Abstract
Vibrio parahaemolyticus is a pathogenic marine bacterium that is the main causative agent of bacterial seafood-borne gastroenteritis in the United States. An increase in the frequency of V. parahaemolyticus-related infections during the last decade has been attributed to the emergence of an O3:K6 pandemic clone in 1995. The diversity of the O3:K6 pandemic clone and its serovariants has been examined using multiple molecular techniques including multilocus sequence analysis, pulsed-field gel electrophoresis, and group-specific PCR analysis. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has become a powerful tool for rapidly distinguishing between related bacterial species. In the current study, we demonstrate the development of a whole-cell MALDI-TOF MS method for the distinction of V. parahaemolyticus from other Vibrio spp. We identified 30 peaks that were present only in the spectra of the V. parahaemolyticus strains examined in this study that may be developed as MALDI-TOF MS biomarkers for identification of V. parahaemolyticus. We detected variation in the MALDI-TOF spectra of V. parahaemolyticus strains isolated from different geographical locations and at different times. The MALDI-TOF MS spectra of the V. parahaemolyticus strains examined were distinct from those of the other Vibrio species examined including the closely related V. alginolyticus, V. harveyi, and V. campbellii. The results of this study demonstrate the first use of whole-cell MALDI-TOF MS analysis for the rapid identification of V. parahaemolyticus.
Recent food-borne illness outbreaks have emphasized the need for rapid, robust, and low-cost methods for microbial identification. Vibrio parahaemolyticus is one of several Vibrio species that cause human infection and occur in coastal estuarine and marine environments worldwide. V. parahaemolyticus causes gastroenteritis, wound infections, and septicemia upon exposure to contaminated water or contaminated undercooked seafood. In the United States, V. parahaemolyticus is the leading causative agent of bacterial seafood-borne gastroenteritis (8). Gastroenteritis-associated V. parahaemolyticus strains typically possess one or both of the thermostable direct hemolysin genes (tdh and trh); however, recent studies have indicated the presence of additional virulence-associated genes including two type III secretion systems (6, 7, 26, 28, 33). Following the emergence of the V. parahaemolyticus O3:K6 pandemic clone in 1995, there has been a rise in the number of reported V. parahaemolyticus-associated infections each year, making this species a pathogen of increasing concern (8, 11). The V. parahaemolyticus pandemic clone was first isolated from outbreaks in Asia in 1995 with the O3:K6 serotype and has since emerged with additional serotypes (30). The worldwide spread of the V. parahaemolyticus O3:K6 clone is a recognized international public health issue that requires the use of standardized methods for global monitoring and surveillance such as pulsed-field gel electrophoresis (PFGE) (22, 34).
Initial isolation of V. parahaemolyticus is often conducted by culturing strains on thiosulfate citrate bile salts sucrose (TCBS) growth medium (15, 23). TCBS is used to selectively enrich for Vibrio spp. from cooccurring non-Vibrio strains; however, TCBS cannot differentiate V. parahaemolyticus from closely related species such as Vibrio harveyi and Vibrio campbellii. Additional molecular analyses are required to positively distinguish V. parahaemolyticus from other, closely related Vibrio species. These methods include group-specific PCR (4), multiplex PCR (38), multilocus sequence analysis (MLSA) (9, 17), comparative gene arrays (43), and whole-genome arrays (18). Often, several of these techniques are employed to distinguish V. parahaemolyticus from closely related Vibrio spp. and to provide greater resolution for discriminating among the pandemic clones (17, 18, 27). The development of a rapid method to distinguish V. parahaemolyticus from other Vibrio species including Vibrio pathogens would greatly aid the identification of strains involved in disease outbreaks when time is critical.
Recent studies have shown that whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) is a powerful tool for the rapid identification of bacteria including Streptococcus spp. (44), Salmonella strains (14), Mycobacterium spp. (35), Arthrobacter spp. (42), Listeria spp. (2), Burkholderia spp. (41), and other diverse nonfermenting clinical bacteria (12, 29). These studies have demonstrated the use of whole-cell MALDI-TOF MS analysis to generate highly reproducible and unique profiles to differentiate these bacterial strains at the species and subspecies levels. Whole-cell MALDI-TOF MS involves growing bacteria under standardized conditions and preparing cells for analysis by washing them to remove residual medium components, followed by resuspension of cells in a matrix that allows protein ionization. The cell-matrix suspension is then spotted onto a MALDI plate, each spot is ionized with a laser, and the ionizable proteins migrate based on their size resulting in the different peak sizes (kDa) in the MALDI-TOF MS spectra. Bacteria are typically grown overnight; however, the specific growth conditions and medium type must be determined and replicated to avoid condition-dependent differences in MADLI-TOF MS spectra (42). The method for preparation of the cells consists of only a few steps, and the protein ionization and generation of the spectra take several seconds. Whole-cell MALDI-TOF MS analysis can thus quickly provide accurate and reproducible generation of bacterial fingerprints that may be analyzed for the presence of biomarker peaks representative of a species or clonal group (2, 25, 35, 41, 44).
In the current study, we have developed a method for whole-cell MALDI-TOF MS identification of V. parahaemolyticus. MALDI-TOF MS analysis was used to differentiate V. parahaemolyticus from nine other Vibrio spp. (V. campbellii, V. cholerae, V. fischeri, V. fluvialis, V. harveyi, V. vulnificus, V. alginolyticus, V. mimicus, and V. mediterranei) and to identify potential V. parahaemolyticus-specific biomarker peaks. The objectives of this study were to determine whether MALDI-TOF MS analysis is reliable for (i) distinguishing V. parahaemolyticus from closely related Vibrio spp. and (ii) detecting variation among the V. parahaemolyticus pandemic clones. Furthermore, we analyzed whether strains that have undergone single gene deletions will have unique fingerprints resulting from changes in their ionizable proteins. This is the first study to use whole-cell MALDI-TOF MS analysis to generate reproducible and unique fingerprints that may be used to rapidly identify Vibrio spp. and to distinguish V. parahaemolyticus from related vibrios.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The V. parahaemolyticus strains examined in this study were isolated from illness-related human and food samples of diverse outbreaks by the Centers for Disease Control and Prevention in Atlanta, GA (Table 1), and are referred to as clinical strains (26, 28). The V. parahaemolyticus environmental strains analyzed were isolated from sediment, water, and oyster samples from Georgia, Florida, and North Carolina using TCBS as previously described (1, 10, 19) (Table 1). V. parahaemolyticus, V. fischeri (36), V. harveyi, V. alginolyticus, V. mimicus, V. vulnificus, V. mediterranei (20), V. campbellii (20), V. fluvialis (20), and V. cholerae were grown in heart infusion broth supplemented with 2% NaCl and 2% agar for plates. Escherichia coli (5) was grown in Luria-Bertani (LB) broth. All strains were grown for 24 h at 30°C with shaking.
TABLE 1.
