Abstract
The multicopper oxidase laccase is widespread in fungi and has great industrial importance. One puzzle regarding laccase production in the basidiomycetous yeast Cryptococcus neoformans is that it is inhibited by high temperature (e.g., 37°C). In this paper, we report identification of a nitrogen metabolite repression-related gene, TAR1, which is responsible for laccase repression. Disruption of TAR1 results in a significant increase in the level of LAC1 mRNA at 37°C. The putative protein Tar1 shares a moderate level of similarity with the nitrogen metabolite repressors Nmr1 and NmrA from Neurospora crassa and Aspergillus nidulans, respectively. Likewise, Tar1 has a negative role in the utilization of nitrate. Furthermore, the structure of Tar1 is unique. Tar1 lacks the long C-terminal region of Nmr1 and NmrA. It contains the canonical Rossmann fold motif, GlyXXGlyXXGly, whereas Nmr1 and NmrA have variable residues at the Gly positions. Interestingly, the promoter region of TAR1 contains three TTC/GAA repeats which are likely the heat shock factor (Hsf) binding sites, implying that Hsf has a role in laccase inhibition. TAR1 mediation of temperature-associated repression of LAC1 suggests a novel mechanism of laccase regulation and a new function for Nmr proteins. Our work may be helpful for industry in terms of promotion of laccase activity.
The copper-containing polyphenol oxidase laccases are widespread in fungi (6). These enzymes have a broad array of industrial and environmental applications (7, 23). For instance, laccases produced by the white rot fungi have ecological significance in the decomposition of plant litter (7, 15). Laccases have also been used for a long time in the removal of lignin from wood fibers in the pulp and paper industry (4) and in the transformation of environmental pollutants, such as trichlorophenol, dyes like orange G and amaranth, and herbicides (3, 28). Lately, laccases have drawn the attention of scientists due to their potential use in producing the biofuel ethanol from lignocellulose biomass (5). Efforts to make novel organic molecules or biomaterials with laccases have also been reported (17, 22).
Despite its practical importance, our knowledge regarding the molecular basis of laccase expression in fungi is limited. The basidiomycete Cryptococcus neoformans provides an excellent model to investigate the regulation of expression of fungal laccases (34). C. neoformans is ubiquitous in soil and is enriched in bird droppings (13), and it has been identified as one of the major fungal pathogens in human immunodeficiency virus-infected patients (13). This fungus exhibits abundant laccase activity, which has been recognized as a major virulence trait (25). Cryptococcal laccase is encoded mainly by the LAC1 gene despite the fact that there are two paralogs in the genome (18, 34). Laccase is the only enzyme responsible for the formation of the high-molecular-weight pigment melanin in the presence of an appropriate polyphenolic substrate (e.g., norepinephrine [NE]) (30). Disruption of LAC1 alone results in elimination of enzymatic activity, melanin biosynthesis, and attenuated virulence in a mouse cryptococcosis model (18, 25, 34). Previous studies showed that several environmental factors affect the production of cryptococcal laccase. A high glucose concentration (e.g., 2%) is a strong inhibitor of laccase expression (19). Glucose depletion initiates laccase expression, which involves the Gα-cyclic AMP-protein kinase A signal pathway (1). The trace element copper is an inducer of laccase activity (33). An elevated temperature (e.g., 37°C) has been shown to be a negative regulator (14), although the optimal temperature for enzymatic activity is 37°C (29). Unfortunately, the mechanism of temperature-associated laccase repression remains unknown.
We previously created a mutant library using a random insertional mutagenesis strategy (34). Mutant DK58 produced dark colonies at 37°C in the presence of NE. In this paper, we report further characterization of the gene that was disrupted in this mutant. This gene, designated TAR1 (temperature-associated repressor; GenBank accession no. FJ379298), putatively is an open reading frame (ORF) exhibiting homology (∼32% identity) to ORFs encoding fungal Nmr1 and NmrA repressors in Aspergillus nidulans and Neurospora crassa, respectively (2, 31). We show here that TAR1 plays negative roles in temperature-mediated laccase expression and in nitrogen metabolism. The structural and functional uniqueness of Tar1 is also described.
MATERIALS AND METHODS
Strains and media.
