Abstract
Previous work, using solubilization of yeast cell walls by carboxymethylation, before or after digestion with β(1-3)- or β(1-6)glucanase, followed by size chromatography, showed that the transglycosylases Crh1p and Crh2p/Utr2p were redundantly required for the attachment of chitin to β(1-6)glucan. With this technique, crh1Δ crh2Δ mutants still appeared to contain a substantial percentage of chitin linked to β(1-3)glucan. Two novel procedures have now been developed for the analysis of polysaccharide cross-links in the cell wall. One is based on the affinity of curdlan, a β(1-3)glucan, for β(1-3)glucan chains in carboxymethylated cell walls. The other consists of in situ deacetylation of cell wall chitin, generating chitosan, which can be extracted with acetic acid, either directly (free chitosan) or after digestion with different glucanases (bound chitosan). Both methodologies indicated that all of the chitin in crh1Δ crh2Δ strains is free. Reexamination of the previously used procedure revealed that the β(1-3)glucanase preparation used (zymolyase) is contaminated with a small amount of endochitinase, which caused erroneous results with the double mutant. After removing the chitinase from the zymolyase, all three procedures gave coincident results. Therefore, Crh1p and Crh2p catalyze the transfer of chitin to both β(1-3)- and β(1-6)glucan, and the biosynthetic mechanism for all chitin cross-links in the cell wall has been established.
The fungal cell wall protects the cell against internal turgor pressure and external mechanical injury. To fulfill these functions, it must be endowed with a resilient structure. Presumably, the cell wall strength is largely due to the cross-links that bind together its components, mainly polysaccharides, giving rise to a tightly knit mesh (6, 11-13). Interestingly, the cross-links must be created outside the plasma membrane, because most of the polysaccharides are extruded as they are synthesized at the membrane; therefore, they do not exist inside the cell. This posits a thermodynamic problem, because there are no obvious sources of energy in the periplasmic space. About 20 years ago we proposed that the free energy may come from existing bonds in the polysaccharide chains and that the new cross-links may be originated by transglycosylation, thus creating a new linkage for each one that is broken (5).
Ascertaining the mechanism of cross-link formation seemed a worthwhile endeavor, both because of the theoretical implications and because the cell wall is a proven target for antifungal compounds; therefore, more knowledge about its synthesis can be of practical interest. For this type of investigation to proceed, it was necessary to devise some method for the quantitative analysis of cell wall cross-links. We developed such a procedure for the evaluation of the proportion of cell wall chitin that is free or bound to β(1-3)- or β(1-6)glucan (4). In this methodology, chitin was specifically labeled in vivo with [14C]glucosamine; cell walls were isolated, and their proteins were eliminated by alkali treatment. The insoluble residue was solubilized by carboxymethylation and analyzed by size fractionation chromatography. By treating the cell walls with different glucanases before carboxymethylation and comparing the chromatographic profiles, we were able to determine the amount of chitin bound to the different glucans, as well as the fraction that was free (4). Armed with this procedure, we could now analyze the cell wall of different mutants that appeared to be candidates for cross-links defects. In this way we found that the two putative transglycosylases Crh1p and Crh2p were redundantly required for the formation of the chitin-β(1-6)glucan linkage. A double mutant crh1Δ crh2Δ had no chitin attached to β(1-6)glucan, although it still contained apparently normal amounts of chitin-β(1-3)glucan complex (7). Further work supported the notion that Crh1p and Crh2p function as transglycosylases, transferring portions of chitin chains to glucan (8). This confirmed our earlier hypothesis.
With the initial intention of finding easier and faster methods, I devised two novel procedures for cell wall analysis. One is based on the affinity between β(1-3)glucan chains, the other on the conversion of chitin in situ into its deacetylated product, chitosan, followed by extraction of the chitosan with acetic acid before or after treatment with specific glucanases. With a wild-type strain, both procedures gave similar results to those of the carboxymethylation-chromatography technique. However, in the double mutant crh1Δ crh2Δ all of the chitin appeared to be free with both new methods. Further investigation showed that the older procedure led to erroneous results for the double mutant, because of the presence of a small amount of chitinase in the β(1-3)glucanase preparation used. After reconciling the results, I conclude that Crh1p and Crh2p are necessary for the formation of cross-links between chitin and either β(1-6) or β(1-3)glucan.
MATERIALS AND METHODS
Strains used and growth conditions.
The strains used are listed in Table 1. The growth medium was YEPGal/Raf (2% peptone, 1% yeast extract, and 2% each of galactose and raffinose). The growth temperature was 30°C, except for strain 4250-3B, which was grown at 25°C. The experimental results depicted in Fig. S1 and S2 in the supplemental material were carried out at more than one temperature, as described previously (references 4 and 7, respectively).
TABLE 1.
Strains used in this study
In vivo labeling and carboxymethylation-chromatography procedure.
In vivo labeling of chitin with [14C]glucosamine was the same for all procedures described here and was carried out as described elsewhere (4). The carboxymethylation-chromatography method was performed as previously described (4).
Preparation of [14C]β(1-6)glucan and [14C]carboxymethyl-β(1-3)glucan.
FY001 cell growth and labeling with [14C]glucose was carried out essentially as described for [14C]glucosamine labeling (4), with the following differences. Cells were grown in YEPD, centrifuged, and suspended in 20 ml of a similar medium, but containing only 0.5% glucose, at a density of ∼1.5 × 107 cells/ml. [14C]glucose (0.1 mCi [3.7 MBq], 250 mCi/mmol; MP Biomedicals) was added, and the yeast was allowed to grow at 30°C for ∼2.5 generations. Cells were harvested, and cell walls were prepared and treated with alkali as described previously (4).
