Abstract
Saccharomyces cerevisiae is exposed to freeze-thaw stress in commercial processes, including frozen dough baking. Cell viability and fermentation activity after a freeze-thaw cycle were dramatically decreased due to freeze-thaw injury. Because this type of injury involves complex phenomena, the injury mechanisms are not fully understood. We examined freeze-thaw injury by indirect gene expression analysis during postthaw incubation after freeze-thaw treatment using DNA microarray profiling. The results showed that genes involved in the homeostasis of metal ions were frequently contained in genes that were upregulated, depending on the freezing period. We assessed the phenotype of deletion mutants of the metal ion homeostasis genes that exhibited freezing period-dependent upregulation and found that the strains with deletion of the MAC1 and CTR1 genes involved in copper ion homeostasis exhibited freeze-thaw sensitivity, suggesting that copper ion homeostasis is required for freeze-thaw tolerance. We found that supplementation with copper ions during postthaw incubation increased intracellular superoxide dismutase activity and intracellular levels of reactive oxygen species were decreased. Moreover, cell viability was increased by supplementation with copper ions. These results suggest that insufficiency of copper ion homeostasis may be one of the causes of freeze-thaw injury.
Yeast (Saccharomyces cerevisiae) cells are exposed to various environmental stresses such as freeze-thaw, high-temperature, osmotic, and air-drying stresses during commercial processes. Freeze-thaw stress is important in the bread-making process, because frozen dough baking has become a major technology (3). Frozen dough baking improves labor conditions for bakers and enables them to provide fresh baked goods for consumers (3). Because frozen dough baking involves freeze-thaw treatment, it exposes yeast cells to freeze-thaw stress, which leads to a significant decrease in the fermentation ability and viability of yeast cells (called “freeze-thaw injury”) (8). The freeze-thaw injury of yeast cells depends on many factors, including freezing periods, freezing temperature, and the physiology of yeast cells (2, 5, 16, 17). Clarification of the changes in the cell physiology of yeast cells caused by freeze-thaw stress is important, because bakers are eager to extend the shelf life of frozen dough. In this study, we attempted to determine the changes in yeast cell physiology due to freeze-thaw injury by indirect gene expression analysis, which is described below.
Freezing subjects yeast cells to low temperature, ice crystal formation in the cells, and dehydration from the cells. This causes both physical damage to cellular components, such as the cell wall, membrane, and proteins, and formation of reactive oxygen species (ROS) (13, 15). Superoxide anions and free radicals are generated in yeast cells during the freeze-thaw process (18), and ROS generation during the thawing process is increased, depending on the freezing period (5, 18). Oxidative stress generated by freeze-thaw treatment enhances the damage to cellular components (9, 25). Because modulation of intracellular levels of ROS after freeze-thaw treatment is required to protect against toxicity, ROS scavenging systems such as glutathione, catalase, and superoxide dismutase (SOD) are believed to be important for freeze-thaw tolerance of yeast cells (1, 2, 17). In particular, copper/zinc SOD (Cu/Zn SOD), which plays a role in oxygen radical detoxification, is necessary to confer full tolerance to freeze-thaw injury (18). Heavy metal ions, such as iron ions and copper ions, are important transition metals for the detoxification of oxygen radicals in yeast cells (6).
Although there have been several studies on the mechanisms of freeze-thaw injury (5, 17, 18, 24), the mechanisms are complex and have not yet been fully elucidated. We therefore examined freeze-thaw injury by indirect gene expression analysis, which was conducted during postthaw incubation after freeze-thaw treatment using DNA microarray profiling. Indirect gene expression analysis may be advantageous for such an examination, because changes of gene expression may reflect the physiology of the freezing-state cell. We hypothesized that the genes involved in freeze-thaw injury may upregulate during postthaw incubation. To elucidate the physiological changes during freeze-thaw injury, we carried out indirect gene expression analysis using yeast cells after they had been frozen for different periods. The upregulated genes in the indirect gene expression analysis were extracted by clustering methods. We found that the genes involved in metal ion homeostasis were specifically upregulated. The importance of the genes extracted by the clustering was confirmed by phenotypic analysis using the deletion strains of the extracted genes. We also showed, by physiological analysis, that insufficiency of copper ion homeostasis causes freeze-thaw injury.
MATERIALS AND METHODS
Yeast strains, media, and viable cell counts.
