Abstract
The EIItMan phosphotransferase system (PTS) permease encoded by the mpt operon is the principal glucose transporter in Listeria monocytogenes. EIItMan participates in glucose-mediated carbon catabolite repression (CCR) and downregulation of virulence gene expression, and it is the receptor for class IIa bacteriocins. The regulation of this important protein and its roles in gene control were examined using derivatives of strain EGD-e in which the mpt operon or its regulatory genes, manR and lmo0095, were deleted. Real-time reverse transcription-PCR analysis showed that the mpt mRNA level was 10- and 100-fold lower in the lmo0095 and manR deletion strains, respectively. The manR mRNA level was higher in the mpt deletion mutant in medium lacking glucose, possibly due to disruption of a regulatory process that normally downregulates manR transcription in the absence of this sugar. Analysis of the mpt deletion mutant also showed that EIItMan participates to various degrees in glucose-mediated CCR of PTS operons. CCR of the lmo0027 gene, which encodes a β-glucoside PTS transporter, required expression of EIItMan. In contrast, genes in two mannose PTS operons (lmo0024, lmo1997, and lmo2002) were repressed by glucose even when EIItMan was not synthesized. A third mannose PTS operon, mpo, was not regulated by glucose or by the level of EIItMan. Finally, the mRNA levels for five genes in the prfA virulence gene cluster were two- to fourfold higher in the mpt deletion mutant. The results show that EIItMan participates to various extents in glucose-mediated CCR of PTS operons and makes a small, albeit significant, contribution to downregulation of virulence gene transcription by glucose in strain EGD-e.
The EIItMan phosphotransferase system (PTS) permease encoded by the mannose permease two (mpt) operon is the principal glucose transporter in Listeria monocytogenes (9, 36, 39). EIItMan plays a central role in class IIa bacteriocin resistance (9, 16, 30, 31, 33, 41), carbon catabolite repression (CCR) (1, 42), and possibly regulation of virulence gene expression (10, 18, 21, 22, 25, 36) in Listeria species. The mpt operon contains three genes, which encode the IIABMan (mptA), IICMan (mptC), and IIDMan (mptD) subunits of the transporter (Fig. 1). Seminal studies of class IIa bacteriocin resistance showed that the mpt promoter is controlled by the σ54 sigma factor (8, 17, 33) and the σ54-associated activator, ManR (encoded by lmo0785) (9, 42). In addition, transcription is controlled by the response regulator protein, ResD (encoded by lmo1948) (21), and, at least in Listeria innocua, by the Crp-Fnr homolog, Lin0142 (20, 41), the ortholog of which in L. monocytogenes is Lmo0095. The role of Lmo0095 in mpt control in L. monocytogenes has not been studied.
FIG. 1.
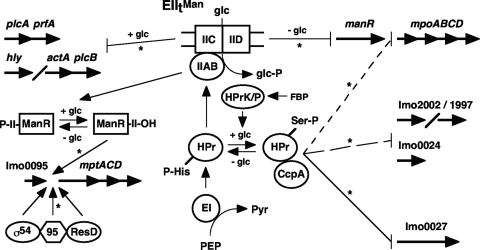
Model for EIItMan regulation of PTS operon and virulence gene transcription in L. monocytogenes. The EIItMan PTS permease transports and phosphorylates glucose (glc) using a phosphate group (P) donated from phosphoenolpyruvate (PEP) and transferred to the permease via the enzyme I (EI) and histidine protein (HPr)-His-P components of the core PTS (center). EIItMan participates in CcpA-independent downregulation of mpt transcription in the absence of glucose through inactivation of ManR by phosphorylation of its PRD-II (lower left). The dephosphorylated species of ManR that predominates in glucose medium activates mpt transcription, as do the σ54, Lmo0095 (95), and ResD proteins. In addition, EIItMan appears to contribute to downregulation of mpt transcription in the absence of glucose by an unknown mechanism that decreases manR transcription (upper right). EIItMan also appears to participate to various extents in CcpA-dependent regulation of other PTS operons in glucose medium (lower right). Downregulation of lmo0027 (solid line) requires EIItMan expression, whereas downregulation of lmo0024, lmo1997, and lmo2002 does not require EIItMan expression (lower dashed line). EIItMan-dependent regulation of the mpo operon (upper dashed line) was not observed in this study but has been reported elsewhere. Lastly, the downregulation of prfA and other virulence genes by EIItMan in glucose medium is indicated at the upper left. Regulatory processes examined in this study are indicated by asterisks. Abbreviations: Pyr, pyruvate; FBP, fructose 1,6-bisphosphate; II, ManR PRD-II. Other gene and protein abbreviations are explained in the text.
The regulation of gene expression by glucose occurs by similar processes in Bacillus subtilis and L. monocytogenes. Namely, transport and metabolism of glucose are coupled to both catabolite control protein A (CcpA)-dependent and CcpA-independent mechanisms of gene control (1, 3, 6, 7, 11, 12, 42) (Fig. 1). In CcpA-dependent CCR, the metabolism of glucose leads to an increase in the level of fructose 1,6-bisphosphate, which is an activator of the regulatory enzyme, HPr kinase/phosphorylase (HPrK/P) (11, 12). HPrK/P phosphorylates the HPr core kinase of the PTS at a regulatory serine residue, activating it for binding to CcpA (7, 11, 12). Subsequently, the HPr-Ser-P/CcpA complex binds to control sites known as cre sequences (catabolite repression elements) in genes involved in catabolism of alternative carbon sources, inhibiting transcription (3, 11, 12). Because EIItMan is the principal glucose transporter in L. monocytogenes, it is thought to play a central role in CcpA-dependent CCR (1). In CcpA-independent CCR, the activities of regulatory proteins that control the transcription of catabolic operons are modulated by PTS transporter-mediated phosphorylation of histidine residues in structural domains called PTS regulation domains (PRDs) (12, 23, 37). It has been proposed that phosphorylation of ManR PRD-II by the EIItMan transporter in the absence of glucose inactivates ManR and downregulates mpt transcription in L. innocua (42) and presumably in L. monocytogenes (Fig. 1).
