Abstract
So far, only scarce and rather diffuse information is available on bacteriophages infecting members of the genus Bifidobacterium. In the current study, we investigated the genetic organization, phylogenetic relationships, and, in some cases, transcription profiles and inducibility of 19 prophage-like elements present on the individual chromosomes of nine bifidobacterial strains, which represent six different species.
Prophage-like elements in microbial genomes represent one of the main contributors of mobile DNA, also known as the mobilome (9), and are the main reason for bacterial interstrain variability (5). The genomic era has allowed the discovery of a very large number of prophage-like elements which have been instrumental in understanding bacterial genome evolution. Like mobile DNA elements, phage DNA may also function as a vector for lateral gene transfer between bacteria. Prophage DNA may thus be a major target for the selective forces that shape the population structure of a bacterial species. The prophage contribution to the total DNA bacterial genome is highly variable: in Actinobacteria, it can represent up to 8% of the total chromosomal DNA (25), but phages may also be absent, as seems to be the case for the largest actinobacterial genome so far sequenced, that of Streptomyces coelicolor A3, as well as the actinobacterial pathogens Mycobacterium leprae and Mycobacterium bovis (7, 25). Interestingly, the genomes of actinophages are clearly mosaic, with regions of obvious sequence similarity interspersed with segments that appear to be unrelated, suggesting that extensive horizontal genetic exchange (or shuffling) is common (11). Within Actinobacteria, bifidobacteria have until recently been considered to be free from phage attacks. However, genome analyses of Bifidobacterium longum subsp. longum NCC2705, B. longum subsp. longum DJO10A, and Bifidobacterium breve UCC2003 have provided evidence that phage infections do take place in this genus (26). Nevertheless, the real extent of prophage distribution in bifidobacterial genomes or their biological role is still unknown. In the present study, we addressed these information gaps by expanding our knowledge on bifidobacterial prophages through analysis of nine bifidobacterial genome sequences, which allowed the identification and comparative analysis of prophage-like elements with respect to their gene content, inducibility, transcription profile, and comparison to other bifidobacterial sequences.
Prophage content of nine bifidobacterial genomes.
As previously described (24, 26, 27), genes encoding integrases and/or cI-type repressors can be used as markers to identify prophages in bacterial genomes. Recently, the genome sequences of Bifidobacterium dentium Bd1, B. dentium ATCC 27678, B. longum subsp. infantis ATCC 15697, B. longum subsp. infantis CCUG 52486, B. bifidum 317B, B. bifidum NCIMB 41171, B. adolescentis L2-32, B. catenulatum DSM 16992, and B. animalis subsp. lactis AD011 genomes (21; M. Ventura, F. Turroni, V. Giubellini, A. Zomer, F. Bottacini, E. Foroni, and D van Sinderen, unpublished data) (NCBI source) have become available for such purposes. Screening for integrase- and cI-type repressor-encoding genes in these genomes revealed the presence of 19 apparently complete or partial prophage-like elements. These putative prophage sequences occupy various genomic positions (names and genome positions are given in Fig. 1) and represent a significant portion of each of the relevant bifidobacterial chromosomes. The currently available bifidobacterial genome sequences contain 22 prophage-like elements, amounting to 3.02% of the Bifidobacterium pangenome content (Fig. 1). Prophages may contribute significantly to the individuality of bacterial strains (4). In the case of bifidobacteria, the relative contribution of prophages to strain-specific DNA varies between species. In the case of B. longum subsp. infantis, whole-genome comparison indicates that prophages are one of the major contributors to variability. In contrast, prophages do not appear to contribute considerably to strain variability in B. dentium, as shown by comparative genomic hybridization using 10 B. dentium strains (Ventura et al., unpublished; also, see below).
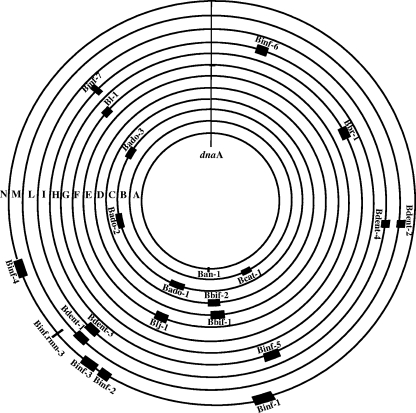
FIG. 1.