The Saccharomyces cerevisiae wild-type strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) was used in this study. The Δsit1, Δfre1, Δfre4, Δfre7, Δctr1, Δree1, Δmac1, and Δpro1 deletion strains derived from BY4741 were obtained from EUROSCARF (the European Saccharomyces cerevisiae Archive for Functional Analysis).
YPD medium (1% yeast extract [Difco Laboratory, Detroit, MI], 2% peptone [Difco Laboratory], and 2% glucose) was used for cultivation of the yeast cells unless otherwise noted. SD medium without copper (2% glucose, 0.17% yeast nitrogen base without Cu and Fe [Bio 101, Vista, CA], 0.5% ammonium sulfate, amino acid, and 1 μM ferric chloride) supplemented with various concentrations of copper ions (0 μM, 1 μM, or 10 μM CuSO4 alone or 10 μM CuSO4 plus 100 μM copper chelator bathocuproinedisulfonic acid disodium salt [BCS]) was used for measurement of the intracellular ROS level and SOD activity.
Viable numbers of yeast cells were determined using YPD medium solidified with 2% agar. Cell suspensions were plated onto the agar plates with appropriate dilution and grown for 2 days at 30°C to allow colony formation, and viable cell numbers were expressed as CFU. Viability was represented by the ratio of viable cell numbers after freeze-thaw treatment to viable cell numbers before freeze-thaw treatment. The viability of cells before freeze-thaw treatment was taken as 100%.
Freeze-thaw treatment condition.
The yeast cells were grown at 30°C with shaking at 200 rpm until the log growth phase (optical density at 600 nm [OD600] of 1) in YPD medium. The cells were harvested by centrifugation, washed in distilled water, and resuspended in an equal volume of distilled water at room temperature. An aliquot of 5 ml was then taken from the cell suspensions and used as a negative control (nonfrozen control). The remaining cell suspensions were divided into 5-ml aliquots and frozen at −30°C for 24 h, 48 h, 72 h, or 96 h. The frozen cell suspensions were then thawed at 30°C in a water bath for 10 min.
Measurement of intracellular levels of ROS.
The intracellular levels of ROS were measured using the oxidant-sensitive probe 2′,7′-dichlorofluorescein diacetate (DCFDA; Molecular Probes, Eugene, OR). After freeze-thaw treatment, the yeast cells were collected and resuspended in YPD medium, and then the cell culture was incubated at 30°C for 60 min (postthaw incubation). Thirty minutes before the end of postthaw incubation, DCFDA (final concentration at 10 μM) was added to the cell culture. The cells were then washed, resuspended in 200 μl of distilled water, and disrupted with glass beads in a FastPrep FP100A instrument (Bio 101) for 40 s at the speed setting of 6.5. Cell extracts (50 μl) were mixed with 450 μl of distilled water, and the fluorescence was measured at λemission = 538 nm (λexcitation = 485 nm) using a SpectraMax Gemini XS microplate spectrofluorometer (Molecular Devices, Sunnyvale, CA). The values of λemission = 538 nm were normalized using the viable cell numbers. The fluorescence intensity of the nonfrozen control cells was relatively taken as 100%.
DNA microarray analysis of indirect gene expression.
We performed DNA microarray analysis of indirect gene expression as follows. The yeast cells were exposed to freeze-thaw treatment for 0, 24, or 48 h, and then the cells were subjected to postthaw incubation for 60 min at 30°C in YPD medium. The RNA samples used for DNA microarray analysis were extracted from the cells after postthaw incubation by a method described previously (23). Affymetrix Yeast Genome 2.0 arrays (Affymetrix, Santa Clara, CA) were used as the DNA microarrays. Poly(A)+ RNA was enriched from total RNA using an Oligotex dT30 Super mRNA purification kit (Takara Bio, Ohtsu, Japan). The quality of the poly(A)+ RNA was confirmed using a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). For cDNA synthesis and cRNA synthesis and labeling, One-cycle target labeling and control reagents (Affymetrix) were used according to the manufacturer's instructions. Biotinylated cRNA was fragmented and used as a probe. The probe was hybridized to a DNA microarray at 45°C for 18 h. The arrays were washed and stained using a GeneChip fluidics station 400 (Affymetrix), and scanning was carried out using a GeneChip scanner (Affymetrix). All experiments were performed in triplicate with independently grown cells.