Recent transcriptome studies have identified a number of PTS operons that are regulated by glucose and other sugars in L. monocytogenes (19, 25, 36). However, the role of EIItMan in CCR in glucose medium has been examined for relatively few genes. One PTS permease whose expression appears to be strongly inhibited by EIItMan-based CcpA-dependent CCR is the β-glucoside PTS permease encoded by the lmo0027 gene (15, 16, 36) (Fig. 1). Another highly expressed PTS operon of the mannose structural family, mpo (mannose permease one) encoding EIIoMan, also may be controlled by EIItMan-initiated CcpA-dependent CCR since its transcription is 12-fold higher in an mptA insertion mutant (2). Interestingly, the mpo operon has a near-consensus σ54 promoter sequence (2). Because the manR gene is located immediately upstream, it is possible that mpo is controlled by ManR. The L. monocytogenes EGD-e genome contains genes encoding subunits of potentially two other PTS permeases of the mannose structural family (14). The regulation of these genes by glucose, EIItMan, and ManR has not been examined.
EIItMan also may be involved in glucose-mediated downregulation of the activity of the master regulator of virulence gene expression in L. monocytogenes, PrfA (positive regulatory factor A) (21, 36). PrfA controls a cluster of genes that encode virulence factors, such as listeriolysin O (hly) and two phospholipase C enzymes (plcA and plcB), which allow internalized cells to escape from the phagocytic vacuole of a mammalian host cell. PrfA also regulates transcription of its own gene (prfA) and the actA gene, which encodes a protein (ActA) that is important for intracellular movement and cell-to-cell spread (13, 24, 34). Recent studies have indicated that CcpA and HPrK/P are not involved in the regulation of PrfA activity and virulence gene transcription by carbohydrates (3, 25). Instead, PrfA activity is negatively correlated with the levels of the dephosphorylated species of PTS transporters for glucose and other PTS sugars (36). The dephosphorylated forms of the transporters accumulate when their substrates are being transported into the cell (Fig. 1). Virulence gene expression also has been reported to be elevated in an mpt deletion mutant (21). However the contribution of EIItMan to virulence gene downregulation was not quantified in this study. Because PTS operon expression and transport of glucose and other PTS sugars decrease in PrfA overexpression strains, it is possible that PrfA activity is controlled by its direct interaction with PTS permeases, such as EIItMan (22).
In this study, we examined the roles of EIItMan, ManR, and Lmo0095 in the regulation of mpt, mpo, and two other PTS operons of the mannose structural family. We also quantified the effects of mpt deletion on expression of genes in the prfA virulence gene cluster. The experiments increase our understanding of the roles played by EIItMan in the regulation of gene expression by glucose in L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains.
All experiments were performed with strains derived from L. monocytogenes EGD-e (ATCC BAA-679) (14) obtained from the American Type Culture Collection (Manassas, VA). Escherichia coli DH5α was used as the host for cloning. DH5α was grown at 37°C in Luria-Bertani (LB) medium supplemented with 0.2% d-glucose and 100 μg/ml ampicillin when it was transformed with pKSV7 (35) and derived plasmids.
Construction of deletion mutant strains.
Experiments were conducted using EGD-e and three deletion mutant strains designated EGD-e Δmpt1, EGD-e ΔmanR1, and EGD-e Δlmo0095-1. The wild-type copies of the genes were replaced by in-frame deletion genes introduced into the EGD-e chromosome by homologous recombination using plasmid pKSV7 (35, 42). Deletion genes were constructed by splice-by-overlap extension PCR (42) using EGD-e genomic DNA as the template and the primers listed in Table 1. Twelve to 18 amino acid codons at the 5′ and 3′ ends of the genes were preserved and fused together in the constructs. In the EGD-e Δmpt1 strain, all three genes in the mpt operon (mptA, mptC, and mptD) were deleted. The sequences of the deletion genes in all strains were confirmed by DNA sequencing.
TABLE 1.
Primers used for construction of deletion mutant strainsa
| Strain | Primer | Direction | Sequence | No. of amino acidsb |
|---|---|---|---|---|
| EGD-e Δlmo0095-1 | lmo0095-A-BamHI | F | CGGGATCCTTTGCGATGAAGTCGTTGCATT | 15 N terminal, 15 C terminal |
| lmo0095-B | R | GGTCGAGTCAGTAACCAAGGTGTCAACAAATCTTCTTTGACCAATTGATGG | ||
| lmo0095-C | F | ACACCTTGGTTACTGACTCGACC | ||
| lmo0095-D-SalI | R | ACGCGTCGACCAAATTCACCGTGAGTTGCGAG | ||
| EGD-e Δmpt1 | lmo0096/8-A-KpnI | F | GCGGGTACCAACGAACTAGTCAATGAAGGTG | 12 N terminal, 15 C terminal |
| lmo0096/8-B | R | CCAACTATACCAACTACGAATAGAAATTCACCGTGAGTTGCGAGGA | ||
| lmo0096/8-C | F | CTATTCGTAGTTGGTATAGTTGG | ||
| lmo0096/8-D-SphI | R | GCGGCATGCAGCAAATGTGTATGTGCCGTTG | ||
| EGD-e ΔmanR1 | manR-A-BamHI | F | CGGGATCCGCCAGCTTGTAATTCTCTACC | 18 N terminal, 18 C terminal |
| manR-B | R | GATATCGTACAAATAACATAGCTTAGTGGAAGGATTTTCTAA | ||
| manR-C | F | CTATGTTATTTGTACGATATC | ||
| manR-D-SalI | R | CCGTCGACGTAATAAACTCTATGTCC |
F, forward; R, reverse. The restriction sites incorporated into primers for cloning are indicated by bold type. The regions of the “B” primers that are complementary to the “C” primers used for splice-by-overlap extension PCR are underlined. Primers were designed using the L. monocytogenes EGD-e genome sequence available at the ListiList server (http://genolist.pasteur.fr/ListiList).
Amino acid codons remaining at the 5′ and 3′ ends of deletion genes. For the mpt deletion strain, the 12 N-terminal amino acids encoded by mptA were fused to the 15 C-terminal amino acids encoded by mptD.
