Abstract
Nisin is a natural bacteriocin produced commercially by Lactococcus lactis and widely used in the food industry as a preservative because of its broad host spectrum. Despite the low productivity and troublesome fermentation of L. lactis, no alternative cost-effective host has yet been found. Bacillus subtilis had been suggested as a potential host for the biosynthesis of nisin but was discarded due to its sensitivity to the lethal action of nisin. In this study, we have reevaluated the potential of B. subtilis as a host organism for the heterologous production of nisin. We applied transcriptome and proteome analyses of B. subtilis and identified eight genes upregulated in the presence of nisin. We demonstrated that the overexpression of some of these genes boosts the natural defenses of B. subtilis, which allows it to sustain higher levels of nisin in the medium. We also attempted to overcome the nisin sensitivity of B. subtilis by introducing the nisin resistance genes nisFEG and nisI from L. lactis under the control of a synthetic promoter library.
One of the biggest challenges the food industry faces is the preservation of food to extend shelf life while ensuring high standards of food safety and quality. Food preservation is made even more difficult by consumer interests that dictate minimal use of chemically synthesized additives and stringent legislation that restricts the use of approved preservatives. These concerns have led to an increased interest in the use of natural bacteriocins as alternative preservatives. One of the most commercially significant bacteriocins produced by some strains of Lactococcus lactis subsp. lactis is nisin (4).
Nisin is a small, antibacterial peptide that belongs to the group A lantibiotics, with activity against a wide variety of gram-positive pathogenic and food spoilage bacteria. Like other lantibiotics, nisin is synthesized as an inactive precursor peptide (prenisin) that undergoes several steps of processing. Mature nisin is an extracellular peptide that contains modified amino acid residues resulting in highly stable thioether bridges (7, 13, 23). Since 1988, nisin has been approved by the Food and Drug Administration (FDA), and results from toxicity studies carried out with nisin levels in excess of those used in foods show that it can be considered nontoxic and safe (15, 16, 18, 39). The nisin mode of action involves the formation of pores in the cytoplasmic membranes of sensitive bacteria, as well as the inhibition of cell wall synthesis, and this dual killing mode makes nisin an effective preservative (11, 12, 19, 21, 45, 48). In L. lactis, protection against the dual killing action of nisin is produced by four self-protection peptides, NisFEG (ABC transporter complex) and NisI (transmembrane protein) (36).
The biggest limitation of nisin application is the high cost of production due to low nisin production rates during fermentation of L. lactis (42). Industrial-scale production of nisin is carried out in batch fermentations where synthesis of nisin starts in the mid-exponential growth phase, reaches a maximum in late exponential phase, and stops once the cells go into stationary phase. Nisin production levels are thus dependent on the growth rate and cell density, both of which are comparatively low for nisin-producing L. lactis cultures (14, 17, 32, 50). The concomitant production of lactic acid also poses a problem, as it results in medium acidification below the optimum pH required for nisin production. Also, occasional problems of phage infections during production and apparent exhaustion of the ability to increase productivity by classical mutagenesis are observed for L. lactis (9, 38, 46).
In order to achieve higher productivity and avoid the troublesome fermentation of L. lactis, the engineering of alternative cost-effective nisin-producing strains is an option. Bacillus subtilis provides one such alternative. Some isolates are themselves known to produce the lantibiotic subtilin (which is related to nisin by divergent evolution) and the nisin-unrelated sublancin 168 (3, 30, 34, 51). More importantly, B. subtilis allows easy and inexpensive large-scale fermentation with high growth rates and high cell densities (33, 37, 47). Nevertheless, a remaining challenge in engineering B. subtilis for this purpose is its sensitivity to nisin (22).
In this study, we have focused on two strategies to obtain nisin resistance in B. subtilis 168. We applied transcriptome and proteome analyses to identify B. subtilis genes that are likely to contribute to inherent nisin resistance. Knockout of the identified genes increased the nisin sensitivity, and overexpression allowed B. subtilis to sustain higher levels of nisin in the medium. We also attempted to overcome the nisin sensitivity of B. subtilis by introducing the nisin immunity genes nisFEG and nisI from L. lactis. These genes were placed under the control of a synthetic promoter library (SPL) (40), which allowed us to optimize their expression in order to achieve greater resistance to nisin.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli NM522 (for gene cloning) and strains of B. subtilis were propagated in Luria-Bertani (LB) broth (Difco) with shaking or on LB agar at 37°C. The applied LB medium contains 4 g/liter NaCl, unless otherwise stated. Chromosomal DNA from a nisin-producing strain of L. lactis was kindly provided by Danisco A/S and used as the genetic source for the nisin immunity genes. Antibiotics were used at the following concentrations: ampicillin, 100 mg/ml; erythromycin, 5 mg/ml; and kanamycin, 5 mg/ml. Gene induction was achieved by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Purified nisin was also provided by Danisco A/S. Nisin stock solutions were made in 0.01 M HCl.
Gene cloning.
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of competent E. coli cells were carried out as described by Sambrook et al. (35). B. subtilis was transformed as described by Anagnostopoulos and Spizizen (1). PCR was carried out as described by van Dijl et al. (44). The nucleotide sequences of primers used for PCR are listed in Table 1, and the plasmids used in this study are listed in Table 2.
TABLE 1.