For the preparation of β(1-6)glucan, 300 μl of cell wall suspension (420,000 cpm) was incubated overnight at 37°C on a rotator with 15 μl of Zymolyase 100T (10 mg/ml in 1 M Tris-HCl [pH 7.5]) and 0.02% sodium azide as a preservative. The tube was centrifuged 5 min at 16,000 × g. The supernatant was dialyzed overnight against 300 ml of water in a Pierce cassette with a 3,500-molecular-weight cutoff. About 120,000 cpm were recovered in 0.4 ml of dialysate.
For the preparation of carboxymethylated β(1-3)glucan, 150 μl of labeled cell walls (210,000 cpm) was incubated overnight at 37°C on rotator with 8 μl of 1 M sodium acetate (pH 5) and 10 μl (∼0.27 μg of protein) of recombinant β(1-6)glucanase (3). The tube was centrifuged 5 min at 16,000 × g. The pellet was carboxymethylated as described previously for the carboxymethylation of cell walls (4). About 114,000 cpm were recovered in 0.85 ml of carboxymethylated material. Portions of each polysaccharide containing 10,000 cpm were used for tests with the curdlan procedure.
Curdlan method.
Curdlan was purchased from Carbomer (San Diego, CA). A 2% suspension is prepared in 0.05 M citrate (pH 3.5). Of this suspension, 0.3 ml was placed in a 2-ml microcentrifuge tube with a conical bottom. The tube was heated at 56°C in a dry bath for 10 min; 0.3 ml of the citrate buffer was added, and the tube was shaken in a Mini-Beadbeater (Biospec Products) for 60 s and then cooled to room temperature. An additional 0.4 ml of citrate buffer was added, and the tube was shaken in the Mini-Beadbeater for 20 s. 14C-labeled samples (10,000 cpm) of carboxymethylated cell walls, either untreated or predigested with recombinant β(1-6)glucanase (3), as previously described (4), were added to the tubes with curdlan gel, prepared as outlined above. The tubes were shaken in the Mini-Beadbeater for 20 s and then placed in a rotator at room temperature for 30 min, followed by centrifugation for 5 min at 16,000 × g. Supernatants were transferred to other tubes, and their volume and radioactivity were measured. The pellets were washed twice with 0.7 ml of the citrate buffer, and the washings were pooled and counted. The total radioactivity in the first supernatant plus washings was calculated. The washed pellets were suspended in 0.5 ml of the citrate buffer, transferred to a scintillation vial, and counted. The radioactivity in each supernatant (plus the washings) was calculated as a percentage of the total radioactivity recovered. The radioactivity of the supernatant from untreated cell walls yielded the amount of free chitin, whereas that from walls pretreated with β(1-6)glucanase corresponds to the free chitin plus the chitin bound to β(1-6)glucan. The difference between the two values represents the chitin bound to β(1-6)glucan. The chitin linked to β(1-3)glucan is given by the difference between the total sum of radioactivity recovered in the fractions and the radioactivity of the supernatant (plus the washings) from walls treated with β(1-6)glucanase. As a control, one can also use walls pretreated with β(1-3)glucanase, where all of the radioactivity should be found in the supernatant.
Chitosan method.
All centrifugations below are for 5 min at 16,000 × g. To screw-cap 1.5-ml conical microcentrifuge tubes, 100 μl of [14C]glucosamine-labeled cell walls (cell walls not treated with alkali; see reference 4 and 30 μl of alkali-treated carrier cell walls (4) were added. The tube was centrifuged, and the supernatant was discarded. The cell walls were suspended in 100 μl of 50% sodium hydroxide. This and all following suspensions were done in a small amount of liquid with a thin (2-mm-diameter) glass rod. The remainder of the liquid was used to wash the glass rod into the suspension (vortexing does not work well with these pellets). The tube, with the screw cap only partially adjusted, was then placed in a small beaker, which was put inside a larger beaker covered with aluminum foil. The assembly was autoclaved for 20 min at 121°C. After cooling, the tube was centrifuged, and the supernatant was discarded. The pellet was suspended in 150 μl of 10 M potassium acetate, and 50 μl of a 10% suspension of Sephadex G-10 was added as the carrier. After centrifugation, the pellet was washed once with 200 μl of 10 M potassium acetate and once with 200 μl of 0.02 M sodium hydroxide. The chitosan that originated from the free chitin was then extracted by suspending the pellet in 200 μl of 10% acetic acid and placing the tube on a rotator at room temperature. All extractions with acetic acid were performed for a minimum of 3 h or overnight, according to convenience. After rotation, the tube was centrifuged, the supernatant was saved, and the pellet was washed with 200 μl of water that was added to the first supernatant. The radioactivity of the combined fraction represents the free chitin (chitosan). The pellet was suspended in 185 μl of water, and 5 μl of 1 M sodium acetate (pH 5) and 10 μl of recombinant β(1-6)glucanase (3) were added. After 3 h of incubation at 37°C on rotator, the tube was centrifuged. The supernatant was saved and counted. The pellet was extracted with 200 μl of 10% acetic acid as described above, followed by centrifugation. The supernatant was then saved, and the pellet was washed with 200 μl of water, which was added to the first supernatant. The radioactivity of this fraction was added to that of the β(1-6)glucanase digestion supernatant to yield the amount of chitin bound to β(1-6)glucan. The pellet was suspended in 185 (or 165) μl of water, and 5 μl of 1 M potassium phosphate (pH 6.3) and 7.5 μl of Zymolyase 100T (Associates of Cape Cod) at 10 mg/ml in 1 M Tris-HCl (pH 7.5) (or 30 μl of chitinase-free zymolyase [see below for the preparation]) were added. The mixture was incubated on a rotator at 37°C for 3 h and then centrifuged. The supernatant was saved and counted. The radioactivity represented the small chitin attached to β(1-3)glucan (see, however, Results for correction). The pellet was extracted with 200 μl of 10% acetic acid, as described above. After centrifugation, the radioactivity of the supernatant was determined. This corresponded to the large chitin bound to β(1-3)glucan.