RNA isolation and mRNA quantitation.
The transcript levels for PTS operon and regulatory genes were measured for strains grown in LB medium supplemented or not supplemented with 0.2% d-glucose. Cultures were grown at 30°C and harvested at an optical density at 600 nm (OD600) of ∼0.5. The levels of the mptA and mpoA mRNAs also were measured for strains grown at 37°C in brain heart infusion (BHI) medium (which contained 0.2% glucose) until the OD600 was ∼1.0. The effects of mpt and lmo0095 deletion on virulence gene transcription were measured for cultures grown in BHI medium containing 0.2% activated charcoal (Sigma-Aldrich, St. Louis, MO) (BHI/C) supplemented or not supplemented with 25 mM d-glucose. Cultures were grown at 37°C and harvested when the OD600 was ∼1.0.
Total cellular RNA was isolated using the RNeasy mini kit method (Qiagen, Valencia, CA) (41, 42). The only change in the previously described RNA isolation and quantitation procedures was that cells were broken using the FastRNA Pro Blue lysing matrix and a Fast Prep-24 cell disruption apparatus (MP Biomedicals, Irvine, CA) prior to RNA isolation. mRNA levels were measured by real-time reverse transcription-PCR using the iQ SYBER green Supermix kit protocol (Bio-Rad, Hercules, CA) and the primer sets listed in Table 2. 16S rRNA was used as the internal standard for all measurements (38). The mRNA values that are reported below are the averages obtained for four or five independent RNA preparations. The 2008 online version of the Relative Expression Software Tool (REST) program (29) was used to calculate changes in mRNA levels and to determine whether changes were significant.
TABLE 2.
Primers used for real-time reverse transcription-PCR measurement of mRNA levelsa
| Gene | Direction | Sequence |
|---|---|---|
| lmo0024 | F | AGGAATGGATATGGCGATTG |
| R | CGGCAATTGATCCAAAAATC | |
| lmo0027 | F | GAACCAGCGATTTACGGTGT |
| R | GACCGAAGATTCCAAGTCCA | |
| lmo0095 | F | GCGGAAATGATTTTGGTGAA |
| R | TCGTAGACACCGAGCATTTG | |
| lmo0096 (mptA) | F | AGGTGTTCGCGTTAAACCAG |
| R | ACGAATTCGATTTTGCCATC | |
| lmo0200 (prfA) | F | AACCAATGGGATCCACAAGA |
| R | CCCGTTCTCGCTAATACTCG | |
| lmo0201 (plcA) | F | CCATTAGGCGGAAAAGCATA |
| R | CAGGTAGAGCGGACATCCAT | |
| lmo0202 (hly) | F | TTAGCTTGGGAATGGTGGAG |
| R | ATTTCGGATAAAGCGTGGTG | |
| lmo0204 (actA) | F | GGAAAGCCATAGCATCATCG |
| R | AGCATCCGCAACTGACTCTT | |
| lmo0205 (plcB) | F | AGCAAATGCCTGTTGTGATG |
| R | TTATCCGCGGACCAACTAAG | |
| lmo0784 (mpoA) | F | TCGGCAAACAGGACAATGTA |
| R | CCGCCAAATAAATCAACCAT | |
| lmo0785 (manR) | F | AGGTGAAACTGGTCGGATTG |
| R | TCTAGCACGCTAGCAAACGA | |
| lmo1997 | F | GAATCCGCGGAAAACTTACA |
| R | CATTAAACGGAGTGCCACCT | |
| lmo2002 | F | AGAGGCGGCAAAACTAGTCA |
| R | CTTCAACCGATTCGACAACA | |
| 16S rRNA | F | AAGCAACGCGAAGAACCTTA |
| R | TGCACCACCTGTCACTTTGT |
F, forward; R, reverse. Primers were designed using the L. monocytogenes EGD-e genome sequence available at the ListiList server (http://genolist.pasteur.fr/ListiList).
Database searches.
Searches for cre sites in the EGD-e genome were conducted using the Pattern Recognition search tool at the ListiList server (http://genolist.pasteur.fr/ListiList). The input sequence used in the searches was TGTTTACGTTTACA, which is the putative autoregulatory cre site located 60 bp upstream of the ccpA gene in L. monocytogenes 10403S (3). The ccpA cre sequence has only one deviation (underlined) from the B. subtilis consensus cre sequence (TGWNANCGNTNWCA, where W is A or T and N is any residue) that has been experimentally established (11, 40). The location and sequence of the ccpA cre site are identical in EGD-e. In the search, six base mismatches were allowed, and this generated a total of 105,675 matches in the genome. The list of matches was examined to find potential cre sites with coordinates located near PTS permease genes.
RESULTS AND DISCUSSION
Regulation of the mpt operon by Lmo0095 and ManR.
We previously determined that the mpt operon in L. innocua Lin11 is positively regulated by the Lin0142 protein (41). The putative ortholog of this protein in L. monocytogenes EGD-e is Lmo0095. Like the lin0142 gene, lmo0095 is located immediately upstream of mptA (lmo0096) (Fig. 1). To determine if Lmo0095 is the functional equivalent of Lin0142, we deleted the lmo0095 gene to obtain the EGD-e Δlmo0095-1 strain and examined the effects on mpt transcription. We also assessed the relative contribution of manR to mpt regulation using the EGD-e ΔmanR1 mutant. While manR has been shown to positively regulate mpt in L. monocytogenes (9), the reduction in mpt expression that occurs in the absence of ManR has not been reported.
The mptA transcript levels in the lmo0095 deletion strain were lower than those in EGD-e (Table 3), indicating that Lmo0095 positively regulates mpt transcription (Fig. 1). The mptA mRNA levels were ∼10-fold lower in LB medium lacking glucose and ∼2-fold lower in medium containing this sugar. In comparison, the mptA transcript levels were ∼100-fold lower in the manR deletion strain regardless of whether glucose was present in the medium. Taken together, the results indicate that while mpt transcription is activated more strongly by ManR than by Lmo0095, Lmo0095 nonetheless is required for full activation of transcription of the operon in glucose medium. mRNA quantitation also revealed that the levels of the lmo0095 and manR transcripts in EGD-e are not changed by addition of glucose to the medium.