Distribution of prophage-like elements and their size relative to the genomes of their bifidobacterial host. Shown are the relative genomic positions of prophage-like elements in the genome of B. animalis subsp. lactis AD011 (ring A), B. catenulatum DSM 16992 (ring B), B. adolescentis L2-32 (ring C), B. bifidum NCIMB 41171 (ring D), B. bifidum 317B (ring E), B. longum subsp. longum DJO10A (ring F), B. longum subsp. longum NCC2705 (ring G), B. breve UCC2003 (ring H), B. longum subsp. infantis CCUG 52486 (ring I), B. dentium ATCC 27678 (ring L), B. dentium Bd1 (ring M), and B. longum subsp. infantis ATCC 15697 (ring N). A central genomic marker (dnaA) is also indicated.
Genome analyses of the identified prophage-like elements.
Database matches (see Table S1 in the supplemental material) allowed a tentative subdivision of the 19 identified prophage-like elements into functional modules (see Table S1 in the supplemental material). Notably, the genomic and modular organization of large parts of the identified prophage genomes resembled that of lambdoid phages (4) and that of the previously described B. longum subsp. longum prophage Bl-1 (28). Bioinformatic analyses of the prophage genomes revealed homology to (pro)phage genomes of high-G+C gram-positive bacteria (e.g., Mycobacterium and Rhodococcus) as well as low-G+C gram-positive bacteria (e.g., enterobacteria and Lactococcus) (see Table S1 in the supplemental material). Furthermore, such analyses revealed the presence of so-called lysogenic conversion genes (LCG), which are presumed to be derived from bacterial DNA and contribute to the ecological fitness of the lysogen. For example, the B. adolescentis L32-2 prophage-like elements Bado-1 and Bado-2 contained genes predicted to encode a type I restriction-modification system, enzymes involved in polyketide biosynthesis, and a protein containing an immunoglobulin-like domain (see Table S1 in the supplemental material). Furthermore, the genome of the prophage-like element Bado-3 contains a gene (ORF1378) encoding a protein similar to a retro-type reverse transcriptase also identified in many Proteobacteria phages (e.g., Bordetella bacteriophages) as well as in the B. longum subsp. longum DJO10A prophage like-element Blj-1 (see Table S1 in the supplemental material). Accordingly, the presence of such a gene constitutes a mechanism for rapid adaptation to allow host range changes (8). In addition, the Binf-4 genome contains a tRNA (carrying a CCA anticodon) which may contribute to the translation capabilities of B. longum subsp. infantis ATCC 15697 and thus the ecological fitness of the lysogen (4).
Among the prophage-like elements identified in the genome of the two B. dentium strains, Bdent-1 and Bdent-2 show very high sequence similarity and conserved synteny with Bdent-3 and Bdent-4, respectively (see Fig. S1 and Table S1 in the supplemental material). In addition, extensive homology was detected between particular sections of the B. dentium prophage-like sequences and predicted or proven structural modules of phages infecting member of the Firmicutes (e.g., Lactococcus) (Fig. 2). The high level of mosaicism displayed by these prophage-like elements may be explained by multiple module exchange events between phages infecting Firmicutes and Actinobacteria. Such events could have been facilitated by the common ecological origin of B. dentium and certain members of the Firmicutes. Recent sequencing data have indeed identified apparent hybrid phages that contain genetic segments from different phage genera (4) and phage families (3, 19).
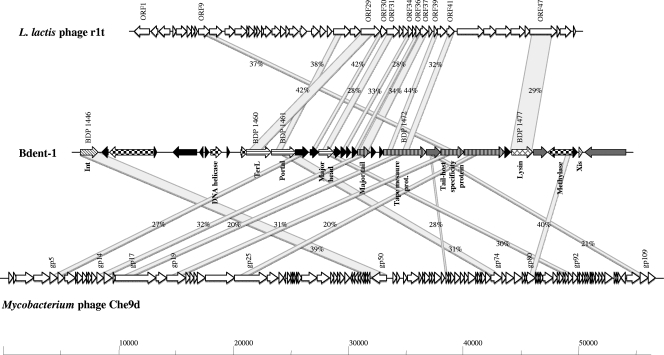
FIG. 2.
Comparative genome maps of a typical bifidobacterial (pro)phage, B. dentium Bd1 prophage-like element Bdent-1, and alignment of the genetic maps of mycobacteriophage Che9d and lactococcal phage r1t. Similar genes are linked by shading, and percentages are amino acid similarities. Predicted functions of encoded proteins identified are indicated. The modular structure of the genomes is indicated by different patterns, indicating their predicted functions: diagonal stripes, lysogeny; large checkerboard, DNA replication; horizontal stripes, DNA packaging and head; narrow vertical stripes, tail and tail fiber; cross-hatching, lysis modules; grey, similar to bacterial protein; black, hypothetical protein.
