Abstract
The transcription dynamics of subunit A of the key gene in methanogenesis (methyl coenzyme M reductase; mcrA) was studied to evaluate the relationship between process rate (methanogenesis) and gene transcription dynamics in a peat soil ecosystem. Soil methanogen process rates were determined during incubation of peat slurries at temperatures from 4 to 37°C, and real-time quantitative PCR was applied to quantify the abundances of mcrA genes and transcripts; corresponding transcriptional dynamics were calculated from mcrA transcript/gene ratios. Internal standards suggested unbiased recovery of mRNA abundances in comparison to DNA levels. In comparison to those in pure-culture studies, mcrA transcript/gene ratios indicated underestimation by 1 order of magnitude, possibly due to high proportions of inactive or dead methanogens. Methane production rates were temperature dependent, with maxima at 25°C, but changes in abundance and transcription of the mcrA gene showed no correlation with temperature. However, mcrA transcript/gene ratios correlated weakly (regression coefficient = 0.76) with rates of methanogenesis. Methanogen process rates increased over 3 orders of magnitude, while the corresponding maximum transcript/gene ratio increase was only 18-fold. mcrA transcript dynamics suggested steady-state expression in peat soil after incubation for 24 and 48 h, similar to that in stationary-phase cultures. mcrA transcript/gene ratios are therefore potential in situ indicators of methanogen process rate changes in complex soil systems.
In recent years, the analysis of bacterial gene expression in environmental samples has developed as an important approach for evaluating the in situ metabolic state of physiologically active microbial cells. Most studies of transcriptional activity have been restricted to environments with low concentrations of coextractable enzymatic inhibitors, organic radicals, or nucleases (21, 53, 56), possibly due to technical issues associated with the low efficiency of extraction and rapid degradation of environmental mRNA. More recently, attempts have been made to quantify the transcription of functional genes and correlated physiological processes in complex soil communities (40, 42). The properties of peat soils are particularly detrimental to high-quality nucleic acid extraction because of acidity (pH 3.8), high water content (80 to 90%), high concentrations of coextractable organic matter, high capacity for complexing nucleic acids (10), and high concentrations of stable free radicals (52) and phenolics (59). Not surprisingly, therefore, the transcriptional dynamics of the key microbial functional genes in peat ecosystems have not yet been investigated.
In peat soils, methane is produced by strictly anaerobic Euryarchaeota, mainly from CO2, H2, and acetate (13, 22, 27, 36, 37). The key functional enzyme in methane generation is methyl coenzyme M reductase (MR), which catalyzes the reduction of one methyl group bound to coenzyme M, releasing CH4 (57). MR is assumed to be specific for methanogens, and evolutionary phylogenies established from sequence analysis of the mcrA gene (which encodes subunit A of MR isoenzyme I) and of 16S rRNA genes are congruent, implying its conserved nature (30, 33).
Characterization of peat methanogen communities by analysis of the diversity of mcrA or 16S rRNA gene sequences (18, 23, 36, 55) is often carried out in conjunction with assessment of the methanogenic potential under laboratory conditions (12-14, 24) and/or field measurement of net methane emission rates (3, 19, 20, 22). Although molecular analysis of methanogen communities generates valuable information and possible correlation of sequence types with ecological niches (23, 35), this approach does not address the dynamics of methanogenesis, for example, during natural drying and rewetting cycles (26). Similarly, analysis of methanogen potential under laboratory conditions may be hampered by distortion of the natural processes leading to methanogenesis.
Recently, Cadillo-Quiroz and colleagues (6) quantified methanogens in peat soil by using real-time quantitative PCR (qPCR) amplification of 16S rRNA genes, and Colwell and colleagues (8) quantified the abundance of mcrA genes in marine sediments, extrapolating data to estimate in situ CH4 production rates. In situ methanogenic activity can potentially be assessed more directly by quantifying the expression of the functional genes encoding MR (47), while quantification of both genes and gene transcripts allows calculation of transcript/gene ratios (51, 61). Such ratios emulate transcript abundance per cell ratios that have been compared with growth and activity in methanogen pure-culture studies (31, 38, 43, 48), potentially allowing in situ analysis of growth state and activity.
The aim of this study was to assess the validity of this approach by determining the relationship of functional gene transcriptional activity and process rates in an unamended peat soil. Methane production rates were determined for peat soil slurries incubated at different temperatures and were compared with the transcriptional dynamics of the mcrA gene by reverse transcription-qPCR (RT-qPCR) and qPCR. A further aim was to assess DNA and mRNA recovery from peat and its influence on qPCR and RT-qPCR data, thereby enabling correction of biases with internal-standard recovery efficiencies.
MATERIALS AND METHODS
Peat origin and incubation.
Peat samples were collected from an ombrotrophic upland blanket peat at Lake Vyrnwy, North Wales, United Kingdom, in November 2007. Vegetation was dominated by immature mown Scottish heather (Calluna vulgaris) interspersed with cotton grass (Eriophorum spp.) on a peat moss (Sphagnum spp.) lawn. The water table was close to the peat surface. Soil samples were taken to a depth of 20 cm after removal of vegetation and roots. Peat was broken up into pieces of approximately 500 cm3 and stored in plastic bags at 4°C until further processing. pHH2O was 3.8 (standard error [SE], 0.05; n = 3). Water content, determined as weight loss after drying at 100°C for 48 h, was 82% (wt/vol) (SE, 0.30; n = 3). Samples were stored at 4°C for up to 6 months prior to analysis.
For preparation of peat slurries, large pieces of peat, stored for 2 to 6 months, were broken up, homogenized by blending (Philips HR1372/90 hand blender, United Kingdom), and gravimetrically adjusted to a water content of 90% (wt/vol). Homogenized peat was evacuated three times in an anaerobic jar at 900 Pa (Eppendorf 5301 diaphragm pump, Cambridge, United Kingdom), flushed with 99.9% N2 to establish anoxic conditions, and incubated at 4°C for 24 h to allow equilibration of residual traces of oxygen with the gas phase. Homogenized anoxic peat (15 g) was dispensed into 100-ml serum bottles in a glove box under constant N2 flow and sealed with butyl rubber septa. Serum bottles were again purged with N2 and incubated in quadruplicate at 4, 10, 15, 25, 32, and 37°C to assess temperature-dependent rates of CH4 production and mcrA transcription. To analyze mcrA transcription at constant CH4 production rates, triplicate peat slurries containing soil that had been stored for either 4 months or 6 months were incubated at 25°C for 16 days or 4 days, respectively, and were sampled with low or high frequency, respectively.
Measurement of methane production rates.
Methane production was assessed by direct injection of 0.5 ml of headspace gas, sampled using a 1-ml disposable syringe, into a Hewlett Packard 5890 series II gas chromatograph (Agilent Technologies, Stockport, United Kingdom) equipped with a flame ionization detector (FID-GC). Gases were separated with a 1.5-m Porapak Q 80:100 mesh column (Sigma-Aldrige, Poole, United Kingdom) with N2 as the carrier gas at a flow rate of 30 ml min−1. The GC injector, oven, and FID temperatures were 100°C, 50°C, and 350°C, respectively. Rates of methane production g−1 peat (dry weight) were calculated by linear regression of increases in CH4 concentration with time, using at least three consecutive measurements with a regression coefficient (r2) of >0.9. Temperature trial peat slurries were incubated for 6 days and analyzed every 2 days. Time course experiment results were analyzed after 0, 1, 2, 3, 5, 7, and 16 days or after 0, 6, 12, 24, 48, 72, and 96 h for peat stored for 4 or 6 months, respectively.