Primers used in this study
| Primer (relevant plasmid) | Sequencea |
|---|---|
| Amplification primers for gene inactivation fragments (pMUTIN2) | |
| mutyvqI+ | GACGCGGAATTCCGGCGGATTCTTATTAATTG |
| mutyvqI− | TTATACGGGATCCCGGAAGCCGGTTCAGGATC |
| mutyvqH+ | GACGCGGAATTCCCGATATGGAAAGCGACATCGC |
| mutyvqH− | TTATACGGGATCCCGCGCTTCCACATCGTCTGC |
| mutyvqG+ | GACGCGGAATTCCGAAGTGACATATCGGCAGGC |
| mutyvqG− | GACGCCGGGATCCCGGCTTACATTAGCATCACCGC |
| mutyvqF+ | GACGCGGAATTCCGCGTTTGCCGCATTTCTGATTTACGC |
| mutyvqF− | TTATACGGGATCCCGACACTTTTACCCGGCGC |
| mutyvqE+ | GACGCGGAATTCCAGCGTGACCGTCGGTTTCGC |
| mutyvqE− | GACGCCGGGATCCCGGTCCAAAAGCTCCGTAAGGC |
| mutyvcR+ | GACGCGGAATTCCGATTATGGGGCCGTCGGGAAGC |
| mutyvcR− | GACGCCGGGATCCCGGCGTAACCAACAAAATCGTCGC |
| mutyvcS+ | GACGCGGAATTCCCAGGCTGAAGGATCATGCACGC |
| mutyvcS− | GACGCCGGGATCCCGGTGACAGACTGTCAAAGC |
| mutyxaH+ | GACGCGGAATTCCGCCGCCGCAGACATACTG |
| mutyxaH− | TTATACGGGATCCCGGTGGATCCGCTGTTC |
| verifypMUTIN+ | ATAATTCTACACAGCCCAGTCCAGACTATTCGG |
| Amplification primers for gene overexpression (pHT315) | |
| pHTyvqIH+ | GACGCGGATCCAAAAGGAGAATGATAAAAATGAAAATAAACAAGAAAACAATAG |
| pHTyvqIH− | GACGCCCCGGGTTATTCATTTGCCGCTTTTGTCTGGTC |
| pHTyvqGFE+ | GACGCGGATCCAAAAGGAGAATGATAAAAATGGTCATTGAGTCGGATAGCAAGG |
| pHTyvqGFE− | GACGCCCCGGG TCAATCAATAATACTCGAATCACGTTC |
| pHTyvcRS+ | GACGCGGATCCAAAAGGAGAATGATAAAAATGAACGTGTTGCAAACAACGAACC |
| pHTyvcRS− | GACGCCCCGGGTTACATACGCTGAAGAACAGC |
| pHTyxaGH+ | GACGCCCCGGGAAAAGGAGAATGATAAAAATGAAAACATTATGTACACATTC |
| pHTyxaGH− | GACGCCCCGGGCTAGACTTTTGTTTTCTTTGCAAT |
| verifypHT+ | AGCGGATAACAATTTCACACAGGA |
| Amplification primers for nisin immunity genes and lacZ (pSac-Kan) | |
| nisFEG+ | TCGGAAGATCTTCTCCCCGCGGGGAGCCGGCGTTCGAAGGAACTACAAAATAAATTATAAGGAGGCACTCAAAATGCAGGTAAAAATTCAAAATCTTTCTAAAAC |
| nisFEG− | ATGCCCCTAGGTCTAATCTTTTTTTTAGATAATGCTACAAG |
| nisI+ | GACCTCTAGCTAGCTAGAGGAGGGAAGAGGAAAGTGGCCTTAATAGGGATAACAGGTTTATCAGG |
| nisI− | CGGGAAGATCTTCGACGTCGTTTCCTACCTTCGTTGCAAGCTTAAAATC |
| lacZ+ | ATGGCGGGGTACCCCTAACCTAACTAAAGGTGGTGAACTACTGTG |
| lacZ− | AGCTCCCTAGGGGTTATTATTATTTTTGACACC |
| Amplification primers for synthetic promoter in front of nisin immunity genes | |
| SP+ | ATGCGTCCCCGCGGGGATCCCCCGGGGGACAGTGATGGGTCCAGAAGGTGCGGCATCG |
| SP− | GCATGGCCGGCATTATANNNNNNNNNNNNNNNNNTGTCAANNNNNNNNNNNNNNNAAACGCAATATGATGCAGTCCCTGCCCTTTC |
Restriction sites are underlined.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristicsa | Reference |
|---|---|---|
| pMUTIN2 | Ampr Eryr | 43 |
| pMEH1 | ΔyvqI::pMUTIN2 Ampr Eryr | This study |
| pMEH2 | ΔyvqH::pMUTIN2 Ampr Eryr | This study |
| pMEH3 | ΔyvqF::pMUTIN2 Ampr Eryr | This study |
| pMEH4 | ΔyvqE::pMUTIN2 Ampr Eryr | This study |
| pMEH5 | ΔyvqG::pMUTIN2 Ampr Eryr | This study |
| pMEH6 | ΔyvcR::pMUTIN2 Ampr Eryr | This study |
| pMEH7 | ΔyvcS::pMUTIN2 Ampr Eryr | This study |
| pMEH8 | ΔyxaH::pMUTIN2 Ampr Eryr | This study |
| pHT315 | pUC19 derivative; Ampr Eryr | 2 |
| pMEH9 | yvqIH::pHT315 Ampr Eryr | This study |
| pMEH10 | yvqGFE::pHT315 Ampr Eryr | This study |
| pMEH11 | yvcRS::pHT315 Ampr Eryr | This study |
| pMEH12 | yxaGH::pHT315 Ampr Eryr | This study |
| pSac-Kan | pUC18 derivative; Ampr Kanr | 28 |
| pMEH4s | SP4-nisFEGI-lacZ::pSac-Kan Ampr Kanr | This study |
| pMEH5s | SP5-nisFEGI-lacZ::pSac-Kan Ampr Kanr | This study |
| pMEH13s | SP13-nisFEGI-lacZ::pSac-Kan Ampr Kanr | This study |
| pMEH46s | SP46-nisFEGI-lacZ::pSac-Kan Ampr Kanr | This study |
Ampr, ampicillin resistant; Eryr, erythromycin resistant; Kanr, kanamycin resistant; SP4, SP5, SP13, and SP46, synthetic promoters from the SPL.