The prophage Bdent-2 sequence is flanked on each side by nearly identical sequences that represent the likely attL and attR sites, and its predicted integration site, a tRNAMet gene, which is reconstituted after phage integration, is similar to that found for several other bifidobacterial prophages (28), reflecting a common target for integration and excision.
Comparative genomics of bifidobacterial prophages.
A systematic dot plot analysis using the nineteen Bifidobacterium prophage genome sequences described here, as well as the three previously described bifidobacterial prophage-like elements (28), revealed only limited DNA similarities (Fig. 3a). These prophages share DNA sequence homology in a patchwise fashion across the nonstructural gene clusters, possibly as a consequence of modular genetic exchanges between phages that share the same ecological niche and related hosts. Moreover, the B. dentium prophages show a high pairwise similarity (see above), which was not observed for the majority of the other bifidobacterial prophages. Exceptions to this were Binf-7 (which is highly similar to B. longum subsp. longum NCC2705 prophage Bl-1 [28]), the two B. bifidum prophage-like elements, and the B. adolescentis L32-2 Bado-3 prophage genome, which shows considerable sequence similarity and genome synteny with the previously described B. longum subsp. longum Blj-1 prophage genome (28) (Fig. 3a). The general organization of the Bado-3 genome shows a one-to-one synteny to similarly sized genes in the genome of the Blj-1 prophage-like element. Such a high level of similarity (greater than 94% at DNA level) indicates that these prophage-like elements must have been capable of infecting bifidobacteria belonging to different species. In contrast, the B. animalis subsp. lactis Ban-1 prophage does not reveal significant homology with any other bifidobacterial prophage sequence.
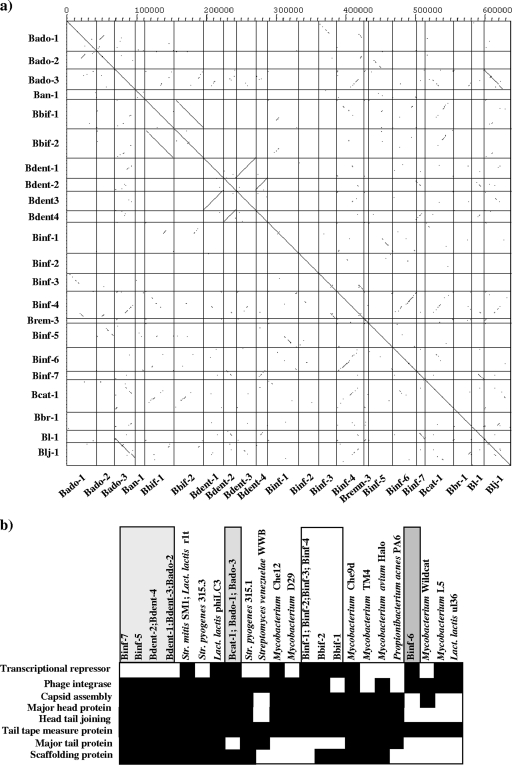
FIG. 3.
Comparative genomic analyses of bifidobacterial prophage-like sequences. (a) Dot plot matrix calculated for the complete genomes of Bado-1, Bado-2, Bado-3, Bbif-1, Bbif-2, Ban-1, Bent-1, Bent-2, Bent-3, Bend-4, Binf-1, Binf-2, Binf-3, Binf-4, Binf-5, Binf-6, Binf-7, Binf-rmn3, Bl-1, Blj-1, Bbr-1, and Bcat-1. The top x axis provides a scale in kilobases; the right y axis and the bottom x axis identify the prophage genomes that were compared in the corresponding square. The dot matrix was calculated using Dotter (22). The comparison window was 50 bp, and the stringency was 30 bp. (b) Bifidobacterial grouping based on the module occurrence and the DNA similarities across the various bifidobacterial prophages. Each column represents a phage or prophage sequence, and each row represents a protein family that is part of a module.
Analysis of the modular organization of the bifidobacterial prophage-like elements Binf-7, Bado-1, Bado-2, Bado-3, and, as previously reported, Blj-1 (28) shows that these prophage-like elements harbor an integrase gene and/or a repressor gene very close to the predicted host lysis module. Such a genomic architecture is atypical for temperate phages of gram-positive bacteria (6) and may suggest the existence of a new lineage of temperate phage.