Determination of conversion of chitin to chitosan with nitrous acid.
Treatment of chitosan with nitrous acid converts glucosamine to anhydromannose, which separates itself from the polymer and dissolves in water (1). Because the glucosamine here was labeled with 14C, it is possible to measure the percent conversion of chitin to chitosan from the amount of radioactivity solubilized by nitrous acid. After autoclaving in 50% sodium hydroxide and washing with potassium acetate and 0.02 M sodium hydroxide as described above, the pellet was suspended in 100 μl of 0.2 M hydrochloric acid, followed by 100 μl of 50 mM sodium nitrite. The mixture was rotated for 3 h at room temperature and centrifuged. The radioactivity of the supernatant fluid represents the amount of chitin converted to chitosan, although it is not an exact measure, because if some short tracks of chitin are still acetylated, they would be liberated as water-soluble chitin oligosaccharides.
Sephacryl S-300 chromatography.
The same Sephacryl S-300 column (80 cm long, 51 cm in diameter) was used throughout. For the previously published procedure (4), the same buffer (50 mM Tris-HCl [pH 7.5] containing 150 mM sodium chloride and 0.03% sodium azide) was used. For acetic acid extracts from the chitosan procedure, 10% acetic acid, containing 1 M sodium chloride, was used to equilibrate the column. The high sodium chloride concentration was needed to prevent the attachment of chitosan to the Sephacryl bed.
Elimination of chitinase activity from zymolyase.
Chitin for this purpose was purified from the commercial product (Sigma-Aldrich) by suspending 1 g in 100 ml of dimethylacetamide containing 6% lithium chloride. After overnight stirring at room temperature, the suspension was centrifuged to eliminate the insoluble residue. The supernatant fluid was added to an equal volume of water in a mechanical homogenizer and homogenized for 3 min. After centrifugation for 5 min at 3,000 × g, the chitin pellet was washed five times with water and suspended in water.
This chitin was used to prepare a 1-ml bed volume column in a 7-mm-diameter tube. The column was equilibrated with 0.1 M potassium phosphate (pH 6.3). A 10-mg portion of Zymolyase 100T was dissolved in 1 ml of 1 M Tris-HCl (pH 7.5). The solution was dialyzed overnight, with one change, against 200 ml of 0.1 M potassium phosphate (pH 6.3). A precipitate that formed during dialysis was centrifuged off for 5 min at 16,000 × g. The supernatant was applied to the chitin column described above. The emerging fluid and a 1-ml wash with 0.1 M potassium phosphate (pH 6.3) were pooled. About 70% of the β(1-3)glucanase activity was recovered, as measured with reduced laminarin as the substrate and the Park and Johnson method for reducing sugar (16).
Measurement of chitinase activity.
Chitinase was measured by viscosimetry, with the falling-sphere method, where the viscosity of a solution is determined by the time taken by a sphere to travel down a tube containing the solution. I used a home-made viscosimeter, with a 0.2-ml pipette as the tube and a 1-mm-diameter glass bead as the sphere.
The substrate for the chitinase reaction, carboxymethylchitin, was prepared from commercial chitin (Sigma-Aldrich) by a minor modification of Hirano's procedure (9). The reaction mixture contained carboxymethylchitin (1.5 mg), enzyme, and 0.1 M potassium phosphate (pH 6.3) in a total volume of 450 μl. Incubation was at 37°C for 15 to 30 min, and the reaction was stopped by placing tubes in a boiling water bath for 5 min.
RESULTS
Curdlan method.
The procedure we previously devised (4) allows the quantitative determination of the three forms of chitin in the cell wall, free chitin, chitin-β(1-3)glucan, and chitin-β(1-6)glucan-β(1-3)glucan, but it is rather cumbersome because it requires, in addition to the carboxymethylation step, several chromatographic separations, followed by lengthy calculations. Searching for a simpler methodology, I tried to take advantage of the affinity of β(1-3)glucan chains for each other, due to hydrogen bonds, which accounts for the insolubility of several polysaccharides with this structure (17). One of them is curdlan, which has the unusual property of forming a gel when heated at 56°C in an aqueous suspension (14). This gel can be disrupted by suspending it in buffer, followed by vigorous shaking. Preliminary experiments showed that 80 to 90% of the radioactivity of 14C-labeled carboxymethylated β(1-3)glucan could be adsorbed by the curdlan gel, whereas none of [14C]β(1-6)glucan (not carboxymethylated) was picked up by the gel. This showed that the curdlan gel was able to adsorb specifically chains of the same structure, even when modified by attached acetic acid residues. It seems probable that the adsorption of the carboxymethylated β(1-3)glucan is not complete because of impurities in the preparation. Based on these results, one would expect that, when presented with the mixture of polysaccharides produced by carboxymethylation of the 14C-labeled cell walls (Fig. 1A), curdlan should be able to adsorb all of them, except for the free chitin. In fact, when the experiment was carried out, ca. 40% of the radioactivity remained in the supernatant, an amount similar to that of free chitin, as determined by the chromatographic method (Table 2). Pretreatment of the cell wall with β(1-6)glucanase increased the amount of radioactivity in the supernatant by ca. 15% of the total, which would correspond to the chitin bound to β(1-6)glucan, which was again similar to that calculated by the chromatographic procedure (Table 2). The amount of chitin attached directly to β(1-3)glucan can be calculated by difference with the total (Fig. 1 and Table 2). Furthermore, when the cell walls were treated with β(1-3)glucanase before carboxymethylation, none of the radioactivity was adsorbed by curdlan, as expected. For the calculations, this incubation with β(1-3)glucanase is not necessary. One limitation of the curdlan method is that it does not discriminate between the large and small chitin attached to β(1-3)glucan that were previously identified (4). Aside from this limitation, the data yielded by the curdlan gel procedure, which does not require chromatography, were similar to those of the previous method. However, when the curdlan method was applied to the crh1Δ crh2Δ double mutant, a surprising result was obtained: whereas a normal amount of chitin bound to β(1-3)glucan had been found by the chromatographic procedure (7), here 100% of the radioactivity remained in the supernatant, as if all of the chitin were free. Because the curdlan method was new and not thoroughly explored, it was first thought that the unexpected result might come from a quirk of the procedure, perhaps as a result of the high molecular weight of chitin in the double mutant. It seemed, however, that in order to understand the discrepancy it would be necessary to develop a third procedure, based on a different principle from the other two.