Nucleic acid extraction.
Peat slurry microcosms were sampled destructively immediately after GC analysis, and samples were frozen immediately in liquid nitrogen and stored at −80°C. Variation between batches during qPCR was avoided by restricting analysis to samples generating the maximum number of 96 qPCR reactions per single assay (including standards and controls). Thus, analysis was performed on slurries from peat stored for 2 months and incubated at 4, 15, 25, and 32°C, on slurries from peat stored for 4 months and sampled at 0, 1, 3, 5, and 16 days (for time course experiments), and on slurries from peat stored for 6 months and sampled at 0, 6, 12, 24, 48, and 72 h. Extractions were performed in duplicate per biological replicate as described previously (16, 41), with some modifications. Peat samples were semidefrosted on ice, and 0.5 g was transferred into prechilled, 2-ml screw-cap Blue Matrix Ribolyser tubes (Hybaid, Ashford, Middlesex, United Kingdom) preloaded with 0.5 ml cetyltrimethylammonium bromide extraction buffer for total nucleic acid extraction. Immediately before addition of cold Tris-buffered phenol and chloroform, 3.3 × 107 cells of Escherichia coli K-12 (NCIMB 10218) suspended in 10 μl of the RNA preservative RNAlater (Ambion, Applied Biosystems, Warrington, United Kingdom) was added to samples from the time course slurries constructed from peat stored for 4 months to provide an internal calibration standard for qPCR assays (see below). Disruption of cells was performed twice for 20 s each time, with intermittent cooling in a Hybaid Ribolyser (Thermo Scientific, Basingstoke, United Kingdom) at speed 4. After initial separation of the aqueous and organic phases, the aqueous phases were transferred to MaXtract high-density gel barrier tubes (Qiagen, Crawley, United Kingdom) and an additional phenol-chloroform extraction, followed by chloroform extraction, was carried out according to the manufacturer's instructions. Precipitation of nucleic acids was carried out for 2 h on ice with the addition of precipitation carrier linear acrylamide (final concentration, 15 μg ml−1; Ambion). Tubes were kept on ice at all times during extraction, and extracts were subdivided into two aliquots for preparation of DNA and RNA templates. DNA and RNA templates were further purified by using a NucleoSpin Extract II silica spin column purification system (Macherey Nagel, Düren, Germany) and an RNeasy minikit (Qiagen), respectively, in accordance with the manufacturers' protocols for high-yield purification. The latter included 10 μl β-mercaptoethanol (≥99.0%; Sigma-Aldrich) ml−1 RLT lysis/binding buffer. DNA residues were removed from RNA templates by digestion with 2 U Turbo DNase (Ambion) for 30 min at 37°C, with the addition of 40 U of the RNase inhibitor RNasin (Promega, Southampton, United Kingdom). DNase-digested RNA extracts were further purified by phenol-chloroform extraction followed by chloroform extraction in gel barrier tubes. Finally, RNA extracts were precipitated with twice the aqueous phase volume of 100% ethanol, 500 mmol ammonium acetate (Sigma-Aldrich), and 15 μg ml−1 linear acrylamide. Salts were removed by washing twice with 75% ethanol, and RNA was redissolved in 12.5 μl RNase-free H2O. DNA and RNA purity and concentrations were established by Nanodrop spectroscopy. DNA aliquots were adjusted to 2 ng μl−1 for qPCR, and RNA aliquots were adjusted to the highest possible common concentration.
Reverse transcription and qPCR.
cDNA was generated from 5 μl of RNA extract with random hexamer primers at a final concentration of 37.5 ng μl−1 with Superscript II reverse transcriptase (Invitrogen, Paisley, United Kingdom) in accordance with the manufacturer's instructions. The reverse transcription reaction mixture also included T4 gene 32 protein (25 ng μl−1; MP Biomedicals, Cambridge, United Kingdom), linear acrylamide (100 ng μl−1), bovine serum albumin (200 ng μl−1; Ambion), and 20 U of the RNase inhibitor RNasin. Generated cDNA was diluted 1 in 5 prior to use in RT-qPCR. For negative controls, aliquots of each RNA extract were pooled in batches of 10 extracts and diluted in proportion to the RT protocol. Performing RT-qPCR on negative controls confirmed the amplification of RNA templates free of DNA contamination. An additional control contained the RT mixture without the template to ensure that all reagents were free of contaminants.
mcrA DNA and mRNA gene fragment copy numbers were quantified by qPCR and RT-qPCR with the ML primer pair (24, 33). All qPCR reactions were carried out in 25 μl reactions with 5 μl of template DNA or cDNA added to 20 μl qPCR reaction mixture containing 2.5 U AccuSure polymerase (Bioline, London, United Kingdom), 1× AccuSure reaction buffer including 2 mmol MgSO4, 500 μmol of each dNTP and SYBR green I (10,000× concentration; Invitrogen) at a final concentration of 0.2×. To facilitate strand separation, dimethyl sulfoxide (5% final concentration; Sigma-Aldrich), betaine (0.65 M; Sigma-Aldrich), and 7-deaza-2′-deoxy-guanosine-5′-triphosphate (50 μmol; Roche Diagnostics, Burgess Hill, United Kingdom) were added to the qPCR master mix (39), and linear acrylamide (100 ng μl−1) and bovine serum albumin (200 ng μl−1) were added to increase enzymatic efficiency. The ML forward and reverse primers were each used at a 1.2 μM concentration. mcrA qPCR was carried out using a hot-start protocol, with an initial denaturation step of 15 min at 95°C and then 40 cycles of denaturation for 30 s at 95°C, annealing for 45 s at 55°C, and extension for 45 s at 68°C, followed by fluorescence quantification at the end of a 79°C step for 8 s to allow dissociation of possible primer dimers and unspecific amplification products. qPCR was finalized by an extension step at 68°C for 10 min and subsequent melt curve analysis for confirmation of amplicon specificity by verification of identical, clearly defined melting peaks and by amplicon size analysis using standard agarose gel electrophoresis. Standard curves for calibration of mcrA qPCR were created using triplicate 10-fold dilution series covering 7 orders of magnitude from 10 to 1 × 107 gene copies per qPCR reaction during each run. The standard consisted of a 782-bp PCR amplicon spanning the mcrA region of Methanosarcina barkeri Schnellen 1947 (DSM 804) created with primers M. barkeri-forward (CTCCATGGGTGAAATGCTTC) and M. barkeri-reverse (AAAGTTGAAGGGCAGGAGGT). M. barkeri primer sequences were aligned to the mcr operon sequence of Methanosarcina barkeri fusaro (NCBI accession no. NC_007355), using the eprimer3 primer prediction software program (49). qPCR reactions were performed in duplicate per DNA or cDNA template and were carried out using a MyIQ5 real-time PCR detection system (Bio-Rad, Hemel Hempstead, United Kingdom). DNA- and cDNA-associated qPCR-determined mcrA copy numbers were expressed as quantities ng−1 nucleic acid, g−1 soil (dry weight), or g−1 soil (dry weight) normalized to internal-standard-recovery efficiency (see below). A survey of available genomic sequences demonstrated the presence of one or two mcrA coding regions, but calculations assumed one mcrA gene copy per cell. Additionally, qPCR amplification efficiency of peat samples was assessed by subjecting serial dilutions of DNA and cDNA to mcrA qPCR with the ML primer pair as described above. Efficiency was calculated as the slope of linear regression of derived copy numbers against the dilution factor.