The existence of such a relatively large number of DNA homology groups suggests that they represent separate evolutionary lineages that have followed distinct evolutionary paths within their host genus, Bifidobacterium. The reason why bifidobacterial prophages seem to exhibit a much broader genetic diversity than those found in other bacteria (e.g., Streptococcus) may be that bifidobacterial prophages are under a much higher selective pressure to diversify within the “species” or to accept alien phage modules from the environment. We speculate that the latter process is in line with the high selective pressure to which their hosts are exposed in their natural ecological niche (e.g., the gastrointestinal tract of mammals) (5, 13).
Bifidobacterial prophage-like element induction.
In order to determine whether the prophage-like elements identified in the genomes of bifidobacteria are cryptic or active phage elements, their inducibility was tested by exposing bifidobacterial cultures to hydrogen peroxide (28). Such analyses were performed targeting 14 of the 19 identified prophage like-elements (Binf-1, Binf-2, Binf-3, Binf-4, Binf-remn-3, Bbif-1, Bado-1, Bado-2, Bado-3, Ban-1, Bdent-1, Bdent-2, Bdent-3 and Bdent-4 phages), since not all bifidobacterial strains were available to us (Supplementary data). To detect excised and circularized phage genomes, PCR products should be obtained with primers targeting the left- and right-most part of prophage genomes and running outwards from the prophages (Tables 1 and 2). As assessed by PCR, no spontaneous induction of prophage-like elements was detected during growth of bifidobacterial cultures in de Man-Rogosa-Sharpe (MRS) medium (data not shown). In contrast, amplicons of 500 bp, 650 bp and 1,000 bp were obtained following hydrogen peroxide treatment or following growth in basal medium mucin (BMM), indicating that a circularized form of each of these bacteriophages was obtained (see Fig. S2a and S2b in the supplemental material). Notably, induction of Bdent-2 was only observed when the culture was grown in BMM medium, which reflects the environmental conditions of the oral cavity (2). When these 500-bp, 650-bp, and 1,000-bp amplicons, which are expected to contain the Binf-4, Bdent-2, and Ban-1 attP sites, respectively, were sequenced and aligned with the deduced Binf-4, Bdent-2, and Ban-1 attL, attR, and/or attB sites, a common core region was identified for each prophage (see Fig. S2 in the supplemental material), presumably representing the sequences where DNA strand exchange takes place during phage genome integration into or excision out of the bacterial chromosome. However, attempts to detect viable Bdent-2, Binf-4, and Ban-1 bacteriophages using plaque assays did not reveal the presence of clear plaques, which may be due to the lack of a suitable indicator strain or because no intact phage particles are produced.
TABLE 1.
Identification of putative attB regions of prophage-like elements
| Prophage-like element | attB sequence |
|---|---|
| Bdent-4 | TGCTCTAACCGACTGAGCTAAC |
| Ban-1 | GGGGTTCAATCCCCCGCGGCTCCAC |
| Binf-1 | TGCCCACATTTTGCCCACATTTT |
| Binf-5 | GCAAGTTGAAGGACGC |
| Bcat-1 | AGCACGGTTTATA |
| Binf-4 | GGGTTCGAGTCCCACCGGAGGCACCTT |
TABLE 2.
Identification of the attP regions of prophage-like elements
| Prophage-like element | attP sequence | PCR primer | PCR primer sequence | PCR amplicon size (bp) |
|---|---|---|---|---|
| Binf-4 | GGGTTCGAGTCCCACCGGAGGCACCTT | Binf4-attP1 | 5′-GACGGTTGGCTGTCGGTC-3′ | 500 |
| Binf4-attP2 | 5′-GATCAGGCCGCGACGACGAC-3′ | |||
| Ban-1 | GGGGTTCAATCCCCCGCGGCTCCAC | Blac-attP1 | 5′-GCTCAGGCGCACTCATG-3′ | 1,000 |
| Blac-attP2 | 5′-GTGGACGTGCAGAACAA-3′ | |||
| Bdent-2 | TGCTCTAACCGACTGAGCTAAC | Bdent2-attP1 | 5′-GTCTCGTCTTAATGGTGCTC-3′ | 650 |
| Bdent2-attP2 | 5′-CTCCACCTCCAACGGTTTC-3′ |
No PCR product indicative of prophage excision was obtained with primers corresponding to the prophage genomes of Binf-1, Binf-2, Binf-3, Binf-rmn3, Bado-1, Bado-2, Bado-3, Bdent-1, and Bbif-1 in the culture supernatants following hydrogen peroxide treatment. Although the majority of these prophage-like elements are not capable of excision from the host chromosome under experimental conditions, we cannot rule out the possibility that when present in their natural habitat, these prophage-like elements can be induced or require a helper phage to become active, as was suggested for the cryptic phages in Lactococcus lactis (24).