Statistical analysis after data acquisition and normalization of expression data was performed using GeneSpring version 7.3.1 (Agilent Technologies) based on the gene expression data from triplicate independent experiments. After data transformation to GeneSpring, per-chip normalization to the 50th percentile was performed, and per-gene normalization to the control samples (nonfrozen samples) was applied to the per-chip normalized data.
We performed K-means clustering analysis on genes that had upregulated expression when their freezing periods were extended (21). The number of iterations was 100, and a similarity measurement was performed using the Pearson correlation coefficients. The genes whose expressions were upregulated depending on freezing periods and that showed upregulation of twofold or more in a 48-h sample compared with the nonfrozen control sample were extracted. To clarify the functions of the extracted genes, we classified the genes by function using FunSpec (19) based on the data in the Munich Information Center for Protein Sequences (MIPS; http://mips.gsf.de/). A probability of the intersection between a given gene list and any given functional category occurring by chance was calculated by FunSpec. The probability is represented by P values, and the P value is a good statistical indicator of overrepresentation of genes in a given functional category (19).
Evaluation of the freeze-thaw sensitivity of gene deletion strains.
We evaluated the freeze-thaw sensitivities of BY4741 and its Δsit1, Δfre1, Δfre4, Δfre7, Δctr1, Δree1, Δmac1, and Δpro1 derivative deletion strains. BY4741 was used as a positive control, and the Δpro1 deletion mutant, which is known to be highly sensitive to freeze-thaw stress (2, 14), was used as a negative control. These strains were grown at 30°C with shaking at 200 rpm until they reached the log growth phase (OD600 of 1) in YPD medium. The yeast cells were exposed to freeze-thaw treatment for 24 or 48 h by the method described above. After the freeze-thaw treatment, the numbers of viable cells in the suspension were determined. Four parameters were used in the data analysis of the freeze-thaw sensitivity of the deletion mutants: AM, defined as the viable cell numbers of deletion strains after freeze-thaw treatment; AW, defined as the viable cell numbers of the wild-type strain after freeze-thaw treatment; BM, defined as the viable cell numbers of deletion strains before freeze-thaw treatment; and BW, defined as the viable cell numbers of the wild-type strain before freeze-thaw treatment. The freeze-thaw sensitivity was evaluated using the value of (AM/AW)/(BM/BW). Mutants that exhibited an (AM/AW)/(BM/BW) of ≤0.5 were defined as hypersensitive to freeze-thaw treatment.
Assessment of the copper supplementation effect on intracellular SOD activity and ROS level after freeze-thaw treatment.
We assessed the effects of supplementation with copper ions on the SOD activity and ROS level as follows. After freeze-thaw treatment, the cells were collected and resuspended in SD medium containing various concentrations of copper ions (0 μM, 1 μM, or 10 μM CuSO4 alone or 10 μM CuSO4 plus 100 μM copper chelator BCS), and the cell culture was then incubated at 30°C for 60 min. SD medium was used in this assay because the copper ion concentration in the culture medium can be controlled during postthaw incubation. After incubation, intracellular SOD activity and the intracellular levels of ROS were measured.
Measurement of intracellular SOD activity.
The intracellular SOD activity was assayed using the SOD assay kit-WST (Dojindo Molecular Technologies, Gaithersburg, MD) according to the manufacturer's instructions. One unit of SOD activity was defined as the amount of enzyme required to inhibit water-soluble tetrazolium salt reduction by 50%. The unit of intracellular SOD activity was normalized as the viable cell numbers. The unit of intracellular SOD activity before the cells were frozen was taken as 100%.
Assessment of the copper supplementation effect on cell viability after freeze-thaw treatment.
We assessed the effects of supplementation with copper ions on viability by measuring the viable cell numbers after freeze-thaw treatment. The cell suspensions with appropriate dilution after freeze-thaw treatment were spread onto YPD or SD agar medium supplemented with various concentrations of copper ions (0 μM, 1 μM, or 10 μM CuSO4 alone or 10 μM CuSO4 plus 100 μM BCS). Viable cell numbers were measured as described above, except that the SD agar plate was incubated for 3 days at 30°C to allow colony formation.
Statistical analysis in the assessment of the copper supplementation effect.
We evaluated the statistical significance of differences in ROS level, SOD activity, and viabilities in the assessment of copper-supplementation effect by Student's t test with two-tailed distributions using Microsoft Office Excel 2007.