Internal standards and dynamic range.
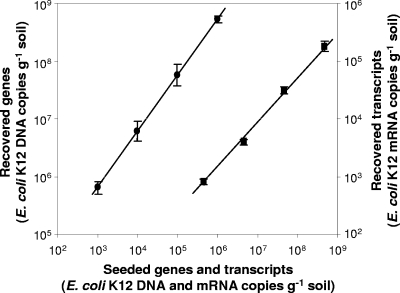
Slurry cDNA and DNA were also used to quantify recovery of an E. coli K-12 internal standard from slurries constructed from peat stored for 4 months. Coding sequences of E. coli K-12 substrain MG1655 (NCBI accession no. U00096) were subjected to BLASTn (1) searches against a database containing the genomic sequences of 12 different methanogens (NCBI accession no. NC_009515, NC_007955, NC_009635, NC_009135, NC_009634, NC_008942, NC_009051, NC_003551, NC_003552, NC_007355, NC_007681, and NC_000916) and three further environmental microbial genomes (NCBI accession no. NC_004757.1, NC_007614.1, and NC_002977). Ten E. coli K-12 coding sequences with no discernible sequence similarity were randomly selected as putative targets for qPCR internal standard assays. For each selected sequence, primers spanning 700 to 1,600 bp and 150 to 250 bp nested within were generated with eprimer3 for production of qPCR calibration standards and qPCR primer pairs, respectively. E. coli K-12 qPCR primers were screened for background PCR amplification with DNA and cDNA templates generated from unspiked peat, as described above, and for analysis of qPCR amplification efficiency on generated qPCR standards and on DNA and cDNA generated from mid-log-phase cultures of E. coli K-12. qPCR primers targeting 196 bp of E. coli coding sequence b0606, gene ahpF, demonstrated good product yield with E. coli K-12 DNA and cDNA templates, with no detectable PCR products on peat DNA and cDNA templates. To quantify internal-standard recovery, DNA and cDNA generated from the time course experiment and constructed from peat stored for 4 months were subjected to E. coli K-12 qPCR using E. coli K-12 qPCR primers b0606-qPCR-f (CGCATGACGTTGACTGAAAT) and b0606-qPCR-r (AGGATCTGACCACCAAAACG), each with the 1,554-bp E. coli K-12 b0606 amplicon as a calibration standard. E. coli K-12 b0606-f (TGCTCGACACAAATATGAAAAC) and b0606-r (TGGTGCGAATCAGGTAGTCA) were used to generate E. coli K-12 b0606 qPCR standards. To calculate E. coli K-12 RNA recovery efficiencies, E. coli K-12 cell suspensions were also subjected to nucleic acid extraction and reverse transcription as described above. E. coli K-12 cell suspensions were produced from mid-log-phase cultures grown for 6 h in Luria-Bertani medium. Pellets obtained by centrifugation of 2.5 ml culture at 10,000 × g for 15 min were resuspended in 1 ml of the RNA preservative RNAlater to generate internal standards or in 1 ml 4% formalin in phosphate-buffered saline for cell counts determined by DAPI (4′,6-diamidino-2-phenylindole) fluorescence-based counting (46) of triplicate samples, indicating 3.3 × 109 (SE, 1.9 × 108) cells ml−1. E. coli K-12 cell suspensions in RNAlater were combined and aliquoted to guarantee equal cell distribution and frozen at −80°C until further use. E. coli K-12 qPCRs were carried out as described above on peat slurry DNA and cDNA templates, E. coli K-12 cell suspensions, and qPCR standards and controls with a 0.4 μM final concentration of E. coli K-12 qPCR primers. The E. coli K-12 qPCR efficiencies were 84 to 90%, with linear regression coefficients (r2) of 0.993 to 0.997. To access the dynamic range of extraction and reverse transcription (42), serial dilutions of E. coli K-12 cell suspensions ranging from 3.3 × 108 to 3.3 × 105 cells per 10 μl were added in triplicate to 0.5 g of homogenized peat as described above, and nucleic acid extractions, qPCR, and RT-qPCR were carried out as described above.
RESULTS
Methane production.
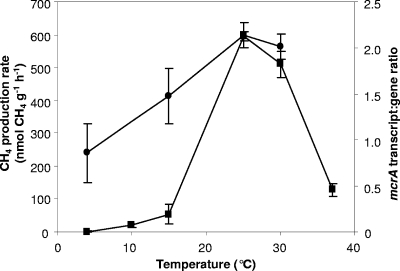
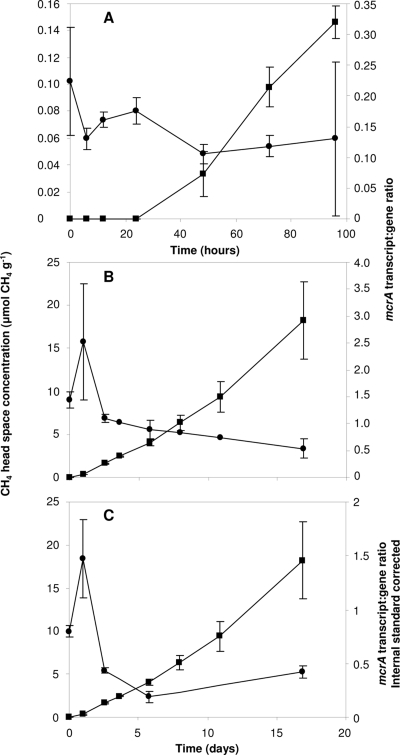
CH4 was produced in all peat slurries, with rates spanning 3 orders of magnitude, from 1.1 to 595 nmol g−1 h−1 (Fig. 1 and 2). In the temperature experiment, CH4 production rates were highest at 25 and 30°C and there were no significant differences in rates between these two temperatures (P > 0.05), suggesting adaptation to temperate climate. Rates were lowest at 4°C, where increases in methane concentration were close to the detection limit of FID-GC. The temperature-dependent rate increase from 4 to 15°C was approximately 6.2 nmol g−1 h−1 °C−1, with substantial increases to 15 and 25/30°C. The methane production rates in both time course experiments at 25°C (Fig. 2A and B) were significantly lower (42.5 [SE, 4.6] nmol g−1 h−1 and 2.0 [SE 0.2] nmol g−1 h−1 for the slurries constructed from peat stored for 4 and 6 months, respectively), equivalent to those at 15°C and 4°C in the temperature experiment, with significant lag periods (24 and 48 h, respectively) (Fig. 2A).
FIG. 1.