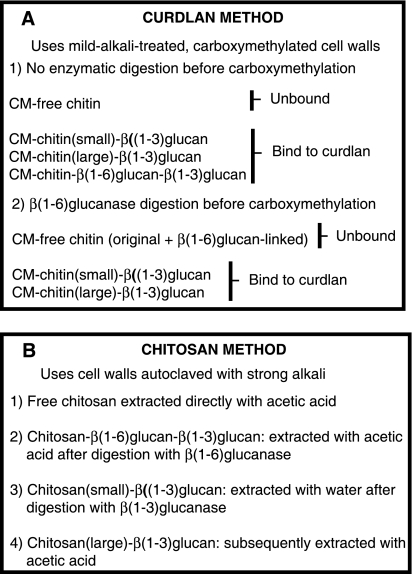
FIG. 1.
Outline of the two novel procedures for determination of chitin cross-links. In both cases chitin was specifically labeled in vivo with [14C]glucosamine and estimated in the various fractions by radioactivity measurement. For details of the procedures, see Materials and Methods. (A) Curdlan method. Free chitin was determined from the unbound radioactivity (step 1). This value was subtracted from the unbound radioactivity (step 2) to obtain the chitin attached to β(1-6)glucan. The chitin bound to β(1-3)glucan can be calculated by subtracting the unbound value (step 2) from the total or by determining the bound radioactivity (step 2). (B) Chitosan method. Each step 1 to 4 was performed on the insoluble residue remaining from the preceding step. The radioactivity of each extract was measured.
TABLE 2.
Comparison of different procedures for the measurement of cross-links between chitin and other polysaccharides in strain FY001
| Cross-link | % of total chitin in each fraction as determined by: |
||
|---|---|---|---|
| Carboxymethylation and chromatography | Curdlan method | Chitosan method | |
| Free chitin | 43 | 41 | 31 |
| Chitin → β(1-6)glucan | 16 | 15 | 25 |
| Chitin → β(1-3)glucan | 41 | 44 | 44 |
Chitosan method.
In the search for a third procedure for the analysis of chitin cross-links, I turned to chitosan, the deacetylated analog of chitin. Chitin is totally insoluble in aqueous solvents, because of the strong hydrogen bonds between the >N-H groups of one chain and the >C=O groups of an adjacent chain. The >C=O groups are no longer present in chitosan, thus eliminating the hydrogen bonds, and this compound is soluble in dilute acetic acid, with protonation of the now free amino groups. It seemed that if one could convert all of the cell wall chitin to chitosan in situ, it should now be possible to extract the free chitosan (coming from free chitin) with acetic acid (Fig. 1B). Subsequently, one could treat the cell walls with β(1-6)glucanase, followed by a similar acetic acid extraction, to obtain the chitosan bound to β(1-6)glucan (Fig. 1B). This would leave in the insoluble residue only the chitosan attached to β(1-3)glucan, which could be removed by digestion with β(1-3)glucanase. The problem here was the deacetylation of chitin, which must be done without breaking other bonds. It would be desirable to accomplish this operation with a mild and specific method. Chitin deacetylases were considered as possible agents. However, these enzymes act very sluggishly on native chitin (2). Trials with a Cryptococcus deacetylase (kindly provided by V. Bouriotis) led to a conversion of only 10% of the cell wall chitin to chitosan, as measured with the nitrous acid procedure (see Materials and Methods). The alternative was treatment of the cell walls with concentrated (50%) sodium hydroxide at high temperature. Although usually this treatment is carried out for many hours, No et al. (15) found that 20 to 30 min is sufficient if the reaction is carried out in the autoclave. This was the chosen treatment (see Materials and Methods). Observation in the microscope showed that, despite the harsh treatment, the cell walls, although shrunken, maintained their shape. If they were washed with water, they tended to disintegrate. However, a wash with high salt (10 M potassium acetate) stabilized and somehow clumped the walls, which could then be washed with water. Nitrous acid liberated more than 90% of the radioactivity from the washed cell walls, showing that chitin had been effectively converted to chitosan.