Microarray data accession number.
Microarray data from the present study have been deposited in the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) under series accession no. GSE15461.
RESULTS
Design of the experimental conditions for indirect gene expression analysis.
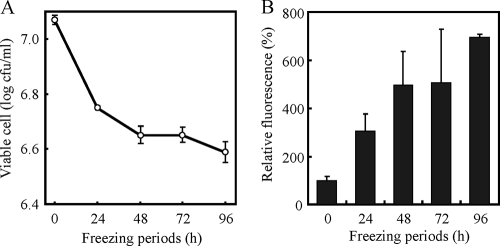
To design the experimental conditions for indirect gene expression analysis, which is an analysis during postthaw incubation, we monitored the viable cell numbers (Fig. 1A) and intracellular levels of ROS (Fig. 1B), because viable cell numbers and ROS levels are considered to be suitable markers for the degree of freeze-thaw injury. The yeast cell suspensions were frozen for 0, 24, 48, 72, or 96 h, and viable cell numbers in the cell suspensions after thawing were monitored (Fig. 1A). The viable cells decreased linearly during the first 48 h of freeze-thaw treatment and remained relatively constant thereafter. These data suggest that drastic changes in cell physiology occurred during these 48 h.
FIG. 1.
Changes in viable cell numbers (A) and intracellular levels of ROS (B) of strain BY4741 after freeze-thaw treatment. In panel A, viable cell numbers were measured after freeze-thaw treatment for 24 to 96 h. In panel B, the intracellular levels of ROS were measured after postthaw incubation preceded by freeze-thaw treatment for 24 to 96 h. The fluorescence intensity of the cells before freeze-thaw treatment was relatively set as 100%. The values are the means and standard deviations of results from three independent experiments.
After freezing, the cells were thawed, and postthaw incubation was then conducted for 60 min. We then measured the intracellular ROS levels after postthaw incubation (Fig. 1B) and found that the levels had increased depending on extension of the freezing periods. In particular, the ROS levels were drastically increased during the first 48 h of freezing, which correlated well with changes in the viable cell numbers. Based on the results of viable cell numbers and intracellular ROS levels, we estimated that drastic changes in cell physiology occurred within 48 h of when the cells were frozen. Therefore, we decided to analyze the gene expression of cells that were treated with freezing for either 24 or 48 h.
Indirect gene expression analysis using a DNA microarray after postthaw incubation.
To examine the changes in cell physiology during freezing, we carried out indirect gene expression analysis using yeast cells after 24 or 48 h of freeze-thaw treatment followed by postthaw incubation at 30°C for 60 min. RNAs were extracted from the cells obtained from the postthaw incubation, and DNA microarray analysis was performed. It is conceivable that genes upregulated by freeze-thaw stress were involved in recovery from the freeze-thaw-injured state in this analysis. To extract genes that were upregulated by freeze-thaw stress, we performed K-means clustering analysis. We found that 65 genes were upregulated in a manner that depended on the freezing period, and these genes showed upregulation of twofold or more in the 48-h sample compared with the nonfrozen control samples.
To characterize the extracted genes, we performed functional classification of the genes using FunSpec based on the data from MIPS (Table 1). We found that the category “homeostasis of metal ions” had the lowest P value, strongly suggesting the physiological relevance of this category of function to recovery from the freeze-thaw-injured state. This category contained genes involved in copper ion transport (CTR1, FRE1, FRE7, and REE1) and iron ion transport (SIT1, FRE1, FRE4, and FRE7). Other than “homeostasis of metal ions,” the category “cellular import” was extracted with relatively low P values. Genes involved in copper ion transport and iron ion transport were included in the “cellular import” category, overlapping with the genes included in the “homeostasis of metal ions” category. These results indicated that the transport mechanisms of heavy metal ions, such as copper ions and iron ions, may be specifically activated during postthaw incubation after freeze-thaw treatment. The transport mechanisms of metal ions may be involved in the recovery process from the freeze-thaw-injured state.
TABLE 1.