Mean peat slurry CH4 production rates and corresponding mean mcrA transcript/gene ratios during incubation of slurries for 6 days at different temperatures. ▪, methane production rate (μmol CH4 g−1 [dry weight] h−1); •, mcrA transcript/gene ratios, calculated from copy numbers ng−1 nucleic acid. Error bars represent the standard deviation from the mean for four replicates.
FIG. 2.
Mean mcrA transcript/gene ratios and corresponding CH4 slurry headspace concentrations during time course incubations at 25°C. Results are shown for slurries constructed from peat stored for 6 months (A) or 4 months (B, C). (A, B) mcrA transcript/gene ratios obtained from copy numbers ng−1 nucleic acid. (C) Ratios calculated from E. coli K-12 DNA and mRNA internal-standard-recovery-corrected mcrA abundances. ▪, methane production rate (μmol CH4 g−1 [dry weight]); •, mcrA transcript/gene ratio. The high SE at the last time point in the time course experiment with peat stored for 6 months was caused by a single mcrA transcript abundance 1 order of magnitude higher than other values. The error bars represent the standard deviation from the mean for three replicates.
Molecular analysis.
Total DNA and RNA yields were high in all experiments, with DNA concentrations within the range of yields reported for soils with high organic content (5). Raw extracts after initial precipitation were faintly discolored, with low A260/A280 ratios, indicating residues of coextracted humic acids. Additional purification with silica spin columns increased ratios to >1.7 for total RNA and DNA extracts. Nucleic acid extraction yields and RNA/DNA ratios varied significantly (P < 0.05) between the three experiments (Table 1). Mean DNA yields from temperature experiment peat slurries were approximately 10-fold higher than those from slurries of peat stored for 6 months, but total RNA/DNA yield ratios were higher in peat stored for 4 months at 4°C and highest in peat stored for 6 months. Amplification of qPCR calibration standards was linear (r2 > 0.994) in all qPCR assays from 102 to 106 copies of the template per reaction, and qPCR efficiency in all assays was between 85% and 95%, allowing confident enumeration of >10 mcrA copies ng−1 template. The peat sample qPCR efficiencies calculated from the slope of linear regressions of template DNA and cDNA dilution series were within the same range, suggesting unbiased enumeration.
TABLE 1.
Temperature- and time-dependent total mcrA transcript and gene abundances, total DNA and RNA yields, and mcrA transcript and gene detection limits
| Parameter | Mean (SE) per exptl data set for: |
||
|---|---|---|---|
| Peat stored for 4 mo (n = 15) | Peat stored for 6 mo (n = 21) | Effect-of-temp expt (n = 16) | |
| mcrA transcript abundance/ng nucleic acid | 6.6 × 103 (1.7 × 103) | 3.9 × 102 (8.5 × 101) | 6.1 × 102 (9.7 × 101) |
| mcrA transcript abundance/g soil | 6.7 × 107 (1.2 × 107) | 1.1 × 107 (3.9 × 106) | 8.5 × 106 (1.4 × 106) |
| mcrA gene abundance/ng nucleic acid | 4.5 × 103 (4.3 × 102) | 2.4 × 103 (2.9 × 102) | 3.3 × 102 (4.6 × 101) |
| mcrA gene abundance/g soil | 1.7 × 108 (4.2 × 107) | 3.6 × 107 (3.9 × 106) | 3.4 × 107 (3.9 × 106) |
| Internal-standard-corrected mcrA transcript abundance/g soil | 4.8 × 108 (8.1 × 107) | ||
| Internal-standard-corrected mcrA gene abundance/g soil | 9.1 × 108 (1.5 × 108) | ||
| Detection limit for mcrA transcript abundance/g soil | 1.9 × 103 | 6.5 × 103 | 3.1 × 103 |
| Detection limit for mcrA gene abundance/g soil | 3.2 × 104 | 1.5 × 104 | 1.5 × 105 |
| Total DNA yield (μg/g) | 32 (2.0) | 15 (0.7) | 152 (12.0) |
| Total RNA yield (μg/g) | 26 (0.8) | 14 (0.8) | 17 (1.0) |
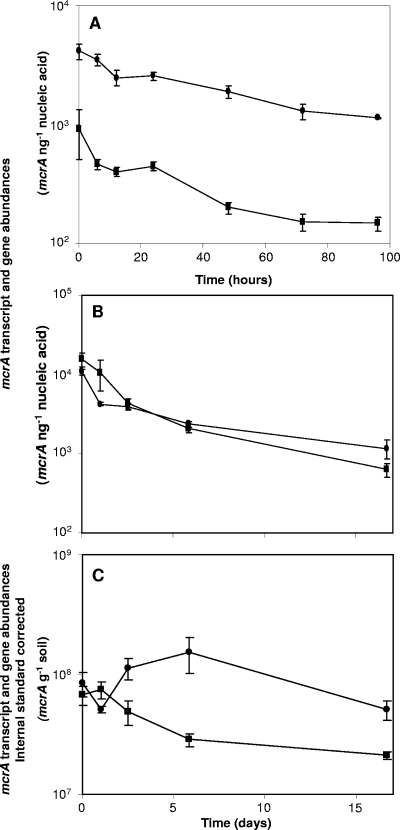
mcrA amplicons were generated during qPCR and RT-qPCR from all extracts, and qPCR specificity was confirmed by fragment size evaluation with conventional agarose gel electrophoresis. To assess possible qPCR inhibition, RT-qPCR and qPCR were carried out on serial dilutions (10−1 to 10−3) of total RNA and DNA extracts, and the proportionality of the resulting mcrA copy numbers to the dilution factors was confirmed (50). Comparison of SEs of technical and biological replicates suggested greater biological than technical variance for DNA extractions but greater technical variance for mRNA extractions. However, differences in mean SE were not significant. The mean copy numbers across all samples were 2.4 × 103 and 2.5 × 103 ng−1 nucleic acid, equivalent to 2.9 × 108 and 1.4 × 108 g−1 soil, for DNA and cDNA, respectively. mcrA gene and transcript copy numbers increased three- and fourfold, respectively, with increasing temperature and were highest at 25°C, corresponding to highest CH4 production rates. However, mcrA gene (2.3 × 102 ng−1 nucleic acid) and transcript (2 × 102 ng−1 nucleic acid) abundances were significant in slurries incubated at 4°C, where rates were close to the detection limit. During time course experiments, gene and transcript abundances decreased with incubation time, such that the highest mcrA gene and transcript copy numbers were detected in slurries with low or absent discernible CH4 production rates (Fig. 2 and 3). The initial mcrA transcript and gene abundances for the time course experiment conducted with peat stored for 4 months were 25- and 10-fold higher, respectively, than those observed after incubation for 16 days and 6- and 4-fold higher, respectively, than those observed after 96 h in the experiment conducted with peat stored for 6 months (Fig. 3).
FIG. 3.