TABLE 3.
Levels of mRNAs for mpt operon and regulatory genes in EGD-e and deletion mutant strains
| Strain | Medium | Levels of mRNAsa |
||
|---|---|---|---|---|
| lmo0095 | manR | mptA | ||
| EGD-e | LB | 1.0 | 1.0 | 1.0 |
| LB + Glc | 1.3 (0.7-2.9) | 2.4 (1.5-4.0) | 8.3 (3.6-16.8)b | |
| EGD-e Δlmo0095-1 | LB | 0 | 1.2 (0.7-2.1) | 0.09 (0.05-0.2)b |
| LB + Glc | 0 | 2.0 (1.0-4.9) | 3.9 (2.4-7.1)b | |
| EGD-e ΔmanR1 | LB | 0.9 (0.5-1.9) | 0 | 0.01 (0.004-0.02)b |
| LB + Glc | 0.8 (0.5-1.4) | 0 | 0.01 (0.004-0.01)b | |
| EGD-e Δmpt1 | LB | 1.2 (0.5-3.1) | 4.8 (2.1-14.1)b | 0 |
| LB + Glc | 1.1 (0.6-2.1) | 2.2 (1.2-4.7) | 0 | |
The average levels of mRNA for each gene were normalized to the level obtained for strain EGD-e grown in the absence of glucose. Values were calculated based on measurements obtained for four or five independent RNA preparations. Standard errors calculated by using the REST statistics program are indicated in parentheses.
The value differs significantly (P < 0.05) from the value obtained for the reference strain.
Similar 100-fold reductions in mpt transcription were observed for an L. innocua Lin11 manR deletion mutant (42). However, disruption of lin0142 expression by transposon Tn917 insertion in L. innocua Lin11 resulted in inhibition of mpt transcription that was greater than that observed here for EGD-e Δlmo0095-1 (41). This could have been because mpt transcription is more dependent on the Lmo0095 ortholog, Lin0142, in L. innocua or because the transposon somehow inhibits the mpt promoter. It is unlikely that the higher level of mpt mRNA observed in the lmo0095 deletion mutant was due to residual mpt transcription originating from the lmo0095 promoter. This is because a rho-independent terminator (14) is preserved downstream of the lmo0095 deletion gene and the lmo0095 and mptA coding sequences are separated by 297 bp.
Regulatory relationships between lmo0095, manR, and mpt transcription.
Levels of mRNAs for the lmo0095 and manR genes were measured in the strains mentioned above and in the EGD-e Δmpt1 mutant in which the mptACD genes are deleted (Table 3). No significant changes in lmo0095 mRNA levels were detected in the manR or mpt deletion strains compared to EGD-e, indicating that neither ManR nor EIItMan regulates lmo0095, at least under the conditions examined. While the manR transcript level was not affected by deletion of lmo0095, it was ∼5-fold higher in the mpt deletion strain grown in the absence of glucose. It was unchanged when the mpt deletion strain was grown in glucose medium. A similar increase (∼3-fold) in the manR transcript level in medium lacking glucose was observed for the L. innocua Lin11 Tn917 insertion mutant mentioned above, in which transcription of mpt is undetectable (41).
These observations suggest that manR transcription is regulated by EIItMan-dependent repression in the absence of glucose (Fig. 1) and by an EIItMan-independent process in glucose-containing medium. Because no differences in the manR mRNA levels in the strains grown in glucose medium were observed, the data do not provide insight into how manR transcription is controlled under these conditions. However, the results obtained for the EGD-e Δmpt1 strain grown in medium lacking glucose are consistent with a control mechanism in which the phosphorylated form of EIItMan is involved. This form of the transporter normally predominates in the absence of glucose and is not present in the mpt deletion mutant. If this proposal is correct, then the data obtained for the lmo0095 deletion mutant suggest that the relatively small amount of the phosphorylated species of EIItMan that remains in this strain is sufficient to keep manR transcription repressed in the absence of glucose. We have considered the possibility that ManR controls the transcription of its own gene and that transcription normally is repressed in medium lacking glucose due to phosphorylation of ManR PRD-II by EIItMan (42). However, this type of control would require that the manR gene is regulated by a σ54 promoter, and a σ54 promoter sequence does not appear to be present upstream of this gene.
Regulation of the mpo operon.
The mpo operon contains four genes, mpoA (lmo0784), mpoB (lmo0783), mpoC (lmo0782), and mpoD (lmo0781), which encode the IIAMan, IIBMan, IICMan, and IIDMan subunits of the EIIoMan permease, respectively (14) (Fig. 1). This operon, like mpt, is one of the most strongly expressed PTS operons in L. monocytogenes (36). The mpo operon is located just downstream of the manR gene (lmo0785), and mpoA is preceded by a putative −24/−12 σ54 promoter sequence (TGGCACAGTTTTTGCG) which is similar to the L. monocytogenes σ54 consensus sequence (TGGCACGGAACTTGCA) (1). For these reasons, we determined whether ManR regulates mpo transcription. We also determined whether EIItMan and Lmo0095 control the operon.
The mpoA mRNA level was unchanged in the EGD-e Δlmo0095-1 and EGD-e ΔmanR1 mutants (Table 4), indicating that transcription of the mpo operon is not controlled by Lmo0095 or ManR, at least under the conditions studied. Consistent with a recent report (36), we observed that mpo transcription is not induced by addition of glucose to the medium. However, we were unable to confirm that in our mpt deletion mutant the level of mpoA mRNA increases when EIItMan is not expressed (2). Because the original analysis of the effects of EIItMan expression on mpo transcription was performed using cultures grown at 37°C in BHI medium, we also obtained measurements under these conditions (Table 4). However, again, no changes in mpo expression were observed in the deletion strains in which either mptA mRNA was absent (EGD-e Δmpt1) or mptA mRNA expression was ∼5-fold (EGD-e Δlmo0095-1) or ∼1,000-fold (EGD-e ΔmanR1) lower. These results contradict previous findings suggesting that mpo transcription is downregulated by EIItMan-mediated CcpA-dependent CCR. In agreement with our findings, other investigators have found that mpo expression does not increase in ccpA and hprK (which encodes HPrK/P) mutants (25). It is unclear why our results differ from the results obtained previously, which also were obtained using a mutant prepared using strain EGD-e (2). The differences may stem from sequence polymorphisms in the two strains or may be due to the types of mpt mutants that were examined. In this regard, an mptA insertion mutant was used in the previous study.
