tips has been reported to increase protein production and secretion in some cases (14, 106), whereas no correlation has been found in others (14). Thus, no generally accepted model exists so far that can be used as basis for rationally optimizing the morphology of filamentous fungi with respect to protein secretion and their rheological behavior in a bioreactor. In order to improve the morphological features of filamentous fungi in industrial processes, much more basic knowledge is required to obtain a deeper insight into the molecular networks regulating fungal morphology.
The formation of highly polarized hyphae is a defining attribute of filamentous growth. Various protein complexes (e.g., the polarisome, exocyst, and Arp2/3 complex), cytoskeletal elements, the Spitzenkörper, lipid rafts, and signaling molecules (GTPases, calcium, and cyclic AMP) are essential for establishing and maintaining polarized growth (for reviews, see references 8, 43, 44, 92, and 102). Basically, it is thought that secretory vesicles delivering proteins and cell wall material to the hyphal apex are transported along microtubules to the Spitzenkörper (8). The Spitzenkörper is a vesicle-rich region present in the apexes of fungal hyphae and defines the center and direction of growth (34, 35, 39, 108). The Spitzenkörper is thought to be newly formed at sites of spore germination and branch formation and is visible only in rapidly growing hyphal tips. The Spitzenkörper consists of vesicles (chitosomes, calcium-containing vesicles, and other vesicles of unknown content), proteins (F-actin, tubulin, formins, and calmodulin), and ribosomes and is viewed as a switching station from microtubule-based to actin microfilament-based vesicle transport (8-10, 20, 39, 45, 80, 92, 97, 103). Actin microfilaments focus in the center of the Spitzenkörper and organize vesicle transport to the plasma membrane (8, 19). The polarisome is a multiprotein complex adjacent to the Spitzenkörper and is thought to play a key role in the nucleation of actin microfilaments and in governing maximal polar growth rate (44, 55, 66, 102, 111). In vivo studies with strains of Neurospora crassa showed that three different chitin synthases concentrate in the center of the Spitzenkörper (80). Besides these known fungal polarity determinants, a kinase complex which is conserved throughout eukaryotic evolution mediates spatial control of cell growth by regulating the actin cytoskeleton. This complex, named TORC2, has been shown to be essential for the determination of cell polarity in Saccharomyces cerevisiae, Dictyostelium discoideum, and mammalian cells (47). However, such a role of TORC2 has not yet been established in any filamentous fungus.
Hyphal branching leads to mycelial development. Usually, branches arise from basal regions (lateral branching); however, new branches can also be formed by tip splitting (apical branching). The physiological details of the process of apical branching have been studied in vegetative hyphae of A. niger, using the temperature-sensitive hyperbranching ramosa-1 mutant (77, 79). Four short-term events were identified: (i) cytoplasmic contraction thought to be triggered by a transient alteration of the cytoskeleton, (ii) retraction of the Spitzenkörper, (iii) disappearance of the Spitzenkörper accompanied by a sharp reduction in the hyphal elongation rate, and (iv) de novo formation of two Spitzenkörper giving rise to two apical branches. Similar events were also observed in other fungi, including wild-type A. niger, Neurospora crassa, and Trichoderma atroviride (78), suggesting a common mechanism for the generation of apical branches in filamentous fungi. It is believed that apical branching results from abnormal accumulation of vesicles at the tip and/or from increased tip-directed transport of vesicles, which exceeds the capacity of the leading tip. To accommodate the abnormal accumulation of the vesicles, the tip divides into two new branches (42).
Only little is known about the molecular basis of apical branching in filamentous fungi. In N. crassa, the act1 mutant, in which actin is positioned subapically instead of apically, displays increased tip splitting (101). Furthermore, the spray mutant (with altered intracellular calcium distribution) and the frost mutant (with disturbed manganese homeostasis) showed excessive apical-branching phenotypes (15, 90). In a large-scale genetic screen for morphological mutants of N. crassa, more proteins whose mutations led to hyphal tip splitting were identified (87), e.g., Ypk1, an ortholog of the yeast and nematode TORC2 effector proteins Ypk1p and SGK-1, and Cdc24, an activator protein of the GTPase Cdc42 (49, 50, 87). A requirement for Cdc42 in tip splitting was also supported by observations of the filamentous yeast Ashbya gossypii (85). Other proteins reported to be specifically required for apical branching in A. gossypii are the protein kinase AgCla4p, the paxillin-like protein AgPxl1p, and the polarisome components AgSpa2p and AgBni1p (5, 55, 56, 85). Congruently, mutations in the AgBni1p ortholog SepA evoked increased apical branching in Aspergillus nidulans (89).
In order to understand the molecular basis for apical branching in A. niger, we have identified and characterized the genetic locus affected in the ramosa-1 mutant. Here, we report that a single point mutation in the rmsA gene (An02g04280) is responsible for the mutant phenotype of the ramosa-1 mutant. The RmsA protein is homologous to the Avo1p/Sin1 protein, which is conserved from yeast to humans and, as a component of the TORC2 complex, is involved in regulating actin cytoskeleton polarity (104, 110). We show that RmsA serves as a functional equivalent of Avo1p and has a pivotal role in polarity maintenance in A. niger. We furthermore present for the first time a transcriptomic fingerprint of apical branching, which enabled us to obtain valuable mechanistic insights into the signaling machinery controlling morphogenesis of A. niger.
MATERIALS AND METHODS
Strains, culture conditions, and molecular techniques.
The A. niger and S. cerevisiae strains used in this study are listed in Table 1. Escherichia coli strain XL1-Blue served as the host for all plasmid work. General cloning procedures in E. coli were done as described by Sambrook and Russel (82). A. niger strains were cultivated in minimal medium (13) containing 1% glucose as a carbon source or in complete medium, consisting of minimal medium supplemented with 1% yeast extract and 0.5% Casamino Acids; 10 mM uridine was added when required.
TABLE 1.
Strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| A. niger | ||
| N402 | Wild type | Lab collection |
| AB4.1 | pyrG− | 98 |
| MA70.15 | ΔkusA pyrG− | 65 |
| T312 | Wild type | 77 |
| ramosa-1 mutant | Mutant of T312 obtained after UV mutagenesis | 77 |
| 14, 17 | Heterokaryotic strain, ΔrmsA | This work |
| S. cerevisiae | ||
| 26474 | Mata/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 YOL078w/Yol078w::Kanr | Research Genetics |
| ARY1 | 26474 containing pGal1-RmsA | This work |
| ARY2 | 26474 containing pGal1-Avo1 | This work |
| ARY1.1B | Segregant from ARY1, ΔYol078w, pGal-RmsA | This work |
| ARY2.4B | Segregant from ARY2, ΔYol078w, pGal-Avo1 | This work |
| ARY2.4C | Segregant from ARY2, pGal-Avo1 | This work |
Fermentation medium (FM) is composed of 0.75% glucose, 0.45% NH4Cl, 0.15% KH2PO4, 0.05% KCl, 0.05% MgSO4, 0.1% salt solution (13), and 0.003% yeast extract. The pH of FM was adjusted to 3. S. cerevisiae strains were cultivated in yeast extract-peptone (YP) medium containing either 1% glucose or 1% galactose as a carbon source. Transformation of A. niger strains was conducted as described earlier (75). S. cerevisiae genetic methods as described by Guthrie and Fink (40) were used.
Bioreactor cultivation.
Freshly harvested conidia (5 × 109) from strain T312 and the ramosa-1 mutant were used to inoculate 5 liters of FM. Cultivations were performed in a BioFlo3000 bioreactor (New Brunswick Scientific), where the temperature, pH (set to 3), and agitation speed were controlled online using the program NBS Biocommand. The cultivation program followed four consecutive phases: (i) 24°C, agitation speed of 250 rpm, and headspace aeration for 13 h; (ii) 37°C, agitation speed of 750 rpm, and sparger aeration for 4 h; (iii) 25°C, agitation speed of 750 rpm, and sparger aeration for 3 h; and (iv) 37°C, agitation speed of 750 rpm, and sparger aeration for 1 h. Mycelial samples were taken after certain time points for microarray and microscopic analyses (see below).
Identification and cloning of rmsA.
The rmsA gene was identified by complementation of the temperature-sensitive phenotype of the ramosa-1 mutant using the cosmid library pAOpyrGcosArp containing genomic DNA of wild-type A. niger (kindly provided by F. Schuren and P. Punt, TNO Nutrition). The complementing plasmid isolated was named pRamosa-13. Restriction analysis revealed that pRamosa-13 contained a 9-kb NcoI fragment as well as a 3.4-kb ClaI/NcoI fragment that were each fully capable of complementing the ramosa-1 mutant phenotype. The 9-kb NcoI fragment and the 3.4-kb ClaI/NcoI fragment were cloned into pUC21 (100), giving pRamosa-18 and pRamosa-19, respectively. Double-stranded DNA sequencing of pRamosa-18 revealed that the 9-kb insert harbored a 2,683-bp-long open reading frame (ORF) that was designated rmsA.
Deletion of rmsA.
To construct an rmsA deletion plasmid, pRamosa-18 was digested with NotI and KpnI to obtain a 6-kb fragment containing the 5′ region of the rmsA gene. A 0.7-kb fragment containing the 3′ flanking region of the rmsA gene was obtained using a XhoI/NotI double restriction of pRamosa-19. A 2.5-kb KpnI/SalI fragment containing the Aspergillus oryzae pyrG gene was obtained from pAO4-13 (29). Three-way ligation of these fragment resulted in the disruption plasmid pΔrmsA. Before transformation into A. niger strain AB4.1 or MA70.15, pΔrmsA was linearized using BglII. Disruption of the rmsA gene in A. niger was analyzed by Southern blot analysis. Genomic DNAs of putative ΔrmsA strains and a wild-type strain were isolated and digested with ClaI. As a probe, a 718-bp XhoI/NcoI rmsA fragment that was labeled by the random-priming method using [α-32P]dATP was used. Hybridizations were carried out at 65°C.
The heterokaryon rescue technique (71) was used to show that rmsA encodes an essential protein. Conidiospores from primary rmsA transformants in the MA70.15 (ΔkusA) background were analyzed for growth on selective medium (lacking uridine). Conidiospores from transformants that did not grow on selective medium (16 out of the 23 primary transformants) were considered potential heterokaryons, which was confirmed by Southern blot analysis. Southern blot analysis of transformants that produced viable conidiospores showed that they had the rmsA deletion construct integrated ectopically.
Complementation of ΔAVO1 in S. cerevisiae.
Plasmid pYES2 (Invitrogen) was used for the expression of AVO1 (Yol078w) and rmsA under the control of the inducible GAL1 promoter. The 3,531-bp fragment containing the AVO1 gene was amplified using primers pYol078P1 and pYol078P2 with S. cerevisiae genomic DNA as a template and cloned using SstI/XbaI restriction sites into pYES2, giving plasmid pGal-Avo1. The 2,535-bp fragment containing the rmsA cDNA was amplified using primers pRamosa23 and pRamosaP24 with genomic cDNA of A. niger (N402) (kindly provided P. Punt, TNO Quality of Life) as a template and cloned into pYES2 using SstI and XbaI, yielding plasmid pGal-RmsA. The AVO1/Yol078w heterozygote deletion strain in the BY4743 background was obtained from Research Genetics and transformed with pGal-Avo1 or pGal-RmsA using the URA3 selection marker. Transformants were grown selectively, induced for sporulation, and subjected to tetrad analysis as described previously (40).