The cell walls were subjected to the series of alternative extractions and enzymatic digestions outlined above (see Materials and Methods). Note that in this case one can theoretically discriminate between small and large chitin attached to β(1-3)glucan, because the former should be solubilized in water by β(1-3)glucanase action, whereas the latter should require subsequent acetic acid extraction (see, however, below). The results showed a reasonable agreement with those of the chromatographic procedure and of the curdlan method (Table 2). When the procedure was applied to the crh1Δ crh2Δ mutant, again practically all of the chitosan was in the first acetic acid extract, as if all of the original chitin were free. The coincidence of this result with the corresponding one of the curdlan method, which is based on an entirely different principle, suggested that the carboxymethylation-chromatography procedure should be reinvestigated.
What was wrong with the carboxymethylation-chromatography method.
In an attempt to understand the cause of the different results with the curdlan and chitosan methods, compared to the older procedure, I incubated carboxymethylated cell walls of the crh1Δ crh2Δ mutant with the β(1-3)glucanase preparation (Zymolyase 100T) and applied both the untreated and the enzyme-digested preparation to a Sephacryl S-300 column (Fig. 2A). Note that the experiment was done in the reverse order than the standard procedure, where the enzymatic treatment precedes the carboxymethylation. Despite this change, the zymolyase digestion resulted in a pronounced shift to the right of the radioactive peak, as found previously with the published method. It was reasoned that, if this effect was due to glucanase action, it should also be produced by a recombinant β(1-3)glucanase. The commercial preparation Quantazyme contains such a protein, expressed from a gene of the same organism from which zymolyase is obtained, Oerskovia xanthineolytica (Arthrobacter luteus). However, Quantazyme incubation had no effect on the chromatographic profile of the radioactive material (Fig. 2A). This result strongly suggested that zymolyase contained, in addition to β(1-3)glucanase, some other enzymatic activity that was responsible for the observed shift. Based on the data from the curdlan and the chitosan procedures, the carboxymethylated cell walls of the crh1Δ crh2Δ mutant should contain only carboxymethylated chitin, which is a good substrate for chitinases. Accordingly, zymolyase caused a time-dependent and enzyme concentration-dependent decrease in viscosity of authentic carboxymethylchitin (Fig. 2B). I successfully eliminated the chitinase activity from the zymolyase preparation by filtering the latter through a chitin column (see Materials and Methods and Fig. 2B). When this chitinase-free preparation was used to digest crh1Δ crh2Δ cell walls before carboxymethylation, the chromatographic profile generated was the same as for untreated cell walls (Fig. 3A) or for cell walls treated with β(1-6)glucanase, which was previously shown to have no effect on cell walls of the double mutant (7). The effect of untreated zymolyase is shown for comparison (Fig. 3B). Clearly, now all three procedures coincide in indicating that in the double mutant all of the chitin is free; therefore, Crh1p and Crh2p are required for the formation of cross-links between chitin and both β(1-3)- and β(1-6)glucan.
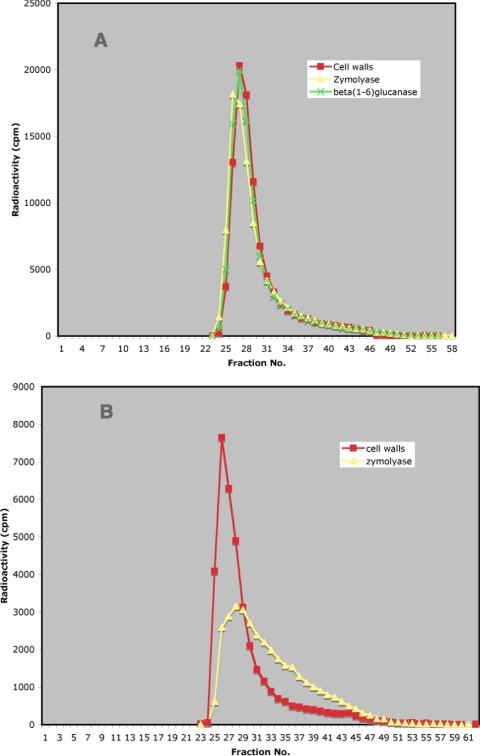
FIG. 2.
(A) Chromatography of crh1Δ crh2Δ carboxymethylated cell walls on a Sephacryl S-300 column, either untreated or after digestion with zymolyase or Quantazyme. The void volume in this and all subsequent Sephacryl columns is at fractions 26 to 27. Also, in all Sephacryl columns the radioactivity represents chitin labeled in vivo with [14C]glucosamine. (B) Determination of chitinase in zymolyase by viscosimetry. Travel time is the time taken by a glass bead to travel the length of a 0.2-ml pipette filled with the solution to be tested. Incubation time is the time of incubation with zymolyase previous to the measurement of viscosity. Symbols: •, 7.5 μl of zymolyase, 10 mg/ml; □, 15 μl of zymolyase, 10 mg/ml; ○, 60 μl of purified zymolyase [contains an amount of β(1-3)glucanase activity similar to that of 15 μl of crude zymolyase]. The broken line indicates the travel time with water as the test solution.
FIG. 3.
Chromatography on Sephacryl S-300 of crh1Δ crh2Δ carboxymethylated cell walls after the original walls were either left untreated or digested with different enzymes. (A) Cell walls, either untreated or digested with either purified zymolyase or recombinant β(1-6)glucanase. (B) Cell walls, either untreated or after digestion with crude zymolyase.
Reevaluation of previous results.
In view of the results with the crh1Δ crh2Δ double mutant, it was important to verify whether the use of chitinase-containing zymolyase had caused other errors. Cell walls of the wild-type FY001 were analyzed with the chitinase-free zymolyase, both with the carboxymethylation-chromatography and with the chitosan procedure (Table 3). The results did not differ greatly from those of Table 2. The chromatographic profiles obtained after incubation with either untreated or purified zymolyase (Fig. 4A and B) do show some difference, but it does not amount to much. On the other hand, chromatography of the fraction made water soluble by zymolyase digestion showed a pronounced decrease in the small chitin fraction, which was reduced in about half, when the purified enzyme was used (Fig. 4C). This fraction disappeared completely in the crh1Δ crh2Δ double mutant when the purified zymolyase was used (see below in Fig. 6).