TABLE 4.
Levels of mRNAs for mannose PTS genes in EGD-e and deletion mutant strains
| Strain | Medium | Levels of mRNAsa |
|||||
|---|---|---|---|---|---|---|---|
| lmo0024 | lmo0027 | mpoA | lmo1997 | lmo2002 | mptA | ||
| Strains grown in LB medium | |||||||
| EGD-e | LB | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| LB + Glc | 0.2 (0.1-0.4)b | 0.02 (0.01-0.08)b | 0.8 (0.3-2.1) | 0.4 (0.2-0.7)b | 0.2 (0.1-0.5)b | ||
| EGD-e Δlmo0095-1 | LB | ND | 1.4 (0.7-3.2) | 1.4 (0.6-2.8) | ND | ND | |
| LB + Glc | ND | 1.6 (0.4-6.9) | 1.4 (0.7-3.4) | ND | ND | ||
| EGD-e ΔmanR1 | LB | 0.5 (0.2-1.1) | 3.1 (1.2-7.1)b | 1.1 (0.4-4.0) | 1.3 (0.7-2.7) | 1.2 (0.5-2.7) | |
| LB + Glc | 0.04 (0.02-0.1)b | 2.3 (1.1-4.8) | 0.7 (0.3-1.6) | 0.2 (0.1-0.3)b | 0.2 (0.1-0.4)b | ||
| EGD-e Δmpt1 | LB | 0.6 (0.3-1.4) | 1.4 (0.6-3.5) | 0.6 (0.2-1.4) | 1.1 (0.7-2.3) | 1.6 (0.7-2.9) | |
| LB + Glc | 0.1 (0.04-0.3)b | 6.2 (2.3-20.0)b | 0.6 (0.2-1.2) | 0.4 (0.2-0.6)b | 0.4 (0.2-0.8)b | ||
| Strains grown in BHI medium | |||||||
| EGD-e | BHI | 1.0 | 1.0 | ||||
| EGD-e Δlmo0095-1 | BHI | 0.6 (0.2-1.5) | 0.2 (0.05-0.5)b | ||||
| EGD-e ΔmanR1 | BHI | 1.7 (1.0-2.8) | 0.001 (0.0-0.001)b | ||||
| EGD-e Δmpt1 | BHI | 0.6 (0.3-1.3) | 0 | ||||
The average mRNA levels for each gene were normalized to the level obtained for strain EGD-e grown in LB medium lacking glucose or for EGD-e grown in BHI medium. Values were calculated based on measurements obtained for four or five independent RNA preparations. Standard errors calculated by using the REST statistics program are indicated in parentheses. ND, not determined.
The value differs significantly (P < 0.05) from the value obtained for the reference strain.
Regulation of lmo0027.
To verify that our deletion strains are not somehow defective in exhibiting relief of CCR for catabolic operons such as mpo, we compared the effects of glucose on expression of the lmo0027 gene, which encodes a β-glucoside-specific PTS permease (14), in EGD-e and the deletion mutants. This gene is strongly upregulated in class IIa bacteriocin-resistant mutants in which EIItMan expression is reduced (15, 16). In addition, lmo0027 transcription is 16-fold greater in an rpoN (σ54) deletion mutant, most likely due to a partial reduction in EIItMan expression (∼5-fold lower for the IIABMan subunit) (1). Based on these observations, it has been proposed that EIItMan downregulates lmo0027 via CcpA-dependent CCR in glucose medium (1, 16) (Fig. 1).
In general agreement with previous studies, we observed that the lmo0027 transcript levels in the lmo0095, manR, and mpt deletion mutants grown in glucose medium were upregulated approximately 80-, 115-, and 310-fold, respectively, compared to the level in EGD-e. We further observed that the lmo0027 mRNA level was ∼50-fold lower in EGD-e in the presence of glucose. Two potential cre sites located 170 (TGCAACCGTTTTCT) and 78 (TGTATGCGTGGAGT) nucleotides upstream of the lmo0027 ATG codon that might be involved in CCR were identified. These sequences contain the 5′ TG and central CG dinucleotide residues that are highly conserved in the B. subtilis cre consensus sequence (4, 11, 40). They differ from the consensus sequence at two and four positions (underlined bases), respectively. Taken together, the results are consistent with the proposal that lmo0027 is regulated by EIItMan-dependent CCR in glucose medium. They further confirm that glucose-mediated CCR is relieved for at least some catabolic operons in the EGD-e Δmpt1 strain and other deletion strains.
Currently, it is unclear why the level of lmo0027 mRNA in the mpt deletion strain in glucose medium was higher than the level in medium lacking glucose. This result cannot simply be attributed to relief of CcpA-mediated CCR, since if this were the case, the mRNA level would be expected to increase only to the level observed in medium lacking glucose. The results suggest that lmo0027 is controlled by different mechanisms depending on whether glucose is present in the medium.
Regulation of other mannose PTS permease genes.