Microarray and Northern analyses.
Culture broth (400 ml each) obtained from the above-described bioreactor cultivations were quickly harvested via filtration, and mycelial samples were immediately frozen using liquid nitrogen. Total RNA extraction, RNA quality control, labeling, Affymetrix microarray chip hybridization, scanning, and signal calculation (P, present; M, marginal; A, absent) were performed as previously described (67). Microarray analyses for T312 and the ramosa-1 mutant were performed on cells obtained from three independent bioreactor cultivations (biological triplicate).
Expression data were analyzed using the program GeneSpring 7.3. (Agilent Technologies). For normalization, default settings were used (50th percentile per chip, median per gene). Genes were defined as being differentially expressed if their expression levels varied at least 1.5-fold in the ramosa-1 samples compared to T312 and if the difference was statistically significant (Student's t test, P value cutoff of 0.05).
Northern analyses using 5 μg RNA from each strain were performed as described earlier (67). For hybridizations, PCR amplicons which were obtained by using the respective primer pairs as listed in Table S1 in the supplemental material were used. For 18S rRNA hybridization, a 2-kb BglII fragment from plasmid pMN1 (18) was used as probe.
Staining procedures and microscopy.
Actin immunostaining and calcofluor white (CFW) and DAPI (4′,6′-diamidino-2-phenylindole) staining of T312 and ramosa-1 germlings were performed as described previously (41, 66, 68). For actin immunostaining, the protocol used included two modifications as described earlier (68): lysing enzyme from Sigma was used for cell wall digestion, and the monoclonal antibody against actin was obtained from MP Biomedicals. For detection of intracellular reactive oxygen species (ROS), a protocol based on nitroblue tetrazolium (NBT) staining (51) was slightly modified as follows. Germlings of both strains were incubated in 50 mM sodium phosphate buffer containing 0.5 mg/ml NBT for 1 h, washed once each with 100% methanol and sterile water, and immediately subjected to microscopy. Microscopic pictures were captured using an Axioplan 2 instrument (Zeiss) equipped with a DKC-5000 digital camera (Sony). Both light (using differential interference contrast settings) and fluorescence (using green fluorescent protein or DAPI settings) images were captured with 40× or 100× objectives. Images were processed using Adobe Photoshop 6.0 (Adobe Systems Inc.).
Bioinformatics.
Responsive genes of the ramosa-1 mutant were functionally classified into FunCat categories as described previously (81). In order to perform gene set enrichment analysis (www.broadinstitute.org/gsea/) (93), expression values were computed using robust multiarray analysis (48). Gene sets based on FunCat categories were generated on a genome-wide scale, whereby the proportion of false positives was controlled by calculating the false discovery rate (< 0.05) according to the method of Benjamini and Hochberg (12). In silico analysis of putative transcription factor binding sites localized in the 1,000-bp upstream regions of A. niger genes was performed using an in-house-developed Perl script named the transcription factor binding site finder (TFBSF). The upstream regions were extracted for 13,750 out of 14,165 ORFs using the annotated genome sequence of A. niger strain CBS 513.88 (73). Upstream regions of 415 ORFs could not be identified due to contig borders or incorrectly determined ORFs (missing start codon). The upstream regions of the differentially expressed genes of the ramosa-1 mutant were searched for the presence of putative binding sites recognized by 25 known transcription factors from Aspergillus or Trichoderma species (see Table S3 in the supplemental material). Furthermore, these sequences were screened for the presence of common but so-far-undescribed putative binding motifs using the motif-based sequence analysis tool (MEME [6] at http://meme.nbcr.net/meme4_1/intro.html).
To determine significant over- or underrepresentation of binding sites, the background distribution of the identified motifs was determined via bootstrapping. For the up- and downregulated sets of genes, 500,000 bootstraps were performed separately. Out of the available 13,750 extracted upstream regions, random sets of equal size as the two sets of interest were selected and the presence of putative binding sites was determined. The average results for these bootstraps were interpreted as the background distribution. The probabilities of an over- or underrepresentation of putative binding sites of a transcription factor of equal or greater extent compared to the requested set was interpreted as the P value.
Microarray data accession number.
The microarray data have been deposited at GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE17641.
RESULTS
The apical-branching phenotype of the ramosa-1 mutant is reversible.
Previous studies have characterized the phenotype of the ramosa-1 mutant and its respective wild-type strain T312 using mature vegetative hyphae. Hyphae of the ramosa-1 mutant developed apical branches when subjected to a restrictive temperature (34 to 40°C), whereas at a permissive temperature (23°C), the ramosa-1 phenotype was similar to that of the wild-type strain (79). In this study, we questioned whether this observation is also valid for young germlings of the ramosa-1 mutant. In order to ensure controlled and equal growth conditions, we cultivated spores of both strains in a bioreactor using a defined temperature program (see below). As followed by the dissolved oxygen tension, equal growth behavior was observed for the ramosa-1 mutant and T312 for the first 17 h of cultivation (data not shown).
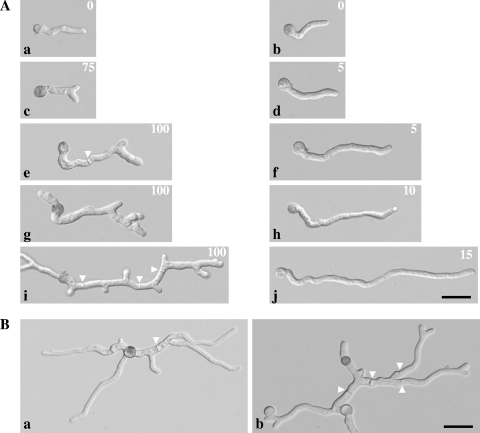
First, spores of both strains were inoculated at 24°C and allowed to germinate for 13 h. After initial swelling, the majority of the spores (>90%) of both strains formed unbranched germ tubes and displayed indistinguishable phenotypes (Fig. 1A, panels a and b). After this phase, the temperature was set to the restrictive temperature of 37°C for a period of 4 h. Within the first hour, already 75% of ramosa-1 germlings had started to develop apical branches, whereas germlings of T312 continued elongating with no branching (Fig. 1A, panels c and d). During further cultivation at 37°C, the majority of T312 germlings grew in an apical fashion, and lateral branches were only rarely observed (Fig. 1A, panels f, h, and j). In contrast, all germlings of the ramosa-1 mutant displayed apical branches, and newly formed tips branched again (usually one of the two apical tips branched apically), suggesting that the ability of ramosa-1 germlings to maintain stable polarity axes is limited to a short period, after which new polarity axes become established via apical branching. Cultivation at the restrictive temperature also produced increased septation and subapical branches next to the septa (Fig. 1A, panels e, g, and i). However, when the temperature was shifted back to the permissive temperature (24°C) and kept there for the next 3 h of cultivation, ramosa-1 germlings regained their ability to elongate without producing apical branches; i.e., they maintained stable polarity axes (Fig. 1B, panel a). This observation indicated that the morphogenetic program of the ramosa-1 mutant can be manipulated by simply altering the ambient growth temperature. Indeed, after an additional upshift to 37°C for 1 hour, new polarity axes became established (Fig. 1B, panel b). Thus, establishment and (re)maintenance of new polarity axes can be induced or repressed in ramosa-1 germlings.
FIG. 1.
Manipulation of polarized growth in the ramosa-1 mutant. (A) Conidia obtained from the ramosa-1 and T312 strains were cultivated under controlled conditions and subjected to differential interference contrast microscopy. Pictures for at least 100 germlings were taken for each time point and the percentage of branched germlings calculated (number at upper right). Young unbranched germlings are visible for both strains after 13 h of growth at 24°C (a and b). After a temperature upshift to 37°C for 1 h, 75% of ramosa-1 germlings show apical branches (c), compared to only 5% of T312 germlings (d). For the next 3 hours at 37°C, 100% of ramosa-1 germlings developed additional apical and subapical branches (e, g, and i) before growth ceased (i). In contrast, the majority of T312 germlings grew in a polar fashion (f, h, and j). (B) Polar growth of ramosa-1 germlings corresponding to panel i resumed after the temperature was downshifted to 24°C for 3 h (a). A subsequent temperature upshift to 37°C for 1 h resulted in the formation of new apical branches (b). Septa are indicated by arrowheads. Bars, 10 μm.
RmsA shows homology to Avo1p/Sin1 proteins.
To identify the genetic locus responsible for the mutant phenotype of the ramosa-1 mutant, a complementation approach was followed. When cultivated on solid medium at 37°C, the ramosa-1 mutant has a drastically reduced radial colony growth rate and forms small, very compact colonies lacking conidiophores and conidia (79). A cosmid library was transformed to the ramosa-1 mutant, and transformants were selected based on their ability to grow and conidiate at 37°C (data not shown). The isolation and subsequent sequencing of the complementing plasmid pRamosa-18 resulted in the identification of a single ORF, designated rmsA.
The 2,683-bp-long ORF is interrupted by two introns, the positions of which were confirmed by sequencing of the corresponding cDNA. As depicted in Fig. S1 in the supplemental material, the deduced 838-amino-acid sequence of RmsA shows a high degree of homology to the Avo1p/Sin1 protein family. In Saccharomyces cerevisiae, Avo1p (adheres voraciously to TOR2) has been shown to be an interactor with the TOR (target of rapamycin) protein complex (TORC2), which is necessary for the polarization of the actin cytoskeleton (61). The Schizosaccharomyces pombe rmsA ortholog Sin1 was originally reported to interact with the mitogen-activated protein (MAP) kinase StyI (105); however, most recently, it has also been described as an essential component of TORC2 (46).
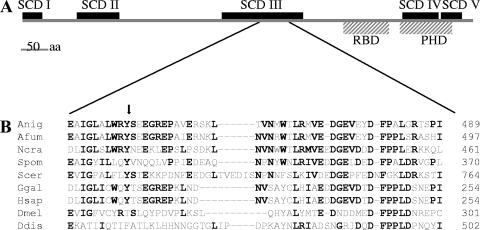
Avo1p/Sin1 proteins, in general, are highly conserved in metazoan species and fungi (104). Five regions with considerable identity (SCDs I to V) have been identified in Avo1p/Sin1 orthologous proteins (104) and are also present in the RmsA protein (SCD I, amino acids [aa] 1 to 39; SCD II, aa 104 to 180; SCD III, aa 369 to 517; SCD IV, aa 702 to 767; SCD V, aa 768 to 816) (Fig. 2A). Schroder et al. have additionally identified two putative domains within most Avo1p/Sin1 orthologs, namely, a Raf-like Ras binding domain and a pleckstrin homology domain (86), which were also identified in RmsA (Ras binding domain, aa 594 to 677; pleckstrin homology domain, aa 699 to 789) (Fig. 2A). Sequence comparison of the rmsA genes in T312 and the ramosa-1 mutant revealed a single point mutation, resulting in an exchange of a highly conserved tyrosine for asparagine at aa 447 (Fig. 2B). Remarkably, this mutation localized within region SCD III, which shows greatest conservation in metazoa and fungi (104).
FIG. 2.