Bacterial strains examineda
| Bacterial species | Strain | Yr of isolation | Serotype | Location | Sample type | Reference or source |
|---|---|---|---|---|---|---|
| V. parahaemolyticus | RIMD2210633 | 1996 | O3:K6 | Japan | Clinical | 26 |
| K1223 | 2004 | O3:K6 | CO | Clinical | 28 | |
| F9083 | 2002 | O3:K6 | AZ | Clinical | 28 | |
| K4435 | 2006 | O3:K6 | GA | Clinical | 28 | |
| ATCC 17802 | 1965 | O3:K6 | Japan | Clinical | 23 | |
| F8023 | 2001 | O4:K12 | GA | Clinical | 28 | |
| K1461 | 2004 | O4:K12 | MA | Clinical | 28 | |
| F5052 | 1997 | O4:K12 | WA | Clinical | 28 | |
| K3566 | 2006 | O4:K63 | LA | Clinical | 28 | |
| K4250 | 2006 | O4:K63 | NY | Clinical | 28 | |
| K4981 | 2007 | O1:Kuk | GA | Clinical | 28 | |
| K5067 | 2007 | O1:K56 | SD | Clinical | 28 | |
| K5330 | 2007 | O5:Kuk | TX | Clinical | 28 | |
| 22702 | 1998 | O5:Kuk | GA | Sediment | 10 | |
| SG176 | 2006 | O5:Kuk | GA | Water | 19 | |
| SG258 | 2006 | O1:Kuk | GA | Sediment | 19 | |
| AF2 | 2006 | O4:K9 | FL | Oyster | 19 | |
| AF67 | 2006 | O5:K37 | FL | Sediment | 19 | |
| J-C2-15 | 1998 | O1:Kuk | NC | Sediment | 1 | |
| S-M2-3-B3 | 1998 | O5:K17 | NC | Sediment | 1 | |
| V. alginolyticus | 2439-03 | 2003 | NA | HI | Clinical | C. Tarr |
| 05-2442 | 2005 | NA | AR | Clinical | C. Tarr | |
| 07-2432 | 2007 | NA | FL | Clinical | C. Tarr | |
| V. vulnificus | 08-2485 | 2008 | NA | LA | Clinical | C. Tarr |
| V. cholerae | O395 | NA | O1 | NA | Clinical | B. Hammer |
| V. mimicus | 08-2437 | 2008 | NA | IN | Clinical | C. Tarr |
| V. harveyi | ATCC BAA-1116 | 1993 | NA | NA | Ocean | B. Bassler |
| V. fischeri | ES114 | NA | NA | HI | Ocean | 36 |
| V. campbellii | 09022 | 1998 | NA | SC | Ocean | 20 |
| V. fluvialis | 0908 | 1998 | NA | SC | Ocean | 20 |
| V. mediterranei | 23023 | 1998 | NA | SC | Ocean | 20 |
| E. coli | K-12 strain MG1655 | NA | NA | NA | Ocean | 5 |
Abbreviations: NA, not available; CO, Colorado; AZ, Arizona; GA, Georgia; MA, Massachusetts; WA, Washington; LA, Louisiana; NY, New York; SD, South Dakota; TX, Texas; FL, Florida; NC, North Carolina; HI, Hawaii; AR, Arkansas; IN, Indiana; SC, South Carolina.
Serotyping.
The serotypes of the V. parahaemolyticus clinical and environmental strains were determined as previously described (13, 15). A slide agglutination test was performed using anti-O and anti-K antisera (Denka; Seiken Corp., Tokyo, Japan).
MALDI-TOF MS.
The MALDI-TOF MS spectra were generated on an Applied Biosystems Voyager DE STR MALDI-TOF mass spectrometer with Data Explorer software operated in the linear, delayed-extraction, and positive-ion modes. The laser (N2, 337 nm) intensity was set above the ion generation threshold. The low-mass gate was m/z 2,400 with a delay time of 750 ns. The accelerating voltage was 25,000 V, and the grid voltage was 95% of the accelerating voltage. The mass spectra were acquired by accumulating a total of 300 laser shots over the m/z range 2,500 to 15,000. The resulting mass spectra were calibrated using the doubly (m/z 6,181) and singly (m/z 12,361) charged ions of the cytochrome c internal standard.
Vibrio strains were grown for 24 h in 3 ml of heart infusion broth with shaking (200 rpm) at 30°C. Following incubation, 1 ml of each culture was transferred to a microcentrifuge tube and centrifuged for 2 min at 13,400 × g. The supernatant was decanted, and cells were washed once in 1 ml of 0.85% NaCl and twice in 1 ml of 50% ethanol with centrifugation in between each of the three washes for 2 min at 13,400 × g. The tubes containing the cell pellets were weighed, and the cells were resuspended in 1% trifluoroacetic acid (TFA) to yield a final concentration of 0.2 mg/μl of cells in TFA. Equal volumes of the TFA bacterial suspension and the optimized MALDI-TOF MS matrix (10-mg/ml sinapinic acid in 50% acetonitrile, 50% water, 0.1% TFA, and 2 pmol/μl of the internal standard cytochrome c) were mixed in a microcentrifuge tube, and then 1.5 μl of this mixture was spotted in triplicate on a stainless steel MALDI-TOF MS sample plate. Samples were allowed to air dry before being loaded in the mass spectrometer. MALDI-TOF MS spectra were generated for each strain by performing three runs on three independently grown cultures.
MALDI-TOF MS data analysis.
The protein peaks analyzed were those that possessed peak areas of 0.5% or greater relative to the most intense peak. A total of three separate runs for each strain analyzed were combined to generate a composite list of peaks. The peaks that had similar upper and lower mass bounds were considered to be a single peak and were averaged to account for the peak variation between runs for each strain. Peaks not present in two or more independently generated spectra for each strain were excluded from the analysis. A composite spectrum was generated for each strain by excluding peaks that were not replicated. A total of 132 peaks that were present in one or more strains were examined by cluster analysis. A binary matrix of 0 and 1 was assigned for each peak to indicate the presence or absence of a peak for cluster analysis. The similarity matrix was analyzed using PAST (v.1.34) (http://folk.uio.no/ohammer/past/doc1.html) as previously described (44). The Dice coefficient with single linkages was used for cluster analysis of MALDI-TOF MS data.
Identification of potential biomarker peaks.
Representative peaks that were present in the MALDI-TOF MS profiles of multiple species and strains for possible use as biomarkers for species and strain level identification were identified using the ExPASy TagIdent tool (16). We examined the presence of peaks within the m/z range of singly and doubly charged cytochrome c that may be due to matrix or positive ion adducts with Na+, Mg2+, K+, and Ca2+. Peaks differing between the ΔopaR strain and RIMD2210633 MALDI-TOF MS spectra and the ΔmutS strain and ATCC 17802 spectra were examined by comparison with predicted proteins of the RIMD2210633 genome using TagIdent (16). The ΔopaR and ΔmutS mutants were constructed as previously described (19).