TABLE 3.
Comparison of different procedures for the measurement of cross-links between chitin and other polysaccharides in strain FY001 with the use of chitinase-free zymolyase
| Cross-link | % of total chitin in each fraction as determined bya: |
||
|---|---|---|---|
| Carboxymethylation and chromatography | Curdlan method | Chitosan method | |
| Free chitin | 43 | 41 | 36 |
| Chitin → β(1-6)glucan | 21 | 15 | 22 |
| Large chitin → β(1-3)glucan | 26 | 27 (33) | |
| Small chitin → β(1-3)glucan | 11 | 16 (10) | |
| Total chitin → β(1-3)glucan | 37 | 44 | 43 |
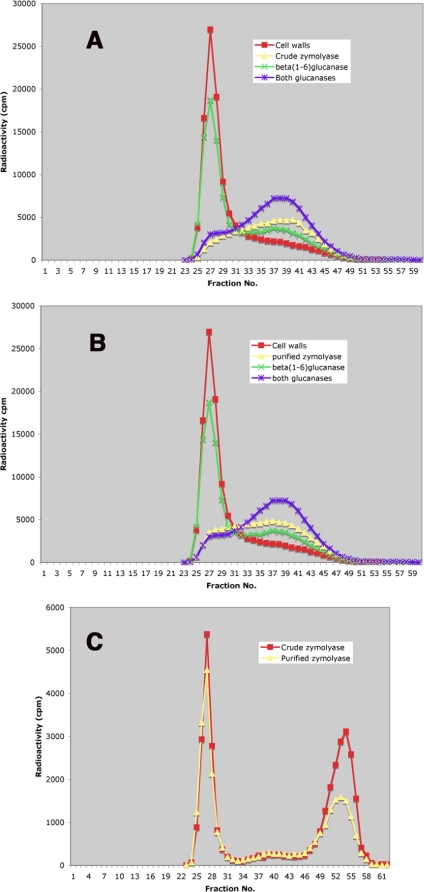
FIG. 4.
Chromatography on Sephacryl S-300 of different cell wall fractions from a wild-type strain (FY001). (A) Carboxymethylated cell walls, either undigested or after incubation with different enzymes, including crude zymolyase. (B) Same as panel A, but purified zymolyase was used. (C) Material solubilized in water by digestion of FY001 cell walls with either crude or purified zymolyase. The first peak corresponds to a chitin-β(1-6)glucan complex (4), and the second peak corresponds to small chitin that was previously linked to β(1-3)glucan (4).
FIG. 6.
Distribution of chitin in the wild type and in different crh mutants, measured by either carboxymethylation-chromatography (A) or the chitosan procedure (B). In both cases, purified zymolyase was used.
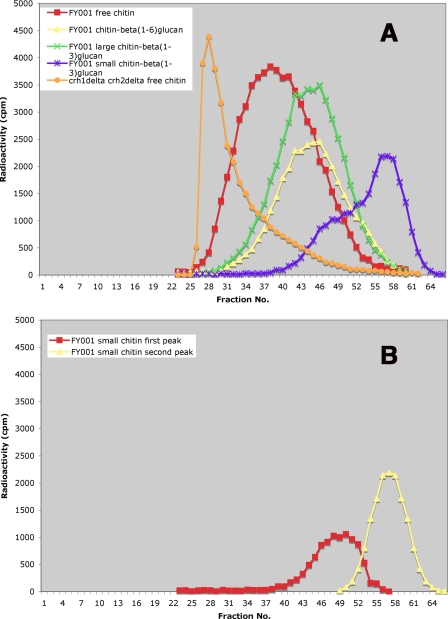
Another aspect of the small chitin fraction deserves comment. Whereas in the carboxymethylation-chromatography method most chitin fractions are together and can only be distinguished by subtracting one curve from another, in the chitosan procedure they are isolated separately. Although each one can be evaluated simply by counting radioactivity, it was of interest to examine their chromatographic profile (Fig. 5A). The free chitin (chitosan) is clearly the one with higher molecular weight, whereas the large chitin attached to β(1-3)glucan and the chitin linked to β(1-6)glucan are smaller and roughly of the same size. The small chitin linked to β(1-3)glucan is heterogeneous, consisting of at least two fractions. Only the later fraction elutes at a position comparable to that of the small chitin in Fig. 4C. This chitin seems to be smaller, which is expected because it lacks the acetyl groups as well as the carboxymethyl residues. The presence of the higher-molecular-weight fraction may result from the greater solubility of chitosan in water relative to chitin. The contribution of each peak to the small chitin can be calculated by plotting the first half of the second peak as symmetrical to the second half and obtaining the first peak by subtraction (Fig. 5B). The first peak can then be added to the value of the large chitin bound to β(1-3)glucan (numbers in parentheses in Table 3). This brings the values for the small chitin closer for the two methods. This correction was applied to all of the data presented here for small chitin-β(1-3)glucan, after carrying out the corresponding chromatographic separations. Similar chromatography of solubilized chitosan from the crh1Δ crh2Δ double mutant showed that the corresponding chitin, which suffered no transfer to glucans, has a higher molecular weight, as would have been predicted (Fig. 5A).
FIG. 5.