Evolutionary development of prophages in bifidobacteria.
With the increasing number of available bacteriophage genome sequences, it has become clear that genetic mosaicism is common to all regions of the phage genome, which has spurred much debate within the Prokaryote Virus Subcommittee of the International Committee on Taxonomy of Viruses regarding the expansion of taxonomy criteria beyond the traditionally used genome type and phage tail and head/capsid morphologies. Several proposals for virus classification have been put forward, mostly based on sequence information (17, 20). Recently, a novel way to investigate phage evolutionary developments was developed, which involves the assignment of reticulate relationships through the construction of a weighted graph where nodes represent phages (14). We performed such a reticulate representation of the evolutionary development of phage sequences using the (pro)phage database with inclusion of the bifidobacterial prophage-like sequences. Figure 4a shows the subgraph defined by the bifidobacterial prophage-like elements and their relatives in the (pro)phage database, revealing the existence of three main phylogenetic clusters that are connected to each other as well as to other phage clusters (see Fig. S3 in the supplemental material for a direct representation of the reticulate groups). These include group I (labeled cl_25) formed by Bdent-1, Bdent-3 and Binf-5; group II (cl_4), represented by Bdent-2, Bdent-4, Bado-2, Bado-3, B-cat1, Bbif-1, Bbif-2, Binf-1, Binf-2, Binf-4, Binf-6, Binf-7, and Binf-remn3; and group III (cl_38), comprising Binf-3 and Bado1. The prophage-like element Ban-1 is not connected to any (pro)phage in the data set, so it does not appear in the graph, and we propose that this be considered the sole representative of a separate group, designated group IV. Notably, group I appears to be an outgroup, as members of this group are not very closely related to any known phage except for Brevibacterium phage BFk20 and the Streptomyces venezuelae phage VWB (Fig. 4a; also, see Fig. S3 in the supplemental material). Furthermore, Binf-5 is phylogenetically close to phages infecting members of the Firmicutes (e.g., L. lactis, Streptococcus thermophilus, and Lactobacillus casei) (Fig. 4a; also, see Fig. S3 in the supplemental material). This may therefore represent evidence pointing to an ancient exchange of DNA sequences between low- and high-G+C-content bacteria. The largest group (cl_4) appears to constitute a transition cluster between Firmicutes- and Actinobacteria-infecting phages (Fig. 4a). Furthermore, we compared the evolutionary development of these prophage sequences and that of their bacterial hosts, using a classical molecular marker such as the 16S rRNA gene sequences, which revealed significant discrepancies (data not shown). The phylogenetic tree of the bacterial hosts points to a clear bacterial speciation between B. longum subsp. infantis, B. adolescentis, B. dentium, and B. bifidum taxa. In contrast, in the reticulate classification scheme of phages, the prophage sequences of B. longum subsp. infantis, B. adolescentis, B. dentium, and B. bifidum were shown to be of a mixed evolutionary origin.
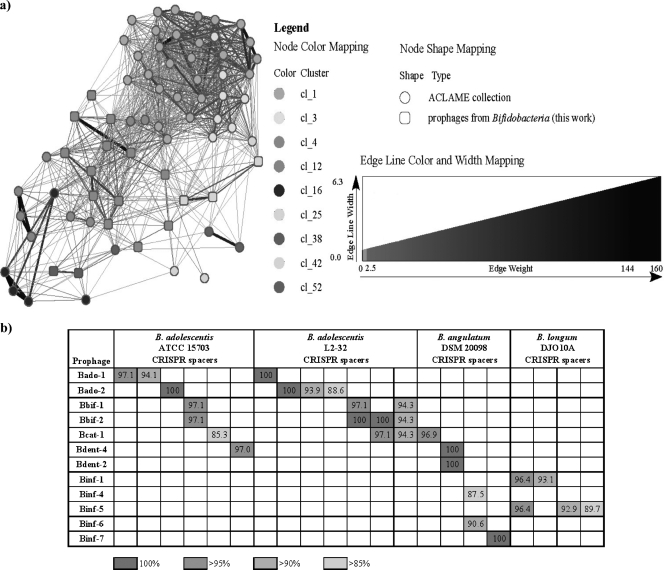
FIG. 4.