We also examined the roles of glucose, EIItMan, and ManR in control of three other genes that may encode components of mannose PTS permeases (Fig. 1). Two of these genes (lmo1997 and lmo2002) are part of a group of genes that appear to encode a complete set of mannose PTS subunits, including lmo1997 (which encodes a IIAMan subunit), lmo2000 (which encodes a IIDMan subunit), lmo2001 (which encodes a IICMan subunit), and lmo2002 (which encodes a IIBMan subunit). Although lmo1997 is not contiguous with the other genes, all four genes may occur in a single transcriptional unit (14). The remaining gene, lmo0024, encodes an isolated IIDMan subunit and is immediately downstream and probably in a transcriptional unit containing three fructose PTS structural family genes. These genes are lmo0021 (which encodes a IIAFru subunit), lmo0022 (which encodes a IIBFru subunit), and lmo0023 (which encodes a IICFru subunit).
mRNA analyses showed that lmo0024, lmo1997, and lmo2002 are significantly repressed (∼3- to 5-fold) by glucose in EGD-e (Table 4). These results are consistent with transcriptome analyses performed with strain EGD, which showed that the levels of expression of lmo2000, lmo2001, and lmo2002 are three- to fivefold higher in medium containing glycerol than in medium containing glucose (36). Similarly, the level of expression of lmo0021, which is linked to lmo0024, is threefold higher in EGD in glycerol medium than in glucose medium. The combined results indicate that the PTS permeases encoded by these genes are unlikely to transport glucose.
Interestingly, the levels of mRNAs for these three genes were not different in the mpt deletion mutant and EGD-e in either medium (Table 4). This behavior differs from that of lmo0027, which was strongly upregulated in the EGD-e Δmpt1 strain grown in glucose medium. Nonetheless, we propose that EIItMan may normally contribute to glucose-mediated CcpA-dependent CCR of these genes (Fig. 1). When EIItMan is not expressed, as in the mpt deletion strain, CCR may continue to be exerted due to the uptake of glucose by another PTS permease or even a non-PTS transporter in the strain (28). In this regard, glucose transport is mediated exclusively by PTS permeases in strain EGD (36), but it is carried out by both PTS and proton motive force-dependent systems in L. monocytogenes Scott A (5). In support of this idea, we observed that addition of glucose to the medium greatly increased the growth rate and cell yield for the mpt deletion strain, indicating that this strain still transports and metabolizes glucose efficiently. In addition, several potential cre sites occur in the lmo0024, lmo1997, and lmo2002 gene regions, and transcriptome studies have indicated that lmo2001 is upregulated in ccpA and hprK mutants (25). In summary, the observed differences in the EIItMan dependence of gene regulation may be due to variations in the level of the HPr-Ser-P/CcpA complex formed when cells take up glucose using different transporters and in the amount of the complex that is required to bind to the cre sites of the genes. It should be noted that the level of the HPr-Ser-P/CcpA complex present in B. subtilis depends on the concentration of fructose 1,6-bisphosphate (11). The concentration of this metabolite may vary based on the rate at which glucose is transported into and metabolized by cells.
Lastly, transcription of lmo0024, lmo1997, and lmo2002 does not appear to be controlled by ManR, because the levels of mRNAs for these genes were unaffected in the EGD-e ΔmanR1 strain in the absence of glucose (Table 4). If ManR positively controlled these genes, decreases in mRNA levels due to manR deletion may be detectable in the absence of glucose, as under these conditions ManR-activated transcription would not be counteracted by CCR. A summary of the effects of deletion mutations on PTS gene regulation is shown in Table 5.
TABLE 5.
Summary of PTS permease gene regulation
| Gene | Encoded protein functiona | Regulatory effectb |
|||
|---|---|---|---|---|---|
| Glucose | EIItMan deletion | ManR deletion | Lmo0095 deletion | ||
| lmo0024 | IIDMan subunit of PTS permease | Down | None | None | ND |
| lmo0027 | EIIBgl permease (IIABCBgl) | Down | Up | Up | Up |
| lmo0096 (mptA) | IIABMan subunit of EIItMan permease | Up | NA | Down | Down |
| lmo0784 (mpoA) | IIAMan subunit of EIIoMan permease | None | None | None | None |
| lmo1997 | IIAMan subunit of PTS permease | Down | None | None | ND |
| lmo2002 | IIBMan subunit of PTS permease | Down | None | None | ND |
The putative functions of PTS permease genes are based on the annotations at the ListiList server (http://genolist.pasteur.fr/ListiList).
ND, not determined. NA, not applicable.
Virulence gene expression increases in the mpt deletion mutant.
Early work showed that virulence gene activity in L. monocytogenes is downregulated by readily metabolized PTS sugars, such as glucose (3, 26, 27, 32). More recent studies have demonstrated that PrfA activity and virulence gene expression are negatively correlated with the level of the nonphosphorylated species of PTS permeases in cells (18, 19, 22, 25, 36) (Fig. 1). While a reciprocal relationship between mpt and virulence gene expression was noted in some of these studies, changes in virulence gene expression were not directly measured using a strain in which mpt is inactivated. In another study, in which the mpt operon was inactivated, increases in virulence gene activity were not quantified (21). For these reasons, we measured the levels of mRNAs for virulence genes in the prfA gene cluster in the mpt deletion strain. We also measured the levels of mRNAs for virulence genes in the lmo0095 deletion mutant, in which the EIItMan level is partially reduced, to determine how sensitive virulence gene expression is to the level of this permease.
For these studies, mRNA quantitation was performed with strains grown in BHI/C medium and BHI/C medium supplemented with 25 mM glucose. These conditions are optimal for detecting the repressive effects of glucose on virulence gene expression in L. monocytogenes (21, 32). Interestingly, addition of glucose to BHI/C medium did not decrease the levels of mRNAs for the prfA, plcA, hly, actA, and plcB virulence genes in EGD-e or the lmo0095 and mpt deletion mutants compared to the levels measured in BHI/C medium alone (data not shown). This result is consistent with a recent study comparing virulence gene repression by glucose in strains EGD and EGD-e, in which it was found that repression is strong in EGD and minimal in EGD-e (36). (Note that virulence gene expression nonetheless was strongly elevated by charcoal for all strains in both media [data not shown].) However, the levels of mRNAs for all five virulence genes were ∼2- to 4-fold higher in the mpt deletion mutant than in EGD-e when the strains were grown in BHI/C medium supplemented with glucose (Table 6). Derepression was not observed in the lmo0095 deletion mutant, which suggests that the reduction in EIItMan expression that occurs in this strain (∼5-fold) (Table 4) is not sufficient to alter virulence gene expression.