PFGE.
PFGE of the V. parahaemolyticus clinical and environmental strains was performed as previously described (22, 34). Bacterial strains embedded in agarose plugs were digested with 50 U of concentrated SfiI (40 U/μl) and 40 U of NotI (Roche, Mannheim, Germany). Restricted DNA fragments were separated by gel electrophoresis using 0.5× Tris-borate-EDTA buffer at 14°C for 18 h in a CHEF Mapper electrophoresis system (Bio-Rad Laboratories, Hercules, CA) with initial and final switch times of 10 s and 35.03 s, respectively. PFGE patterns were analyzed with BioNumerics software v.5.01 (Applied-Maths, Kortrijk, Belgium). The patterns were compared using the Dice coefficient with a band position tolerance and optimization of 1.5%, and clustering was performed using the unweighted pair group method with arithmetic averages. Salmonella enterica serovar Braenderup H9812 restricted with 50 U of XbaI (Roche, Mannheim, Germany) was used as a control strain for gel normalization.
MLSA.
MLSA of seven housekeeping genes PCR amplified from V. parahaemolyticus clinical and environmental strains was performed using previously developed primers that contained the M13 forward and reverse primers to be used for sequencing (17). PCR amplification was performed with NEB Phusion High Fidelity polymerase with the GC buffer. All PCR amplicons were gel purified on a 0.7% SeaKem LE agarose gel (Lonza, Switzerland) containing ethidium bromide. The expected products were excised from the gel and extracted using a Sigma GenElute gel extraction kit (Sigma-Aldrich, St. Louis, MO). Sequencing was performed using M13 universal primers at the Georgia Institute of Technology Genomics Core Facility.
Sequence alignments and a concatenation of the MLSA data were performed in MEGA (v.4.1) (24). A neighbor-joining tree was constructed from the concatenated nucleotide sequences with the Kimura two-parameter correction model and 1,000 bootstrap replications. The concatenated tree was constructed from recA (642-nucleotide [nt]), gyrB (618-nt), pntA (414-nt), pyrC (531-nt), dtdS (498-nt), dnaE (587-nt), and tnaA (470-nt) nucleotide sequences.
Nucleotide sequence accession numbers.
The seven housekeeping gene sequences (recA, gyrB, pyrC, pntA, dtdS, dnaE, and tnaA) for each V. parahaemolyticus strain examined in this study were deposited in GenBank under the accession numbers FJ577370 to FJ577500. The recA sequences of K1223 and 22702 were previously submitted under the accession numbers EU652262 and EU018456, respectively. The nucleotide sequences of the other vibrios and the E. coli strain examined in the housekeeping gene analysis were available in GenBank.
RESULTS
Whole-cell MALDI-TOF MS identification of Vibrio species.
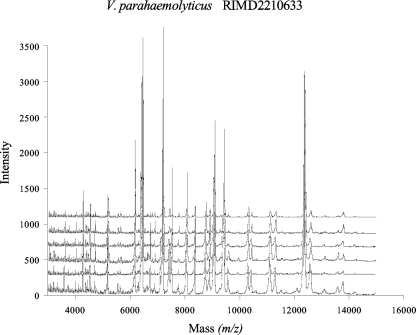
MALDI-TOF MS fingerprinting of whole-cell lysates produced spectra that were highly reproducible for the pandemic O3:K6 V. parahaemolyticus strain RIMD2210633 (Fig. 1). The spectra shown in Fig. 1 represent six individual runs of V. parahaemolyticus RIMD2210633 that were generated from different overnight cultures grown on different days. Identical prominent peaks were detected for the six different spectra generated from V. parahaemolyticus RIMD2210633; however, the peak intensity was variable for each of the cultures analyzed (Fig. 1).
FIG. 1.
MALDI-TOF MS spectra of V. parahaemolyticus RIMD2210633 showing the reproducibility of spectra. Each MALDI-TOF MS spectrum represents an independently generated data set.
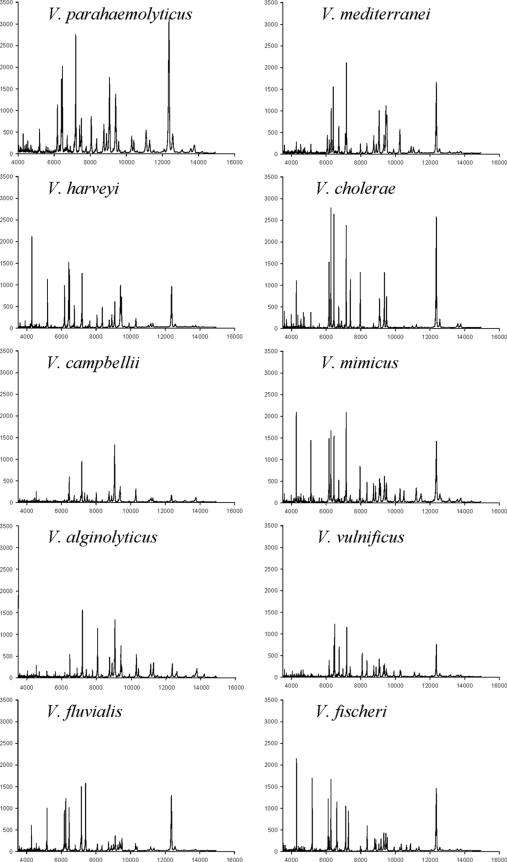
MALDI-TOF MS fingerprinting of phylogenetically diverse Vibrio strains using whole-cell lysates revealed spectra that could be used to distinguish V. parahaemolyticus from other Vibrio species including the closely related V. alginolyticus, V. harveyi, and V. campbellii (Fig. 2). Two additional species examined, previously identified by phylogenetic analysis as being related to V. fluvialis-V. furnissii and V. mediterranei (20), exhibited different spectra. In addition, unique and distinguishable spectra were generated for the distantly related species V. vulnificus, V. mimicus, V. cholerae, and V. fischeri. The spectra generated for each of the seven Vibrio spp. and E. coli revealed unique and reproducible MALDI-TOF MS fingerprints.
FIG. 2.
Whole-cell MALDI-TOF MS spectra of 10 Vibrio spp. including the V. parahaemolyticus O3:K6 clonal pandemic strain RIMD2210633. The mass intensity is on the y axis, and the x axis indicates the mass-to-charge ratio (m/z). Each spectrum is a representative run from a total of three independent runs performed for each strain.
MALDI-TOF MS analysis of V. parahaemolyticus clinical and environmental strains.