Sample preparation for transcriptome and proteome analyses.
Before performing the transcriptome and proteome analyses of nisin-stressed B. subtilis 168 cells, mRNAs and proteins were isolated from cells grown in LB medium with 10 g/liter NaCl at 37°C with the addition of five doses of 50 ng/ml of nisin. The first addition of nisin was performed when cells reached an optical density at 600 nm (OD600) of 0.2, and four subsequent additions were made at 5-min intervals. Samples for RNA and protein purification were taken 10 min after the last nisin addition.
DNA microarray.
Three independent RNA preparations (biological triplicates) from each sample were isolated according to the protocol from the Promega SV RNA isolation kit. The RNA samples were labeled and hybridized to B. subtilis 168 trpC glass slide microarrays purchased from Eurogentec. Chromosomal DNA was isolated according to the protocol from Qiagen by using a genomic-tip 100/G, and 2 μg of DNA was labeled overnight at 37°C in a mixture containing 5 μl 10× deoxynucleoside triphosphate mix, 3 μl Cy3-dCTP (Amersham), and 1 μl Klenow enzyme (Roche). Labeled cDNA was prepared from 16 μg RNA by using StrataScript reverse transcriptase (Stratagene) for the incorporation of Cy5 dye (Amersham). Labeled DNA and cDNA were purified using a QIAquick purification kit (Qiagen) and dried before being resuspended in a mixture of 8 μl E. coli tRNA, 16 μl 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2.6 μl 1 M HEPES (pH 7.0), 2.6 μl 10% sodium dodecyl sulfate (SDS), and 10.7 μl Denhardt solution. Samples were heated for 3 min in a boiling water bath, cooled at room temperature for 5 min, and centrifuged at maximum speed for 2 min to remove any solid particles from the hybridization mixture. This mixture was put onto the microarray slide, sealed with a coverslip in a hybridization chamber, and incubated overnight at 63°C. Following hybridization, microarray slides were washed briefly in prewarmed (63°C) 2× SSC-0.1% SDS to remove the coverslip and then washed twice for 5 min in each of the following buffers: (i) 1× SSC (room temperature) and (ii) 0.2× SSC (room temperature). Microarray slides were dried by centrifugation at 1,200 rpm for 5 min before being scanned by a GenePix 4000A scanner (Axon Instruments, Inc.). Fluorescent spots and the local background intensities were identified and quantified using BlueFuse software (BlueGnome, Oxford, United Kingdom). To compensate for unequal levels of dye incorporation, data centering to zero was performed for each block. The data were analyzed using GeneSpring software. We considered genes to be differentially expressed if they displayed ≥2-fold changes. Array data are available in the supplemental material.
2D gel electrophoresis.
B. subtilis cells grown with and without the addition of five doses of 50 ng/ml of nisin were harvested by centrifugation (5,000 × g for 15 min). They were washed with Tris-buffered saline, pH 7.5, and lysed by four 1-min beatings with glass beads (0.10 to 0.11 μm [product no. G4649; Sigma]) in a lysis buffer containing 50 mM Tris, pH 7.5, 0.3% SDS, 0.2 M dithiothreitol, 3.3 mM MgCl2, 16.7 μg/ml RNase, and 1.67 U/ml DNase. Following lysis, the extract was kept on ice for 30 min before being centrifuged at 14,000 × g for 20 min. Protein concentrations in the samples were determined in triplicate using the Bradford protein assay with bovine serum albumin as a standard (6). Proteomic analyses of the cell extracts, including two-dimensional (2D) gel electrophoresis, imaging, spot picking, digestion, and matrix-assisted laser desorption ionization-time of flight analysis, were carried out as described by Holmes et al. (20) using 100 to 125 μg protein per immobilized pH gradient strip.
Construction of B. subtilis knockout strains.
Individual gene knockouts in B. subtilis were constructed by inserting a 300- to 500-bp internal fragment of yvqI, yvqH, yvqG, yvqF, yvqE, yvcS, yvcR, or yxaH into the integrational plasmid pMUTIN2 (43). The pMUTIN2 insertions into the chromosome, leading to single-crossover gene disruption, were confirmed by PCR amplification of a DNA fragment by primers which hybridize upstream of the genes and within pMUTIN2.
Construction of B. subtilis overexpression strains.
The genes found to be overexpressed in B. subtilis during nisin stress are grouped together in four small operons: yvqIH, yvqGFE, yvcRS, and yxaGH. Even though yxaG was not identified as being overexpressed in the DNA array experiments, we chose to overexpress it in case it should be needed for full yxaH activity. All four gene fragments were cloned into the shuttle vector pHT315 (2) and examined by sequencing. Transformation of B. subtilis was confirmed by PCR amplification of a DNA fragment using primers which hybridize upstream of the genes and within pHT315.
Construction of B. subtilis SPL upstream of the nisin immunity genes.
The open reading frames of nisFEG and nisI were PCR amplified from L. lactis chromosomal DNA and ligated together. The nisFEGI gene fragment was sent for sequencing to verify the correct sequence before it was cloned into the integration vector pSac-Kan, which had been cured of a SacII restriction site by site-directed mutagenesis (28). The reporter gene lacZ from E. coli was fused to the end of the nisFEGI gene construct in order to perform β-galactosidase assays to determine the strength of the SPL, which was cloned in front of the nisin immunity genes. The SPL fragment was inserted in front of the nisFEGI-lacZ gene construct by using a degenerated reverse primer (Table 1) amplifying a fragment of 500 bp of noncoding DNA from B. subtilis, as described by Solem and Jensen (40). E. coli was transformed with the SPL-nisFEGI-lacZ ligation mixture, and resultant transformants were pooled. The chromosome of B. subtilis was then transformed at the sacA locus with plasmid DNA from the pooled transformants. Transformants were first selected based on their blue coloration on LB agar plates containing the appropriate antibiotics and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and thereafter examined for nisin sensitivity and β-galactosidase activity.