C. neoformans serotype A strain H99 (= ATCC 208821) (20) was used as the wild type in this study. H99FOA was a uracil auxotrophic mutant of H99 that was used as a recipient for making the second targeted disruption mutant designated the Δtar1 mutant. The Δtar1C strain was a reconstituted strain in which a wild-type copy of TAR1 was transformed into Δtar1. YPD medium (2% glucose, 2% Bacto peptone, 1% yeast extract) was used for routine growth of C. neoformans. Low-glucose (0.1%) asparagine (Asn) salt agar or liquid medium (0.1% asparagine, 0.3% KH2PO4; pH 5.2) was used for melanin production in the presence of the laccase substrate NE (100 mg/liter) (29).
Measurement of laccase enzymatic activity.
The laccase enzymatic activity assay was carried out as described previously (29). Briefly, yeast cells grown overnight in 25 ml YPD were pelleted and washed three times with sterile water. Cells were then resuspended in 200 μl sterile double-distilled H2O, plated onto Asn salt agar plates or inoculated into liquid Asn medium containing 0.1% glucose, and incubated at 37°C or 30°C for laccase induction. A total of 107 cells were resuspended in 0.05 M sodium phosphate (pH 6.5) for the NE oxidation assay, in which the optical density at 475 nm was determined. One unit of enzymatic activity was defined as an increase of 0.001 U of absorbance at 475 nm in 30 min. The assay was carried out in triplicate, and the error was expressed as the standard deviation.
RNA preparation and Northern blotting.
To extract total RNA, cells were induced in liquid Asn medium as indicated in the figure legends for each assay at 30°C or 37°C for 8 h. Cells were collected, and total RNA was isolated by using the protocol provided with a BIOZOL total RNA extraction reagent kit (BIOER Technology Co., Ltd., Hangzhou, China). Northern blotting was conducted by following the manufacturer's instructions for an N+-Magaprobe nylon transfer membrane (GE Osmonics Inc., MN). The membrane was probed with a 0.4-kb PCR fragment of LAC1 and a 0.5-kb ACT1 PCR fragment (see below). The probe was labeled with [α-32P]dCTP using a random primer DNA labeling kit (TaKaRa Biotech Co. Ltd., Dalian, China) according to the manufacturer's instructions.
Targeted disruption of TAR1 by homologous recombination.
To create a second targeted TAR1disruption mutant, a disruption construct was first made. Two primers, 58-S and 58-A, were used to amplify a 1.0-kb fragment of TAR1 with H99 genomic DNA as the template. The sequences of 58-S and 58-A were 5′CTTGTGCAGTTCCGACGG and 5′GAGCTGCATATACGGCATG, respectively. The selectable marker, a 1.2-kb cryptococcal Ura5 gene, was inserted into the TAR1 fragment at a unique NdeI site to obtain a disruption cassette (see Fig. 2A). The disruption cassette (length, 2.2 kb) was amplified by PCR using the same primers, and approximately 10 μg of the PCR product was transformed into H99FOA by a biolistic procedure with the Bio-Rad PDS-1000/He particle delivery system (Bio-Rad Laboratories, Inc., CA). For Δtar1 mutant screening, a PCR procedure was employed with a pair of primers, Ura5P3 and 58-T3a, which gave rise to a 1.0-kb PCR product in positive candidates, while no products were amplified with the wild type. The sequences of Ura5P3 and 58-T3a were 5′GAGTACGGTGTTCAGAGGTCT and 5′TTGTCGGAAGTACTGCTCC, respectively. 58-T3a was located 300 bp downstream of the disruption construct. The PCR products were sequenced by TaKaRa Biotech Co. Ltd. (Dalian, China) for verification. Two positive candidates, TX16 and TX30, were obtained for Southern analysis.
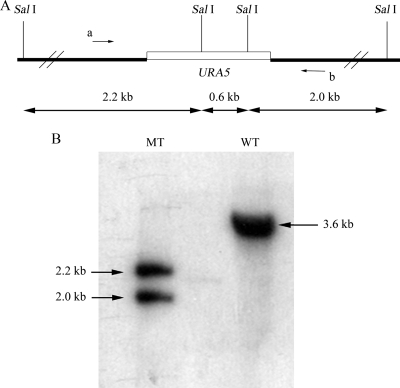
FIG. 2.
(A) Diagram of the predicted disruption at the TAR1 locus. SalI cutting sites are indicated. Arrows a and b indicate primers 58-S and 58-A, respectively. (B) Southern blotting to confirm the disruption of TAR1. Two SalI restriction bands (∼2.2 and 2.0 kb) were detected by probing with the wild-type 1.0-kb fragment amplified with primers 58-S and 58-A. The wild type had only one 3.6-kb band, as shown in panel A. Lane MT, mutant; lane WT, wild type.