Alignment of RmsA and its ortholog proteins. (A) Schematic representation of the domains present in RmsA. (B) A region within the SCD III domain showing highest homology between the orthologous proteins. Amino acids that are identical in at least four species are highlighted in bold. The tyrosine residue that has been exchanged in RmsA present in the ramosa-1 mutant is indicated by an arrow. Anig, Aspergillus niger RmsA (GenBank accession number An02g04280); Afum, Aspergillus fumigatus Sin1 (XP_755553); Ncra, Neurospora crassa Sin1 (XP_322410); Spom, Schizosaccharomyces pombe Sin1 (NP_59470); Scer, Saccharomyces cerevisiae Avo1 (NP_014563); Hsap, Homo sapiens MAPKAP1 (BC002326); Ggal, Gallus gallus Sin1 (AF153127); Ddis, Dictyostelium discoideum RipA (XP_638477); Dmel, Drosophila melanogaster (AAF58247).
Deletion of the rmsA gene is lethal.
To investigate the consequences of a loss of function of the rmsA gene in A. niger, a gene deletion vector (pΔrmsA) was constructed, in which an internal part of the rmsA ORF was replaced by the pyrG selection marker from A. oryzae. The wild-type strain AB4.1 (pyrG−) was repeatedly transformed with pΔrmsA; however, all attempts to detect any transformants deleted for rmsA failed, suggesting that the gene is, similarly to its S. cerevisiae counterpart AVO1, essential. This assumption was confirmed by using strain MA70.15 (ΔkusA, pyrG−) as the recipient strain. This strain has been shown to be valuable for the generation of balanced heterokaryons in primary transformants (65). Southern blot analysis of primary transformants obtained with strain MA70.15 proved the heterokaryotic nature of selected transformants; i.e., the hybridization patterns were congruent with the presence of the wild-type and rmsA-deleted genes, respectively (see Fig. S2 in the supplemental material). Conidia derived from these heterokaryotic transformants failed to grow under selective conditions (medium lacking uridine), supporting the conclusion that the rmsA gene is essential in A. niger. To test whether the rmsA deletion was essential at different temperatures or could be suppressed by high-osmolarity medium, spores from the heterokaryotic ΔrmsA 17 strain were plated at different growth temperatures (25°C, 32°C, and 37°C) or in high-osmolarity medium (1.2 M sorbitol or 0.6 M KCl). None of these conditions allowed growth on selective medium, indicating that these conditions could not rescue the rmsA deletion.
RmsA complements the AVO1 null phenotype of S. cerevisiae.
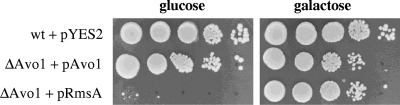
The RmsA ortholog Avo1p is an essential protein in S. cerevisiae (61). In order to get a first insight into the function of RmsA, a heterozygous AVO1::kanMX4/AVO1 S. cerevisiae strain was transformed with a plasmid that contained the rmsA wild-type gene under the control of the S. cerevisiae GAL1 promoter (plasmid pGal-RmsA). As controls, the recipient strain was transformed with the S. cerevisiae AVO1 gene under the control of the same promoter (plasmid pGal-Avo1) and with the empty plasmid (pYES2). Transformed diploid cells were allowed to sporulate, and haploid spores were dissected on galactose plates. The analysis confirmed that AVO1 is an essential gene, as only two viable spores per tetrad were obtained after dissecting avo1/AVO1 heterozygote stains containing the empty pYES2 plasmid. Dissection of the avo1/AVO1 diploid strain containing pGal-Avo1 resulted in four viable spores, and subsequent analysis showed that expression of AVO1 from the GAL1 promoter could rescue the avo1 deletion on both galactose and glucose media (Fig. 3). The rmsA gene was also fully competent in rescuing the loss of AVO1 function, indicating that RmsA is a functional homolog of Avo1p. However, complementation was obtained only on galactose medium, indicating that high levels of rmsA are required for full complementation. S. cerevisiae wild-type strains derived from dissecting the avo1/AVO1 heterozygote diploid strain containing the plasmid pGal-RmsA or pGal-Avo1, respectively, did not result in any altered phenotype compared to the wild-type situation (data not shown), suggesting that overexpression of Avo1p or RmsA is not detrimental to S. cerevisiae.
FIG. 3.
Complementation of AVO1 deletion with the rmsA gene in S. cerevisiae. Cell suspensions of S. cerevisiae strains were spotted onto solid YP-glucose or YP-galactose medium and inoculated for 3 days at 28°C. Cells were spotted in a 10-fold dilution series starting with ∼1 × 105 cells as the highest concentration. Note that the GAL1 promoter allows leaky expression under noninduced conditions (glucose). wt, wild type.
Mutation of RmsA results in actin and chitin depolarization.
Avo1p has been described to be essential for the maintenance of TORC2 integrity in S. cerevisiae (109). TORC2 is a protein complex that is required for actin polarization, thereby mediating spatial control of cell growth, as well as for positive regulation of the cell wall integrity (CWI) pathway via activation of Rom2p, a GDP/GTP exchange factor for Rho1p (28, 84). We assumed that if a similar complex existed in A. niger and had the same function, actin localization and cell wall organization would be disturbed in the ramosa-1 mutant when grown at 37°C due to a nonfunctional (or partially functional) RmsAY447N protein.
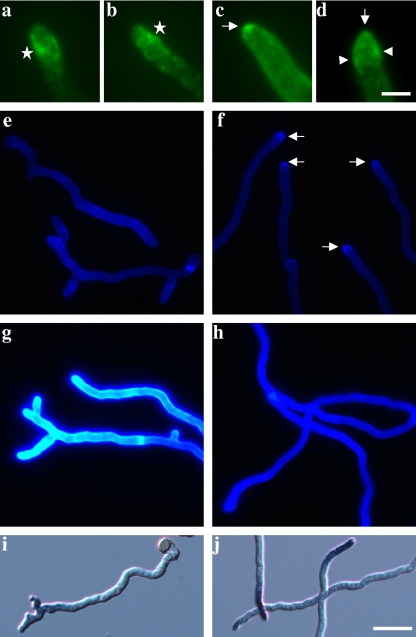
We thus visualized actin via immunofluorescence and chitin via CFW staining in ramosa-1 and T312 germlings shifted for 4 h to the restrictive temperature (similar to those in panels i and j in Fig. 1A). As depicted in Fig. 4, actin localization differed in ramosa-1 germlings compared to the wild-type situation. Actin patches were highly clustered at the apex, i.e., at the extreme tip, in 42 out of 49 hyphal tips analyzed from wild-type germlings (85.7%) (Fig. 4c). In seven hyphal tips (14.3%), we could observe a faint actin spot at the apex together with actin patches behind the apex (Fig. 4d). These patches presumably form a cortical actin ring corresponding to the actin collar most recently observed in mature hyphae of A. nidulans by time-lapse microscopy (94). In comparison, only 21 out of 68 hyphal tips (30.4%) from the ramosa-1 mutant displayed a considerable congregation of actin patches at the hyphal apex (similar to that shown in Fig. 4c), and in only three of these could we detect a basal actin collar (similar to that in Fig. 4d). In the majority of ramosa-1 tips analyzed (69.6%), however, actin patches were scattered randomly along the subapex (Fig. 4a and b), resembling the actin delocalization effect of the actin-depolymerizing agent cytochalasin A in A. nidulans (94) and the localization pattern of actin in the act1 mutant of N. crassa (101).
FIG. 4.
Phenotypic analysis of ramosa-1 and T312 germlings. Samples were taken after cultivation for 13 h at 24°C, followed by cultivation for 4 h at 37°C. Microscopic pictures were taken for at least 50 germlings from each strain, and representative pictures are shown. (a to d) Actin immunostaining of the ramosa-1 mutant (a and b) and T312 (c and d). Polarized actin localization is indicated by arrows, an actin collar by arrowheads, and depolarized actin patches by stars. (e to h) CFW staining of the ramosa-1 mutant (e and g) and T312 (f and h). Pictures for panels e and f were taken using an automatic exposure time, and pictures for panels g and h were taken using a 100-ms exposure time. Polarized chitin localization as visible in panel f is indicated by arrows. The increase in CFW fluorescence intensity in panel g suggests enhanced chitin levels at ramosa-1 cell walls. (i and j) Microscopic images of the ramosa-1 mutant (i) and T312 (j) stained with NBT. The presence of ROS is reflected by blue hyphal tips. Bars, 5 μm for panels a to d and 10 μm for panels e to j.
Actin polarization at the hyphal tip has been shown to be required for polarized chitin synthesis in A. nidulans (95). This observation might also hold true for A. niger, as chitin was found to be accumulated at the hyphal apex in wild-type germlings (Fig. 4f). This cap-like distribution, however, was not present in ramosa-1 germlings (Fig. 4e), suggesting that loss of actin polarization in the ramosa-1 mutant distorts chitin synthesis at the hyphal tip. Remarkably, the overall CFW fluorescence was found to be much stronger for ramosa-1 germlings than for T312 germlings (Fig. 4g and h), possibly hinting at a higher chitin abundance in the cell walls of the ramosa-1 mutant, which is in accordance with the previous finding that disappearance of the Spitzenkörper during apical branching is accompanied by considerable cell wall thickening (77).
No differences between ramosa-1 and wild-type germlings were observed when they were subjected to DAPI staining. The nuclear distributions, i.e., a regular spacing between nuclei, were similar in both strains (data not shown). Likewise, staining for ROS, described to be required for polarized growth of plant organs (22) and fungi (88), did not reveal any obvious discrepancies between the two strains. A tip-localized accumulation of ROS was found for the ramosa-1 mutant and T312 (Fig. 4i and j).
Gene expression profiling in the ramosa-1 mutant induced for apical branching.
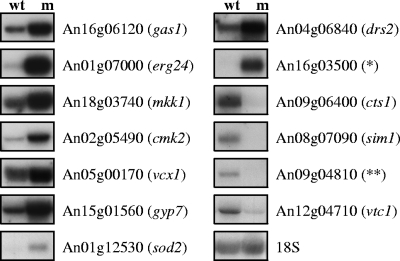
Microarray analyses were performed using RNA samples obtained from each three bioreactor runs (biological triplicate) of the ramosa-1 mutant and T312 (negative control). To analyze early events in apical branching, germlings (corresponding to panels c and d in Fig. 1A) were harvested 1 h after the temperature upshift to 37°C. Microarray data were processed as described in Materials and Methods, and genes showing at least 1.5-fold-higher or -lower expression in the ramosa-1 mutant were evaluated as being differentially expressed. Expression of 136 genes out of 14,165 A. niger genes was modulated, and 109 thereof displayed increased expression levels in the ramosa-1 strain. A comprehensive list of all differentially expressed genes is depicted in Table S2 in the supplemental material. Gene orthologs which have been shown to be important for polar growth in N. crassa (52, 87) are indicated there. In order to validate the changes in gene expression, Northern analyses for 13 selected genes were performed using the same RNA samples as utilized for the microarray analyses. As shown in Fig. 5, the Northern data are in good agreement with the microarray data; i.e., genes showing high or low levels of differential expression in the ramosa-1 mutant displayed comparable signals of intense or modest upregulation (or downregulation) in the Northern experiment.
FIG. 5.