Mean mcrA transcript and gene abundances during time course incubations at 25°C. Results are shown for slurries constructed from peat stored for 6 months (A) and 4 months (B, C). (A, B) mcrA abundances normalized to quantities ng−1 nucleic acid. (C) mcrA abundances g−1 soil (dry weight), corrected by E. coli K-12 internal-standard recovery. ▪, transcript abundance; •, gene abundance. The means and SEs for last three time points for panels B and C were used to calculate relationships between rate and ratio. Error bars represent the standard deviation of the mean for four replicates.
mcrA transcript/gene ratios.
mRNA transcript/gene ratios were highest at 25°C, increasing on average 2.5-fold from 4°C to 25°C (Fig. 1), with a maximum 4-fold increase. During the time course experiment with peat stored for 4 months, the initial ratio was relatively high (1.2× mean) and increased by day 2 (2× mean) and then decreased (0.7× mean). Subsequent decreases were not significant (P > 0.05), indicating steady-state transcriptional activity (Fig. 2B). Variation in ratios was greatest during early stages of incubation with peat stored for 6 months, with the highest ratios at zero hour (1.5× mean). Again, ratios were more consistent at 48 to 96 h, with less variation (0.7 to 0.85× mean) (Fig. 2A). Mean transcript/gene ratios were similar in temperature and time course experiments with peat stored for 4 months (1.6 and 1.5, respectively) but lower with peat stored for 6 months (0.15), which also showed the lowest CH4 production rate (Fig. 1 and 2A and B).
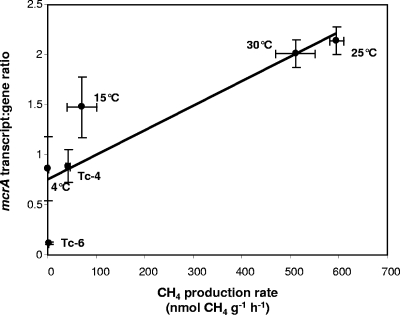
Data were analyzed further by assuming that temperature experiment mcrA transcript/gene abundance ratios and time course ratios during the later time points (based on the mean ratio from at least three consecutive data points with the least variation from the mean) reflected steady-state conditions. mcrA transcript/gene ratios from all experiments increased with corresponding CH4 production rates over an 18-fold range, implying dependence of process rate on transcription ratio (Fig. 4), with a slope of 0.26, an intercept of 1.9, and a regression coefficient of 0.79.
FIG. 4.
Mean slurry CH4 production rates and corresponding mean mcrA transcript/gene ratios. The line shows linear regression (slope = 0.0025; intercept = 0.75; regression coefficient = 0.79). Ratios and rates were compiled from the temperature experiment and both time course experiments. Ratios were calculated from mcrA transcript and gene abundance ng−1 nucleic acid. The error bars represent the standard deviation from the mean for three replicates. Labels denote relevant incubation conditions: Tc-6 represents the time course for slurries constructed from 6-month-old peat; Tc-4 represents the time course for 4-month-old peat; “4°C” represents the 4°C temperature trial, etc.
Calibration of mcrA mRNA and DNA abundances by internal standards.
To assess potential bias in determining gene and transcript abundances, extraction efficiencies were determined by calculating recovery for E. coli K-12 cells added to peat stored for 4 months. qPCR of E. coli K-12 cell suspensions yielded 1.4 × 106 DNA copies μl−1 (SE, 5.1 × 105), 50% lower than DAPI fluorescence-determined cell counts (3.3 × 106 μl−1), and 1.1 × 104 cDNA copies μl−1. Recovery of E. coli K-12 cell suspensions added to peat soil in a dilution series over 4 orders of magnitude (3.3 × 105 to 3.3 × 108) was linear (Fig. 5) (r2 > 0.99) for both E. coli K-12 genes and transcripts from 1.4 × 105 to 1.4 × 108 and 1.1 × 103 to 1.1 × 106 for DNA and cDNA, respectively. Slopes of linear regressions indicated recovery efficiencies of 62% for DNA and 92% for cDNA, suggesting unbiased analysis of transcript dynamics. However, recoveries from time courses with peat stored for 4 months were lower and varied between individual samples. The mean extraction efficiencies were 25% (SE, 7.2) and 14% (SE, 1.6) for DNA and cDNA, respectively. Applying the individual standard recovery correction factors to the respective mcrA transcript and gene abundances for this experiment increased the means to 9.1 × 108 and 4.8 × 108 mcrA copies g−1 soil, respectively. This reduced the mean mcrA transcript/gene ratio to 0.7, while maintaining a similar range of ratios from minimum to maximum (3.4-fold). However, temporal changes in ratios obtained from internal-standard-corrected values gave results that were similar to, if somewhat higher than, those normalized to quantities ng−1 nucleic acid (Fig. 2B and C and 3B and C).
FIG. 5.
Dynamic range of DNA and mRNA recovery after addition of 3.3 × 105 to 3.3 × 108 E. coli K-12 cells to peat before extraction. The slope of linear regression indicates the combined efficiency of extraction and qPCR or RT-qPCR for DNA and mRNA, respectively. The efficiencies were 62 and 93% for DNA and RNA, respectively. ▪, DNA; •, mRNA. Error bars represent the standard deviation from the mean; some error bars are smaller than the symbols.
DISCUSSION
Influence of extraction efficiency on measurement of transcription dynamics.
Recently, Nicolaisen and colleagues (42) evaluated mRNA extraction for assessing transcription dynamics in an agricultural soil. Our findings support their observations that rapid processing and maintenance of RNase-free and constantly cold conditions are necessary for extraction of high-quality environmental mRNA. Additional purification steps were required for peat, due to the high content of coextractable humic acids, but the proposed inclusion of antioxidants in the extraction buffer (7, 28) did not increase mcrA transcript or gene abundances (data not shown).
Variation between batches was minimized by analysis of sample numbers that could be processed simultaneously. The recovery efficiencies for extractions with added E. coli K-12 cell suspensions were constant over 4 orders of magnitude, and efficiency was high (>90% for mRNA), suggesting maximal recovery or possibly overestimation by qPCR (Fig. 5). All extractions were carried out using the same protocol and reagent lot by one person. Nevertheless, internal-standard-recovery efficiencies varied between batches of extractions and between individual extractions. In addition, the E. coli K-12 extraction efficiencies for DNA and mRNA obtained from long-term-incubation samples were significantly lower than the efficiencies for serial dilutions but were within the range of reported cell recoveries from soil (e.g., 31% DNA recovery from agricultural soil [44]), suggesting sensitivity to minor, unrecognized changes. Little is known about mRNA extraction efficiencies from environmental samples, but previous studies suggest that recoveries were significantly higher than reported previously (34, 53). Plasmids could be used as internal DNA standards, rather than cells (9), and T7 polymerase-amplified amplified RNA could also be used to calculate recovery efficiencies for mRNA. The addition of E. coli K-12 cell suspensions in RNA preservative RNAlater to peat prior to extraction allowed quantification of DNA and mRNA recovery efficiencies for each extraction. Efficiencies are thus less likely to be affected by differences in extraction of intracellular and extracellular nucleic acids. However, compared to that of transcript/gene ratios normalized to nucleic acid levels, the influence of recovery efficiencies was low because of similar DNA and mRNA recovery efficiencies. Confirmation of transcription dynamics by two independent normalization methods thus increases confidence in the validity of mRNA analysis in soil. Nevertheless, maximum ratios corrected for recovery efficiency were also significantly underestimated in comparison to what was found for stationary-phase Methanococcus vannielii cells (17). The mcrA gene abundances were similar to those obtained for an acidic peatland soil in which methanogen rRNA genes constituted approximately 55% of the total archaeal abundance (6), suggesting similar dominance of methanogens in the investigated peat soils.