Southern blotting for verification of Δtar1.
Cryptococcal genomic DNA was prepared for Southern blotting as described previously (33). Approximately 8 μg DNA from each strain was digested with SalI, which cut Ura5 twice. The probe was the same fragment of TAR1 used for the disruption cassette (1.0 kb). Digested DNA was subjected to 0.8% agarose gel electrophoresis, and blotting was carried out by following the manufacturer's instructions for an N+-Magaprobe nylon transfer membrane (GE Osmonics Inc., MN). The DNA probe was labeled with [α-32P]dCTP using a random primer DNA labeling kit (TaKaRa Biotech Co. Ltd., Dalian, China).
RT-PCR for LAC1 and TAR1 transcripts.
To determine the LAC1 mRNA level by reverse transcription-PCR (RT-PCR) (see Fig. 5), primers LAC-A and LAC-S, whose sequences were 5′-TACAACTTTCCCCGACCTC and 5′-GATGGAGAAGGTGAGCGTC, respectively, were used for RT-PCR, which resulted in a 0.4-kb product. For determination of the TAR1 transcription level, primers TAR-A and TAR-S, whose sequences were 5′-GATGTCAGGATAGCCCAC and 5′-ATCTCCCTCCGCTTTCTC, respectively, were used to amplify TAR1 cDNA, which resulted in a 700-bp product. In both cases ACT1 mRNA was used as the internal standard. The primers used for ACT1 mRNA amplification were 5′CGCTATCCTCCGTATCGATCTTGC and 5′CTGCTGGAAGGTAGACAAAGAGG, which generated a 0.5-kb PCR product. The RT-PCR was performed using a TaKaRa BcaBEST RNA PCR kit (TaKaRa Biotech Co. Ltd., Dalian, China). The initial quantity of the total RNA template in each sample was determined with a spectrometer and neutralized to 0.5 μg/μl. The PCR was performed for 21 cycles to the mid-log phase as determined previously. An aliquot (5 μl) of PCR products was subjected to electrophoretic separation in 2.0% denaturing agarose gels. A fast and sensitive procedure to determine the abundance of LAC1 mRNA relative to that of the endogenous standard ACT1 mRNA was performed using a specific charge-coupled device imaging system and specific software, Quantity One (ChemiDoc, XRS; Bio-Rad, CA) (10, 12). Briefly, gels were stained with ethidium bromide, and images were recorded using the charge-coupled device imaging system and transillumination. The cDNA band recordings were evaluated with the software mentioned above. The mRNA abundance was measured in triplicate, and the standard variation was also calculated.
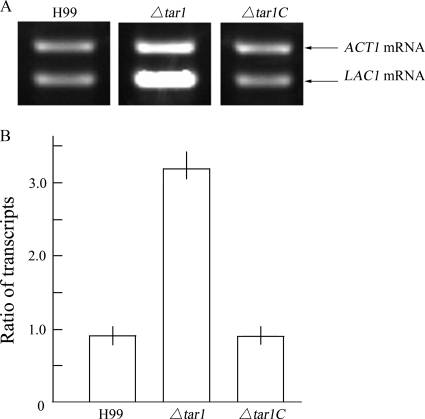
FIG. 5.
Increased LAC1 expression in the Δtar1 mutant. (A) Derepression of LAC1 transcription in the mutant. RT-PCR amplification showed that the level of LAC1 mRNA was increased dramatically in the Δtar1 mutant compared to H99 and the Δtar1C strain. Cells were induced at 37°C for 12 h in liquid Asn medium. (B) Ratio of LAC1 mRNA to ACT1 mRNA for each strain. The amount of LAC1 mRNA was 3.35-fold larger than the amount of ACT1 mRNA in the Δtar1 mutant, while the ratios of the transcripts were 0.84 ± 0.12 and 0.81 ± 0.17 for H99 and the Δtar1C strain, respectively, verifying that LAC1 transcription was depressed in the Δtar1 mutant.
RESULTS
Temperature-dependent laccase production in C. neoformans.