TABLE 6.
Levels of mRNAs for virulence genes in EGD-e and deletion mutant strains
| Strain | Levels of mRNAsa |
||||
|---|---|---|---|---|---|
| prfA | plcA | hly | actA | plcB | |
| EGD-e | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| EGD-e Δlmo0095-1 | 2.3 (0.7-5.9) | 2.6 (0.8-7.8) | 2.3 (0.5-10.0) | 2.7 (0.8-7.3) | 2.8 (0.8-8.6) |
| EGD-e Δmpt1 | 2.4 (1.3-3.9)b | 4.4 (2.3-8.9)b | 2.9 (1.6-5.7)b | 4.1 (2.2-9.5)b | 3.1 (1.8-5.6)b |
Strains were grown in BHI/C medium supplemented with 25 mM glucose. The average mRNA levels for each gene were normalized to the level obtained for the EGD-e reference strain. Values were calculated based on measurements obtained for four or five independent RNA preparations. Standard errors calculated by using the REST statistics program are indicated in parentheses.
The value differs significantly (P < 0.05) from the value obtained for EGD-e.
Summary and conclusions.
It has been determined that the Lmo0095 protein positively regulates mpt transcription in L. monocytogenes EGD-e. While Lmo0095 is not as important as ManR in regulating this operon, it contributes significantly to expression of the EIItMan transporter, particularly in medium lacking glucose. The lmo0095 deletion strain is >1,000-fold more resistant to the class IIa bacteriocin pediocin AcH than EGD-e is. This indicates that resistance to class IIa bacteriocins in L. monocytogenes could result from spontaneous inactivation of the lmo0095 gene. We also determined that manR transcription is downregulated by an EIItMan-dependent mechanism in medium lacking glucose. It is proposed that this is a physiologically important process that functions in addition to phosphorylation control of ManR activity (42) to minimize synthesis of EIItMan in medium lacking glucose (Fig. 1).
EIItMan was found to contribute to various extents to CCR of PTS operons in glucose medium. CCR of the lmo0027 gene was strongly dependent on the expression of EIItMan, whereas CCR of the lmo0024, lmo1997, and lmo2002 mannose PTS permease genes was not. We propose that the dependence of these genes on EIItMan for CCR varies because they differ with respect to the concentration of the HPr-Ser-P/CcpA complex that is needed for binding to their cre sites. Possibly, the level of the HPr-Ser-P/CcpA complex in EGD-e Δmpt1 is sufficient to occupy cre sites that regulate the mannose PTS permease genes but not lmo0027.
Finally, the experiments showed that the EIItMan permease contributes to a small but significant degree to downregulation of virulence gene transcription in glucose medium. Given the interest in using class IIa bacteriocins to control the growth of L. monocytogenes in foods, the basis for this relationship and the effects of mpt inactivation on virulence in bacteriocin-resistant mutants should be more fully investigated. In this regard, a resD mutant, in which mpt expression is greatly reduced and virulence gene activity is elevated in glucose medium, did not exhibit increased invasiveness and growth in cell culture lines (21). However, experiments have not been carried out with a mpt deletion mutant per se, and it is not known if the upregulation of virulence genes that occurs in this genetic background results in greater infectivity.
Acknowledgments
This work was supported by grant R21 AI065623 to K.W.M. from the National Institute of Allergy and Infectious Diseases. We also acknowledge financial support from the University of Wyoming Agricultural Research Station.
Footnotes
Published ahead of print on 4 September 2009.
REFERENCES
- 1.Arous, S., C. Buchrieser, P. Folio, P. Glaser, A. Namane, M. Hebraud, and Y. Hechard. 2004. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology 150:1581-1590. [DOI] [PubMed] [Google Scholar]
- 2.Arous, S., K. Dalet, and Y. Hechard. 2004. Involvement of the mpo operon in resistance to class IIa bacteriocins in Listeria monocytogenes. FEMS Microbiol. Lett. 238:37-41. [DOI] [PubMed] [Google Scholar]
- 3.Behari, J., and P. Youngman. 1998. A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes. J. Bacteriol. 180:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, S.-K., and M. H. Saier, Jr. 2005. Regulation of sigL expression by the catabolite control protein CcpA involves a roadblock mechanism in Bacillus subtilis: potential connection between carbon and nitrogen metabolism. J. Bacteriol. 187:6856-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen, D. P., and R. W. Hutkins. 1994. Glucose uptake by Listeria monocytogenes Scott A and inhibition by pediocin JD. Appl. Environ. Microbiol. 60:3870-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, D. P., A. K. Benson, and R. W. Hutkins. 1998. Cloning and expression of the Listeria monocytogenes Scott A ptsH and ptsI genes, coding for HPr and enzyme I, respectively, of the phosphotransferase system. Appl. Environ. Microbiol. 64:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, D. P., A. K. Benson, and R. W. Hutkins. 1999. Mutational analysis of the role of HPr in Listeria monocytogenes. Appl. Environ. Microbiol. 65:2112-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalet, K., C. Briand, Y. Cenatiempo, and Y. Hechard. 2000. The rpoN gene of Enterococcus faecalis directs sensitivity to subclass IIa bacteriocins. Curr. Microbiol. 41:441-443. [DOI] [PubMed] [Google Scholar]
- 9.Dalet, K., Y. Cenatiempo, P. Cossart, The European Listeria Genome Consortium, and Y. Hechard. 2001. A σ54-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263-3269. [DOI] [PubMed] [Google Scholar]
- 10.Deutscher, J., R. Herro, A. Bourand, I. Mijakovic, and S. Poncet. 2005. P-Ser-HPr—a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim. Biophys. Acta 1754:118-125. [DOI] [PubMed] [Google Scholar]
- 11.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutscher, J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87-93. [DOI] [PubMed] [Google Scholar]