Analysis of prophage phylogenetic relationships (a) and similarities of CRISPR spacers and prophage-like element sequences (b). (a) Reticulate relationships between Bado-1, Bado-2, Bado-3, Bbif-1, Bbif-2, Ban-1, Bent-1, Bent-2, Bent-3, Bend-4, Binf-1, Binf-2, Binf-3, Binf-4, Binf-5, Binf-6, Binf-7, Binf-rmn3, and Bcat-1 prophage-like elements and other sequenced phages and prophages (phages and prophages not directly linked in the graph to the bifidobacterial prophages have been excluded for clarity). The main clusters where the (pro)phages are classified are mapped onto the nodes with different shading, and the relevance of the links is represented with a gradient of grey intensity and width. (b) DNA similarities of CRISPR spacers and prophage-like element sequences.
An alternative, novel approach to achieve taxonomic assessment of bacteriophages is based on the genomic analysis of a single or multiple phage modules to allow a rational hierarchical classification system (18). This phage taxonomy system employs comparative genomic analysis of the integrase gene, repressor-encoding genes, and elements of the DNA packaging-head gene cluster, such as the capsid assembly proteins, scaffolding proteins (3), and/or other structural-protein-encoding genes. This approach was undertaken to analyze the taxonomy of bifidobacterial prophages, only in this case the modules were not selected a priori but were the result of cooccurrence patterns of protein-encoding genes across the various phage genomes. The combinational nature of these prophages, in particular their intermediate position between phages from Firmicutes and Actinobacteria, is readily seen; e.g., modules 3 and 5 are present in certain bifidobacterial prophages and mycobacteriophages, whereas modules 1 and 4 relate the bifidobacterial prophages to different groups of phages infecting streptococci (module 4), staphylococci (module 1), and lactic acid bacteria (modules 1 and 4) (see Fig. S4 in the supplemental material). Based on the profile of modules as well as the DNA similarities exhibited by the prophage-like elements, four distinct prophage groups can be delineated (Fig. 3b). These include Binf-7, Binf-5, Bdent-1, Bdent-2, Bdent-3, Bdent-4, and Bado-2 for group A, Binf-1, Binf-2, Binf-3, Binf-4, Bbif-1, and Bbif-2 for group B, Bcat-1, Bado-1, and Bado-3 for group C, and Binf-6 for group D (Fig. 3b). The first group in particular contains prophages that seem to have followed a similar evolutionary development (see above). However, in contrast to the reticulate classification scheme, phage classification based on comparative analyses of phage modules focuses on only a part of a phage genome and does not consider the full genome sequences, thus yielding a somewhat restricted evolutionary image.
Sequence similarities between bifidobacterial prophages and CRISPR spacers.
CRISPRs (clustered regularly interspaced short palindromic repeats) are a diverse family of DNA repeats widely distributed among archaea and bacteria. A CRISPR is a defense mechanism which provides immunity against phages and plasmids in a sequence-specific manner (1, 15, 23). Since CRISPR spacers provide a historical record of interactions between hosts and viruses, we investigated whether bifidobacterial prophage sequences showed similarity to CRISPR spacers found in bifidobacterial genomes. We therefore investigated sequence similarity between 19 prophage sequences and Bifidobacterium CRISPR spacers, which revealed 29 matches (with identity values over 85%) between 12 prophages and 21 CRISPR spacers (Fig. 4b; also, see Table S2 in the supplemental material), including 8 fully identical matches. The observation of perfect matches between presumed prophage sequences and bifidobacterial CRISPR spacers strongly suggests that phages with these sequences once infected the sequenced strains. The 12 prophages originated from five distinct species, while the 21 CRISPR spacers were derived from three species, and within-species matches were observed for B. adolescentis and B. longum. Multiple hits were observed within a single prophage sequence on several occasions, including identical hits (Fig. 4b), indicating that phages containing such sequences were and perhaps still are prevalent. The conclusion from these data is that various bifidobacteria seem to rely on the dynamic CRISPR system to counter the selective pressure applied by the apparent large diversity of phages. The observed Bifidobacterium CRISPR polymorphism (12) and the extensive prophage genetic shuffling are likely to be correlated and illustrate extensive interplay between bifidobacterial populations and their infecting phages.
Transcriptome analysis of bifidobacterial prophage-like elements.