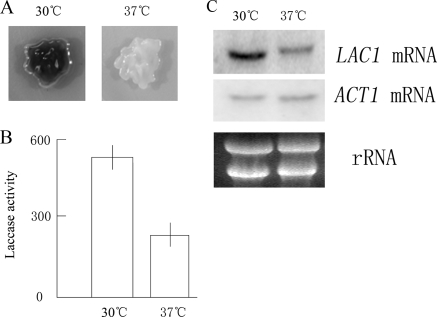
It has been established that both melanin biosynthesis and laccase activity are inhibited at 37°C, although the optimal temperature for laccase activity is 37°C (Fig. 1A and B) (14, 29). As shown in Fig. 1A, the melanin production by the yeast cells on NE-containing plates at 37°C was significantly less than that at 30°C. The laccase enzymatic activity was 239 ± 8.4 U at 37°C, compared to 530 ± 2.4 U at 30°C, as shown in Fig. 1B. Northern blotting confirmed that the level of LAC1 transcription decreased remarkably at 37°C (Fig. 1C). Thus, laccase repression by temperature takes place at the transcriptional level in C. neoformans.
FIG. 1.
Repression of melanin biosynthesis, laccase activity, and LAC1 transcription at an elevated temperature. (A) Melanin biosynthesis in H99 decreased significantly and a lighter colony formed at 37°C. Fresh H99 cells were plated on NE-containing Asn agar and incubated overnight at 37°C or 30°C as indicated. (B) Inhibition of laccase enzymatic activity in cells grown at 37°C. The activity of cells incubated at 37°C was 239 ± 8.4 U, and the activity of cells incubated at 30°C was 530 ± 2.4 U. Cells were induced for 8 h in liquid Asn salt medium. (C) Northern blotting showing the repression of LAC1 transcription under the conditions described above for panel B. Ten micrograms of total RNA was loaded in each lane. ACT1 mRNA served as a control.
Identification of TAR1 responsible for laccase repression at 37°C.
An insertional mutant, DK58, overproduced melanin at 37°C on NE-containing Asn agar (33). The gene disrupted in DK58 was recovered by using a plasmid rescue approach. Sequencing revealed that the transformed vector pBS-Ura5 was integrated 70 nucleotides upstream of the putative ATG codon of an ORF with an unknown function. The gene was designated TAR1. To verify that disruption of TAR1 was responsible for the laccase derepression phenotype in DK58, a second targeted knockout mutant, the Δtar1 mutant, was created via homologous recombination with wild-type strain H99FOA (ura5−). Two Δtar1 mutant candidates, TX16 and TX30, were obtained from nearly 2,000 transformants by using PCR screening. Southern blotting verified that TX16 was an authentic Δtar1 disruption mutant (Fig. 2B). The targeted knockout mutant TX16, designated the Δtar1 mutant, was used in this study. The Δtar1 mutant exhibits a derepression phenotype for laccase and melanin production (Fig. 3A). A reconstituted strain was subsequently generated by transforming a wild-type copy of TAR1 into the Δtar1 mutant. The complemented strain, designated the Δtar1C strain, exhibited laccase repression at 37°C (Fig. 3A).
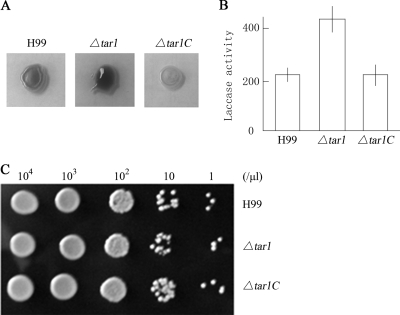
FIG. 3.
Derepression of melanin biosynthesis and laccase activity in the Δtar1 mutant. (A) The Δtar1 mutant apparently produced more melanin than wild-type strain H99 and the complemented Δtar1C strain at 37°C after 36 h. (B) The laccase activity of the Δtar1 mutant was 413 ± 15 U at 37°C after 36 h of incubation on Asn agar, while the laccase activities of H99 and the Δtar1C strain were 238 ± 13 and 235 ± 17 U, respectively. (C) Disruption of TAR1 has no effect on growth. The strains exhibited similar growth rates. Three-microliter portions of cell suspensions (cell concentrations are indicated at the top) were dropped on NE-containing Asn agar and incubated overnight at 37°C.
It should be noted that the Δtar1C strain and the wild type did not produce the same amount of melanin on plates for an unknown reason. Thus, we measured the laccase enzymatic activity of 107 cells grown at 37°C with a liquid assay (29). The results of this assay are presented in Fig. 3B. The laccase activity of 107 Δtar1 mutant cells was 413 ± 15 U, whereas the laccase activities of 107 cells of H99 and the Δtar1C mutant were 239 ± 13 U and 235 ± 17 U, respectively. The enzymatic activity of the Δtar1 mutant was significantly higher than that of either H99 or the Δtar1C mutant, verifying the negative role of TAR1 in laccase production. Since growth kinetics may have caused the increase in laccase activity in the Δtar1 mutant, we compared the growth of the three strains on Asn agar. As shown in Fig. 3C, the strains had similar growth rates, eliminating the possibility that a mutation in TAR1 caused a growth defect at the restrictive temperature. Taken together, the results described above demonstrate that TAR1 acts as a repressor in laccase production in response to high temperature.