β-Galactosidase experiments.
In order to determine the strengths of the different synthetic promoters, a β-galactosidase assay was performed using a modified Miller protocol (29). The B. subtilis culture was grown in LB medium at 37°C until the OD600 reached 0.5. Then a 2-ml aliquot of cells was harvested and resuspended in 2 ml of Z-buffer (6 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0), and the OD600 was recorded. A sample of 100 to 800 μl of the cell suspension was lysed for 5 min at 30°C with 200 μl of the lysozyme stock (Z-buffer with 2.5 mg/ml lysozyme), and Z-buffer was added to obtain a total volume of 1 ml. The lysed sample was mixed with 8 μl of 10% Triton X-100 and 100 μl of 4-mg/ml ONPG (o-nitrophenyl-β-d-galactopyranoside) and incubated at 30°C until the reaction mixture turned yellow. The reaction was stopped by adding 1 ml of 0.5 M Na2CO3, and the time was noted. The OD420 and OD550 were recorded, and the specific β-galactosidase activity was calculated as follows: 1,000[OD420 − 1.75(OD550)]/[time (in minutes)·OD600 sample volume (in milliliters)].
Growth experiment with nisin addition.
B. subtilis wild-type, knockout, and overexpression strains and strains harboring the nisin immunity genes were grown in LB medium with the appropriate antibiotics at 37°C. Nisin was added either in five doses of 50 ng/ml to cultures at an OD600 of 0.2 (as described above) or in a single dose of 600 to 900 ng/ml to cultures at an OD600 of 0.5. The growth was monitored until the cells entered stationary phase, after which the experiments were terminated.
RESULTS
Identification of genes involved in nisin resistance in B. subtilis 168.
B. subtilis cells, despite producing their own lantibiotics, are sensitive to nisin. However, we supposed that there might be general cellular mechanisms for coping with membrane stress that, if optimized, could contribute to improving nisin tolerance. To identify such factors, B. subtilis cells were stressed during growth by the addition of 50 ng/ml of nisin five times at 5-min intervals, starting when cultures reached an OD600 of 0.2. Global gene expression patterns of the stressed B. subtilis cells were compared to those of the control without nisin addition by using spotted DNA microarrays. The data presented here are the means of results for three independent biological replicates. A total of eight differentially expressed genes were identified, all of which were upregulated during nisin stress. Some of these genes have been associated previously with membrane stress in B. subtilis (25, 27, 31). No downregulated genes were found (Table 3). We attempted to confirm the transcriptome data by a proteomic approach, in which a selection of proteins overproduced under nisin stress were picked from the gels and identified by matrix-assisted laser desorption ionization-time of flight analysis. This strategy allowed us to confirm the upregulation of two genes already identified by the transcriptome analysis, yvqH and yvcR (Table 3). In order to understand the individual impacts of the genes found by transcriptome and proteome analyses, all eight genes were respectively knocked out and overexpressed. The resulting strains were compared to the wild type with respect to nisin resistance.
TABLE 3.
Characteristics of all genes significantly overexpressed in nisin-stressed B. subtilis cells
| Gene namea | Change in expression (n-fold) | P value | Function or description of gene product |
|---|---|---|---|
| yvqI | 30.1 | 0.00005 | No known function |
| yvqH | 30.7 | 0.00005 | Similar to hypothetical proteins from B. subtilis |
| yvqG | 2.6 | 0.0002 | No known function |
| yvcS | 5.2 | 0.008 | Similar to ABC transporter (permease) |
| yvqF | 2.4 | 0.01 | No known function |
| yvcR | 4.2 | 0.01 | Similar to ABC transporter (ATP-binding protein) |
| yxaH | 3.3 | 0.01 | Similar to hypothetical proteins |
| yvqE | 2.2 | 0.02 | Similar to two-component sensor histidine kinase |
Genes marked in bold correspond to genes identified by 2D gel spot picking.
Inactivation of genes induced by nisin stress leads to increased nisin sensitivity.
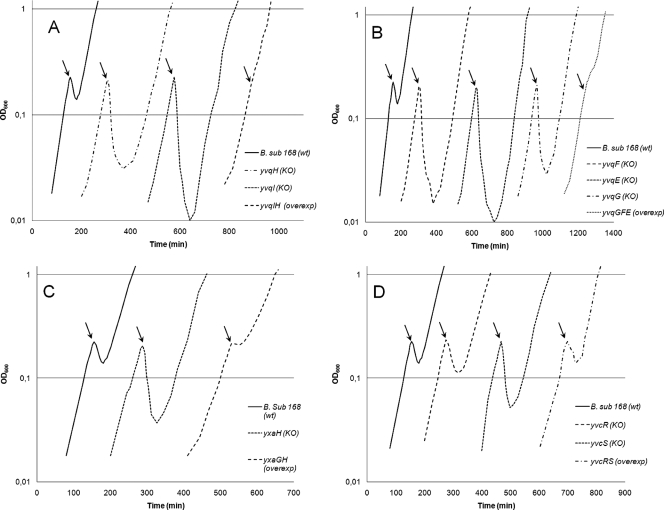
In this study, yvqI, yvqH, yvqG, yvqF, yvqE, yvcS, yvcR, and yxaH were inactivated individually to determine the effects on nisin sensitivity. It was expected that the knockout strain would show greater nisin sensitivity than the wild type if the inactivated gene played a role in the inherent nisin resistance of B. subtilis. The strains were tested under the same conditions used for DNA array experiments. As shown in Fig. 1, all knockout strains presented an altered growth phenotype compared to that of the wild-type strain. The largest changes are seen when yvqI or yvqH has been inactivated (Fig. 1A). Both the yvqI and yvqH knockout strains are considerably more sensitive to nisin than the wild type, which correlates well with the fact that yvqI and yvqH were found to be the two most upregulated genes in the transcriptome analysis. Furthermore, the strains with inactivated yvqF, yvqE, and yvqG also showed considerably greater nisin sensitivity than the wild type, although the transcription of these genes was activated 10-fold less than that of yvqI and yvqH (Fig. 1B). The inactivation of yxaH also led to increased nisin sensitivity (Fig. 1C), whereas the smallest changes were detected in strains with yvcR or yvcS inactivated (Fig. 1D).