DNA microarray data of 2,143 B. dentium Bd1 open reading frames as well as almost all identified B. animalis subsp. lactis AD011 prophage genes were used to identify prophage genes that are transcribed during the lysogenic state. The DNA microarray data analyses revealed that most prophage genes were not transcribed when the B. dentium cultures were grown in MRS medium, with the exception of a small number of weakly expressed genes that are predicted to form part of the lysogeny module (e.g., repressor cI). In contrast, a large number of prophage genes were transcribed in B. dentium Bd1 cultures grown in BMM (see Fig. S1 in the supplemental material). Notably, only genes from Bdent-2 were transcribed, whereas Bdent-1 appeared to be completely silent. Transcribed genes of the Bdent-2 prophage genome correspond to the predicted excisionase gene, DNA packaging module (e.g., portal protein and prohead protease), and many prophage genes encoding hypothetical proteins (e.g., BDP_586 and BDP_587) (see Fig. S1 in the supplemental material). Previous studies have shown that orthologs of the latter phage modules are expressed only during the lytic lifestyle or when a prophage is activated (24). In contrast, no transcription of the presumed lysogeny module was observed. Notably, not all the genes encompassing the predicted structural module of Bdent-2 are transcribed, and this may explain why these DNA sequences are not capable of producing viable phage particles. No transcription was detected for any gene of the Ban-1 prophage-like element, except for a very weak level of transcription of the cI-like gene (data not shown).
Conclusions.
The presence of 19 prophage-like sequences and their similarities to CRISPR loci and double-stranded DNA phages spanning a broad phylogenetic range of host bacteria clearly demonstrates that bifidobacteria do regularly encounter these viral parasites. With respect to prophage-host interactions, our data reveal interesting parallels between bifidobacteria and other gut commensals, as reflected by a similar genetic organization and by the fact that prophages are a quantitatively important element of mobile DNA and constitute a sizable part of strain-specific DNA (26).
Bifidobacterial prophage-like elements exhibit similarities to phages infecting both Firmicutes and Actinobacteria, somehow representing an evolutionary bridge between these phage groups that may have arisen as a result of a shared habitat. If one considers the fact that bifidobacteria belong to the phylum Actinobacteria, which evolved from low-G+C-content ancestors (10), one can speculate that they have been infected by bacteriophages in relatively distant as well as recent times.
Horizontal exchange plays a crucial role in the evolution of bacterial genomes (16), and the rampant mosaicism of phage genomes, generated by exchange of genome modules between phages, illustrates the powerful creativity of this process. The significance of the modular exchange between bifidobacterial prophages is unknown, but such events may arguably be held responsible for the genetic variability of bifidobacterial phages, including their ability to infect different bifidobacterial strains and species. Our data contain clear evidence of cross-species infection, which may be the consequence of such modular exchanges, resembling that described for streptococcal/lactococcal phages (18).
Prophages in bifidobacteria may be of significant importance to their hosts. As described for prophages identified in other genomes (4), bifidobacterial prophages are expected to contribute to genome variability and have been postulated to contribute genes known as LCG, which increase the fitness of the lysogenic host. Interestingly, our analyses indicate the occurrence of LCG in some of the bifidobacterial prophages, encompassing genes which may provide an ecological advantage to the lysogen and thus guarantee a more efficient colonization of the human intestine by the bacterial host.
In vivo experiments will be needed to determine if the prophage-like elements which appear to be noninducible can in fact be excised from the host chromosome. Such findings will be important to show bifidobacterial genome stability, particularly in strains that are used as probiotics, whose health-promoting activity and safety may otherwise be affected due to the loss of genetic material. Furthermore, prophages may be used to develop new genetic tools for bifidobacteria, such as gene transfer and integration of knock-out systems.
The results obtained in this study depict a scenario where, in contrast to other closely related taxa (e.g., Mycobacterium), bifidobacteria are under great selection pressure from bifidobacterial (pro)phages. These apparently extensive interactions between bifidobacteria and phages substantiate the notion that CRISPRs and (pro)phages drive the coevolution of both phage and host genomes and furthermore emphasize the idea that the population and diversity of bifidobacteria, important members of the gut microbiota, may fluctuate due to bacteriophage-mediated extermination.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the prophage-like element sequences are as follows: Binf-1, NC_011593; Binf-2, NC_011593; Binf-3, NC_011593; Binf-4, NC_011593; Binf-5, NZ_ABQQ00000000; Binf-6, NZ_ABQQ00000000; Binf-7, NZ_ABQQ00000000; Binf-remn-3, NC_011593; Bbif-1, GQ141189; Bbif-2, NZ_ABQP00000000; Bado-1, NZ_AAXD00000000; Bado-2, NZ_AAXD00000000; Bado-3, NZ_AAXD00000000; Bcat-1, NZ_ABXY00000000; Ban-1, NC_011835; Bdent-3 and Bdent-1, NZ_ABIX00000000); and Bdent-4 as well as Bdent-2, NZ_ABIX00000000.