TAR1 encodes an Nmr family repressor of nitrogen utilization.
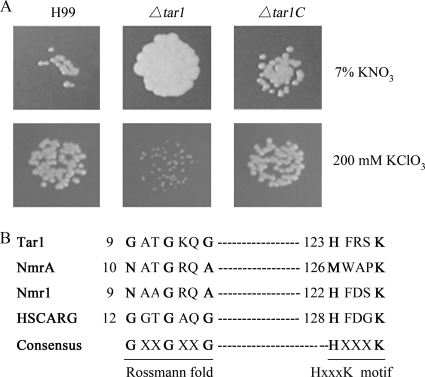
The cloned TAR1 gene and the complete cDNA (GenBank accession no. FJ379297) form a 1.16-kb ORF with three introns. This ORF putatively encodes a peptide with 288 amino acids. Tar1 shares 32% identity with the equivalent N-terminal part of the fungal Nmr repressors Nmr1 and NmrA. Previous studies found that Nmr proteins are negative regulators of nitrogen metabolite repression (2, 16). We wondered whether Tar1 had a negative role in nitrogen metabolism in C. neoformans as well. We used a couple of plate assays to demonstrate the repressive effect of TAR1 on the utilization of potassium nitrate (KNO3) (2). Utilization of a secondary nitrogen source, such as potassium nitrate, is an indication of derepression of the nitrate reductase gene (21, 31). As shown in Fig. 4A, the Δtar1 mutant cells grew significantly faster than the wild-type H99 cells and the reconstituted Δtar1C mutant cells in the presence of 7% KNO3, suggesting that there was derepression of nitrate metabolism in the Δtar1 mutant. Potassium chlorate (KClO3) is a toxic analog of KNO3, and its toxicity is also used to indicate activation of the nitrate reductase gene. As shown in Fig. 4A, 200 mM KClO3 exhibited high toxicity for the Δtar1 mutant. In conclusion, the cryptococcal TAR1 gene negatively regulates nitrogen metabolism in C. neoformans.
FIG. 4.
(A) Enhanced utilization of potassium nitrate by the Δtar1 mutant. H99 and the Δtar1C strain produced petite colonies in the presence of 7% KNO3 as the only nitrogen source (upper panels). The Δtar1 mutant showed high sensitivity to 200 mM KClO3 under nitrogen-limiting conditions (5 mM ammonium tartrate). A total of 103 cells in suspension were dropped onto Asn salt agar with 2% glucose at 30°C. (B) Alignment of the Rossmann fold for fungal Nmr proteins and the human HSCARG protein. Cryptococcal Tar1 and HSCARG have a typical Rossmann motif conserved in the members of the family, while Nmr1 and NmrA have variations in the sequence. An unknown HXXXK motif (where X is any amino acid residue) was also found in Tar1. The numbers in the sequences indicate the positions of the motifs in each protein.
Increased level of LAC1 mRNA in the Δtar1 mutant at 37°C.
To explore the mechanism underlying TAR1 modulation of laccase production, we utilized an RT-PCR approach to determine the level of LAC1 mRNA in the mutant. The methods used for preparation of total RNA and RT-PCR amplification are described in Materials and Methods. As shown in Fig. 5A, the level of LAC1 mRNA was significantly higher in the Δtar1 mutant than in H99 and the Δtar1C mutant, confirming that TAR1 acts on the steady level of LAC1 mRNA at 37°C. The complemented Δtar1C strain exhibited temperature-dependent inhibition of LAC1 transcription. For further verification, we determined the relative abundance of LAC1 mRNA compared to the abundance of ACT1 mRNA with a nonradioactive RT-PCR protocol which was shown to be as sensitive as radioactive RT-PCR (10, 12). The relative abundance was expressed as the ratio of LAC1 mRNA to ACT1 mRNA (Fig. 5B). In the Δtar1 mutant, the level of LAC1 mRNA was 3.35-fold higher than the level of ACT1 mRNA, whereas the levels of these two mRNAs were approximately the same in both H99 (ratio, 0.84 ± 0.12) and the complemented strain (ratio, 0.81 ± 0.17). This result confirmed again that TAR1 acts as a repressor in LAC1 transcription.