FIG. 1.
Growth phenotypes of knockout (KO) and overexpression (overexp) strains of B. subtilis (B. sub.): strains with knockouts of yvqI and yvqH and overexpression of yvqIH (A); strains with knockouts of yvqF, yvqE, and yvqG and overexpression of yvqGFE (B); strains with knockout of yxaH and overexpression of yxaGH (C); and strains with knockouts of yvcS and yvcR and overexpression of yvcRS (D). wt, wild type. Cell growth was measured at 600 nm, and at an OD600 of 0.2, 50-ng/ml aliquots of nisin were added five times at 5-min intervals. The black arrows indicate the time of the first nisin addition. The experiments were carried out in triplicate, and one representative curve for each strain is shown.
Overexpression of nisin-induced genes leads to increased nisin tolerance.
To determine if the effects on the knockout strains could be reversed to obtain some level of nisin resistance, strains overexpressing the nisin-induced genes were constructed. For the overexpression experiments, the eight genes have been grouped together into four small operons, yvqIH, yvqGFE, yvcRS, and yxaGH, according to their normal genetic organization in B. subtilis. Despite the fact that the function of yxaH is unknown, it has been shown to be expressed as part of the yxaGH operon and may therefore act with yxaG in a cooperative manner, which is the reason for including yxaG in this study (49). The resultant overexpression strains were tested under the same conditions as the knockout strains, and their growth phenotypes are shown in Fig. 1. The strain overexpressing yvqIH is clearly more resistant than the wild type (Fig. 1A). Overexpression of yvqGFE produces a similar effect (Fig. 1B), whereas the overexpression of both yxaGH (Fig. 1C) and yvcRS (Fig. 1D) does not seem to overcome the lethal action of the added nisin. Overexpression of yxaGH is beneficial to some extent, as the growth is stalled for approximately 25 min during the addition of nisin, after which it continues at a 40% decrease in the specific growth rate. The increased nisin tolerance of strains with overexpressed yvqIH and yvqGFE correlates with the sensitivity of the strains with the same genes inactivated, although the yvqGFE genes were expressed considerably less than yvqIH. The fact that the two overexpression strains exhibited increased nisin resistance clearly indicates their potential in alleviating the nisin-induced stress during tentative nisin production in B. subtilis.
Introduction of nisin immunity genes under the control of an SPL results in increased nisin tolerance.
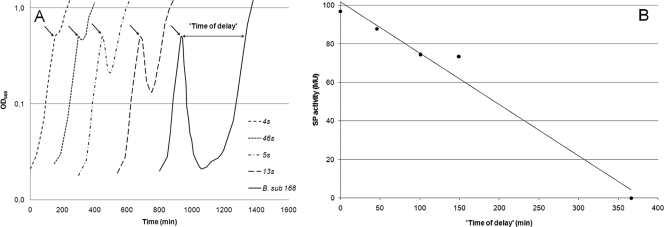
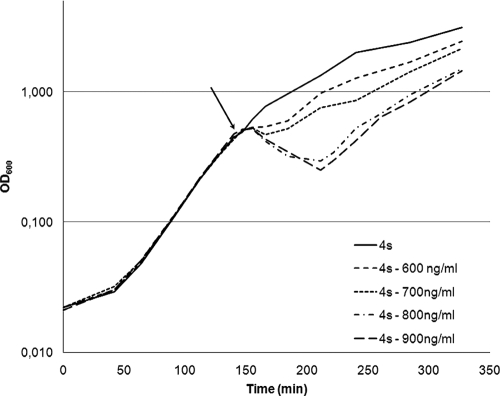
Prior to the introduction of the nisin immunity genes from L. lactis into B. subtilis 168, an SPL was fused upstream of the nisFEGI-lacZ gene construct. With the use of an SPL, it is possible to modulate the gene expression and screen for the optimal window of expression in a given situation. In this case, we correlated the strengths of our promoters (measured as β-galactosidase activities) to nisin sensitivity. Samples for β-galactosidase assays were taken at an OD600 of 0.5, after which 600 ng/ml of nisin was added to the culture (which resulted in a reduction of the OD) and the time of delay was calculated as the time from the addition of nisin until the cells reached an OD600 of 0.5 again. As depicted in Fig. 2A, the tested transformants showed different times of delay, varying from 0 min to around 300 min for the wild type. In Fig. 2B, the β-galactosidase activities (in Miller units) are plotted against the times of delay, and these data suggest a direct correlation between these two properties. High β-galactosidase activity (indicating a strong synthetic promoter) corresponds to a short time of delay and thereby also to greater resistance against nisin. A strain named 4s has been identified as the most resistant candidate in this screening. The influence of four different nisin concentrations (600, 700, 800, and 900 ng/ml) on the growth of the 4s strain was studied, and the growth of 4s in the presence of these concentrations was compared to growth without nisin addition. The results (Fig. 3) show that the addition of 600 ng/ml of nisin has only a small effect on the growth of 4s compared to the effect on wild-type B. subtilis, which has a drop in OD600 of 96% when 600 ng/ml is added (Fig. 2A). The addition of even higher nisin concentrations to the culture of 4s had much less dramatic effects than those on wild-type B. subtilis.