Northern analysis of selected genes differentially expressed in the ramosa-1 mutant. Five micrograms of RNA was loaded onto each lane (wt, T312; m, ramosa-1 mutant) and hybridized with different probes as indicated. Control hybridization with 18S RNA confirmed equal loading. Gene names given in parentheses refer to the closest S. cerevisiae homolog (Table 3), *, unclassified protein selected from Table S2 in the supplemental material; **, predicted high-affinity glucose transporter (Table 3).
Functional classification of all responsive genes of the ramosa-1 mutant into FunCat categories (81) revealed that the category with the largest number of differentially expressed genes is that of genes involved in metabolism (Table 2). Apical branching of the ramosa-1 mutant has been reported to be accompanied by substantial reduction of the elongation rate of the parent hypha prior to appearance of apical branches (77). A possible explanation for this observation might be diminished ATP production as a result of reduced glucose uptake caused by decreased expression of An09g04810 (predicted glucose transporter) and a switch to anaerobic energy generation reflected by increased transcription of genes encoding a pyruvate decarboxylase (An02g06820) and a alcohol dehydrogenase (An02g02060) (Table 3).
TABLE 2.
Functional categories of genes up- or downregulated in the ramosa-1 mutant compared to wild-type strain T312a
| Functional category | No. of genes: |
|
|---|---|---|
| Upregulated | Downregulated | |
| Metabolism | 32 | 11 |
| Amino acid metabolism | 4 | 1 |
| Nitrogen and sulfur metabolism | 2 | 1 |
| Nucleotide metabolism | 1 | 1 |
| Phosphate metabolism | 1 | |
| C compound and carbohydrate metabolism | 18 | 7 |
| Lipid, fatty acid, and isoprenoid metabolism | 6 | 1 |
| Energy | 2 | |
| Cell cycle and DNA processing | 2 | 1 |
| Transcription | 2 | 1 |
| Protein synthesis | ||
| Protein fate | 7 | |
| Cellular transport and transport mechanism | 5 | 2 |
| Cellular communication | 1 | |
| Cell rescue, defense, and virulence | 3 | |
| Regulation of/interaction with cellular environment | 2 | |
| Subcellular localization | 2 | 1 |
| Transport facilitation | 1 | 1 |
| Unclassified proteins | 50 | 10 |
An annotated list of all responsive genes, including the fold change, P value, and classification, can be found in Table S2 in the supplemental material. Statistically enriched FunCat subcategories, determined by using the gene set enrichment analysis tool (93) (false discovery rate, <0.05), are also depicted in Table S2 in the supplemental material.
TABLE 3.
Selected responsive genes in the ramosa-1 mutant, ordered into different processes and functions
| Category and ORF | Genea | Fold changeb | P value | Predicted protein functionc | Closest S. cerevisiae homolog |
|---|---|---|---|---|---|
| Energy generation | |||||
| An02g06820 | 2.62 | 0.024 | Pyruvate decarboxylase | Pdc6 | |
| An02g02060 | 2.15 | 0.039 | Alcohol dehydrogenase | ||
| Amino acid metabolism | |||||
| An11g07960 | (9.74) | 0.025 | Glutaminase | ||
| An17g00910 | 6.03 | 0.004 | γ-Aminobutyrate transaminase | Uga1 | |
| An02g14590 | 3.15 | 0.029 | NAD+-dependent glutamate dehydrogenase | Gdh2 | |
| Oxidative stress-responsive proteins | |||||
| An09g01050 | (22.64) | 0.038 | PAFAH, removal of oxidized membrane phospholipids | ||
| An15g07670 | (10.66) | 0.055 | Tyrosinase involved in melanin synthesis | ||
| An01g06970 | 6.506 | 0.005 | d-Arabinose dehydrogenase | Ara1 | |
| An06g01610 | (3.97) | 0.029 | Heat shock protein | Hsp12 | |
| An15g01840 | 2.49 | 0.038 | Secoisolariciresinol dehydrogenase | ||
| An01g12530 | 2.39 | 0.055 | Manganese superoxide dismutase | Sod2 | |
| Cell wall synthesis and remodeling | |||||
| An16g06120 | gelF | (78.40) | 0.004 | Glycosylphosphatidylinositol-anchored β-1,3-glucanosyltransferase | Gas1 |
| An04g03830 | 18.18 | 0.006 | Glycosylphosphatidylinositol-anchored cell wall protein | ||
| An13g02510 | crhE | 7.49 | 0.003 | Glycosylphosphatidylinositol-anchored chitin transglycosidase | Crh1 |
| An11g06540 | mndA | (4.24) | 0.033 | β-Mannosidase | |
| An03g05560 | 3.74 | 0.008 | Spherulin 4-like cell surface protein | ||
| An18g03740 | mkkA | 3.72 | 0.025 | MAP kinase kinase involved in CWI pathway | Mkk1/2 |
| An16g08090 | dfgE | 3.24 | 0.043 | Glycosylphosphatidylinositol-anchored endo-mannanase | Dfg1 |
| An08g09610 | agnD | 3.17 | 0.016 | α-1,3-Glucanase | |
| An07g05570 | chsA | 2.39 | 0.034 | Chitin synthase class II, similar to ChsA of A. nidulans | Chs1 |
| An14g03910 | 2.26 | 0.052 | α-1,2-Mannosyltransferase | Kre2 | |
| An18g05910 | (2.25) | 0.041 | α-1,2-Mannosyltransferase | Kre2 | |
| An03g05530 | 2.18 | 0.051 | Endo-β-1,4-glucanase | ||
| An04g05550 | 2.61 | 0.037 | Mucin-like protein | Flo11 | |
| An02g11620 | 0.32 | 0.039 | Cell wall protein | ||
| An09g06400 | ctcA | 0.14 | 0.013 | Glycosylphosphatidylinositol-anchored chitinase, similar to ChiA of A. nidulans | Cts1 |
| Phospholipid signalingd | |||||
| An02g080503 | (12.36) | 0.056 | Phosphatidyl synthase, synthesis of phosphatidyl alcohols | ||
| An01g07000 | 10.75 | 0.051 | C14 sterol reductase, ergosterol synthesis | Erg24 | |
| An16g050206 | 7.34 | 0.002 | Inositol hexaki-/heptaki-phosphate kinase, synthesis of IP6/IP7 | Vip1 | |
| An18g064105 | 4.11 | 0.011 | Plasma membrane protein promoting PI4P synthesis | Sfk1 | |
| An15g070404 | 3.77 | 0.026 | Phospholipase D, synthesis of PA | Spo14 | |
| An04g038702 | (3.24) | 0.018 | PA phosphatase, synthesis of DAG | ||
| An11g053301 | 1.95 | 0.051 | Diacylglycerol pyrophosphate phosphatase, synthesis of DAG | Dpp1 | |
| Calcium signaling and homeostasis | |||||
| An02g05490 | 2.97 | 0.028 | Ca2+/calmodulin-dependent protein kinase | Cmk2 | |
| An16g03050 | 2.39 | 0.033 | Ca2+/calmodulin-dependent protein kinase | Cmk2 | |
| An02g06350 | (5.48) | 0.025 | Vacuolar Ca2+/H+ exchanger | Pmc1 | |
| An01g03100 | 4.49 | 0.009 | Vacuolar Ca2+/H+ exchanger | Vcx1 | |
| An05g00170 | 2.15 | 0.054 | Vacuolar Ca2+/H+ exchanger | Vcx1 | |
| Other signaling processes | |||||
| An15g01560 | (3.21) | 0.029 | GTPase-activating protein involved in protein trafficking | Gyp7 | |
| An04g01500 | 2.98 | 0.044 | Putative C2H2 zinc finger transcription factor | ||
| An08g07090 | 0.06 | 0.054 | SUN family protein involved in replication | Sim1 | |
| An16g07890 | 0.04 | 0.037 | Similar to A. nidulans transcription factor RosA | Ume6 | |
| An12g04710 | 0.29 | 0.031 | Negative regulator of Cdc42 | Vtc1 | |
| Transporters | |||||
| An02g04160 | 4.85 | 0.010 | Mitochondrial phosphate translocator | Mir1 | |
| An13g02320 | (4.70) | 0.027 | Vacuolar glutathione S-conjugate ABC transporter | Ycf1 | |
| An09g00930 | 3.44 | 0.055 | Na+/K+-exchanging ATPase alpha-1 chain | ||
| An02g01480 | 3.78 | 0.021 | MFS multidrug transporter | ||
| An16g06300 | 3.18 | 0.013 | Low-affinity Fe(II) transporter | Fet4 | |
| An04g06840 | 2.39 | 0.052 | Ca2+/phospholipid-transporting ATPase | Drs2 | |
| An14g01860 | 0.41 | 0.051 | Mitochondrial carrier protein | Rim2 | |
| An12g05510 | 0.38 | 0.024 | Siderophore-iron transporter | Taf1 | |
| An15g03900 | 0.32 | 0.049 | Vacuolar zinc transporter | Zrc1 | |
| An18g01220 | 0.29 | 0.059 | Allantoin permease | Dal5 | |
| An15g07460 | 0.26 | 0.023 | Oligopeptide transporter | ||
| An09g04810 | 0.06 | 0.011 | High-affinity glucose transporter |
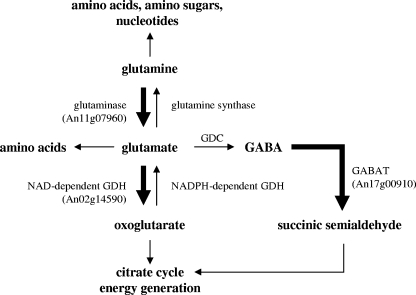
Within the category of metabolism, genes coding for proteins involved in glutamate metabolism (An11g07960 coding for glutaminase and An02g14590 coding for a NAD+-dependent glutamate dehydrogenase) were upregulated (Table 3 and Fig. 6). Most interestingly, deletion of the NADPH-dependent glutamate dehydrogenase results in reduced branching frequencies in A. nidulans and Penicillium chrysogenum (96). Glutamate is one of the main cellular precursors used for the synthesis of other amino acids, suggesting that amino acid starvation might be sensed in the ramosa-1 mutant. Alternatively, glutamate might have been used to fuel the citrate cycle or as a precursor of γ-aminobutyrate, whose further metabolization is important for counteracting oxidative stress in S. cerevisiae and Arabidopsis thaliana (25, 31). The key enzyme of the γ-aminobutyrate shunt, γ-aminobutyrate transaminase, showed enhanced expression in the ramosa-1 mutant (An17g00910 in Fig. 6), which could probably point toward increased ROS production in the ramosa-1 mutant. Supportive of this assumption is upregulation of other A. niger genes which show considerable sequence homology to oxidative stress-responsive and ROS-scavenging proteins from other eukaryotes, e.g., d-arabinose dehydrogenase (An01g06970), platelet-activating factor acetylhydrolase (An09g01050), and manganese superoxide dismutase (An01g12530) (1, 58, 99).
FIG. 6.
Pathways involved in glutamate metabolism. Genes upregulated in the ramosa-1 mutant are indicated with their ORF codes. The directions of the respective enzymatic reactions are indicated with thick arrows. GDH, glutamate dehydrogenase; GDC, glutamate decarboxylase; GABA, γ-aminobutyrate; GABAT, γ-aminobutyrate transaminase.