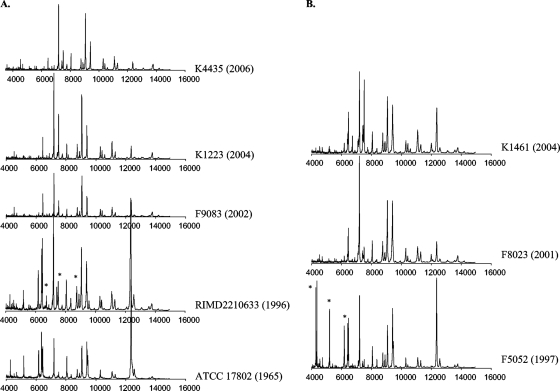
We examined whether MALDI-TOF MS fingerprints can be used to distinguish between V. parahaemolyticus strains including ones belonging to the O3:K6 pandemic clone. We analyzed V. parahaemolyticus O3:K6 clones and the closely related clinical O4:K12 strains compared to diverse clinical and environmental strains. There were visual differences between the MALDI-TOF MS spectra of post-1995 clonal O3:K6 strains (K4435, K1223, F9083, and RIMD2210633) and that of a pre-1995 O3:K6 strain (ATCC 17802) (Fig. 3A). Overall, the spectra of the post-1995 O3:K6 strains were very similar. There were two peaks present in the spectra of the RIMD2210633 pandemic clone isolated from Japan that were not present in the spectra of the post-1995 strains K4435, K1223, and F9083, which were isolated from disease-associated samples in the United States. These peaks are located at approximately m/z 5,000 and 8,500. In addition, there were several prominent peaks within the m/z 7,000 to 9,500 range of the spectra of the post-1995 O3:K6 strains that were absent from the spectrum of the pre-1995 O3:K6 strain.
FIG. 3.
(A) MALDI-TOF MS of V. parahaemolyticus post-1995 clonal pandemic O3:K6 strains (K4435, K1223, F9083, and RIMD2210633) and a pre-1995 nonclonal O3:K6 strain, ATCC 17802. The x axis indicates the mass-to-charge ratio of each peak (m/z). The y axis of each individual plot indicates the peak intensity. The year that each strain was isolated is indicated in parentheses. Peaks present in the RIMD2210633 spectra that were absent in the ATCC 17802 spectra are indicated by asterisks. (B) MALDI-TOF MS of the clonal O4:K12 strains K1461, F8023, and F5052. The x axis indicates the mass-to-charge ratio of each peak (m/z). The y axis of each individual plot indicates the peak intensity. The year that each strain was isolated is indicated in parentheses. Peaks present in one spectrum that were absent in another are indicated by asterisks.
In addition, we examined the differences in the MALDI-TOF MS fingerprints of the closely related O4:K12 clinical strains (K1461, F8023, and F5052) (Fig. 3B). The spectra of the more recently isolated strains F8023 and K1461 were nearly identical, except for a few additional peaks present in the m/z 6,000 to 8,000 range of the K1461 spectrum. In contrast, there were additional peaks detected in the m/z 4,000 to 6,000 range of F5052 that were not present in the spectra of F8023 and K1461. The majority of the peaks were present in the m/z 6,000 to 13,000 range, except for three additional peaks of F5052 that were present in the m/z 4,000 to 5,000 range. There was a peak detected at m/z 10,428 to 10,432 that was present exclusively in the three O4:K12 strains, indicating that this peak may be a biomarker for the closely related O4:K12 strains.
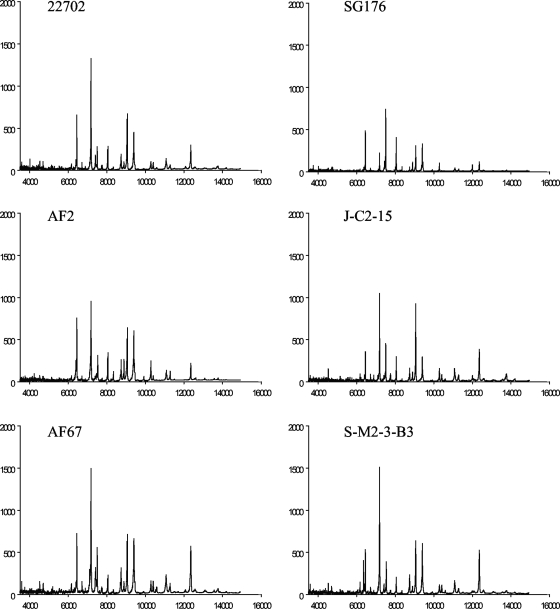
The V. parahaemolyticus environmental strains examined exhibited unique MALDI-TOF MS fingerprints (Fig. 4). There were visual similarities present among strains isolated from the same location such as the J-C2-15 and S-M2-3-B3 strains isolated from North Carolina. Overall, there were many similar peaks present in the spectra generated for the V. parahaemolyticus environmental strains.
FIG. 4.
MALDI-TOF MS of environmental strains isolated from sediment and water samples from coastal Georgia, Florida, and North Carolina. The x axis indicates the mass-to-charge ratio of each peak (m/z). The y axis of each individual plot indicates the mass intensity.
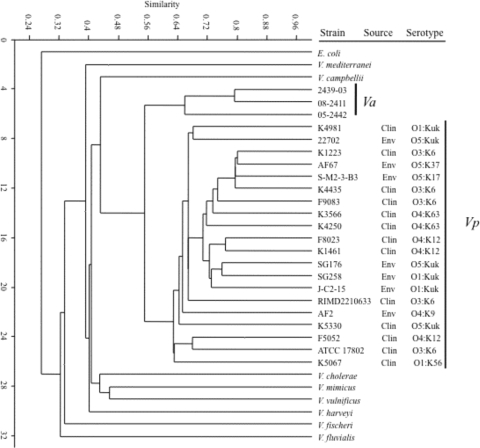
Single linkage cluster analysis performed in PAST with the Dice coefficient was used to generate a dendrogram that showed the relative differences in the MALDI-TOF MS fingerprints of the bacterial strains analyzed (Fig. 5). The V. parahaemolyticus strains formed a distinct group that was separate from the other vibrios examined. The V. parahaemolyticus strains examined formed a distinct group from the other Vibrio spp. and E. coli. The V. parahaemolyticus strains exhibited from 64 to 80% similarity based on binary analysis of the absence or presence of MALDI-TOF MS peaks. The V. parahaemolyticus strains exhibited 56 to 80% similarity relative to the V. alginolyticus strains, which formed a distinct group. In contrast, the MALDI-TOF MS fingerprints of V. parahaemolyticus exhibited 40 to 45% similarity to those of V. harveyi, V. campbellii, V. mimicus, V. vulnificus, V. mediterranei, and V. cholerae and approximately 32% similarity to the fingerprints of V. fluvialis and V. fischeri. Cluster analysis showed that there were different levels of similarity in the MALDI-TOF MS fingerprints from the V. parahaemolyticus strains examined. The V. parahaemolyticus environmental strains SG176 and SG258, which were isolated from coastal Georgia in 2006, exhibited similarity that demonstrated that these strains had unique MALDI-TOF MS fingerprints in comparison with the V. parahaemolyticus environmental strain 22702, which was isolated from coastal Georgia in 1998. The V. parahaemolyticus environmental strain AF67, which was isolated from Florida, exhibited comparable similarities to the environmental strains J-C2-15 and S-M2-3-B3, which were isolated from North Carolina in 1998. Cluster analysis of V. parahaemolyticus O3:K6 and O4:K12 clones examined confirmed that the MALDI-TOF MS fingerprints had different levels of similarity (Fig. 5), as was suggested in a visual comparison of the MALDI-TOF MS fingerprints (Fig. 3). An exception is the O4:K12 strains F8023 and K1461, which exhibited comparable similarity in their MALDI-TOF MS fingerprints both by cluster analysis (Fig. 5) and by visual inspection (Fig. 3).