Functional classification of genes whose expressions were upregulated depending on the freezing period in indirect gene expression analysis
| MIPS functional categorya | P valueb | Gene(s) |
|---|---|---|
| Homeostasis of metal ions | 3.73E−04 | SIT1, FRE1, FRE4, FRE7, CTR1, REE1 |
| Cellular import | 1.83E−03 | SIT1, FRE1, CTR1, HXT5, HXT11 |
| Oxidative stress response | 2.00E−03 | GPX2, HSP12, CTT1, GRE2 |
| Protein folding and stabilization | 2.11E−03 | SSA1, SSA4, HSP26, HSP42, HSP82 |
| Oxidation of fatty acids | 3.29E−03 | POT1, SPS19 |
| Unfolded protein response | 4.58E−03 | SSA1, SSA4, HSP26, HSP42 |
| Stress response | 4.97E−03 | HSP30, HSP82, MRK1, UBI4, DDR2, YJL144W |
| Siderophore-iron transport | 5.91E−03 | SIT1, FRE4 |
| Alcohol fermentation | 6.94E−03 | NDE2, ADH2 |
| Heavy metal ion transport | 7.44E−03 | FRE1, FRE7, CTR1 |
| Nonvesicular endoplasmic reticulum transport | 9.23E−03 | SSA1, SSA4 |
| Glycolysis methylglyoxal bypass | 9.84E−03 | GRE2 |
| Sodium-driven symporter | 9.84E−03 | PHO89 |
| Others | ADR1, ADY2, AGP2, ATO2, BAG7, DSF1, ECM4, FMP16, FMP40, GLC3, GPH1, INO1, JEN1, MOH1, NCA3, NQM1, OTU1, PRM5, PUT1, PYK2, RNR3, RTC2, SHC1, SOL4, SPG1, UGX2, YPS6, YDR034W-B, YER053C-A, YGR110W, YLR149C, YML131W, YMR085W, YNL092W, YNL134C, YNL194C, YNL195C, YOR376W-A |
Genes were classified by gene function using FunSpec based on the categories defined by the MIPS database.
Functional categories with P values of <0.01 are listed.
In the “homeostasis of metal ions” category, we found that the genes whose transcription was regulated by Mac1 were contained at high frequency. Mac1 activates the expression of six genes (CTR1, CTR3, FRE1, FRE7, IRC7, and REE1) under a copper deficiency condition (7, 10), which, taken together with our results, suggests that the import of copper ions may be activated during postthaw incubation and that a deficiency in utilizing copper ions may occur in freeze-thaw-injured cells. We therefore decided to analyze the gene functions with regard to tolerance to freeze-thaw stress of the genes contained in the category “homeostasis of metal ions” and MAC1.
Evaluation of the freeze-thaw sensitivity of deletion mutants of genes involved in homeostasis of metal ions.
To examine the correlation between freeze-thaw tolerance and the function of metal ion homeostasis genes, we evaluated the freeze-thaw sensitivity of deletion strains of the genes contained in the category “homeostasis of metal ions” and the gene encoding transcriptional factor Mac1. BY4741 (positive control), the Δpro1 negative control strain, and the deletion strains were frozen for 24 or 48 h, and freeze-thaw sensitivity was evaluated as residual cell viability. As shown in Fig. 2, the Δctr1 and Δmac1 strains showed hypersensitivity to freeze-thaw stress. The Δctr1 strain in particular showed a higher level of freeze-thaw sensitivity than did the Δpro1 strain, a representative strain hypersensitive to freeze-thaw stress (2, 14). Other strains with FRE1, FRE4, FRE7, REE1, and SIT1 deletions showed sensitivity equivalent to that of BY4741 (data not shown). These data indicated that CTR1 and MAC1 play a critical role in tolerance to freeze-thaw stress in S. cerevisiae. Because CTR1 and MAC1 were involved in copper ion transport, the maintenance of copper ion homeostasis may be important for freeze-thaw tolerance.
FIG. 2.
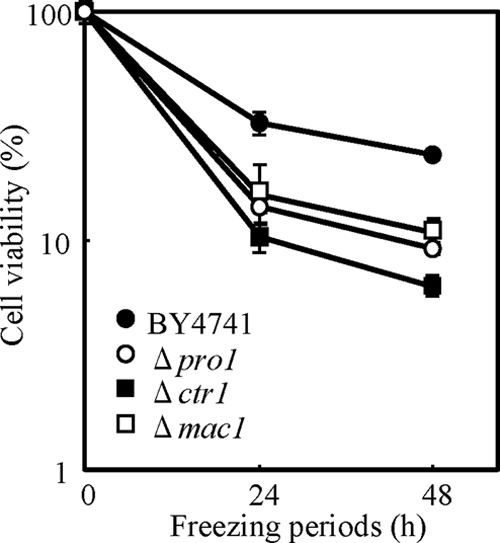
The viabilities of strain BY4741 (as a positive control) and the Δmac1, Δctr1, and Δpro1 (as a negative control) deletion strains were measured after freeze-thaw treatment for 24 or 48 h. The values are means and standard deviations of results from three independent experiments.