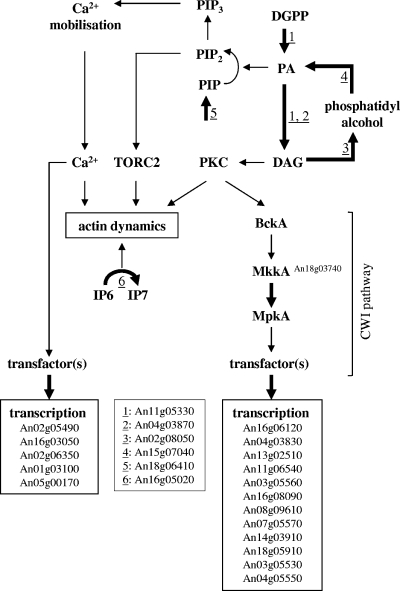
Finally, 13 genes in the category related to metabolism with predicted function in cell wall biosynthesis and integrity were significantly upregulated in the ramosa-1 mutant, whereas two genes were downregulated (Table 3 and Fig. 7), indicating that apical branching is accompanied by considerable cell wall reorganization. Congruently, the mkkA gene (An18g03740), encoding a MAP kinase kinase predicted to function in the CWI signaling pathway of A. niger, showed increased expression. To the group of upregulated cell wall genes belonged a chitin synthase gene (An07g05570), whose increased expression might explain the apparent enhanced chitin level in the ramosa-1 mutant as shown in Fig. 4g.
FIG. 7.
Genes upregulated in the ramosa-1 mutant and their allocation into the processes of cell wall synthesis, (phospho)lipid synthesis, and calcium homeostasis (indicated with thick arrows). For the predicted gene functions, see Table 3. For explanations of the connection of these processes in eukaryotes, see Discussion.
Six genes predicted to function in the synthesis of (phospho)lipid signaling molecules such as phosphatidate (PA) (An02g08050 and An15g07040; reactions 3 and 4 in Fig. 7), phosphatidylinositol-4′-monophosphate (PIP) (An18g06410, reaction 5 in Fig. 7), diacylglycerol (DAG) (An04g03870 and An11g05330, reactions 1 and 2 in Fig. 7), and inositolpyrophosphates (IP) (An16g05020, reaction 6 in Fig. 7) were upregulated in the ramosa-1 mutant (Table 3 and Fig. 7). These molecules play important roles in the regulation of actin polarization, CWI, and calcium signaling in lower and higher eukaryotes (see Discussion and Fig. 7), thus probably reflecting an important influence of phospholipid signaling in the process of apical branching.
Furthermore, genes encoding putative effector proteins of the calcium signaling machinery were upregulated, such as two Ca2+/calmodulin-dependent protein kinases (An02g05490 and An16g03050) and three vacuolar Ca2+ pumps (An02g06350, An01g03100, and An05g00170), hinting at the possibility that the mechanisms underlying apical branching might, beside CWI and phospholipid signaling, also involve the calcium signaling machinery (Fig. 7). In support of this notion is the most recent observation that a strong calcium spike accompanies apical branching in Fusarium oxysporum hyphae (54). Finally, 12 genes putatively encoding transport proteins for ions (Pi, Na+, K+, Fe2+, and Zn2+) and small molecules (phospholipids, amino acids, peptides, and glucose) displayed differential transcription in the ramosa-1 mutant (Table 3), suggesting that (i) apical branching might in general require ion homeostatic and metabolic control systems and/or (ii) the RmsA protein has a function not only in actin polarization but additionally in ion homeostasis and energy metabolism.
Promoter analysis of differentially expressed genes.
In order to unravel potential transcription factors involved in up- or downregulation of the 136 differentially expressed genes, we screened the 1,000-bp upstream regions of all genes for the presence of binding sites established for 25 transcription factors from different Aspergillus and Trichoderma species (using the in-house-developed TFBSF). We also used MEME (6) to screen for the presence of common but new DNA binding motifs. We determined the frequency of occurrence of these motifs in the group of 109 upregulated and 27 downregulated genes and evaluated whether these motifs are statistically significant over- or underrepresented compared to groups of the same size comprising randomly selected genes of A. niger (500,000 bootstrap samples, P < 0.05).
Using these in silico approaches, we were able to identify four motifs with the TFBSF tool and 27 motifs with the MEME tool as being enriched or underrepresented within the upstream regions of genes upregulated in the ramosa-1 mutant. As shown in detail in Table S3 in the supplemental material, the four enriched Aspergillus motifs identified with the TFBSF tool were also among the sites identified by MEME and are binding motifs of the transcription factors AbaA (asexual development and dimorphism) (2, 17, 57), CrzA (calcium signaling, cell wall formation, and polar growth) (33, 91), CreA (carbon catabolite repression and polar growth) (59, 112), and AmdR (amino acid utilization) (27), respectively. Among the set of MEME sites, we could additionally identify one site as a BrlA site (asexual development) (23) and three sites as cyclic AMP-responsive elements involved in protein kinase A signaling (metabolism, polar growth, and dimorphism) (11, 16, 38). Within the set of genes downregulated in the ramosa-1 mutant, three overrepresented sites were identified with the TFBSF tool which were also present among the 10 identified MEME sites (see Table S3 in the supplemental material): Seb1 (osmotic stress response) (74), AnCP/AnCF (CCAAT binding factor involved in, e.g., respiration) (21, 53), and BrlA (asexual development) (23). These data are in good agreement with the microarray and phenotypic data obtained which suggested a complex reorganization in the ramosa-1 mutant involving polar growth control, carbon and nitrogen metabolism, respiration, and calcium homeostasis.
For the majority of the known Aspergillus/Trichoderma transcription factor binding sites analyzed (15), we found a similar frequency of occurrence in genes differentially expressed in the ramosa-1 mutant compared to randomly selected genes (see Table S3 in the supplemental material). Although this does not necessarily preclude these trans factors from any role in the ramosa-1 mutant transcriptional response, it can be speculated that their involvement in regulating early events during apical branching is less important than the involvement of transcription factors showing enriched or underrepresented motifs. To our surprise, however, binding sites for the effector regulator of the CWI pathway RlmA are not significantly overrepresented in genes upregulated in the ramosa-1 mutant, suggesting that (i) RlmA does not have the same prominent function as its S. cerevisiae ortholog Rlm1p (i.e., not all A. niger cell wall-related genes are regulated by RlmA), (ii) CWI activates not only RlmA but also another as-yet-unknown transcription factor(s), or (iii) CWI signaling is not the only pathway responsible for the expression of cell wall-related genes.
DISCUSSION
The ramosa-1 mutant as a model system to study polarity control.
This study shows that the ramosa-1 strain can be considered an excellent model system to study the morphogenetic program of A. niger. All critical steps of hyphal morphogenesis (establishment, maintenance, and loss of maintenance of polar axes) can be altered within a single system by changing the cultivation temperature. Hence, different aspects of fungal polarity can be simultaneously studied in this strain. In addition, this model system can be used to address the question of whether an increase in the number of branches (per total cell mass) will indeed improve protein secretion in A. niger. Using the approach described in this study, the frequency of branching as well as the length of hyphal compartments can easily be adjusted by running defined temperature programs. Determination of the amount of secreted proteins and its relation to the amount of branches per hyphal compartment can be one systematic attempt to answer this question. Furthermore, the ramosa-1 mutant offers the possibility to determine the optimal number of branches per hyphal compartment in order to improve the rheological behavior of A. niger in industrial settings.
A proposed role for RmsA in polarity control.
Only a single point mutation within the rmsA gene is responsible for the mutant phenotype of the ramosa-1 mutant. The Y447N mutation of RmsA seems to have no apparent consequence for the growth of the ramosa-1 mutant when it is cultivated at low temperature (24°C), suggesting that RmsAY447N can still fulfill its cellular function. However, at higher temperature, Y447 seems to be essential for the function of RmsA, and its change to asparagine has profound consequences on the growth of A. niger, such as loss of polarity maintenance, slow growth, and defective asexual development (this work and reference 77). Several scenarios are imaginable to explain this temperature-dependent phenotype: (i) RmsAY447N is stable but lowered in its stability at 37°C and becomes readily degraded, (ii) an interaction of RmsAY447N with other proteins is possible at low temperature but disturbed at higher temperature, or (iii) RmsA is crucial for the survival of A. niger at high but not at low temperature. Our data seem to exclude the third possibility, as deletion of the rmsA gene is already lethal for A. niger when it is cultivated at 25°C.
The lethality of AVO1 deletion in S. cerevisiae can be complemented by rmsA, demonstrating that RmsA is a functional equivalent of Avo1p, a component of the S. cerevisiae TORC2 complex (61). Avo1p is essential for the maintenance of TORC2 integrity (109), a complex that is composed of six proteins, namely Avo1p, Avo2p, Avo3p/Tsc11p, Lst8p, Bit61p, and Tor2p (30, 61, 76). With the exception of Avo2p and Bit61p, sequences with obvious similarity to all S. cerevisiae TORC2 components can be found in the genome of A. niger (see Table S4 in the supplemental material). Respective sequences (except for Avo2p and Bit61p, which appear to be unique to S. cerevisiae) are also present in other eukaryotes ranging from S. pombe over Drosophila to mammals and have at least partially been shown to associate in a TORC2 complex (109). This high degree of evolutionary conservation makes it reasonable to predict that a TORC2-like complex exists in A. niger and that RmsA is a component of it.
TORC2 has an essential function for actin polarization and hence determination of cell polarity in S. cerevisiae, Dictyostelium discoideum, and mammalian cells but not in the fission yeast S. pombe (63, 109). In this study, we have observed a depolarized actin localization at the hyphal tip when the ramosa-1 mutant is cultivated at the restrictive temperature (Fig. 4), strongly suggesting that RmsA, i.e., the proposed A. niger TORC2, carries out a function in cytoskeletal organization. Most interestingly, the disturbed actin localization observed is reminiscent of the subapical localization of actin in the act1 mutant of N. crassa, a mutant that displays increased apical branching (101). Loss of actin polarization in the ramosa-1 mutant (hypothetically due to reduced TORC2 integrity as a consequence of a less functional RmsAY447N) may thus be considered as a key event of apical branching.
Transcriptomic insights into the process of apical branching.
What, then, are the cellular events involved in apical branching? The transcriptomic data obtained in this work are most probably a reflection of two events: (i) the consequences of a defective RmsA/TORC2 function(s) (resulting in loss of polarity and disturbance of other TORC2-controlled processes) and (ii) establishment of two new polarity axes. The function of TORC2 in polar growth control via regulation of actin polarization and/or protein kinase C (PKC) activities is conserved from yeast to mammals (reviewed in reference 49). Additionally, TORC2 has also been reported to be an upstream regulator of another TOR-containing complex (TORC1) in mammals (49). TORC1 is also conserved from yeast to mammals and regulates a myriad of cell growth-related processes (e.g., transcription, translation, and protein turnover) by integrating signals from nutrients, energy status, and stressors. Thus, TORC1 regulates temporal growth, whereas TORC2 regulates spatial growth (49). However, the discovery that mammalian TORC2 acts not only in parallel to but also upstream of TORC1 brings challenge in understanding TORC2 signaling mechanisms and in answering the question posed above. The transcriptomic response of the ramosa-1 mutant includes changes in carbon, nitrogen, (phospho)lipid, and energy metabolism as well as in the stress response and ion homeostasis. As mentioned above, probably not all of these responses are related to the process of apical branching. Below, we propose and discuss a possible involvement of three cellular processes in the process of apical branching—(phospho)lipid signaling, calcium signaling, and CWI signaling—and their suspected connections to TORC2 functions.