FIG. 5.
Cluster analysis of Vibrio spp. examined using the Jaccard similarity coefficient with single linkage associations. The similarity represents the relationship of strains as determined by a binary matrix denoting the presence or absence of peaks in the MALDI-TOF MS fingerprints. Clin, clinical; Env, environmental.
Identification of V. parahaemolyticus strains by PFGE and MLSA.
Analysis of the PFGE patterns revealed the genetic diversity of the 20 V. parahaemolyticus clinical and environmental strains (Fig. 6). Exceptions to this were the grouping of the O3:K6 clonal pandemic strains as well as the O4:K12 strains in the dendrogram, indicating genetic homogeneity within these two serotypes. The PFGE profiles of the remaining clinical and environmental strains did not exhibit relatedness based on serotype or isolation source.
FIG. 6.
Dendrogram comparing SfiI and NotI PFGE patterns of clinical and environmental V. parahaemolyticus strains.
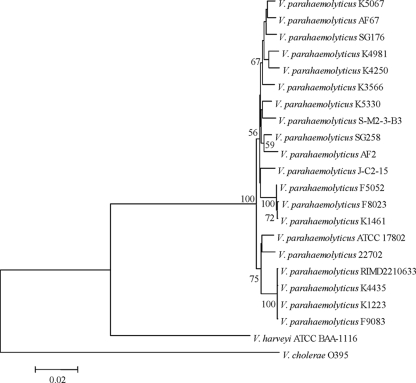
Another molecular method frequently used to identify V. parahaemolyticus is MLSA. To determine whether MALDI-TOF MS analysis of whole-cell lysate is reliable for distinguishing V. parahaemolyticus from other vibrios and for investigating the relatedness of V. parahaemolyticus strains, we compared MALDI-TOF MS analysis with an MLSA of seven housekeeping genes from all V. parahaemolyticus strains examined. There were two closely related groups within the V. parahaemolyticus group, one composed of isolates of the O3:K6 clone and the other consisting of isolates with the O4:K12 serotype (Fig. 7). An exception for the overall concatenation was the presence of strain 22702 in a group with the O3:K6 strains. V. parahaemolyticus 22702 was isolated from a sediment sample from a Georgia salt marsh and has the O5:Kuk serotype. Analysis of the individual gene trees of each of the seven housekeeping genes showed that 22702 was related to other environmental strains for six of the seven genes. In the recA tree V. parahaemolyticus 22702 formed a group with the O3:K6 strains (data not shown). The relatedness of 22702 to the O3:K6 strains was also observed in the seven-gene concatenated tree (Fig. 7).
FIG. 7.
A neighbor-joining tree constructed with the concatenated nucleotide sequences (3,760 nt) of the seven housekeeping genes recA (642 nt), gyrB (618 nt), pntA (414 nt), pyrC (531 nt), dtdS (498 nt), dnaE (587 nt), and tnaA (470 nt) using the Kimura two-parameter correction model and 1,000 bootstrap replications. The nucleotide sequences of all other vibrios and E. coli were available from GenBank. The scale bar represents the number of substitutions per site. Only bootstrap values of ≥50 are shown.
Identification of potential biomarker peaks and differentiation of wild-type and mutant strains by MALDI-TOF MS analysis.
There were a total of 132 conserved peaks that were shared by at least two or more strains or species analyzed. Of the conserved peaks, there were 30 that were detected only in the spectra of V. parahaemolyticus strains (Table 2; see also Table S1 in the supplemental material). In addition, there was a peak present at m/z 6,409 that was detected in the spectra of 15 of the 20 V. parahaemolyticus strains examined as well as V. campbellii, V. mediterranei, and E. coli, suggesting that this peak may be a useful biomarker for the identification of Gammaproteobacteria.
TABLE 2.
Predicted proteins of select whole-cell MALDI-TOF MS peaks
| Peak mass (Da) | Predicted gene | Predicted protein | Species or strain(s) in which present |
|---|---|---|---|
| Select potential V. parahaemolyticus biomarker peaks | |||
| 3,601 | 0 | V. parahaemolyticus | |
| 4,706 | 0 | V. parahaemolyticus, V. alginolyticus | |
| 6,368 | 0 | V. parahaemolyticus | |
| 6,724 | 0 | V. parahaemolyticus | |
| 8,712 | VP3074 | ATP synthase subunit c | V. parahaemolyticus |
| VP2780 | SlyX | ||
| VP0098 | Sec-independent protein translocase protein TatA/E homolog | ||
| 9,933 | VP2817 | Hfq | V. parahaemolyticus |
| VP2331 | 50S ribosomal protein L31 type B | ||
| VP2582 | Hypothetical protein | ||
| VP0160 | RNA polymerase omega subunit RpoZ | ||
| 10,431 | VP1210 | 50S ribosomal protein L25 | V. parahaemolyticus (O4:K12) |
| VP0261 | 30S ribosomal protein S19 | ||
| 11,294 | VP1294 | IHF-α | V. parahaemolyticus, V. harveyi |
| VP1283 | Hypothetical protein | ||
| VP0268 | 50S ribosomal protein L24 | ||
| 12,088 | VPA1222 | Hypothetical protein GIY-YIG catalytic domain | V. parahaemolyticus, V. alginolyticus |
| VP2986 | CyaY | ||
| VP2923 | 50S ribosomal protein L7/L12 | ||
| VP2766 | Met regulon regulatory protein MetJ | ||
| VP2178 | YbaB-like hypothetical protein | ||
| VP0262 | 50S ribosomal protein L22 | ||
| 13,756-13,765 | VP0280 | 30S ribosomal protein S11 | V. parahaemolyticus |
| VP3028 | Protein CrcB homolog | ||
| VP2843 | Fumarate reductase subunit D | ||
| VP2773 | 30S ribosomal protein S12 | ||
| VP2568 | Holo-acyl carrier protein synthase | ||
| VP0004 | RNase P protein | ||
| 14,211 | VP0448 | Hypothetical protein | V. parahaemolyticus, V. alginolyticus |
| VP0283 | 50S ribosomal protein L17 | ||
| RIMD2210633 vs opaR strain | |||
| 6,612 | VP0275 | 50S ribosomal protein L30 | RIMD2210633 |
| 7,155 | VP2529 | Zinc-binding protein | opaR strain |
| VP0265 | 50S ribosomal protein L29 | ||
| 7,414 | VP2335 | Hypothetical protein | RIMD2210633 |
| 9,931 | VP2817 | Hfq | opaR strain |
| VP2331 | 50S ribosomal protein L31 type B | ||
| VP0160 | RNA polymerase omega subunit RpoZ | ||
| 10,416 | VP1210 | 50S ribosomal protein L25 | RIMD2210633 |
| VP0261 | 30S ribosomal protein S19 | ||
| 10,594 | VPA0321 | Hypothetical protein sigma 70 region | RIMD2210633 |
| VPA0286 | GroES protein 2 | ||
| VP2627 | Probable Fe2+-trafficking protein | ||
| VP2619 | YggU-like hypothetical protein | ||
| VP2029 | IHF-β | ||
| 13,093 | VPA0577 | Pterin-4-alpha-carbinolamine dehydratase | RIMD2210633 |
| VP2530 | 50S ribosomal protein L19 | ||
| VP0646 | Hypothetical protein | ||
| 14,209 | VP0448 | Hypothetical protein | RIMD2210633 |
| VP0283 | 50S ribosomal protein L17 | ||