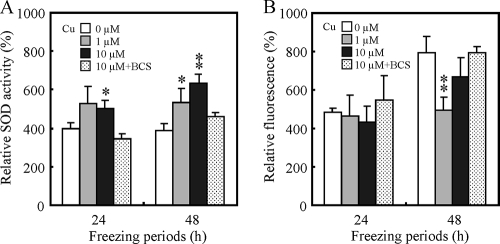
Effects of supplementation with copper ions on intracellular SOD activity and ROS levels.
Based on the results described above, we speculated that insufficiency of copper ion homeostasis was a cause of freeze-thaw injury. We therefore investigated the effects of supplementation with copper ions on intracellular SOD activity and ROS levels, because Cu/Zn SOD, which plays a role in oxygen radical detoxification, requires copper ions for its activity. Intracellular SOD activity and intracellular levels of ROS after postthaw incubation were measured under various copper ion-containing conditions. Figure 3A shows the effects of supplementation with copper ions during postthaw incubation on intracellular SOD activity. The total SOD activities were significantly increased if copper ions were supplemented at the concentration of 1 μM or 10 μM to the medium used for postthaw incubation. Inversely correlated with SOD activity, intracellular ROS levels were decreased by supplementation with copper ions (Fig. 3B). Supplementation with BCS, which is a specific copper chelator, inhibited the effects of copper ions (Fig. 3A and B). These results suggested that copper ions functioned to decrease the degree of freeze-thaw injury by SOD activation and to decrease the ROS level. It is possible that one of the causes of freeze-thaw injury is insufficiency of copper ion homeostasis.
FIG. 3.
Changes in intracellular SOD activity (A) and intracellular levels of ROS (B) in BY4741 by supplementation with copper ions during postthaw incubation. In panel A, the intracellular SOD activities were measured after postthaw incubation preceded by freeze-thaw treatment for 24 or 48 h using SD medium containing various copper ion concentrations. The unit of SOD activity of the cells before freeze-thaw treatment was relatively set as 100%. In panel B, the intracellular levels of ROS were measured after postthaw incubation preceded by freeze-thaw treatment for 24 or 48 h using SD medium containing various copper ion concentrations. The fluorescence intensity of the cells before freeze-thaw treatment was relatively set as 100%. The values are the means and standard deviations of results from three independent experiments. Asterisks and double asterisks indicate that there are significant differences between the values from samples supplemented with no copper and the values from respective samples with P values of <0.05 and <0.01, respectively.
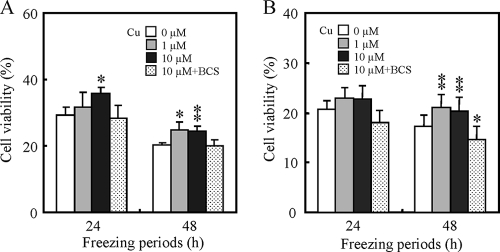
Effects of supplementation with copper ions on the cell viability of freeze-thaw-injured cells.
To evaluate the effects of supplementation with copper ions on cell viability after freeze-thaw treatment, we assessed cell viability using YPD and SD medium supplemented with copper. Figure 4 shows the correlation between cell viability and supplementation of the copper ion concentration in YPD (Fig. 4A) and SD (Fig. 4B) media. Supplementation of CuSO4 in either medium significantly increased the viability of cells frozen for 48 h. These results suggest that supplementation with copper ions was effective in increasing the cell viability of freeze-thaw-injured cells.
FIG. 4.
Effects of supplementation with copper ions on cell viability of BY4741 after freeze-thaw treatment. The cell viabilities after freeze-thaw treatment were measured using YPD (A) and SD (B) agar media containing various copper ion concentrations. The viability of cells before freeze-thaw treatment was relatively set as 100%. The data shown are means of more than triplicate measurements from a representative experiment. Error bars represent the standard deviations of the means. Asterisks and double asterisks indicate that there are significant differences between the viabilities of samples supplemented with no copper and the viabilities of respective samples with P values of <0.05 and <0.01, respectively.