Temperature-dependent upregulation of TAR1 transcription.
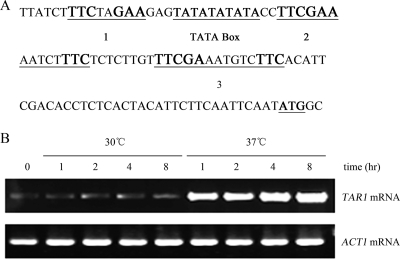
Analysis of the TAR1 promoter revealed three classic cis elements of heat shock transcription factor (Hsf) binding sites (10) (Fig. 6A). The first motif, TCCNNGAA (where N is a variable base), is located 95 bp upstream of the putative ATG codon. The second and third motifs are identical to the gap-type Hsf binding repeat, TCCGAA(N)5TCC, found in baker's yeast (11). This structural property strongly suggests that TAR1 is a temperature-activated gene that is likely activated via heat shock transcription factor. Thus, we carried out an RT-PCR to examine the transcription of TAR1 at both 30°C and 37°C. Interestingly, TAR1 expression was dramatically unregulated at 37°C after the cells were incubated for 1 h or longer (Fig. 6B). In contrast, the TAR1 gene was expressed only at a basal level at 30°C. The level of mRNA of the control ACT1 gene was constant regardless of the temperature. This finding may shed light on the mechanism of laccase repression by TAR1 at higher temperatures.
FIG. 6.
(A) Structural features of TAR1 promoter region. Three putative Hsf binding motifs are indicated by large bold type. Motif 1, TCCNNGAA (where N is a variable base), is located 95 bp upstream of the ATG codon. Motifs 2 and 3 are identical to the gap-type TCCGAA(N)5TCC repeats found in baker's yeast. The position of a TATA box-like element is also indicated. (B) Temperature-dependent regulation of TAR1 transcription determined by RT-PCR amplification. Cells of H99 were incubated in liquid Asn salt medium at 30°C or 37°C for 1, 2, 4, and 8 h as indicated. The TAR1 transcription at 37°C was dramatically enhanced compared to that at 30°C.
DISCUSSION
Opportunistic pathogenic microorganisms, such as C. neoformans, can thrive at a wide range of temperatures. C. neoformans provides a good model to investigate gene regulation by temperature (8, 24, 27). This yeast produces a copper-containing polyphenol oxidase laccase that is inhibited by high temperature (e.g., 37°C), although the repression is puzzlingly incomplete (Fig. 1A and Fig. 3A) (14, 29). In this study, we demonstrated that an Nmr-like TAR1 gene in C. neoformans is responsible for laccase repression at 37°C. Disruption of TAR1 resulted in significant increases in laccase activity and melanin production at the restrictive temperature. A wild-type copy of TAR1 restored the inhibition. At the amino acid sequence level, Tar1 exhibits moderate similarity to Nmr1 of N. crassa and NmrA of A. nidulans. Like Nmr1 and NmrA, Tar1 was implicated in the repression of utilization of secondary nitrogen sources. However, Tar1 has structural features distinct from those of its fungal counterparts. Tar1 is dramatically smaller; it has only 288 amino acids, while Nmr1 and NmrA consist of 488 and 352 amino acids, respectively. Tar1 lacks the long C-terminal part of the fungal Nmr proteins. The latter two proteins share ∼60% amino acid sequence identity. Tar1 has only ∼32% identity to either Nmr1 or NmrA. Nonetheless, Tar1 harbors a canonical Rossmann fold motif, GlyXXGlyXXGly (where X is a variable residue), in its N-terminal region (residues 9 to 15 of Tar1) (Fig. 4B), whereas the proposed motif sequence in NmrA or Nmr1 is AsnXXGlyXXAla based on crystallography data (26, 32). In fact, the closest homolog of Tar1 in the GenBank database is a protein from bacteria (data not shown). The unique structural and functional features suggest that Tar1 encodes a novel repressor.
The molecular targets of Tar1 responsible for laccase repression remain a mystery. Nmr1 and NmrA act by inhibiting a global GATA family transcription activator (31). However, analysis of the promoter region of LAC1 did not find the GATA factor binding repeat HGATAR (where H is A, T, or C) (9), suggesting that a different target of Tar1 is involved in laccase repression. As shown in Fig. 6, transcription of TAR1 itself is dramatically enhanced at 37°C. Thus, the scenario for laccase repression by Tar1 may be as follows. When the temperature increases, Hsf is activated to form homotrimers that bind to the motifs on the TAR1 promoter. TAR1 transcription is upregulated, and more Tar1 protein is synthesized. As a consequence, Tar1 inhibits the activity of an unknown target factor which positively modulates laccase transcription and hence blocks further LAC1 transcription.