| ATCC 17802 vs mutS strain | |||
| 6,287 | VP0082 | Transmembrane protein | ATCC 17802 |
| 6,574 | VP0275 | 50S ribosomal protein L30 | ATCC 17802 |
| VP0186 | 50S ribosomal protein L33 | ||
| 7,167 | VP2546 | CsrA | ATCC 17802 |
| 10,416 | VP1210 | 50S ribosomal protein L25 | ATCC 17802 |
| VP0261 | 30S ribosomal protein S19 | ||
| 12,569 | VPA1414 | Hypothetical protein, ASCH domain | ATCC 17802 |
| VP0273 | 50S ribosomal protein L18 | ||
| VP2098 | Phosphorelay protein LuxU |
Identification of the potential proteins corresponding to the potential biomarker peaks was performed in silico by searching the sequenced V. parahaemolyticus RIMD2210633 genome using the TagIdent tool of ExPASy (16). The majority of the peaks tentatively identified by comparison with the V. parahaemolyticus RIMD2210633 predicted proteome were ribosomal proteins (Table 2). In addition, there were peaks that corresponded to regulatory proteins such as Hfq and RpoZ and numerous hypothetical proteins (Table 2; see also Table S1 in the supplemental material). Frequently, there were multiple predicted proteins corresponding to certain peaks and certain proteins that corresponded to multiple peaks within a certain m/z range. V. parahaemolyticus-specific peaks are shown in Table 2, and all conserved peaks that were present in two or more species are listed in Table S1 in the supplemental material.
To determine whether MALDI-TOF MS analysis is sensitive enough to generate unique fingerprints for V. parahaemolyticus strains that have undergone changes in single genes, we compared the MALDI-TOF MS fingerprints of an in-frame deletion mutant of the quorum sensing regulator opaR and a deletion mutant of the mismatch repair gene mutS to those of the wild-type strains (see Fig. S1 in the supplemental material). The MALDI-TOF MS spectrum of a V. parahaemolyticus RIMD2210633 ΔopaR strain had peaks of lower intensity than those of the wild-type strain (see Fig. S1 in the supplemental material). While most of the peaks were conserved between the RIMD2210633 and ΔopaR strain spectra, there were a few peaks that differed. For example, there were a peak at approximately m/z 6,612 and a peak at m/z 7,414 that were present in the RIMD2210633 spectrum and absent from the ΔopaR strain spectrum (see Fig. S1 in the supplemental material) that were predicted to correspond to a 50S ribosomal protein and a hypothetical protein (Table 2). There were four additional peaks of lower intensity that were present in at least two independently generated spectra of RIMD2210633 that were absent from the opaR strain spectra (Table 2). The additional RIMD2210633 peaks were tentatively assigned as possible hypothetical proteins, 50S and 30S ribosomal proteins, the chaperonin GroES, the integration host factor IHF-β, and a dehydratase (Table 2). There were two peaks present in the ΔopaR strain spectra that were repeatedly absent from the RIMD2210633 spectra at approximately m/z 7,154 and 9,930 (Table 2). The unique ΔopaR strain peaks were assigned as a possible zinc-binding protein, a ribosomal protein, and the regulatory proteins Hfq and RpoZ (Table 2).
There were no visible differences in the spectra of ATCC 17802 and the ΔmutS mutant (see Fig. S1 in the supplemental material); however, there were several low-intensity peaks detected that differed between the fingerprints of ATCC 17802 and the ΔmutS strain. There was a peak present at m/z 6,287 in the spectrum of ATCC 17802 and absent from the ΔmutS strain that corresponded to a transmembrane protein (Table 2). In addition, there were peaks at m/z 6,574, 7,167, 10,416, and 12,568 that were predicted to be 50S ribosomal proteins, CsrA, a hypothetical protein, and the phosphorelay protein LuxU that were detected for ATCC 17802 and not the ΔmutS strain (Table 2).
DISCUSSION
An increase in the frequency of V. parahaemolyticus-associated infections has heightened the need for a method of rapid and reliable identification of V. parahaemolyticus strains (11, 30). Current methods for identification of V. parahaemolyticus involve biochemical tests (15); serotyping, PCR screening, and DNA hybridization analysis of housekeeping genes or disease-associated genes (3, 28, 32, 38, 43); MLSA (9, 17); and PFGE (22, 27, 34). Often, distinction of V. parahaemolyticus from other closely related vibrios requires the use of multiple techniques.
In the current study, we demonstrate the development of a high-throughput whole-cell MALDI-TOF MS technique for the identification of V. parahaemolyticus clinical and environmental strains, which takes less than 2 minutes for each sample spot. In this study, we demonstrate that whole-cell MALDI-TOF MS analysis of V. parahaemolyticus and other Vibrio spp. is a highly reproducible method that is effective for distinguishing between related species as previously demonstrated for other bacteria (37, 44). Comparison of MALDI-TOF MS fingerprints and cluster analysis of the peaks can be used to distinguish V. parahaemolyticus from other vibrios including the closely related V. alginolyticus, V. harveyi, and V. campbellii. Often, these three species are nearly indistinguishable from each other by 16S rRNA gene analysis, requiring analysis of multiple housekeeping genes for accurate species identification (39, 40). We identified a total of 30 peaks that were present only in the spectra of the V. parahaemolyticus strains examined in this study. These peaks could be potential V. parahaemolyticus biomarkers, though further investigation will be needed to validate their use as biomarkers for the rapid detection of V. parahaemolyticus by whole-cell MALDI-TOF MS. In the present study, we normalized the spectra to a threshold that excluded most of the peaks from the several thousand that were generated for each strain, reducing the number of analyzable peaks to a manageable set of approximately 30 to 60 per spectra. Analysis of a larger number of strains and exclusion of outlier peaks would increase the level of similarity and better resolve the relationships among the V. parahaemolyticus strains in the cluster analysis.