FIG. 2.
Growth phenotypes of strains of B. subtilis 168 (B. sub 168) harboring the synthetic promoter-nisFEGI-lacZ construct (A) and characterization of the strengths of the different synthetic promoters (SP) (B). All strains were grown in LB medium at 37°C, and cell growth was measured at 600 nm. The β-galactosidase activity (expressed in Miller units [MU]) is plotted against the time of delay. Samples for the β-galactosidase assay were taken from cultures at an OD600 of 0.5, after which 600 ng/ml of nisin was added to the cultures and the time of delay was calculated as the time from the addition of nisin until the cells reached an OD600 of 0.5 again. The black arrows indicate the time of the nisin addition.
FIG. 3.
Growth profiles of the best candidate, 4s, harboring the nisin immunity genes expressed under the control of a synthetic promoter. All strains were grown in LB medium at 37°C; at an OD600 of 0.5, cultures received four different nisin concentrations (600, 700, 800, and 900 ng/ml), and growth in the presence of nisin was compared to growth without the nisin addition. The black arrow indicates the time of the different nisin additions.
DISCUSSION
One of the biggest challenges and disadvantages in achieving heterologous nisin production in an alternative host like B. subtilis is the sensitivity to the lethal action of nisin. In this study, we pursued two approaches to overcome this sensitivity and possibly reevaluate B. subtilis as a nisin production host. The first was to boost the natural defenses of B. subtilis, and the second was to introduce the L. lactis nisin immunity genes.
We first applied global transcriptome and proteome analyses to define the response of B. subtilis 168 to the antimicrobial action of nisin. Under the tested conditions, eight genes were identified as being overexpressed and, thereby, potentially involved in coping with nisin-induced stress. The most strongly induced genes were yvqI and yvqH, encoding a putative transmembrane protein and an E. coli phage shock protein homologue, respectively (8, 25). The genes yvcR and yvcS showed moderate upregulation during nisin stress, and they are believed to encode a putative ABC transporter system of unknown function (25, 31). The upregulation of yvqH and yvcR was also confirmed by the proteome experiments. Among the responses from the eight overexpressed genes, the lowest transcriptional responses to nisin were from yxaH and the yvqGFE operon. While the precise function of yxaH is unknown, it has been shown previously to be expressed as part of the yxaGH operon, in which yxaG encodes an iron-containing quercetin 2,3-dioxygenase that cleaves flavonoids (49). Encoded by the yvqGFE operon, YvqG exhibits homology to other uncharacterized putative proteins and bears transmembrane helices that are indicative of membrane localization (24). YvqF has conserved domains that are predicted for membrane proteins and shows 70% similarity to a predicted transporter protein, LiaF, from Bacillus cereus. The YvqE protein is a histidine kinase antibiotic response regulator that is part of the YvqCE two-component system. This two-component system is involved in the response to a subset of cell wall-targeting antibiotics that interfere with the lipid II cycle in the cytoplasmic membrane (27).
The response observed when cells of B. subtilis are stressed with nisin is likely to be a general membrane or cell surface stress response rather than a specific response to nisin. This pattern has also been shown in a previous study with whole-genome DNA microarrays for L. lactis IL1403 and L. lactis IL1403 Nisr, in which a 75-fold increase in the wild-type nisin resistance level was reached by consecutively growing the wild-type strain in the presence of increasing nisin concentrations in the medium. This study showed that a large part of the response observed may be due to general stress rather than nisin resistance (25). Measurements of membrane permeabilization in E. coli have also shown that there is a correlation between membrane alteration and nisin sensitivity. Cells are more sensitive toward nisin during membrane stress (9). Boziaris and Adams (5) also showed that cells of Salmonella enterica serovar Enteritidis PT4 and PT7 and Pseudomonas aeruginosa could became nisin sensitive during membrane stress; however, all strains rapidly recovered resistance. Also, the fact that seven of the eight genes, those in the yvqIH, yvqGFE, and yvcRS operons, have previously been found to be induced by different cell wall stresses, including naturally occurring cationic peptides with antibacterial activities, indicates general membrane stress (25, 27, 31). In 2003, Mascher et al. also found that the yvqIH operon shows the most dramatic response when cells of B. subtilis are treated with either vancomycin or bacitracin but without conferring any resistance to either of the two antibiotics (26). They also showed that the disruption of yvqI and yvqH has no effect on the sensitivity of the mutants to any of the antibiotics tested. However, our present results with the eight studied genes indicate measurable effects on B. subtilis nisin tolerance caused by gene inactivation or overexpression. The overexpression of the yvqIH operon gave the highest level of nisin resistance, which correlates with the fact that this operon is also the most strongly expressed during nisin-induced stress. Since B. subtilis never encounters nisin in its natural environment, chances of the bacterium's harboring genes specific for nisin resistance are minimal. Therefore, the results obtained in this study point toward a general stress response induced by the addition of nisin during exponential growth, which can nevertheless be used to some extent toward optimizing a nisin-producing strain.