The synthesis of important (phosho)lipid signaling molecules (PA, DAG, and PIP) and IP seems to be increased during apical branching, as genes encoding the corresponding synthetic enzymes showed enhanced expression (Table 3 and Fig. 7). Although the function of PA has not been studied in detail for filamentous fungi, a recent report demonstrated that reduced production of PA causes polarity defects in A. nidulans (62), which supports our data. Importantly, PA serves as precursor for DAG in S. cerevisiae, which in turn functions as activator of PKC (Pkc1p), a component of the CWI pathway which is localized upstream of the MAP kinase kinase Mkk1/2p (MkkA) (Fig. 7) (for a review, see reference 103). PA, however, has multiple regulatory roles in eukaryotes, such as promotion of actin polymerization and activation of TOR and PI(4)5 kinases to increase conversion from PIP to phosphatidylinositol-4′,5′-bisphosphate (PIP2) (103). PIP2 in turn can stimulate actin polymerization via interaction with actin-interacting proteins (reviewed in reference 24) and promotes actin remodeling by recruiting TORC2-interacting proteins (Slm1p and Slm2p) and different GTPases (e.g., Rho1p and its activator Rom2p) to membrane compartments (3, 4, 83). The GTPase Cdc42 has also been reported to activate mammalian PI(4)5 kinases, thereby increasing local PIP2 levels (reviewed in reference 83). Interestingly, we found a downregulation of An12g04710, which displays similarity to the yeast Vtc1p and Nrf1p proteins, which are negative regulators of Cdc42 (70). Reduced expression of An12g04710 could thus also hint at increased PIP2 synthesis during apical branching. Finally, An16g05020, a gene with homology to the genes encoding the S. cerevisiae Vip1p and S. pombe Asp1p proteins (inositol hexaki-/heptaki-phosphate kinase; reaction 6 in Fig. 7), showed increased expression. Asp1p has been reported to be necessary for the integrity of cortical actin patch organization (32).
Remarkably, PIP2 serves as precursor for phosphatidylinositol-1′,4′,5′-triphosphate (PIP3), which has been described as inducing calcium release from intracellular stores in mammalian cells (reviewed in reference 7). An increase in cytosolic calcium levels has been shown to activate the calcium/calmodulin/calcineurin/Crz1p signaling pathway in S. cerevisiae, which induces calcineurin- or Crz1p-dependent transcription of genes whose protein products are involved in ion homeostasis, cell wall synthesis, and signaling (reviewed in reference 26). All components of this pathway are conserved in A. niger (see Table S5 in the supplemental material). CrzA, the A. nidulans homolog of Crz1p, has also been demonstrated to be positively involved in transcriptional regulation of a chitin synthase gene (chsB) and the vcxA gene, encoding a vacuolar Ca2+ pump (91). Among the genes upregulated in the ramosa-1 mutant, at least five genes whose protein products can be predicted to function in calcium signaling and homeostasis were identified, i.e., two Ca2+/calmodulin-dependent protein kinases and three vacuolar Ca2+ pumps (two of these encode VCX1/vcxA-homologous genes). However, other A. niger genes also might be effector genes of a calcium response, as CrzA binding motifs were significantly overrepresented in genes upregulated in the ramosa-1 mutant (see Table S3 in the supplemental material). Furthermore, a recent finding in S. cerevisiae demonstrated that calcineurin negatively controls TORC2 and that TORC2 negatively regulates calcineurin, a mutual antagonism that is also reflected by the observation that about 50% of the genes which are upregulated during TORC2 inhibition overlap with calcineurin/Crz1p-dependent genes (69). Among this set of overlapping S. cerevisiae genes are also genes showing increased expression in the ramosa-1 mutant (An15g01560/ GTPase Gyp7p, An02g06350/vacuolar Ca2+/H+ exchanger Pmc1p, An02g05490 and An16g03050/protein kinase Cmk2, and An00g07168/aspartic protease) (Table 3; see Table S2 in the supplemental material). Most importantly, this report has evidenced that calcineurin-mediated events cause depolarization of the actin cytoskeleton, whereas TORC2 counteracts this process—opposing control mechanisms that have also been observed in mammalian cells (see reference 69 and references therein). The ramosa-1 transcriptome data hint at the possibility that a similar antagonism might occur in A. niger; i.e., actin depolarization in the ramosa-1 mutant might be a consequence of increased calcium signaling which originates from defective TORC2 signaling.
The actin organization defect in an S. cerevisiae conditional tor2 loss-of-function mutant (tor2ts) can be suppressed by overexpressing CWI pathway components (for a review, see reference 60). Our data demonstrated enhanced transcription of the predicted CWI component mkkA, point toward activation of PkcA by DAG, showed increased expression of cell wall biosynthesis and remodeling genes, and suggested increased chitin deposition in the ramosa-1 mutant (Fig. 4 and Table 3). These responses resemble the connection between TORC2 and CWI signaling in S. cerevisiae, suggesting that reinforced CWI signaling in the ramosa-1 mutant could be an adaptive response to counteract defective RmsAY447N (i.e., TORC2) function.
Finally, the transcriptomic data also point toward increased expression of oxidative stress response genes in the ramosa-1 mutant (Table 3). However, the fact that we could not detect any differences in tip-localized ROS staining in ramosa-1 and wild-type germlings (Fig. 4) suggests that increased ROS production can be sufficiently counteracted by enhanced expression of ROS-scavenging genes in the ramosa-1 mutant. An appealing explanation for the observed induction of oxidative stress genes might be given by a recent report on S. cerevisiae, where it has been demonstrated that TORC2 inhibits the response to oxidative stress (69). In other words, defective TORC2 signaling in the ramosa-1 mutant could mimic the presence of oxidative stress within the cell and might further reflect a similar interconnection between TORC2, oxidative stress, CWI, and calcium signaling pathways in A. niger as reported for S. cerevisiae. Taken together, the transcriptomic fingerprint of the ramosa-1 mutant suggests that at least four signaling cascades might be involved in the process of apical branching: (phospho)lipid, calcium, TORC2, and CWI signaling. In our previous work, we used a pharmacological approach to induce (sub)apical branch formation in A. niger germlings. The transcriptomic profiles obtained likewise suggested a role for (phospho)lipid, TORC2, and CWI signaling during polarity control (67).
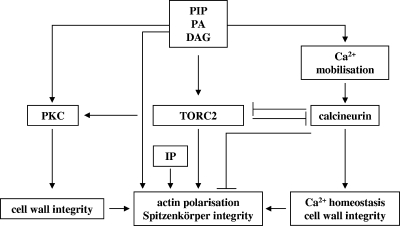
A hypothetic model for cellular events involved in apical branching.
We previously proposed “that the apical branching phenotype in ramosa-1 is triggered by a molecular event that induces a transient alteration in cytoskeleton organization” (77, 78). Our present transcriptomic analysis shows a number of changes that either singly or in combination would bring about the physiological changes that contract the actin cytoskeleton, dislodge the Spitzenkörper, interrupt hyphal elongation, and set the stage for the subsequent formation of two new centers of polarized growth. Based upon transcriptomic, genetic, and phenotypic data obtained in this study and in our previous work (77, 78), we propose the following working model for the process of apical branching in A. niger (Fig. 8). We hypothesize that the actin polarization defect in the ramosa-1 mutant is evoked by a nonfunctional or partially functional RmsAY447N protein, whereas in wild-type cells, the primary trigger for a momentary disruption of actin integrity has yet to be identified. As a consequence of actin depolarization, the Spitzenkörper detaches from the apex and disintegrates, and polarity maintenance becomes lost in the leading hypha. This event is counteracted by increased (phospho)lipid and CWI signaling aiming at actin repolarization and increased cell wall biosynthesis, whereupon two new Spitzenkörper and thereby two new polarity axes become established. The stability of these polarity axes, however, cannot be maintained in the ramosa-1 mutant: increased (phospho)lipid signaling provokes excessive calcium release from internal stores, which results in enhanced calcineurin activity, the hypothetic cause for redepolarization of actin. In wild-type hyphae, however, intact TORC2 signaling antagonizes calcineurin-mediated actin depolarization, thereby ensuring polarity maintenance.
FIG. 8.
A reconstructed model for polarity control and apical branching in A. niger. The model rests on transcriptomic and phenotypic data obtained from ramosa-1 mutant and wild-type A. niger (this work and references 77 and 78), as well as on assumptions deduced from conserved mechanisms in yeast and mammalian systems (see Discussion for references). Basically, TORC2 exerts at least three important cellular functions. (i) TORC2 plays a crucial role in maintaining actin polarization, among other things, via calcineurin inhibition. (ii) TORC2 is essential for cell wall biosynthesis and activates PKC, the initiator kinase of the CWI pathway. (iii) TORC2 inhibits expression of oxidative stress genes under nonstressed conditions (not indicated in the figure). RmsAY447N, however, negatively interferes with A. niger TORC2 function, resulting in actin depolarization, induction of apical branch formation, and derepression of oxidative stress genes. To counteract failure of TORC2, increased synthesis of important (phospho)lipid signaling molecules takes place (reactions 1 to 6) (Table 3 and Fig. 7), resulting in (i) increased Rom2p/Rho1p activation (via PIP2) aiming to repolarize actin, (ii) reactivation of TORC2 (via PIP2), (iii) reinforced PKC activation (via DAG) and thereby amplified expression of cell wall-related genes, and (iv) repolarization of actin (via IP7).
Conclusions.
This work provides the first molecular insights into the nature of apical branching of A. niger and potential RmsA function. The genes belonging to the transcriptomic fingerprint of apical branching constitute a valuable compilation of genes, whose further analysis will unravel their exact contribution to polar growth of A. niger. The apical branching transcriptome of the ramosa-1 mutant allowed us to reconstruct and predict mechanistic details of signaling networks putatively involved in polarity control in A. niger. The exact functions of the presumed networks as well as their interconnections remain to be elucidated in future studies, as do the identities of the A. niger TORC2 complex and its targets.
Supplementary Material
Acknowledgments
We acknowledge Peter Punt and Frank Schuren for providing us with the A. niger cosmid and cDNA libraries. We thank the anonymous reviewers for their helpful suggestions to improve the manuscript.
The research group of C.A.M.J.J.v.d.H. is part of the Kluyver Centre for Genomics of Industrial Fermentation, which is supported by The Netherlands Genomics Initiative.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://ec.asm.org/
REFERENCES
- 1.Amako, K., K. Fujita, C. Iwamoto, M. Sengee, K. Fuchigami, J. Fukumoto, Y. Ogishi, R. Kishimoto, and K. Goda. 2006. NADP(+)-dependent d-arabinose dehydrogenase shows a limited contribution to erythroascorbic acid biosynthesis and oxidative stress resistance in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 70:3004-3012. [DOI] [PubMed] [Google Scholar]
- 2.Andrianopoulos, A., and W. E. Timberlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audhya, A., and S. D. Emr. 2002. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell 2:593-605. [DOI] [PubMed] [Google Scholar]
- 4.Audhya, A., R. Loewith, A. Parsons, L. Gao, M. Tabuchi, H. Zhou, C. Boone, M. Hall, and S. Emr. 2004. Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J. 23:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayad-Durieux, Y., P. Knechtle, S. Goff, F. Dietrich, and P. Philippsen. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113:4563-4575. [DOI] [PubMed] [Google Scholar]
- 6.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 7.Balla, T. 2006. Phosphoinositide-derived messengers in endocrine signaling. J. Endocrinol. 188:135-153. [DOI] [PubMed] [Google Scholar]
- 8.Bartnicki-García, S. 2002. Molecular biology of fungal development, p. 29-58. Marcel Decker Inc., New York, NY.