- 13.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 14.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 15.Gravesen, A., P. Warthoe, S. Knochel, and K. Thirstrup. 2000. Restriction fragment differential display of pediocin-resistant Listeria monocytogenes 412 mutants shows consistent overexpression of a putative β-glucoside-specific PTS system. Microbiology 146:1381-1389. [DOI] [PubMed] [Google Scholar]
- 16.Gravesen, A., M. Ramnath, K. B. Rechinger, N. Andersen, L. Jansch, Y. Hechard, J. W. Hastings, and S. Knochel. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361-2369. [DOI] [PubMed] [Google Scholar]
- 17.Hechard, Y., C. Pelletier, Y. Cenatiempo, and J. Frere. 2001. Analysis of σ54-dependent genes in Enterococcus faecalis: a mannose PTS permease (EIIMan) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology 147:1575-1580. [DOI] [PubMed] [Google Scholar]
- 18.Herro, R., S. Poncet, P. Cossart, C. Buchrieser, E. Gouin, P. Glaser, and J. Deutscher. 2005. How seryl-phosphorylated HPr inhibits PrfA, a transcription activator of Listeria monocytogenes virulence genes. J. Mol. Microbiol. Biotechnol. 9:224-234. [DOI] [PubMed] [Google Scholar]
- 19.Joseph, B., S. Mertins, R. Stoll, J. Schar, K. R. Umesha, Q. Luo, S. Muller-Altrock, and W. Goebel. 2008. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190:5412-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559-592. [DOI] [PubMed] [Google Scholar]
- 21.Larsen, M. H., B. H. Kallipolitis, J. K. Christiansen, J. E. Olsen, and H. Ingmer. 2006. The response regulator ResD modulates virulence gene expression in response to carbohydrates in Listeria monocytogenes. Mol. Microbiol. 61:1622-1635. [DOI] [PubMed] [Google Scholar]
- 22.Marr, A. K., B. Joseph, S. Mertins, R. Ecke, S. Muller-Altrock, and W. Goebel. 2006. Overexpression of PrfA leads to growth inhibition of Listeria monocytogenes in glucose-containing culture media by interfering with glucose uptake. J. Bacteriol. 188:3887-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin-Verstraete, I., V. Charrier, J. Stulke, A. Galinier, B. Erni, G. Rapoport, and J. Deutscher. 1998. Antagonistic effects of dual PTS-catalysed phosphorylation on the Bacillus subtilis transcriptional activator LevR. Mol. Microbiol. 28:293-303. [DOI] [PubMed] [Google Scholar]
- 24.Mengaud, J., C. Geoffroy, and P. Cossart. 1991. Identification of a new operon involved in Listeria monocytogenes virulence: its first gene encodes a protein homologous to metalloproteases. Infect. Immun. 59:1043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mertins, S., B. Joseph, M. Goetz, R. Ecke, G. Seidel, M. Sprehe, W. Hillen, W. Goebel, and S. Muller-Altrock. 2007. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J. Bacteriol. 189:473-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 27.Park, S. F., and R. G. Kroll. 1993. Expression of listeriolysin and phosphatidylinositol-specific phospholipase C is repressed by the plant-derived molecule cellobiose in Listeria monocytogenes. Mol. Microbiol. 8:653-661. [DOI] [PubMed] [Google Scholar]
- 28.Paulsen, I. T., S. Chauvaux, P. Choi, and M. H. Saier, Jr. 1998. Characterization of glucose-specific catabolite repression-resistant mutants of Bacillus subtilis: identification of a novel hexose:H+ symporter. J. Bacteriol. 180:498-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramnath, M., M. Beukes, K. Tamura, and J. W. Hastings. 2000. Absence of a putative mannose-specific phosphotransferase system enzyme IIAB component in a leucocin A-resistant strain of Listeria monocytogenes, as shown by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 66:3098-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramnath, M., S. Arous, A. Gravesen, J. W. Hastings, and Y. Hechard. 2004. Expression of mptC of Listeria monocytogenes induces sensitivity to class IIa bacteriocins in Lactococcus lactis. Microbiology 150:2663-2668. [DOI] [PubMed] [Google Scholar]
- 32.Ripio, M.-T., K. Brehm, M. Lara, M. Suarez, and J.-A. Vazquez-Boland. 1997. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 179:7174-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robichon, D., E. Gouin, M. Debarbouille, P. Cossart, Y. Cenatiempo, and Y. Hechard. 1997. The rpoN (σ54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J. Bacteriol. 179:7591-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scortti, M., H. J. Monzo, L. Lacharme-Lora, D. A. Lewis, and J. A. Vazquez-Boland. 2007. The PrfA virulence regulon. Microbes Infect. 9:1196-1207. [DOI] [PubMed] [Google Scholar]
- 35.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 36.Stoll, R., S. Mertins, B. Joseph, S. Muller-Altrock, and W. Goebel. 2008. Modulation of PrfA activity in Listeria monocytogenes upon growth in different culture media. Microbiology 154:3856-3876. [DOI] [PubMed] [Google Scholar]
- 37.Stulke, J., M. Arnaud, G. Rapoport, and I. Martin-Verstraete. 1998. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 28:865-874. [DOI] [PubMed] [Google Scholar]
- 38.Tasara, T., and R. Stephan. 2007. Evaluation of housekeeping genes in Listeria monocytogenes as potential internal control references for normalizing mRNA expression levels in stress adaptation models using real-time PCR. FEMS Microbiol. Lett. 269:265-272. [DOI] [PubMed] [Google Scholar]
- 39.Vadyvaloo, V., A. Arous, A. Gravesen, Y. Hechard, R. Chauhan-Haubrock, J. W. Hastings, and M. Rautenbach. 2004. Cell-surface alterations in class IIa bacteriocin-resistant Listeria monocytogenes strains. Microbiology 150:3025-3033. [DOI] [PubMed] [Google Scholar]
- 40.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:6238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue, J., I. Hunter, T. Steinmetz, A. Peters, B. Ray, and K. W. Miller. 2005. Novel activator of mannose-specific phosphotransferase system permease expression in Listeria innocua, identified by screening for pediocin AcH resistance. Appl. Environ. Microbiol. 71:1283-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue, J., and K. W. Miller. 2007. Regulation of the mpt operon in Listeria innocua by the ManR protein. Appl. Environ. Microbiol. 73:5648-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]