The second strategy for increasing nisin resistance in B. subtilis attempted in this study was the introduction of the nisin immunity genes nisFEG and nisI from L. lactis. In 2003, Stein et al. performed agar diffusion assays and showed that the expression of the immunity genes in B. subtilis confers a nisin immunity level of up to approximately 30% of that found in the nisin-producing L. lactis strain (41). This result shows the real potential of obtaining a nisin-resistant B. subtilis strain, especially if it is possible to find the optimal expression levels for the immunity genes. We have therefore tried to optimize the expression levels of the nisin immunity genes by using the SPL approach. The promoter libraries obtained by this approach contain promoters with virtually any level of activity, which makes this technology well suited for metabolic optimization (40). In this study, we have constructed an SPL fused to the nisin immunity genes and the lacZ reporter gene from E. coli. During the β-galactosidase activity screening of many of the transformants, we found that there is direct correlation between the acquired nisin immunity and the strength of the synthetic promoter. The strongest nisin tolerance was found in the 4s strain, which showed a 15-fold increase in the nisin resistance level compared to that of wild-type B. subtilis. We were able to challenge the strain with 300 ng/ml of nisin at an OD600 of 0.2 without affecting growth, whereas wild-type B. subtilis could handle only 20 ng/ml of nisin at an OD600 of 0.2 without exhibiting an effect on growth (data not shown). Experiments with higher nisin concentrations in which nisin was added at an OD600 of 0.5 showed that the 4s strain was able to tolerate 600 ng/ml with only a small effect on growth, in contrast to wild-type B. subtilis, which showed a 96% drop in the OD600 when 600 ng/ml of nisin was added. Even though the coordinated expression of nisFEGI with a synthetic promoter in B. subtilis is quite different from the autoregulatory control of nisin immunity in L. lactis, our results demonstrate the possibility of eventually obtaining a strain with nisin immunity levels high enough for the heterologous production of nisin in B. subtilis. Comparing the nisin immunity of the 4s strain with the resistance conferred by the overexpression of yvqIH, which showed the most promising results of all the overexpressed natural genes in B. subtilis, revealed that the effect of the nisin immunity genes exceeds the general stress response by a factor of 10 (data not shown). However, the resistance was permanent in all strains.
In this study, we have reevaluated the use of B. subtilis as a potential host organism for the heterologous production of nisin. We have demonstrated that it is possible to boost the natural defenses of B. subtilis to some extent by overexpressing some of the bacterium's own genes upregulated under nisin stress. Similar to Stein et al. (41), we have also shown that it is possible to circumvent the nisin sensitivity of B. subtilis to some extent by introducing the nisin resistance genes nisFEG and nisI from L. lactis under the control of an SPL. However, the expression of nisFEGI under the control of a synthetic promoter in B. subtilis, like the expression of nisIFEG under the control of the Pspac promoter (41), is quite different from the autoregulatory control of the nisin immunity genes in L. lactis. We did not succeed in increasing nisin resistance in B. subtilis to a level that is sufficient to allow commercial production; however, further optimization and combination of different approaches along the principles set out in this study may bring us closer to that target in the future.
Supplementary Material
Acknowledgments
This work was supported by Danisco A/S, the Technical University of Denmark, and the FOOD graduate school.
We thank Francis Mulholland and Mike Gasson's groups, both at the Institute of Food Research, Colney, Norwich NR4 7UA, United Kingdom, for help with the proteome analysis and for help with the transcriptome analysis, respectively.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, S., and J. N. Hansen. 1988. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J. Biol. Chem. 263:9508-9514. [PubMed] [Google Scholar]
- 4.Bhattacharyya, B. K., and D. Bhattacharjee. 2007. Bacteriocin: a biological food preservative. J. Food Sci. Technol. 44:459-464. [Google Scholar]
- 5.Boziaris, I. S., and M. R. Adams. 2001. Temperature shock, injury and transient sensitivity to nisin in Gram negatives. J. Appl. Microbiol. 91:715-724. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A dye binding assay for protein. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Breukink, E., and B. de Kruijff. 1999. The antibiotic nisin, a special case or not. Biochim. Biophys. Acta 1462:223-234. [DOI] [PubMed] [Google Scholar]
- 8.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao-Hoang, L., P. A. Marechal, M. Le-Thanh, and P. Gervais. 2008. Synergistic action of rapid chilling and nisin on the inactivation of Escherichia coli. Appl. Microbiol. Biotechnol. 79:105-109. [DOI] [PubMed] [Google Scholar]
- 10.Coffey, A., and R. P. Ross. 2002. Bacteriophage-resistance systems in dairy starter strains: molecular analysis to application. Antonie van Leeuwenhoek 82:303-321. [PubMed] [Google Scholar]
- 11.Deegana, L. H., P. D. Cottera, C. Hilla, and P. Rossb. 2006. Bacteriocins: biological tools for bio-preservation and shelf-life extension. Int. Dairy J. 16:1058-1071. [Google Scholar]
- 12.Delves-Broughton, J., P. Blackburn, R. J. Evans, and J. Hugenholtz. 1996. Applications of the bacteriocin, nisin. Antonie van Leeuwenhoek 69:193-202. [DOI] [PubMed] [Google Scholar]
- 13.Delves-Broughton, J. 1998. Nisin. International Dairy Federation document no. 329, p. 9-12. International Dairy Federation, Brussels, Belgium.