Links between process rate and transcript abundance.
The ratio of mRNA transcript/gene abundance has been suggested as a more direct measure of transcriptional activity than absolute abundances (42, 61). Relationships between mcrA transcription and methane production rates in three sets of peat soil slurries were studied. Although transcript abundance did not correlate well with process rate, there was evidence for linear correlation (regression coefficient r2 = 0.79) between methane production rate and transcript/gene ratio, whose use may overcome variation in reproducibility and differences in cell abundance (42, 61). This approach also allows some comparison with pure-culture studies of MR transcription and activity (15, 17, 43, 45). In batch cultures of the mesophile Methanococcus vannielii (17), 180, 200 to 400, and 50 mcrA mRNA transcripts cell−1 were observed during early lag phase, initial exponential growth, and stationary phase, respectively, suggesting increase in MR content during exponential growth and reduction in stationary-phase cells. These values are more than an order of magnitude greater than those obtained with the peat slurries toward the end of the incubations, when physiological conditions are likely to be comparable to those in stationary phase. Analysis of internal-standard recovery (Fig. 5) suggests that DNA and mRNA recovery efficiencies from peat are similar, as found for tfdA transcript and gene abundances in agricultural soil (42). The difference in ratio may therefore result from a higher proportion of dead or inactive methanogens in peat soil, with low levels of mcrA mRNA.
mcrA transcript/gene ratios.
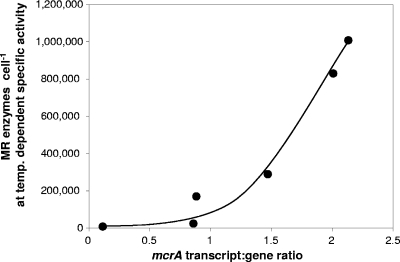
In vivo MR specific activities vary between strains and growth conditions. For example, Methanothermobacter thermautotrophicus, Methanimicrococcus blatticola, and Methanococcus maripaludis have specific activities of 3 to 5, 1.6, and 0.6 μmol CH4 min−1 mg−1 cell protein, respectively, (11, 32, 54). MR content will also vary with cell size (4, 60, 62). Temperature-dependent CH4 production rates were calculated per peat methanogen cell (μmol CH4 min−1 mcrA DNA−1), and temperature-dependent in vitro MR specific activities for the thermophile Methanothermobacter marburgensis (15) were used to estimate the MR cell content for peat methanogens required to achieve observed CH4 production rates (Fig. 6). MR was assumed to constitute approximately 6% of the cell dry weight for M. thermautotrophicus (2), equivalent to approximately 46,000 MR molecules per cell (assuming 300 kDa for MR, a cell size of 0.475 μm in diameter by 5.0 μm in length, and 50% protein content [dry weight] [2, 29, 62]). The recovery efficiency of methanogens was assumed to be 25% during qPCR, and 10% of the total population was assumed to be active, as indicated by internal-standard recovery and low transcript/gene ratios. Calculations using these assumptions predict that the number of MR enzymes required for observed CH4 production rates at 25°C exceeds the maximum number possible, even for larger cells (e.g., 250,000 for Methanosarcina balticum) or that the specific MR activity was ≥35 μmol CH4 mg−1 MR min−1 at 25°C. This suggests that MR specific activities at low temperatures in peat methanogens are significantly higher than those in M. marburgensis and/or that the number of active cells was underestimated by qPCR and extraction efficiency correction. The steep increase in extrapolated MR enzymes with increasing transcript/gene ratios (Fig. 6) further suggests that MR cell content was not maximal, probably due to substrate limitation, at the highest observed CH4 production rates in 25°C and 30°C peat slurries during the temperature experiment, even though the rate at 10°C was higher than those for a Finnish oligotrophic fen (14). These data indicate that the highest possible mcrA transcript/gene ratios were probably not reached during this experiment.
FIG. 6.
Number of MR molecules per cell required to generate the corresponding CH4 production rates per cell (mcrA DNA abundance), calculated using the specific temperature-dependent in vitro activities of M. marburgensis for 4, 15, and 25°C, extrapolated from data reported by Goenrich and colleagues (15) (0.085, 0.28, and 1.6 μmol CH4 min−1 mg−1, respectively). For 30°C, the MR activity of 25°C was used, as CH4 production during the temperature trial was highest at 25 and 30°C.
In peat slurries incubated at 4°C and in time course experiments with peat stored for 6 months, mcrA transcript abundance was significant at CH4 production rates close to the detection limit and were high at the beginning of the time course experiments. The half-life for mcrA at 37°C is 15 min (17). Temperate peat methanogens therefore maintain relatively high mcrA transcript abundances when activity is low or undetectable, as found in the mesophile M. vannielii, but not during early growth phase in the thermophile M. thermautotrophicus (43, 45). Initial time course samples were taken after exposure to ambient temperatures during preparation, which may have induced transcriptional activity in 4°C peat slurries, increasing initial ratios. Peat used for temperature and time course experiments had been stored for 2 and 6 months, respectively, and although gene and transcript abundances were similar, the latter exhibited significantly lower CH4 production rates and a longer lag period before detectable methane production. The low rates may have been due to other aspects of methanogen physiology following extended exposure to low temperatures, substrate limitation, and oxygen exposure but demonstrate a discrepancy between transcriptional and physiological activities. Nevertheless, while physiological activity in the separate experiments and slurries may have been affected by different parameters (temperature, substrate availability, physiological status, and pH change), the correlation of transcript/gene ratios with physiological rates suggests that ratios are indicative of in situ physiological activity, regardless of how it is induced.
In conclusion, the quantification of gene or transcript abundance alone allowed only limited insight into methanogen process dynamics in peat soil. The presence of dead, inactive, and dormant cells suggests that transcript abundance may more accurately reflect the functional activity of the peat methanogen community, and significant correlation was found between transcript/gene ratio and process rate. However, this approach for analysis of in situ function must be used with caution. MR specific activity is strongly temperature dependent (15), regulation of process rate may require transcription of several genes (25, 58), and ratios for natural ecosystems may not be directly comparable with those for pure cultures. Also, the approximation of in situ physiological MR activity and cell content requires detailed information, from pure-culture studies of representative acidophilic temperate peat methanogens (4), on mRNA transcript abundances and related enzyme activities under natural conditions. Additionally, the relationship of physiological rates and transcription per template ratio poses analytical problems for environmental applications. Relatively high “background” ratios equated to low physiological rates, while an 18-fold increase was associated with the highest observed process rates, and interpretation of transcript/gene ratios requires a better understanding of the mRNA regulatory system of the relevant organisms. Nevertheless, quantification of transcript/gene ratios provided a better indication of physiological activity than gene abundance. While this study identifies some potential difficulties in directly relating transcriptional and functional activities, the approach may have particular advantages in comparing in situ functional activities of different physiological groups within a soil community and in determining short-term responses to changes in environmental conditions.