Our work suggests a novel mechanism of regulation of laccase expression in the basidiomycete C. neoformans. The finding that TAR1 is involved in the repression of laccase suggests a new biological function for Nmr proteins. The structural uniqueness suggests that this family of repressors in fungi is evolutionarily diverse.
Acknowledgments
This work was supported in part by Natural Science Foundation of China grant 30770043 and by National Basic Research Program (“973” Program) grant 2007CB707801. We also acknowledge use of the C. neoformans Genome Project, Stanford Genome Technology Center (http://www-sequence.stanford.edu), funded by the NIAID/NIH under cooperative agreement AI47087.
We acknowledge the Duke Center for Genome Technology (http://cneo.genetics.duke.edu) for providing gene sequences related to this work.
Footnotes
Published ahead of print on 4 September 2009.
REFERENCES
- 1.Alspaugh, J. A., R. Pukkila-Worley, T. Harashima, L. M. Cavallo, D. Funnell, G. M. Cox, J. R. Perfect, J. W. Kronstad, and J. Heitman. 2002. Adenylyl cyclase functions downstream of the G-alpha protein GPA1 and controls mating and pathogenicity. Eukaryot. Cell 1:75-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrianopoulos, A., S. Kourambas, J. A. Sharp, M. A. Davis, and M. J. Hynes. 1998. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J. Bacteriol. 180:1973-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asgher, M., H. N. Bhatti, M. Ashraf, and R. L. Legge. 2008. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation 19:771-783. [DOI] [PubMed] [Google Scholar]
- 4.Bajpai, P., A. Anand, and P. K. Bajpai. 2006. Bleaching with lignin-oxidizing enzymes. Biotechnol. Annu. Rev. 12:349-378. [DOI] [PubMed] [Google Scholar]
- 5.Balan, V., L. da Costa Sousa, S. P. Chundawat, R. Vismeh, A. D. Jones, and B. E. Dale. 2008. Mushroom spent straw: a potential substrate for an ethanol-based biorefinery. J. Ind. Microbiol. Biotechnol. 35:293-301. [DOI] [PubMed] [Google Scholar]
- 6.Baldrian, P. 2006. Fungal laccases—occurrence and properties. FEMS Microbiol. Rev. 30:215-242. [DOI] [PubMed] [Google Scholar]
- 7.Blackwood, C. B., M. P. Waldrop, D. R. Zak, and R. L. Sinsabaugh. 2007. Molecular analysis of fungal communities and laccase genes in decomposing litter reveals differences among forest types but no impact of nitrogen deposition. Environ. Microbiol. 9:1306-1316. [DOI] [PubMed] [Google Scholar]
- 8.Chow, E. D., O. W. Liu, S. O'Brien, and H. D. Madhani. 2007. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: nitric oxide stress and body temperature. Curr. Genet. 52:137-148. [DOI] [PubMed] [Google Scholar]
- 9.Gomez, D., I. Garcia, C. Scazzocchio, and B. Cubero. 2003. Multiple GATA sites: protein binding and physiological relevance for the regulation of the proline transporter gene of Aspergillus nidulans. Mol. Microbiol. 50:277-289. [DOI] [PubMed] [Google Scholar]
- 10.Hager, G., E. Eckert, and F. W. Schwaiger. 1999. Semiquantitative analysis of low levels of mRNA expression from small amounts of brain tissue by nonradioactive reverse transcriptase-polymerase chain reaction. J. Neurosci. Methods 89:141-149. [DOI] [PubMed] [Google Scholar]
- 11.Hashikawa, N., N. Yamamoto, and H. Sakurai. 2007. Different mechanisms are involved in the transcriptional activation by yeast heat shock transcription factor through two different types of heat shock elements. J. Biol. Chem. 282:10333-10340. [DOI] [PubMed] [Google Scholar]
- 12.Horikoshi, T., and M. Sakakibara. 2000. Quantification of relative mRNA expression in the rat brain using simple RT-PCR and ethidium bromide staining. J. Neurosci. Methods 99:45-51. [DOI] [PubMed] [Google Scholar]
- 13.Idnurm, A., Y. S. Bahn, K. Nielsen, X. Lin, J. A. Fraser, and J. Heitman. 2005. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753-764. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson, E. S., and H. S. Emery. 1991. Temperature regulation of the cryptococcal phenoloxidase. J. Med. Vet. Mycol. 29:121-124. [DOI] [PubMed] [Google Scholar]