(A) Chromatography on Sephacryl S-300 of different chitosan fractions extracted from FY001 (wild type) or RCA003 (crh1Δ crh2Δ mutant) cell walls with the chitosan procedure. (B) The second peak (yellow) is obtained by drawing a curve symmetrical to its second half, taking fraction 58 as the maximum. The first peak (red) can then be drawn by subtracting the second peak calculated values from the experimental measurements.
The chitin distribution in the wild type and the crh mutants, as obtained with purified zymolyase, did not greatly differ from previous values (Fig. 6), except for the crh1Δ crh2Δ double mutant (compare with Fig. 2 of reference 4) and was similar to the older procedure and to the chitosan method (Fig. 6). Finally, two previously published experiments were repeated with the carboxymethylation-chromatography procedure. One was determination of chitin distribution at the mother-bud neck and in lateral walls (see Fig. S1 in the supplemental material, cf. with Fig. 8 in reference 4). The other was the effect of temperature on the chitin distribution (see Fig. S2 in the supplemental material, cf. with Fig. 6 of reference 7). In both cases, the new results are similar to the previous ones, with only minor quantitative differences.
DISCUSSION
In the work described here, an advance in methodology led to the conclusion that previous results about the attachment of chitin to other cell wall components were partially incorrect in a crh1Δ crh2Δ strain. Based on this result, Crh1p and Crh2p were found to be responsible for the linkage of chitin not only to β(1-6)glucan, as previously reported (7) but also to the main structural component of the cell wall, β(1-3)glucan. As part of this investigation, I developed two new procedures for the quantitative determination of chitin cross-links to other polysaccharides in the yeast cell wall, which may be useful for future research, both in yeast and other fungi. These methods are based on different principles from each other and from the previously devised carboxymethylation-chromatography technique; therefore, they may provide critical information in difficult cases. After the curdlan procedure was devised and used, a literature search showed that the same principle, adhesion between different β(1-3)-linked polysaccharides, had been used for a different purpose, i.e., to study the effect of molecular weight on the action of schizophyllan, a fungal polysaccharide, on the regeneration of yeast protoplasts (10). This confirms the specificity of the polysaccharide interaction. In their work, Hisamatsu et al. (10) used curdlan hard gel, obtained at 120°C, which I found less effective than the soft gel formed at a lower temperature.
Both the curdlan and the chitosan procedures are simpler than the older one because they do not require a chromatographic step. The chitosan method does not even entail carboxymethylation. The whole operation is carried out in a microcentrifuge tube, from which the different extracts are withdrawn, thus minimizing losses. The procedure is much faster than the carboxymethylation-chromatography method, lasting 2 to 3 days instead of about 10. In addition, several samples can be run simultaneously. The correction described above for the small chitin linked to β(1-3)glucan does require one chromatographic separation, but three such chromatographies can be run in 1 day.
Because in the chitosan method each type of chitin (now chitosan) is extracted separately, it is possible to determine the size of the individual fractions (Fig. 5A). Interestingly, the chitins linked to β(1-3)- and β(1-6)glucan show a similar size distribution and are smaller than the free chitin, which suggests that a relatively short fragment of the nascent chitin is transferred by Crh1p and Crh2p to the glucans. This type of experiment also shows that the chitin of the double mutant crh1Δ crh2Δ has a higher molecular weight than any of the chitins from the wild type (Fig. 5), a finding which is in agreement with the expectation that no part of that chitin is transferred to an acceptor.
There is fairly good agreement among the three procedures now available (Table 3, Fig. 6). Some of the differences may arise from the harsh alkali treatment in the chitosan method. It would be desirable to find a milder way to deacetylate chitin, but none is currently available, since the enzymatic treatment with deacetylase proved ineffective, and acid hydrolysis would break polysaccharide bonds. Where all three methods now agree is in the absence of any bound chitin in the cell wall of the crh1Δ crh2Δ mutant. The two new procedures were instrumental in forcing a review of the older one, which resulted in the finding of chitinase contamination in zymolyase. It may be asked whether the possibility of that contamination was entertained earlier. Surprisingly, this was indeed the case. When it was found that zymolyase solubilized in water a large amount of radioactivity from [14C]glucosamine-labeled cell walls (4), it was first suspected that chitinase was responsible. However, Quantazyme, which contains no chitinase, had a similar effect (4) and the solubilized material turned out to be a mixture of a chitin-β(1-6)glucan complex plus “small chitin,” previously attached to β(1-3)glucan. Paradoxically, this result put to rest our suspicion about the presence of chitinase in the zymolyase preparation.
For a β(1-3)glucanase preparation free from chitinase, purification of zymolyase was preferred over use of Quantazyme, for two reasons: first, from previous experience it seemed that zymolyase digests β(1-3)glucan faster and more completely than Quantazyme, perhaps because it contains more than one glucanase. Second, Quantazyme is no longer available commercially, and preparation of the recombinant enzyme would be laborious, whereas purification of zymolyase with a chitin column is very simple and fast. Because the purification is based on substrate affinity, it is very specific. The lack of effect of the purified zymolyase on the chitin of the crh1Δ crh2Δ mutant confirms that the chitinase was responsible for the previous spurious results.
Except for the case of the crh1Δ crh2Δ double mutant, results with the purified β(1-3)glucanase do not differ greatly from those obtained previously. The exception was in the sharp decrease in the small chitin-β(1-3)glucan fraction. Thus, part of that small chitin must come from degradation of chitin by chitinase. It may be asked whether the small chitin-β(1-3)glucan is a separate entity or just represents the tail end of a wide range of fragments of different lengths transferred from nascent chitin to glucan. Analysis of the material generated by the chitosan procedure suggests a separate peak for the small chitin (Fig. 5), but with the evidence at hand this point cannot be settled.