Acknowledgments
This study was supported by funding from the NERC UK Population Biology Network (UKPopNet).
We thank the Royal Society for the Protection of Birds (RSPB) for allowing access to the Lake Vyrnwy special area of conservation.
Footnotes
Published ahead of print on 11 September 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ankel-Fuchs, D., R. Jaenchen, N. A. Gebhardt, and R. K. Thauer. 1984. Functional relationship between protein-bound and free factor F430 in Methanobacterium. Arch. Microbiol. 139:332-337. [Google Scholar]
- 3.Basiliko, N., J. B. Yavitt, P. M. Dees, and S. M. Merkel. 2003. Methane biogeochemistry and methanogen communities in two northern Peatland ecosystems, New York State. Geomicrobiol. J. 20:563-577. [Google Scholar]
- 4.Bräuer, S. L., H. Cadillo-Quiroz, E. Yashiro, J. B. Yavitt, and S. H. Zinder. 2006. Isolation of a novel acidiphilic methanogen from an acidic peat bog. Nature 442:192-194. [DOI] [PubMed] [Google Scholar]
- 5.Bürgmann, H., M. Pesaro, F. Widmer, and J. Zeyer. 2001. A strategy for optimizing quality and quantity of DNA extracted from soil. J. Microbiol. Methods 45:7-20. [DOI] [PubMed] [Google Scholar]
- 6.Cadillo-Quiroz, H., S. Brauer, E. Yashiro, C. Sun, J. Yavitt, and S. Zinder. 2006. Vertical profiles of methanogenesis and methanogens in two contrasting acidic peatlands in central New York State, USA. Environ. Microbiol. 8:1428-1440. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., M. G. Dumont, N. P. McNamara, P. M. Chamberlain, L. Bodrossy, N. Stralis-Pavese, and J. C. Murrell. 2008. Diversity of the active methanotrophic community in acidic peatlands as assessed by mRNA and SIP-PLFA analyses. Environ. Microbiol. 10:446-459. [DOI] [PubMed] [Google Scholar]
- 8.Colwell, F. S., S. Boyd, M. E. Delwiche, D. W. Reed, T. J. Phelps, and D. T. Newby. 2008. Estimates of biogenic methane production rates in deep marine sediments at Hydrate Ridge, Cascadia margin. Appl. Environ. Microbiol. 74:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyne, K. J., S. M. Handy, E. Demir, E. B. Whereat, D. A. Hutchins, K. J. Portune, M. A. Doblin, and S. C. Cary. 2005. Improved quantitative real-time PCR assays for enumeration of harmful algal species in field samples using an exogenous DNA reference standard. Limnol. Oceanogr. 3:381-391. [Google Scholar]
- 10.Crecchio, C., and G. Stotzky. 1998. Binding of DNA on humic acids: effect on transformation of Bacillus subtilis and resistance to DNase. Soil Biol. Biochem. 30:1061-1067. [Google Scholar]
- 11.Ellermann, J., S. Rospert, R. K. Thauer, M. Bokranz, A. Klein, M. Voges, and A. Berkessel. 1989. Methyl-coenzyme-M reductase from Methanobacterium thermoautotrophicum (strain Marburg). Purity, activity and novel inhibitors. Eur. J. Biochem. 184:63-68. [DOI] [PubMed] [Google Scholar]
- 12.Galand, P. E., H. Juottonen, H. Fritze, and K. Yrjälä. 2005. Methanogen communities in a drained bog: effect of ash fertilization. Microb. Ecol. 49:209-217. [DOI] [PubMed] [Google Scholar]
- 13.Galand, P. E., H. Fritze, R. Conrad, and K. Yrjälä. 2005. Pathways for methanogenesis and diversity of methanogenic archaea in three boreal peatland ecosystems. Appl. Environ. Microbiol. 71:2195-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galand, P. E., S. Saarnio, H. Fritze, and K. Yrjälä. 2002. Depth related diversity of methanogen Archaea in Finnish oligotrophic fen. FEMS Microbiol. Ecol. 42:441-449. [DOI] [PubMed] [Google Scholar]
- 15.Goenrich, M., E. C. Duin, F. Mahlert, and R. K. Thauer. 2005. Temperature dependence of methyl-coenzyme M reductase activity and of the formation of the methyl-coenzyme M reductase red2 state induced by coenzyme B. J. Biol. Inorg. Chem. 10:333-342. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennigan, A. N., and J. N. Reeve. 1994. Messenger-RNAs in the methanogenic Archaeon Methanococcus-vannielii—numbers, half-lives and processing. Mol. Microbiol. 11:655-670. [DOI] [PubMed] [Google Scholar]
- 18.Høj, L., R. A. Olsen, and V. L. Torsvik. 2008. Effects of temperature on the diversity and community structure of known methanogenic groups and other archaea in high Arctic peat. ISME J. 2:37-48. [DOI] [PubMed] [Google Scholar]
- 19.Høj, L., R. A. Olsen, and V. L. Torsvik. 2005. Archaeal communities in high Arctic wetlands at Spitsbergen, Norway (78°N) as characterized by 16S rRNA gene fingerprinting. FEMS Microbiol. Ecol. 53:89-101. [DOI] [PubMed] [Google Scholar]
- 20.Høj, L., M. Rusten, L. E. Haugen, R. A. Olsen, and V. L. Torsvik. 2006. Effects of water regime on archaeal community composition in Arctic soils. Environ. Microbiol. 8:984-996. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, D. E., K. P. Nevin, and D. R. Lovley. 2004. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl. Environ. Microbiol. 70:7251-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn, M. A., C. Matthies, K. Küsel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juottonen, H., P. E. Galand, E.-S. Tuittila, J. Laine, H. Fritze, and K. Yrjälä. 2005. Methanogen communities and Bacteria along an ecohydrological gradient in a northern raised bog complex. Environ. Microbiol. 7:1547-1557. [DOI] [PubMed] [Google Scholar]
- 24.Juottonen, H., P. E. Galand, and K. Yrjälä. 2006. Detection of methanogenic Archaea in peat: comparison of PCR primers targeting the mcrA gene. Res. Microbiol. 157:914-921. [DOI] [PubMed] [Google Scholar]
- 25.Kahnt, J., B. Buchenau, F. Mahlert, M. Krueger, S. Shima, and R. K. Thauer. 2007. Post-translational modifications in the active site region of methyl-coenzyme M reductase from methanogenic and methanotrophic archaea. FEBS J. 274:4913-4921. [DOI] [PubMed] [Google Scholar]
- 26.Knorr, K.-H., B. Glaser, and C. Blodau. 2008. Fluxes and 13C isotopic composition of dissolved carbon and pathways of methanogenesis in a fen soil exposed to experimental drought. Biogeosci. Discuss. 5:1457-1473. [Google Scholar]
- 27.Kotsyurbenko, O. R., K.-J. Chin, M. V. Glagolev, S. Stubner, M. V. Simankova, A. N. Nozhevnikova, and R. Conrad. 2004. Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ. Microbiol. 6:1159-1173. [DOI] [PubMed] [Google Scholar]
- 28.Lal, L., R. Sahoo, R. K. Gupta, P. Sharma, and S. Kumar. 2001. RNA Isolation from high-phenolic tea leaves and apical buds. Plant Mol. Biol. Rep. 19:181a-181f. [Google Scholar]