- 14.de Vuyst, L., and E. J. Vandamme. 1992. Influence of the carbon source on nisin production in Lactococcus lactis subsp. lactis batch fermentations. J. Gen. Microbiol. 138:571-578. [DOI] [PubMed] [Google Scholar]
- 15.FDA. 1988. Nisin preparation: affirmation of GRAS status as a direct human food ingredient. FDA regulation 54.6120. Fed. Regist. 53:11247. [Google Scholar]
- 16.Frazer, A. C., M. Sharratt, and J. R. Hickman. 1962. The biological effects of food additives—nisin. J. Sci. Food Agric. 13:32-42. [Google Scholar]
- 17.Guerra, P. N., L. M. Rua, and L. Pastrana. 2001. Nutritional factors affecting the production of two bacteriocins from lactic acid bacteria on whey. Int. J. Food Microbiol. 70:267-281. [DOI] [PubMed] [Google Scholar]
- 18.Hara, S., K. Yakazu, K. Nakakawaii, T. Takenchi, T. Kobayashi, M. Sata, Z. Imat, and T. Shibuya. 1962. An investigation of toxicity of nisin. Tokyo Med. Univ. J. 20:175-207. [Google Scholar]
- 19.Hasper, H. E., N. E. Kramer, J. L. Smith, J. D. Hillman, C. Zachariah, O. P. Kuipers, B. de Kruijff, and E. Breukink. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636-1637. [DOI] [PubMed] [Google Scholar]
- 20.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. Ketley, and J. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243-257. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, S. D., E. Breukink, E. Tischenko, M. A. G. Lutters, B. de Kruijff, R. Kaptein, A. M. J. J. Bonvin, and N. A. J. van Nuland. 2004. The nisin lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat. Struct. Mol. Biol. 11:963-967. [DOI] [PubMed] [Google Scholar]
- 22.Hyde, A. J., J. Parisot, A. McNichol, and B. B. Bonev. 2006. Nisin-induced changes in Bacillus morphology suggest a paradigm of antibiotic action. Proc. Natl. Acad. Sci. USA 103:19896-19901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jack, R. W., J. R. Tagg, and B. Ray. 1995. Bacteriocins of gram-positive bacteria. Microbiol. Rev. 59:171-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of liaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer, N. E., S. A. van Hijum, J. Knol, J. Kok, and O. P. Kuipers. 2006. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 50:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 27.Mascher, T., S. L. Zimmer, T.-A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system liaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Middleton, R., and A. Hofmeister. 2004. New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid 51:238-245. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 30.Paik, S. H., A. Chakicherla, and J. N. Hansen. 1998. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J. Biol. Chem. 273:23134-23142. [DOI] [PubMed] [Google Scholar]
- 31.Pietiainen, M., M. Gardemeister, M. Mecklin, S. Leskela, M. Sarvas, and V. P. Kontinen. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577-1592. [DOI] [PubMed] [Google Scholar]
- 32.Qiao, M., M. J. Omaetxebarria, R. Ra, I. Oruetxebarria, and P. E. J. Saris. 1997. Isolation of a Lactococcus lactis strain with high resistance to nisin and increased nisin production. Biotechnol. Lett. 19:199-202. [Google Scholar]
- 33.Riesenberg, D., and R. Guthke. 1999. High-cell-density cultivation of microorganisms. Appl. Microbiol. Biotechnol. 51:422-430. [DOI] [PubMed] [Google Scholar]
- 34.Rintala, H., T. Graeffe, L. Paulin, N. Kalkkinen, and P. E. J. Saris. 1993. Biosynthesis of nisin in the subtilin producer Bacillus subtilis ATCC6633. Biotechnol. Lett. 15:991-996. [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Saris, P. E. J., T. Immonen, M. Reis, and H. Sahl. 1996. Immunity to lantibiotics. Antonie van Leeuwenhoek 69:151-159. [DOI] [PubMed] [Google Scholar]
- 37.Schallmey, M., A. Singh, and O. P. Ward. 2004. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1-17. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu, H., T. Mizuguchi, E. Tanaka, and S. Shioya. 1999. Nisin production by a mixed-culture system consisting of Lactococcus lactis and Kluyveromyces marxianus. Appl. Environ. Microbiol. 65:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shtenberg, A. J., and A. D. Igmatèv. 1969. Toxicological evaluation of some combinations of food preservatives. Food Cosmet. Toxicol. 8:369-380. [DOI] [PubMed] [Google Scholar]
- 40.Solem, C., and P. R. Jensen. 2002. Modulation of gene expression made easy. Appl. Environ. Microbiol. 68:2397-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein, T., S. Heinzmann, I. Solovieva, and K. Entian. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278:89-94. [DOI] [PubMed] [Google Scholar]
- 42.Tolonen, M., P. E. J. Saris, and M. Siika-aho. 2004. Production of nisin with continuous adsorption to Amberlite XAD-4 resin using Lactococcus lactis N8 and L. lactis LAC48. Appl. Microbiol. Biotechnol. 63:659-665. [DOI] [PubMed] [Google Scholar]
- 43.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 44.van Dijl, J. M., A. de Jong, G. Venema, and S. Bron. 1995. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. J. Biol. Chem. 270:3611-3618. [DOI] [PubMed] [Google Scholar]
- 45.van Heusden, H. E., B. de Kruijff, and E. Breukink. 2002. Lipid II induces a transmembrane orientation of the pore-forming peptide lantibiotic nisin. Biochemistry 41:12171-12178. [DOI] [PubMed] [Google Scholar]
- 46.van Hul, J. S., and W. R. Gibbons. 1997. Neutralization/recovery of lactic acid from Lactococcus lactis: effects on biomass, lactic acid and nisin production. World J. Microbiol. Biotechnol. 13:527-532. [Google Scholar]
- 47.Westers, L., H. Westers, and W. J. Quax. 2004. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim. Biophys. Acta 1694:299-310. [DOI] [PubMed] [Google Scholar]
- 48.Wiedemann, I., E. Breukink, C. van Kraaij, O. P. Kuipers, G. Bierbaum, B. de Kruijff, and H. Sahl. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772-1779. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]
- 50.Yu, P., N. W. Dunn, and W. S. Kim. 2002. Lactate removal by anionic-exchange resin improves nisin production by Lactococcus lactis. Biotechnol. Lett. 24:59-64. [Google Scholar]
- 51.Yuksel, S., and J. N. Hansen. 2007. Transfer of nisin gene cluster from Lactococcus lactis ATCC 11454 into the chromosome of Bacillus subtilis 168. Appl. Microbiol. Biotechnol. 74:640-649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.