Note that the results described here do not invalidate our findings on transfer of chitin to β(1-3)-linked fluorescent oligosaccharides (8). Rather, it may be concluded that the reactions studied there serve as models for chitin transfer not only to β(1-3)-linked side chains of β(1-6)glucan but also to the main chain of β(1-3)glucan.
What is not in doubt is that the glycosyltransferases Crh1p and Crh2p are responsible for the attachment of chitin to both β(1-6)- and β(1-3)glucan, i.e., for all of the wall cross-links in which chitin has been found to be involved. This finding simplifies the situation with regard to chitin cross-links because a single system gives rise to all of them. On the other hand, it is not clear how the system is regulated to favor transfer to one or the other of the acceptors. Previous work, confirmed here, indicated that most of the bound chitin is linked to β(1-3)glucan at the mother-daughter neck, whereas it is attached to β(1-6)glucan in the lateral walls. How this differential localization is achieved remains to be explored. Another consequence of the results reported here is that we know how to abolish all chitin cross-links. This should facilitate studies of the physiological function of the cross-links. Work toward that goal is in progress.
Supplementary Material
Acknowledgments
I thank J. Arroyo, V. Farkas, and P. McPhie for useful discussions and a critical reading of the manuscript and V. Bouriotis for a sample of Cryptococcus chitin deacetylase.
This study was supported by a National Institutes of Health grant (Intramural Research Program, NIDDK).
Footnotes
Published ahead of print on 4 September 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Allan, G. G., and M. Peyron. 1995. Molecular weight manipulation of chitosan. I. Kinetics of depolymerization by nitrous acid. Carbohydr. Res. 277:257-272. [DOI] [PubMed] [Google Scholar]
- 2.Araki, Y., and E. Ito. 1975. A pathway of chitosan formation in Mucor rouxii. Enzymatic deacetylation of chitin. Eur. J. Biochem. 55:71-78. [DOI] [PubMed] [Google Scholar]
- 3.Bom, I. J., S. K. Dielbandhoesing, K. N. Harvey, S. J. C. M. Oomes, F. M. Klis, and S. Brul. 1998. A new tool for studying the molecular architecture of the fungal cell wall: one step purification of recombinant Trichoderma β-(1-6)-glucanase expressed in Pichia pastoris. Biochim. Biophys. Acta 1425:419-424. [DOI] [PubMed] [Google Scholar]
- 4.Cabib, E., and A. Durán. 2005. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J. Biol. Chem. 280:9170-9179. [DOI] [PubMed] [Google Scholar]
- 5.Cabib, E., B. Bowers, A. Sburlati, and S. J. Silverman. 1988. Fungal cell wall synthesis: the construction of a biological structure. Microbiol. Sci. 5:370-375. [PubMed] [Google Scholar]
- 6.Cabib, E., D.-H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679-19682. [DOI] [PubMed] [Google Scholar]
- 7.Cabib, E., N. Blanco, C. Grau, J. M. Rodríguez-Peña, and J. Arroyo. 2007. Crh1p and Crh2p are required for the cross-linking of chitin to β(1-6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 63:921-935. [DOI] [PubMed] [Google Scholar]
- 8.Cabib, E., V. Farkas, O. Kosík, N. Blanco, J. Arroyo, and P. McPhie. 2008. Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J. Biol. Chem. 283:29859-29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirano, S. 1988. Water-soluble glycol chitin and carboxymethylchitin. Method. Enzymol. 161:408-410. [Google Scholar]
- 10.Hisamatsu, M., T. Mishima, K. Teranishi, and T. Yamada. 1997. The correlation between adhesion of schizophyllan to yeast glucan and its effect on regeneration of yeast protoplasts. Carbohydr. Res. 298:117-121. [DOI] [PubMed] [Google Scholar]
- 11.Klis, F. M., A. Boorsma, and P. W. J. De Groot. 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23:185-202. [DOI] [PubMed] [Google Scholar]
- 12.Kollár, R., E. Petraková, G. Ashwell, P. W. Robbins, and E. Cabib. 1995. Architecture of the yeast cell wall. The linkage between chitin and β(1→3)glucan. J. Biol. Chem. 270:1170-1178. [DOI] [PubMed] [Google Scholar]
- 13.Kollár, R., B. B. Reinhold, E. Petráková, H. J. C. Yeh, G. Ashwell, J. Drgonová, J. C. Kaptein, F. M. Klis, and E. Cabib. 1997. Architecture of the yeast cell wall: β(1→6)glucan interconnects mannoprotein, β(1→3)glucan, and chitin. J. Biol. Chem. 272:17762-17775. [DOI] [PubMed] [Google Scholar]
- 14.McIntosh, M., B. A. Stone, and V. A. Stanisich. 2005. Curdlan and other bacterial (1→3)-β-d-glucans. Appl. Microbiol. Biotechnol. 68:163-173. [DOI] [PubMed] [Google Scholar]
- 15.No, H. K., Y. I. Cho, H. R. Kim, and S. P. Meyers. 2000. Effective deacetylation of chitin under conditions of 15 psi/121°C. J. Agric. Food Chem. 48:2625-2627. [DOI] [PubMed] [Google Scholar]
- 16.Park, J. T., and M. J. Johnson. 1949. A submicrodetermination of glucose. J. Biol. Chem. 181:149-151. [PubMed] [Google Scholar]
- 17.Sletmoen, M., and B. T. Stokke. 2008. Higher order structure of (1,3)-β-d-glucans and its influence on their biological activities and complexation abilities. Biopolymers 89:310-321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.