- 29.Loferer-Krössbacher, M., J. Klima, and R. Psenner. 1998. Determination of bacterial cell dry mass by transmission electron microscopy and densitometric image analysis. Appl. Environ. Microbiol. 64:688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lueders, T., K.-J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 31.Luo, H.-W., H. Zhang, T. Suzuki, S. Hattori, and Y. Kamagata. 2002. Differential expression of methanogenesis genes of Methanothermobacter thermoautotrophicus (formerly Methanobacterium thermoautotrophicum) in pure culture and in cocultures with fatty acid-oxidizing syntrophs. Appl. Environ. Microbiol. 68:1173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupa, B., E. L. Hendrickson, J. A. Leigh, and W. B. Whitman. 2008. Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl. Environ. Microbiol. 74:6584-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 34.Meckenstock, R., P. Steinle, J. R. Van Der Meer, and M. Snozzi. 1998. Quantification of bacterial mRNA involved in degradation of 1,2,4-trichlorobenzene by Pseudomonas sp. strain P51 from liquid culture and from river sediment by reverse transcriptase PCR (RT/PCR). FEMS Microbiol. Lett. 167:123-129. [DOI] [PubMed] [Google Scholar]
- 35.Merilä, P., P. E. Galand, H. Fritze, E.-S. Tuittila, K. Kukko-Oja, J. Laine, and K. Yrjälä. 2006. Methanogen communities along a primary succession transect of mire ecosystems. FEMS Microbiol. Ecol. 55:221-229. [DOI] [PubMed] [Google Scholar]
- 36.Metje, M., and P. Frenzel. 2007. Methanogenesis and methanogenic pathways in a peat from subarctic permafrost. Environ. Microbiol. 9:954-964. [DOI] [PubMed] [Google Scholar]
- 37.Metje, M., and P. Frenzel. 2005. Effect of temperature on anaerobic ethanol oxidation and methanogenesis in acidic peat from a northern wetland. Appl. Environ. Microbiol. 71:8191-8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan, R. M., T. D. Pihl, J. Nölling, and J. N. Reeve. 1997. Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum δH. J. Bacteriol. 179:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musso, M., R. Bocciardi, S. Parodi, R. Ravazzolo, and I. Ceccherini. 2006. Betaine, dimethyl sulfoxide, and 7-deaza-dGTP, a powerful mixture for amplification of GC-rich DNA sequences. J. Mol. Diagn. 8:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicol, G. W., S. Leininger, C. Schleper, and J. I. Prosser. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966-2978. [DOI] [PubMed] [Google Scholar]
- 41.Nicol, G. W., D. Tscherko, T. M. Embley, and J. I. Prosser. 2005. Primary succession of soil Crenarchaeota across a receding glacier foreland. Environ. Microbiol. 7:337-347. [DOI] [PubMed] [Google Scholar]
- 42.Nicolaisen, M. H., J. Bælum, C. S. Jacobsen, and J. Sørensen. 2008. Transcription dynamics of the functional tfdA gene during MCPA herbicide degradation by Cupriavidus necator AEO106 (pRO101) in agricultural soil. Environ. Microbiol. 10:571-579. [DOI] [PubMed] [Google Scholar]
- 43.Nölling, J., T. D. Pihl, A. Vriesema, and J. N. Reeve. 1995. Organization and growth phase-dependent transcription of methane genes in two regions of the Methanobacterium thermoautotrophicum genome. J. Bacteriol. 177:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okano, Y., K. R. Hristova, C. M. Leutenegger, L. E. Jackson, R. F. Denison, B. Gebreyesus, D. Lebauer, and K. M. Scow. 2004. Application of real-time PCR to study effects of ammonium on population size of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 70:1008-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pihl, T. D., S. Sharma, and J. N. Reeve. 1994. Growth phase-dependent transcription of the genes that encode the two methyl coenzyme M reductase isoenzymes and N5-methyltetrahydromethanopterin: coenzyme M methyltransferase in Methanobacterium thermoautotrophicum ΔH. J. Bacteriol. 176:6384-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 47.Radl, V., A. Gattinger, A. Chroňa′ková, A. Němcová, J. Čuhel, M. Šimek, J. C. Munch, M. Schloter, and D. Elhottová. 2007. Effects of cattle husbandry on abundance and activity of methanogenic archaea in upland soils. ISME J. 1:443-452. [DOI] [PubMed] [Google Scholar]
- 48.Reeve, J. N., J. Nölling, R. M. Morgan, and D. R. Smith. 1997. Methanogenesis: genes, genomes, and who's on first? J. Bacteriol. 179:5975-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice, P., L. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 50.Ritalahti, K. M., B. K. Amos, Y. Sung, Q. Wu, S. S. Koenigsberg, and F. E. Löffler. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl. Environ. Microbiol. 72:2765-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharkey, F. H., I. M. Banat, and R. Marchant. 2004. Detection and quantification of gene expression in environmental bacteriology. Appl. Environ. Microbiol. 70:3795-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinozuka, T., Y. Enomoto, H. Hayashi, H. Andod, and T. Yamaguchi. 2001. Variation of free radical concentrations of peat humic acid by methylation, p. 83-93. In E. A. Ghabbour and G. Davies (ed.), Humic substances, structures, models and functions. RSC Publishing, Cambridge, United Kingdom.
- 53.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2007. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl. Environ. Microbiol. 73:3612-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sprenger, W. W., J. H. P. Hackstein, and J. T. Keltjens. 2005. The energy metabolism of Methanomicrococcus blatticola: physiological and biochemical aspects. Antonie van Leeuwenhoek 87:289-299. [DOI] [PubMed] [Google Scholar]
- 55.Steinberg, L. M., and J. M. Regan. 2008. Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl. Environ. Microbiol. 74:6663-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steunou, A.-S., D. Bhaya, M. M. Bateson, M. C. Melendrez, D. M. Ward, E. Brecht, J. W. Peters, M. Kühl, and A. R. Grossman. 2006. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl. Acad. Sci. USA 103:2398-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Marjory Stephenson Prize Lecture. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 58.Thauer, R. K., A.-K. Kaster, H. Seedorf, W. Buckel, and R. Hedderich. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579-591. [DOI] [PubMed] [Google Scholar]
- 59.Vaughan, D., and B. G. Ord. 1982. An in vitro effect of soil organic matter fractions and synthetic humic acids on the generation of superoxide radicals. Plant Soil 66:113-116. [Google Scholar]
- 60.von Klein, D., H. Arab, H. Völker, and M. Thomm. 2002. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 6:103-110. [DOI] [PubMed] [Google Scholar]
- 61.Yun, J. J., L. E. Heisler, I. I. Hwang, O. Wilkins, S. K. Lau, M. Hyrcza, B. Jayabalasingham, J. Jin, J. McLaurin, M. S. Tsao, and S. D. Der. 2006. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic Acid Res. 34:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeikus, J. G., and R. S. Wolfe. 1972. Methanobacterium thermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J. Bacteriol. 109:707-715. [DOI] [PMC free article] [PubMed] [Google Scholar]