DISCUSSION
Yeast cells suffer freeze-thaw injury during the freeze-thaw process, and the viability and fermentation ability of yeast cells after thawing are dramatically decreased. Extension of the freezing period is especially damaging to yeast cells (13, 15). Elucidation of the physiological changes during freezing may contribute to the improvement of commercial processes such as frozen dough baking technology.
In this study, we attempted to clarify the physiological changes of yeast cells caused by freeze-thaw injury by indirect gene expression analysis conducted during postthaw incubation after freeze-thaw treatment. We suspected that the changes of gene expression observed in indirect gene expression analysis might reflect the physiology of the freeze-thaw-injured state of yeast cells. The genes involved in causing freeze-thaw injury may be upregulated during postthaw incubation.
We found that the genes involved in the homeostasis of metal ions, such as copper ions and iron ions, were upregulated in indirect gene expression analysis following freeze-thaw treatment, and this is the first report of such a finding. It has been reported that various environmental stresses, such as air-drying, alkaline pH, and acid stresses, are closely associated with metal ion homeostasis in yeast cells (11, 20, 22). Our results suggest the involvement of metal ion homeostasis in freeze-thaw injury.
MAC1 and CTR1 gene deletion strains showed hypersensitivity to freeze-thaw stress. MAC1 and CTR1 genes are involved in copper ion uptake to a cell under a copper-deficient condition (4, 7, 10, 12). Copper ions are an essential trace element for almost all organisms because of their ability to act as a cofactor in a variety of important enzymes for optimal antioxidant defense, including Cu/Zn SOD and cytochrome c oxidase (6, 26). Deficiency of copper ions decreases the viability of cells subjected to oxidative stress (6, 26). However, the deletion strains of metal ion homeostasis genes other than MAC1 and CTR1 included in the “homeostasis of metal ions” category (Table 1) did not show freeze-thaw sensitivity. We speculate that these deletion strains may be insensitive to freeze-thaw stress because of the redundant function of other genes. We confirmed the positive effects of copper ion supplementation on the metabolism of freeze-thaw-injured cells, which requires copper ions. Our results showed that intracellular SOD activity was increased and the intracellular level of ROS was decreased by supplementation with copper ions during postthaw incubation. Furthermore, cell viability after freeze-thaw treatment was increased by copper supplementation. Freeze-thaw injury is closely related to oxidative stress by ROS formation during the freeze-thaw process, and protection from ROS improves cell viability (2, 5, 17, 18). The results described in the present study suggest that an insufficiency of copper ion homeostasis in yeast cells may correlate with suffering a freeze-thaw injury, because copper ions play a role in the intracellular detoxification of the level of ROS.
To the best of our knowledge, the present study is the first report to show the correlation between freeze-thaw injury and copper ion homeostasis. However, we have obtained no evidence that copper ions play a molecular role in freeze-thaw tolerance. Further study is needed to determine the role copper ions play in increasing tolerance to freeze-thaw stress. Our data may accelerate clarification of the mechanisms of freeze-thaw injury and contribute to the commercial application of freeze-thaw processes, including frozen dough baking technology.
Acknowledgments
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry.
Footnotes
Published ahead of print on 11 September 2009.