- 9.Bartnicki-García, S. 2006. Chitosomes: past, present and future. FEMS Yeast Res. 6:957-965. [DOI] [PubMed] [Google Scholar]
- 10.Bartnicki-García, S., F. Hergert, and G. Gierz. 1989. Computer simulation of fungal morphogenesis and the mathematical basis for hyphal tip growth. Protoplasma 153:46-57. [Google Scholar]
- 11.Bencina, M., M. Legisa, and N. D. Read. 2005. Cross-talk between cAMP and calcium signalling in Aspergillus niger. Mol. Microbiol. 56:268-281. [DOI] [PubMed] [Google Scholar]
- 12.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 13.Bennett, J. W., and L. Lasure. 1991. More gene manipulations in fungi. Academic Press, San Diego, CA.
- 14.Bocking, S. P., M. G. Wiebe, G. D. Robson, K. Hansen, L. H. Christiansen, and A. P. Trinci. 1999. Effect of branch frequency in Aspergillus oryzae on protein secretion and culture viscosity. Biotechnol. Bioeng. 65:638-648. [DOI] [PubMed] [Google Scholar]
- 15.Bok, J. W., T. Sone, L. B. Silverman-Gavrila, R. R. Lew, F. J. Bowring, D. E. Catcheside, and A. J. Griffiths. 2001. Structure and function analysis of the calcium-related gene spray in Neurospora crassa. Fungal Genet. Biol. 32:145-158. [DOI] [PubMed] [Google Scholar]
- 16.Borges-Walmsley, M. I., and A. R. Walmsley. 2000. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8:133-141. [DOI] [PubMed] [Google Scholar]
- 17.Borneman, A. R., M. J. Hynes, and A. Andrianopoulos. 2000. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol. Microbiol. 38:1034-1047. [DOI] [PubMed] [Google Scholar]
- 18.Borsuk, P., M. Nagieć, P. Stepień, and E. Bartnik. 1982. Organization of the ribosomal RNA gene cluster in Aspergillus nidulans. Gene 17:147-152. [DOI] [PubMed] [Google Scholar]
- 19.Bourett, T. M., and R. J. Howard. 1991. Ultrastructural immunolocalisation of actin in a fungus. Protoplasma 163:199-202. [Google Scholar]
- 20.Bracker, C. E., J. Ruiz-Herrera, and S. Bartnicki-Garcia. 1976. Structure and transformation of chitin synthetase particles (chitosomes) during microfibril synthesis in vitro. Proc. Natl. Acad. Sci. USA 73:4570-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brakhage, A. A., A. Andrianopoulos, M. Kato, S. Steidl, M. A. Davis, N. Tsukagoshi, and M. J. Hynes. 1999. HAP-like CCAAT-binding complexes in filamentous fungi: implications for biotechnology. Fungal Genet. Biol. 27:243-252. [DOI] [PubMed] [Google Scholar]
- 22.Carol, R. J., and L. Dolan. 2006. The role of reactive oxygen species in cell growth: lessons from root hairs. J. Exp. Bot. 57:1829-1834. [DOI] [PubMed] [Google Scholar]
- 23.Chang, Y. C., and W. E. Timberlake. 1993. Identification of Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, H., B. W. Bernstein, and J. R. Bamburg. 2000. Regulating actin-filament dynamics in vivo. Trends Biochem. Sci. 25:19-23. [DOI] [PubMed] [Google Scholar]
- 25.Coleman, S., T. Fang, S. Rovinsky, F. Turano, and W. Moye-Rowley. 2001. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 276:244-250. [DOI] [PubMed] [Google Scholar]
- 26.Cyert, M. S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143-1150. [DOI] [PubMed] [Google Scholar]
- 27.Davis, M. A., J. M. Kelly, and M. J. Hynes. 1993. Fungal catabolic gene regulation: molecular genetic analysis of the amdS gene of Aspergillus nidulans. Genetica 90:133-145. [DOI] [PubMed] [Google Scholar]
- 28.deHart, A., J. Schnell, D. Allen, J. Tsai, and L. Hicke. 2003. Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol. Biol. Cell 14:4676-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Ruiter-Jacobs, Y. M., M. Broekhuijsen, S. E. Unkles, E. I. Campbell, J. R. Kinghorn, R. Contreras, P. H. Pouwels, and C. A. van den Hondel. 1989. A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae. Curr. Genet. 16:159-163. [DOI] [PubMed] [Google Scholar]
- 30.Fadri, M., A. Daquinag, S. Wang, T. Xue, and J. Kunz. 2005. The pleckstrin homology domain proteins Slm1 and Slm2 are required for actin cytoskeleton organization in yeast and bind phosphatidylinositol-4,5-bisphosphate and TORC2. Mol. Biol. Cell 16:1883-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fait, A., A. Yellin, and H. Fromm. 2005. GABA shunt deficiencies and accumulation of reactive oxygen intermediates: insight from Arabidopsis mutants. FEBS Lett. 579:415-420. [DOI] [PubMed] [Google Scholar]
- 32.Feoktistova, A., D. McCollum, R. Ohi, and K. Gould. 1999. Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 152:895-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fortwendel, J. R., P. R. Juvvadi, N. Pinchai, B. Z. Perfect, J. A. Alspaugh, J. R. Perfect, and W. J. Steinbach. 2009. Differential effects of inhibiting chitin and 1,3-β-d-glucan synthesis in Ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob. Agents Chemother. 53:476-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geitmann, A., and A. M. Emons. 2000. The cytoskeleton in plant and fungal cell tip growth. J. Microsc. 198:218-245. [DOI] [PubMed] [Google Scholar]
- 35.Girbardt, M. 1957. Der Spitzenkörper von Polystictus versicolor. Planta 50:47-59. [Google Scholar]
- 36.Gordon, C. L., V. Khalaj, A. F. Ram, D. B. Archer, J. L. Brookman, A. P. Trinci, D. J. Jeenes, J. H. Doonan, B. Wells, P. J. Punt, C. A. van den Hondel, and G. D. Robson. 2000. Glucoamylase::green fluorescent protein fusions to monitor protein secretion in Aspergillus niger. Microbiology 146:415-426. [DOI] [PubMed] [Google Scholar]
- 37.Grimm, L. H., S. Kelly, R. Krull, and D. C. Hempel. 2005. Morphology and productivity of filamentous fungi. Appl. Microbiol. Biotechnol. 69:375-384. [DOI] [PubMed] [Google Scholar]
- 38.Grosse, C., T. Heinekamp, O. Kniemeyer, A. Gehrke, and A. A. Brakhage. 2008. Protein kinase A regulates growth, sporulation, and pigment formation in Aspergillus fumigatus. Appl. Environ. Microbiol. 74:4923-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grove, S. N., and C. E. Bracker. 1970. Protoplasmic organization of hyphal tips among fungi: vesicles and Spitzenkörper. J. Bacteriol. 104:989-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-933. [PubMed] [Google Scholar]
- 41.Harris, S., J. Morrell, and J. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris, S. D. 2008. Branching of fungal hyphae: regulation, mechanisms and comparison with other branching systems. Mycologia 100:823-832. [DOI] [PubMed] [Google Scholar]
- 43.Harris, S. D., and M. Momany. 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41:391-400. [DOI] [PubMed] [Google Scholar]
- 44.Harris, S. D., N. D. Read, R. W. Roberson, B. Shaw, S. Seiler, M. Plamann, and M. Momany. 2005. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howard, R. J. 1981. Ultrastructural analysis of hyphal tip cell growth in fungi: Spitzenkörper, cytoskeleton and endomembranes after freeze-substitution. J. Cell Sci. 48:89-103. [DOI] [PubMed] [Google Scholar]
- 46.Ikeda, K., S. Morigasaki, H. Tatebe, F. Tamanoi, and K. Shiozaki. 2008. Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle 7:358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoki, K., and K. L. Guan. 2006. Complexity of the TOR signaling network. Trends Cell Biol. 16:206-212. [DOI] [PubMed] [Google Scholar]
- 48.Irizarry, R. A., B. M. Bolstad, F. Collin, L. M. Cope, B. Hobbs, and T. P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacinto, E., and A. Lorberg. 2008. TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 410:19-37. [DOI] [PubMed] [Google Scholar]
- 50.Jones, K. T., E. R. Greer, D. Pearce, and K. Ashrafi. 2009. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 7:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones, M., M. Raymond, Z. Yang, and N. Smirnoff. 2007. NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58:1261-1270. [DOI] [PubMed] [Google Scholar]
- 52.Kasuga, T., and N. L. Glass. 2008. Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot. Cell 7:1549-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato, M., A. Aoyama, F. Naruse, Y. Tateyama, K. Hayashi, M. Miyazaki, P. Papagiannopoulos, M. A. Davis, M. J. Hynes, T. Kobayashi, and N. Tsukagoshi. 1998. The Aspergillus nidulans CCAAT-binding factor AnCP/AnCF is a heteromeric protein analogous to the HAP complex of Saccharomyces cerevisiae. Mol. Gen. Genet. 257:404-411. [DOI] [PubMed] [Google Scholar]
- 54.Kim, H. S., K. Czymmek, and S. Kang. 2009. Development of fluorescent protein-based biosensors for Ca2+ and pH to monitor physiological changes during Arabidopsis thaliana-Fusarium oxysporum interactions. Abstr. 25th Fungal Genetics Conference, p. 559.