In addition, we show that isolates of the V. parahaemolyticus pandemic clone exhibited spectra that had detectable and reproducible differences in the numbers and positions of peaks. This was especially true when comparing more recently isolated strains to older strains. For example, the more recently isolated clonal O3:K6 and O4:K12 strains exhibited MALDI-TOF MS fingerprints different from those of strains isolated in 1996 and 1997 (when the pandemic clones were first detected). In addition, there was one peak at m/z 10,430 that was detected only for the O4:K12 strains analyzed. Analysis of additional clonal O4:K12 strains will be required to ascertain whether this peak and additional peaks can be used as biomarkers for identification of the O4:K12 strains that are frequently associated with disease outbreaks in the United States (8, 30).
Furthermore, differences in the whole-cell MALDI-TOF MS spectra of some of the V. parahaemolyticus strains examined may reflect strain adaptation to a particular geographic location. For example, the MALDI-TOF MS spectra of the V. parahaemolyticus O3:K6 strains RIMD2210633 and ATCC 17802, which were isolated from Japan, exhibited more similarities than the spectra of O3:K6 strains isolated from the United States. In addition, the spectra of the V. parahaemolyticus environmental strains SG176 and SG258, both of which were isolated from coastal Georgia, exhibited many similarities. The analysis of additional V. parahaemolyticus strains isolated from distinct geographical regions and development of biomarker peaks are required to support the use of MALDI-TOF MS for identification of the geographical origin of certain strain types. In this regard, MALDI-TOF MS could be a powerful tool to monitor the emergence and spread of clonal groups as they become more frequent causative agents of disease outbreaks, such as the emergence of the O3:K6 clonal pandemic strain in India in 1995 (31). Overall, the detectable differences among the isolates of the pandemic clone and the closely related O4:K12 strains suggest that MALDI-TOF MS may be a valuable tool to distinguish between isolates of the pandemic clone that were associated with outbreaks during different years and possibly biogeographic regions. Further analysis of a larger number of the V. parahaemolyticus isolates of the pandemic clone would be required in order to develop a database of fingerprints and to validate the usefulness of MALDI-TOF MS for rapidly tracking the emergence and distribution of the disease-associated strains. In addition, the development of a whole-cell MALDI-TOF MS fingerprint database for the rapid identification of diverse vibrios would require the use of a standardized protocol that exhibits interlaboratory reproducibility and consists of an optimized medium type and growth conditions for each species.
In this study, we demonstrate that whole-cell MALDI-TOF MS analysis is sensitive enough to detect only a few genetic changes such as inactivation of a single regulatory gene or DNA repair pathway. Whole-cell MALDI-TOF MS analysis showed variation in the MALDI-TOF MS fingerprints of a single strain following deletion of the quorum sensing regulator opaR or the mismatch repair gene mutS, which results in an increased accumulation of mutations in V. parahaemolyticus (19). Quorum sensing has been shown to regulate type III secretion of V. parahaemolyticus (21), which was shown to translocate effector proteins, resulting in cytotoxicity to eukaryotic cells (33). The ability to monitor changes in global regulatory systems such as quorum sensing that are involved in the pathogenicity mechanism of vibrios would provide additional information on the nature of disease-causing strains.
In this study, we report the first use of whole-cell MALDI-TOF MS analysis as a powerful tool for identification of V. parahaemolyticus strains. Based on the findings of our study, the rapid and reliable generation of whole-cell MALDI-TOF MS fingerprints would be an important tool for the initial identification of V. parahaemolyticus and other Vibrio spp. In addition, we identified potential peaks that could be further developed into biomarkers for detection of V. parahaemolyticus and analysis of differences among disease-causing strains. Further application of this method would involve the construction of a database of whole-cell MALDI-TOF MS fingerprints from V. parahaemolyticus and related disease-causing vibrios that could be referenced for identification of the causative agents of disease outbreaks. Research is ongoing to determine whether the whole-cell MALDI-TOF MS approach described in this study may be used to detect multiple disease-causing vibrios present in food or environmental samples.
Supplementary Material
Acknowledgments
This project was supported in part by GT/CDC seed grant no. 1241326 to P. A. Sobecky and C. A. Bopp. Instrumentation for sequencing was supported by NSF grant DBI-0304606.
We thank B. Hammer for providing the V. cholerae and V. harveyi strains and C. Tarr for providing the V. vulnificus, V. alginolyticus, and V. mimicus strains used in this study.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bagwell, C. E., Y. M. Piceno, A. Ashburne-Lucas, and C. R. Lovell. 1998. Physiological diversity of the rhizosphere diazotroph assemblages of selected salt marsh grasses. Appl. Environ. Microbiol. 64:4276-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbuddhe, S. B., T. Maier, G. Schwarz, M. Kostrzewa, H. Hof, E. Domann, T. Chakraborty, and T. Hain. 2008. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5402-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bej, A. K., D. P. Patterson, C. W. Brasher, M. C. Vickery, D. D. Jones, and C. A. Kaysner. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh, and trh. J. Microbiol. Methods 36:215-225. [DOI] [PubMed] [Google Scholar]
- 4.Bhoopong, P., P. Palittapongarnpim, R. Pomwised, A. Kiatkittipong, M. Kamruzzaman, Y. Nakaguchi, M. Nishibuchi, M. Ishibashi, and V. Vuddhakul. 2007. Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J. Clin. Microbiol. 45:1544-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Boyd, E. F., A. L. V. Cohen, L. M. Naughton, D. W. Ussery, T. T. Binnewies, O. C. Stine, and M. A. Parent. 2008. Molecular analysis of the emergence of pandemic Vibrio parahaemolyticus. BMC Microbiol. 8:110-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burdette, D. L., M. L. Yarbrough, A. Orvedahl, C. J. Gilpin, and K. Orth. 2008. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proc. Natl. Acad. Sci. USA 105:12497-12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2006. Vibrio parahaemolyticus infections associated with consumption of raw shellfish—three states, 2006. MMWR Morb. Mortal. Wkly. Rep. 55:854-856. [PubMed] [Google Scholar]
- 9.Chowdhury, N. R., O. C. Stine, J. G. Morris, and G. B. Nair. 2004. Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J. Bacteriol. 42:1280-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Criminger, J. D., T. H. Hazen, P. A. Sobecky, and C. R. Lovell. 2007. Nitrogen fixation by Vibrio parahaemolyticus and its implications for a new ecological niche. Appl. Environ. Microbiol. 73:5959-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniels, N. A., L. MacKinnon, R. Bishop, S. Altekruse, B. Ray, R. M. Hammond, S. Thompson, S. Wilson, N. H. Bean, P. M. Griffin, and L. Slutsker. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 12.Degand, N., E. Carbonelle, B. Dauphin, J. L. Beretti, M. Le Bourgeois, I. Sermet-Gaudelus, C. Segonds, P. Berche, X. Nassif, and A. Ferroni. 2008. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePaola, A., J. Ulaszek, C. A. Kaysner, B. J. Tenge, J. L. Nordstrom, J. Wells, N. Puhr, and S. M. Gendel. 2003. Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microbiol. 69:3999-4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieckmann, R., R. Helmuth, M. Erhard, and B. Malorny. 2008. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:7767-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliot, E. L., C. A. Kaysner, L. Jackson, and M. L. Tamplin. 1998. Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and other Vibrio spp., p. 9.01-9.27. In U.S. Food and Drug Administration bacteriological analytical manual, 8th ed. AOAC International, Gaithersburg, MD.