Supplementary Material
Acknowledgments
This work was financially supported by an SFI Principal Investigator award (08/IN.1/B1909) to D.V.S. and a Science Foundation Ireland (SFI) CSET award to the Alimentary Pharmabiotic Centre, by the Italian Award for Outstanding Young Researchers scheme “Incentivazione alla mobilita' di studiosi stranieri e italiani residenti all'estero,” and by a Marie Curie Reintegration Grant (MERG-CT-2005-03080) to M.V. The project described was partially supported by NIH-NIGMS T32-GM08799 (D.A.S.).
We thank Carlos Canchaya for helpful discussions and bioinformatic assistance.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH.
Footnotes
Published ahead of print on 4 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw, D. J., K. A. Homer, P. D. Marsh, and D. Beighton. 1994. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology 140:3407-3412. [DOI] [PubMed] [Google Scholar]
- 3.Brüssow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 4.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canchaya, C., G. Fournous, S. Chibani-Chennoufi, M. L. Dillmann, and H. Brussow. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417-424. [DOI] [PubMed] [Google Scholar]
- 6.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Doulatov, S., A. Hodes, L. Dai, N. Mandhana, M. Liu, R. Deora, R. W. Simons, S. Zimmerly, and J. F. Miller. 2004. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature 431:476-481. [DOI] [PubMed] [Google Scholar]
- 9.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, R. S. 2001. The branching order and phylogenetic placement of species from completed bacterial genomes, based on conserved indels found in various proteins. Int. Microbiol. 4:187-202. [DOI] [PubMed] [Google Scholar]
- 11.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath, P., A. C. Coute-Monvoisin, D. A. Romero, P. Boyaval, C. Fremaux, and R. Barrangou. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131:62-70. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence, J. G., W. Hendrix, and S. Casjens. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9:535-540. [DOI] [PubMed] [Google Scholar]
- 14.Lima-Mendez, G., J. Van Helden, A. Toussaint, and R. Leplae. 2008. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25:762-777. [DOI] [PubMed] [Google Scholar]
- 15.Marraffini, L. A., and E. J. Sontheimer. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochman, H., and J. G. Lawrence. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 17.Pride, D. T., T. M. Wassenaar, C. Ghose, and M. J. Blaser. 2006. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proux, C., D. van Sinderen, J. Suarez, P. Garcia, V. Ladero, G. F. Fitzgerald, F. Desiere, and H. Brussow. 2002. The dilemma of phage taxonomy illustrated by comparative genomics of Sfi21-like Siphoviridae in lactic acid bacteria. J. Bacteriol. 184:6026-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage phiP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sela, D. A., J. Chapman, A. Adeuya, J. H. Kim, F. Chen, T. R. Whitehead, A. Lapidus, D. S. Rokhsar, C. B. Lebrilla, J. B. German, N. P. Price, P. M. Richardson, and D. A. Mills. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 105:18964-18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sonnhammer, E. L., and R. Durbin. 1995. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1-10. [DOI] [PubMed] [Google Scholar]
- 23.Sorek, R., V. Kunin, and P. Hugenholtz. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181-186. [DOI] [PubMed] [Google Scholar]
- 24.Ventura, M., A. Zomer, C. Canchaya, M. O'Connell-Motherway, O. Kuipers, F. Turroni, A. Ribbera, E. Foroni, G. Buist, U. Wegmann, C. Shearman, M. J. Gasson, G. F. Fitzgerald, J. Kok, and D. van Sinderen. 2007. Comparative analyses of prophage-like elements present in two Lactococcus lactis strains. Appl. Environ. Microbiol. 73:7771-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ventura, M., C. Canchaya, D. Pridmore, and H. Brussow. 2003. Integration and distribution of Lactobacillus johnsonii prophages. J. Bacteriol. 185:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ventura, M., C. Canchaya, M. Kleerebezem, W. M. de Vos, R. J. Siezen, and H. Brussow. 2003. The prophage sequences of Lactobacillus plantarum strain WCFS1. Virology 316:245-255. [DOI] [PubMed] [Google Scholar]
- 28.Ventura, M., J. H. Lee, C. Canchaya, R. Zink, S. Leahy, J. A. Moreno-Munoz, M. O'Connell-Motherway, D. Higgins, G. F. Fitzgerald, D. J. O'Sullivan, and D. van Sinderen. 2005. Prophage-like elements in bifidobacteria: insights from genomics, transcription, integration, distribution, and phylogenetic analysis. Appl. Environ. Microbiol. 71:8692-8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.