REFERENCES
- 1.Aguilera, J., F. Randez-Gil, and J. A. Prieto. 2007. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol. Rev. 31:327-341. [DOI] [PubMed] [Google Scholar]
- 2.Ando, A., T. Nakamura, Y. Murata, H. Takagi, and J. Shima. 2007. Identification and classification of genes required for tolerance to freeze-thaw stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. FEMS Yeast Res. 7:244-253. [DOI] [PubMed] [Google Scholar]
- 3.Attfield, P. V. 1997. Stress tolerance: the key to effective strains of industrial baker's yeast. Nat. Biotechnol. 15:1351-1357. [DOI] [PubMed] [Google Scholar]
- 4.De Freitas, J. M., J. H. Kim, H. Poynton, T. Su, H. Wintz, T. Fox, P. Holman, A. Loguinov, S. Keles, M. van der Laan, and C. Vulpe. 2004. Exploratory and confirmatory gene expression profiling of mac1Δ. J. Biol. Chem. 279:4450-4458. [DOI] [PubMed] [Google Scholar]
- 5.Du, X., and H. Takagi. 2005. N-Acetyltransferase Mpr1 confers freeze tolerance on Saccharomyces cerevisiae by reducing reactive oxygen species. J. Biochem. 138:391-397. [DOI] [PubMed] [Google Scholar]
- 6.Gaetke, L. M., and C. K. Chow. 2003. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 189:147-163. [DOI] [PubMed] [Google Scholar]
- 7.Gross, C., M. Kelleher, V. R. Iyer, P. O. Brown, and D. R. Winge. 2000. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 275:32310-32316. [DOI] [PubMed] [Google Scholar]
- 8.Hsu, K. H., R. C. Hoseney, and P. A. Seib. 1979. Frozen dough. I. Factors affecting stability of yeasted doughs. Cereal Chem. 56:419-424. [Google Scholar]
- 9.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 10.Jungmann, J., H. A. Reins, J. Lee, A. Romeo, R. Hassett, D. Kosman, and S. Jentsch. 1993. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 12:5051-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawahata, M., K. Masaki, T. Fujii, and H. Iefuji. 2006. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 6:924-936. [DOI] [PubMed] [Google Scholar]
- 12.Kuhlbrandt, W. 2004. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5:282-295. [DOI] [PubMed] [Google Scholar]
- 13.Mazur, P. 1970. Cryobiology: the freezing of biological systems. Science 168:939-949. [DOI] [PubMed] [Google Scholar]
- 14.Morita, Y., S. Nakamori, and H. Takagi. 2003. l-Proline accumulation and freeze tolerance of Saccharomyces cerevisiae are caused by a mutation in the PRO1 gene encoding γ-glutamyl kinase. Appl. Environ. Microbiol. 69:212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muldrew, K., and L. E. McGann. 1990. Mechanisms of intracellular ice formation. Biophys. J. 57:525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura, T., H. Takagi, and J. Shima. 2009. Effects of ice-seeding temperature and intracellular trehalose contents on survival of frozen Saccharomyces cerevisiae cells. Cryobiology 58:170-174. [DOI] [PubMed] [Google Scholar]
- 17.Odani, M., Y. Komatsu, S. Oka, and H. Iwahashi. 2003. Screening of genes that respond to cryopreservation stress using yeast DNA microarray. Cryobiology 47:155-164. [DOI] [PubMed] [Google Scholar]
- 18.Park, J. I., C. M. Grant, M. J. Davies, and I. W. Dawes. 1998. The cytoplasmic Cu, Zn superoxide dismutase of Saccharomyces cerevisiae is required for resistance to freeze-thaw stress. Generation of free radicals during freezing and thawing. J. Biol. Chem. 273:22921-22928. [DOI] [PubMed] [Google Scholar]
- 19.Robinson, M. D., J. Grigull, N. Mohammad, and T. R. Hughes. 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serrano, R., D. Bernal, E. Simon, and J. Arino. 2004. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279:19698-19704. [DOI] [PubMed] [Google Scholar]
- 21.Sherlock, G. 2000. Analysis of large-scale gene expression data. Curr. Opin. Immunol. 12:201-205. [DOI] [PubMed] [Google Scholar]
- 22.Shima, J., A. Ando, and H. Takagi. 2008. Possible roles of vacuolar H+-ATPase and mitochondrial function in tolerance to air-drying stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. Yeast 25:179-190. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka-Tsuno, F., S. Mizukami-Murata, Y. Murata, T. Nakamura, A. Ando, H. Takagi, and J. Shima. 2007. Functional genomics of commercial baker's yeasts that have different abilities for sugar utilization and high-sucrose tolerance under different sugar conditions. Yeast 24:901-911. [DOI] [PubMed] [Google Scholar]
- 24.Terao, Y., S. Nakamori, and H. Takagi. 2003. Gene dosage effect of l-proline biosynthetic enzymes on l-proline accumulation and freeze tolerance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 69:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thannickal, V. J., and B. L. Fanburg. 2000. Reactive oxygen species in cell signaling. Am. J. Physiol. Lung Cell Mol. Physiol. 279:L1005-L1028. [DOI] [PubMed] [Google Scholar]
- 26.Vonk, W. I., C. Wijmenga, and B. van de Sluis. 2008. Relevance of animal models for understanding mammalian copper homeostasis. Am. J. Clin. Nutr. 88:840S-845S. [DOI] [PubMed] [Google Scholar]