- 55.Knechtle, P., F. Dietrich, and P. Philippsen. 2003. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14:4140-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knechtle, P., A. Kaufmann, D. Cavicchioli, and P. Philippsen. 2008. The Paxillin-like protein AgPxl1 is required for apical branching and maximal hyphal growth in A. gossypii. Fungal Genet. Biol. 45:829-838. [DOI] [PubMed] [Google Scholar]
- 57.Kohler, T., S. Wesche, N. Taheri, G. H. Braus, and H. U. Mosch. 2002. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot. Cell 1:673-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kono, N., T. Inoue, Y. Yoshida, H. Sato, T. Matsusue, H. Itabe, E. Niki, J. Aoki, and H. Arai. 2008. Protection against oxidative stress-induced hepatic injury by intracellular type II platelet-activating factor acetylhydrolase by metabolism of oxidized phospholipids in vivo. J. Biol. Chem. 283:1628-1636. [DOI] [PubMed] [Google Scholar]
- 59.Kulmburg, P., M. Mathieu, C. Dowzer, J. Kelly, and B. Felenbok. 1993. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol. Microbiol. 7:847-857. [DOI] [PubMed] [Google Scholar]
- 60.Levin, D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 62.Malavazi, I., M. Savoldi, M. da Silva Ferreira, F. Soriani, P. Bonato, M. de Souza Goldman, and G. Goldman. 2007. Transcriptome analysis of the Aspergillus nidulans AtmA (ATM, Ataxia-Telangiectasia mutated) null mutant. Mol. Microbiol. 66:74-99. [DOI] [PubMed] [Google Scholar]
- 63.Matsuo, T., Y. Otsubo, J. Urano, F. Tamanoi, and M. Yamamoto. 2007. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 27:3154-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McIntyre, M., C. Muller, J. Dynesen, and J. Nielsen. 2001. Metabolic engineering of the morphology of Aspergillus. Adv. Biochem. Eng. Biotechnol. 73:103-128. [DOI] [PubMed] [Google Scholar]
- 65.Meyer, V., M. Arentshorst, A. El-Ghezal, A. C. Drews, R. Kooistra, C. A. M. J. J. van den Hondel, and A. F. Ram. 2007. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 128:770-775. [DOI] [PubMed] [Google Scholar]
- 66.Meyer, V., M. Arentshorst, C. van den Hondel, and A. Ram. 2008. The polarisome component SpaA localises to hyphal tips of Aspergillus niger and is important for polar growth. Fungal Genet. Biol. 45:152-164. [DOI] [PubMed] [Google Scholar]
- 67.Meyer, V., R. A. Damveld, M. Arentshorst, U. Stahl, C. A. van den Hondel, and A. F. Ram. 2007. Survival in the presence of antifungals: genome-wide expression profiling of Aspergillus niger in response to sublethal concentrations of caspofungin and fenpropimorph. J. Biol. Chem. 282:32935-32948. [DOI] [PubMed] [Google Scholar]
- 68.Momany, M. 2001. Molecular and cellular biology of filamentous fungi: a practical approach, p. 119-125. Oxford University Press, Oxford, United Kingdom.
- 69.Mulet, J., D. Martin, R. Loewith, and M. Hall. 2006. Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 281:33000-33007. [DOI] [PubMed] [Google Scholar]
- 70.Murray, J., and D. Johnson. 2000. Isolation and characterization of Nrf1p, a novel negative regulator of the Cdc42p GTPase in Schizosaccharomyces pombe. Genetics 154:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osmani, A. H., B. R. Oakley, and S. A. Osmani. 2006. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protoc. 1:2517-2526. [DOI] [PubMed] [Google Scholar]
- 72.Papagianni, M. 2004. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 22:189-259. [DOI] [PubMed] [Google Scholar]
- 73.Pel, H. J., J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. de Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. Danchin, A. J. Debets, P. Dekker, P. W. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. Driessen, C. d'Enfert, S. Geysens, C. Goosen, G. S. Groot, P. W. de Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. van den Hombergh, C. A. van den Hondel, R. T. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. van der Maarel, R. Meulenberg, H. Menke, M. A. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. van Peij, A. F. Ram, U. Rinas, J. A. Roubos, C. M. Sagt, M. Schmoll, J. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. van de Vondervoort, H. Wedler, H. A. Wosten, A. P. Zeng, A. J. van Ooyen, J. Visser, and H. Stam. 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25:221-231. [DOI] [PubMed] [Google Scholar]
- 74.Peterbauer, C. K., D. Litscher, and C. P. Kubicek. 2002. The Trichoderma atroviride seb1 (stress response element binding) gene encodes an AGGGG-binding protein which is involved in the response to high osmolarity stress. Mol. Genet. Genomics 268:223-231. [DOI] [PubMed] [Google Scholar]
- 75.Punt, P. J., and C. A. van den Hondel. 1992. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 216:447-457. [DOI] [PubMed] [Google Scholar]
- 76.Reinke, A., S. Anderson, J. McCaffery, J. r. Yates, S. Aronova, S. Chu, S. Fairclough, C. Iverson, K. Wedaman, and T. Powers. 2004. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279:14752-14762. [DOI] [PubMed] [Google Scholar]
- 77.Reynaga-Peña, C. G., and S. Bartnicki-Garcia. 1997. Apical branching in a temperature sensitive mutant of Aspergillus niger. Fungal Genet. Biol. 22:153-167. [DOI] [PubMed] [Google Scholar]
- 78.Reynaga-Peña, C. G., and S. Bartnicki-Garcia. 2005. Cytoplasmic contractions in growing fungal hyphae and their morphogenetic consequences. Arch. Microbiol. 183:292-300. [DOI] [PubMed] [Google Scholar]
- 79.Reynaga-Peña, C. G., G. Gierz, and S. Bartnicki-Garcia. 1997. Analysis of the role of the Spitzenkörper in fungal morphogenesis by computer simulation of apical branching in Aspergillus niger. Proc. Natl. Acad. Sci. USA 94:9096-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Riquelme, M., S. Bartnicki-Garcia, J. M. Gonzalez-Prieto, E. Sanchez-Leon, J. A. Verdin-Ramos, A. Beltran-Aguilar, and M. Freitag. 2007. Spitzenkörper localization and intracellular traffic of GFP-labeled CHS-3 and CHS-6 chitin synthases in living hyphae of Neurospora crassa. Eukaryot. Cell 6:1853-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruepp, A., A. Zollner, D. Maier, K. Albermann, J. Hani, M. Mokrejs, I. Tetko, U. Guldener, G. Mannhaupt, M. Munsterkotter, and H. W. Mewes. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32:5539-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, NY.
- 83.Santarius, M., C. H. Lee, and R. A. Anderson. 2006. Supervised membrane swimming: small G-protein lifeguards regulate PIPK signalling and monitor intracellular PtdIns(4,5)P2 pools. Biochem. J. 398:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmidt, A., J. Kunz, and M. Hall. 1996. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. USA 93:13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmitz, H. P., A. Kaufmann, M. Kohli, P. P. Laissue, and P. Philippsen. 2006. From function to shape: a novel role of a formin in morphogenesis of the fungus Ashbya gossypii. Mol. Biol. Cell 17:130-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schroder, W. A., M. Buck, N. Cloonan, J. F. Hancock, A. Suhrbier, T. Sculley, and G. Bushell. 2007. Human Sin1 contains Ras-binding and pleckstrin homology domains and suppresses Ras signalling. Cell Signal. 19:1279-1289. [DOI] [PubMed] [Google Scholar]
- 87.Seiler, S., and M. Plamann. 2003. The genetic basis of cellular morphogenesis in the filamentous fungus Neurospora crassa. Mol. Biol. Cell 14:4352-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Semighini, C., and S. Harris. 2008. Regulation of apical dominance in Aspergillus nidulans hyphae by reactive oxygen species. Genetics 179:1919-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharpless, K. E., and S. D. Harris. 2002. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sone, T., and A. J. Griffiths. 1999. The frost gene of Neurospora crassa is a homolog of yeast cdc1 and affects hyphal branching via manganese homeostasis. Fungal Genet. Biol. 28:227-237. [DOI] [PubMed] [Google Scholar]
- 91.Spielvogel, A., H. Findon, H. N. Arst, L. Araujo-Bazan, P. Hernandez-Ortiz, U. Stahl, V. Meyer, and E. A. Espeso. 2008. Two zinc finger transcription factors, CrzA and SltA, are involved in cation homoeostasis and detoxification in Aspergillus nidulans. Biochem. J. 414:419-429. [DOI] [PubMed] [Google Scholar]
- 92.Steinberg, G. 2007. Hyphal growth: a tale of motors, lipids, and the Spitzenkörper. Eukaryot. Cell 6:351-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102:15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Taheri-Talesh, N., T. Horio, L. Araujo-Bazán, X. Dou, E. Espeso, M. Peñalva, S. Osmani, and B. Oakley. 2008. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 19:1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takeshita, N., A. Ohta, and H. Horiuchi. 2005. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell 16:1961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thykaer, J., K. Rueksomtawin, H. Noorman, and J. Nielsen. 2009. Disruption of the NADPH-dependent glutamate dehydrogenase affects the morphology of two industrial strains of Penicillium chrysogenum. J. Biotechnol. 139:280-282. [DOI] [PubMed] [Google Scholar]
- 97.Torralba, S., and I. B. Heath. 2001. Cytoskeletal and Ca2+ regulation of hyphal tip growth and initiation. Curr. Top. Dev. Biol. 51:135-187. [DOI] [PubMed] [Google Scholar]
- 98.van Hartingsveldt, W., I. E. Mattern, C. M. van Zeijl, P. H. Pouwels, and C. A. van den Hondel. 1987. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 206:71-75. [DOI] [PubMed] [Google Scholar]
- 99.van Loon, A., B. Pesold-Hurt, and G. Schatz. 1986. A yeast mutant lacking mitochondrial manganese-superoxide dismutase is hypersensitive to oxygen. Proc. Natl. Acad. Sci. USA 83:3820-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 101.Virag, A., and A. J. Griffiths. 2004. A mutation in the Neurospora crassa actin gene results in multiple defects in tip growth and branching. Fungal Genet. Biol. 41:213-225. [DOI] [PubMed] [Google Scholar]
- 102.Virag, A., and S. D. Harris. 2006. Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryot. Cell 5:881-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang, G., L. Lu, C. Y. Zhang, A. Singapuri, and S. Yuan. 2006. Calmodulin concentrates at the apex of growing hyphae and localizes to the Spitzenkörper in Aspergillus nidulans. Protoplasma 228:159-166. [DOI] [PubMed] [Google Scholar]
- 104.Wang, S. Z., and R. M. Roberts. 2005. The evolution of the Sin1 gene product, a little known protein implicated in stress responses and type I interferon signaling in vertebrates. BMC Evol. Biol. 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilkinson, M. G., T. S. Pino, S. Tournier, V. Buck, H. Martin, J. Christiansen, D. G. Wilkinson, and J. B. Millar. 1999. Sin1: an evolutionarily conserved component of the eukaryotic SAPK pathway. EMBO J. 18:4210-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wongwicharn, A., B. McNeil, and L. M. Harvey. 1999. Effect of oxygen enrichment on morphology, growth, and heterologous protein production in chemostat cultures of Aspergillus niger B1-D. Biotechnol. Bioeng. 65:416-424. [DOI] [PubMed] [Google Scholar]
- 107.Wösten, H. A., M. S. Moukha, J. H. Sietsma, and J. G. Wessels. 1991. Localisation of growth and secretion of proteins in Aspergillus niger. J. Gen. Microbiol. 137:2017-2023. [DOI] [PubMed] [Google Scholar]
- 108.Wright, G. D., J. Arlt, W. C. Poon, and N. D. Read. 2007. Optical tweezer micromanipulation of filamentous fungi. Fungal Genet. Biol. 44:1-13. [DOI] [PubMed] [Google Scholar]
- 109.Wullschleger, S., R. Loewith, W. Oppliger, and M. Hall. 2005. Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 280:30697-30704. [DOI] [PubMed] [Google Scholar]
- 110.Yang, Q., K. Inoki, T. Ikenoue, and K. L. Guan. 2006. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20:2820-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zheng, X. D., Y. M. Wang, and Y. Wang. 2003. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol. Microbiol. 49:1391-1405. [DOI] [PubMed] [Google Scholar]
- 112.Ziv, C., R. Gorovits, and O. Yarden. 2008. Carbon source affects PKA-dependent polarity of Neurospora crassa in a CRE-1-dependent and independent manner. Fungal Genet. Biol. 45:103-116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.