- 15.Martínez, A. T., M. Speranza, F. J. Ruiz-Dueñas, P. Ferreira, S. Camarero, F. Guillén, M. J. Martínez, A. Gutiérrez, and J. C. del Río. 2005. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 8:195-204. [PubMed] [Google Scholar]
- 16.Marzluf, G. A. 1997. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 61:17-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikolasch, A., and F. Schauer. 2009. Fungal laccases as tools for the synthesis of new hybrid molecules and biomaterials. Appl. Microbiol. Biotechnol. 82:605-624. [DOI] [PubMed] [Google Scholar]
- 18.Missall, T. A., J. M. Moran, J. A. Corbett, and J. K. Lodge. 2005. Distinct stress responses of two functional laccases in Cryptococcus neoformans are revealed in the absence of the thiol-specific antioxidant Tsa1. Eukaryot. Cell 4:202-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurudeen, T. A., and D. G. Ahearn. 1979. Regulation of melanin production by Cryptococcus neoformans. J. Clin. Microbiol. 10:724-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perfect, J. R., D. L. Toffaletti, and T. H. Rude. 1993. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 61:4446-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platt, A., T. Langdon, H. N. Arst, D. Kirk, D. Tollervey, J. M. M. Sanchez, and M. X. Caddick. 1996. Nitrogen metabolite signaling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3′ untranslated region of its mRNA. EMBO J. 15:2791-2801. [PMC free article] [PubMed] [Google Scholar]
- 22.Riva, S. 2006. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 24:219-226. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez Couto, S., and J. L. Toca Herrera. 2006. Industrial and biotechnological applications of laccases: a review. Biotechnol. Adv. 24:500-513. [DOI] [PubMed] [Google Scholar]
- 24.Rosa e Silva, L. K., C. C. Staats, L. S. Goulart, L. G. Morello, M. H. Pelegrinelli Fungaro, A. Schrank, and M. H. Vainstein. 2008. Identification of novel temperature-regulated genes in the human pathogen Cryptococcus neoformans using representational difference analysis. Res. Microbiol. 159:221-229. [DOI] [PubMed] [Google Scholar]
- 25.Salas, S. D., J. E. Bennett, K. J. Kwon-Chung, J. R. Perfect, and P. R. Williamson. 1996. Effect of the laccase gene LAC1 on virulence of Cryptococcus neoformans. J. Exp. Med. 184:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stammers, D. K., J. Ren, K. Leslie, C. E. Nichols, H. K. Lamb, S. Cocklin, A. Dodds, and A. R. Hawkins. 2001. The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 20:6619-6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steen, B. R., T. Lian, S. Zuyderduyn, W. K. MacDonald, M. Marra, S. J. Jones, and J. W. Kronstad. 2002. Temperature-regulated transcription in the pathogenic fungus Cryptococcus neoformans. Genome Res. 12:1386-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wesenberg, D., I. Kyriakides, and S. N. Agathos. 2003. White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol. Adv. 22:161-187. [DOI] [PubMed] [Google Scholar]
- 29.Williamson, P. R. 1994. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. J. Bacteriol. 176:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williamson, P. R., K. Wakamatsu, and S. Ito. 1998. Melanin biosynthesis in Cryptococcus neoformans. J. Bacteriol. 180:1570-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao, X., Y. H. Fu, and G. A. Marzluf. 1995. The negative-acting NMR regulatory protein of Neurospora crassa binds to and inhibits the DNA-binding activity of the positive-acting nitrogen regulatory protein NIT2. Biochemistry 34:8861-8868. [DOI] [PubMed] [Google Scholar]
- 32.Zheng, X., X. Dai, Y. Zhao, Q. Chen, F. Lu, D. Yao, Q. Yu, X. Liu, C. Zhang, X. Gu, and M. Luo. 2007. Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein. Proc. Natl. Acad. Sci. USA 104:8809-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu, X., and P. R. Williamson. 2003. A CLC-type chloride channel gene is required for laccase activity and virulence in Cryptococcus neoformans. Mol. Microbiol. 50:1271-1281. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, X., and P. R. Williamson. 2004. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 5:1-10. [DOI] [PubMed] [Google Scholar]