- 16.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607. In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ.
- 17.González-Escalona, N., J. Martinez-Urtaza, J. Romero, R. T. Espejo, L. Jaykus, and A. DePaola. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, H., H. C. Wong, B. Kan, Z. Guo, X. Zeng, S. Yin, X. Liu, R. Yang, and D. Zhou. 2008. Genome plasticity of Vibrio parahaemolyticus: microevolution of the ‘pandemic group.’ BMC Genomics 9:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazen, T. H., K. D. Kennedy, S. Chen, S. V. Yi, and P. A. Sobecky. 2009. Inactivation of mismatch repair increases the diversity of Vibrio parahaemolyticus. Environ. Microbiol. 11:1254-1266. [DOI] [PubMed] [Google Scholar]
- 20.Hazen, T. H., D. Wu, J. A. Eisen, and P. A. Sobecky. 2007. Sequence characterization and comparative analysis of three plasmids isolated from environmental Vibrio spp. Appl. Environ. Microbiol. 73:7703-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kam, K. M., C. K. Y. Luey, M. B. Parsons, K. L. F. Cooper, G. B. Nair, M. Alam, M. A. Islam, D. T. L. Cheung, Y. W. Chu, T. Ramamurthy, G. P. Pazhani, S. K. Bhattacharya, H. Watanabe, J. Terajima, E. Arakawa, O.-A. Ratchtrachenchai, S. Huttayananont, E. M. Ribot, P. Gerner-Smidt, and B. Swaminathan. 2008. Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: an international multicenter collaborative study. J. Clin. Microbiol. 46:2766-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, T., S. Enomoto, R. Sakazaki, and S. Kuwahara. 1963. A new selective isolation medium for the Vibrio group; on a modified Nakanishi's medium (TCBS agar medium). Nippon Saikingaku Zasshi 18:387-392. [DOI] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 25.Lay, J. O. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172-194. [DOI] [PubMed] [Google Scholar]
- 26.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Urtaza, J., A. Lozano-Leon, A. DePaola, M. Ishibashi, K. Shimada, M. Nishibuchi, and E. Liebana. 2004. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J. Clin. Microbiol. 42:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meador, C. E., M. B. Parsons, C. A. Bopp, P. Gerner-Smidt, J. A. Painter, and G. J. Vora. 2007. Virulence gene- and pandemic group-specific marker profiling of clinical Vibrio parahaemolyticus isolates. J. Clin. Microbiol. 45:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellmann, A., J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. LaSala, M. Kostrzewa, and D. Harmsen. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair, G. B., T. Ramamurthy, S. K. Bhattacharya, B. Dutta, Y. Takeda, and D. A. Sack. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okuda, J., M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panicker, G., D. R. Call, M. J. Krug, and A. K. Bej. 2004. Detection of pathogenic Vibrio spp. in shellfish by using multiplex PCR and DNA microarrays. Appl. Environ. Microbiol. 70:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, K. S., T. Ono, M. Rokuda, M. H. Jang, K. Okada, T. Iida, and T. Honda. 2004. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72:6659-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsons, M. B., K. L. Cooper, K. A. Kubota, N. Puhr, S. Simington, P. S. Calimlim, D. Schoonmaker-Bopp, C. Bopp, B. Swaminathan, P. Gerner-Smidt, and E. M. Ribot. 2007. PulseNet USA standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio parahaemolyticus. Foodborne Pathog. Dis. 4:285-292. [DOI] [PubMed] [Google Scholar]
- 35.Pignone, M., K. M. Greth, J. Cooper, D. Emerson, and J. Tang. 2006. Identification of mycobacteria by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. J. Clin. Microbiol. 44:1963-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauer, S., A. Freiwald, T. Maier, M. Kube, R. Reinhardt, M. Kostrzewa, and K. Geider. 2008. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One 3:e2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarr, C. L., J. S. Patel, N. D. Puhr, E. G. Sowers, C. A. Bopp, and N. A. Strockbine. 2007. Identification of Vibrio isolates by a multiplex PCR assay and rpoB sequence determination. J. Clin. Microbiol. 45:134-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, F. L., D. Gevers, C. C. Thompson, P. Dawyndt, S. Naser, B. Hoste, C. B. Munn, and J. Swings. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, F. L., B. Gomez-Gil, A. T. R. Vasconcelos, and T. Sawabe. 4 May 2007. Multilocus sequence analysis reveals that Vibrio harveyi and V. campbellii form distinct species. Appl. Environ. Microbiol. doi: 10.1128/AEM.00020-07. [DOI] [PMC free article] [PubMed]
- 41.Vanlaere, E., K. Sergeant, P. Dawyndt, W. Kallow, M. Erhard, H. Sutton, D. Dare, B. Devreese, B. Samyn, and P. Vandamme. 2008. Matrix-assisted laser desorption ionisation-time-of-flight mass spectrometry of intact cells allows rapid identification of Burkholderia cepacia complex. J. Microbiol. Methods 75:279-286. [DOI] [PubMed] [Google Scholar]
- 42.Vargha, M., Z. Takáts, A. Konopka, and C. H. Nakatsu. 2006. Optimization of MALDI-TOF MS for strain level differentiation of Arthrobacter strains. J. Microbiol. Methods 66:399-409. [DOI] [PubMed] [Google Scholar]
- 43.Vora, G. J., C. E. Meador, M. M. Bird, C. A. Bopp, J. D. Andreadis, and D. A. Stenger. 2005. Microarray-based detection of genetic heterogeneity, antimicrobial resistance, and the viable but nonculturable state in human pathogenic Vibrio spp. Proc. Natl. Acad. Sci. USA 102:19109-19114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williamson, Y. M., H. Moura, A. R. Woolfitt, J. L. Pirkle, J. R. Barr, M. D. G. Carvalho, E. P. Ades, G. M. Carlone, and J. S. Sampson. 2008. Differentiation of Streptococcus pneumoniae conjunctivitis outbreak isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74:5891-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.