Abstract
Cryptosporidium is an important waterborne protozoan parasite that can cause severe diarrhea and death in the immunocompromised. The current methods used to monitor for Cryptosporidium oocysts in water are the microscopy-based USEPA methods 1622 and 1623. These methods assess total levels of oocysts in source waters, but do not determine oocyst viability or genotype. Recently, propidium monoazide (PMA) has been used in conjunction with molecular diagnostic tools to identify species and assess the viability of bacteria. The goal of this study was the development of a Cryptosporidium PMA-PCR (CryptoPMA-PCR) assay that includes PMA treatment prior to PCR analysis in order to prevent the amplification of DNA from dead oocysts. The results demonstrated that PMA penetrates only dead oocysts and blocks amplification of their DNA. The CryptoPMA-PCR assay can also specifically detect live oocysts within a mixed population of live and dead oocysts. More importantly, live oocysts, not dead oocysts, were detected in raw waste or surface water samples spiked with Cryptosporidium oocysts. This proof-of-concept study is the first to demonstrate the use of PMA for pre-PCR treatment of Cryptosporidium oocysts. The CryptoPMA-PCR assay is an attractive approach to specifically detect and genotype viable Cryptosporidium oocysts in the water, which is critical for human health risk assessment.
Cryptosporidium is a protozoan parasite infecting many animals and humans. Infection occurs following ingestion of oocyst-contaminated food, drinking water, or recreational water. It can also be transmitted from human to human or animal to human (13). Typically, it causes a self-limiting diarrheal disease in immunocompetent individuals but can become severe and even lead to death in the immunocompromised (e.g., AIDS patients, the elderly, and infants) (51). This parasite has been detected in many drinking water sources and is considered an important waterborne contaminant (52). The Cryptosporidium hominis-linked outbreak in Milwaukee, WI, in 1993, which involved over 400,000 infections and at least 54 deaths, is a clear example of a cryptosporidiosis outbreak due to oocyst-contaminated drinking water (24, 27). There are currently 20 valid species and over 40 genotypes of this parasite. The Cryptosporidium species C. canis, C. felis, C. hominis, C. meleagridis, C. muris, C. parvum, and C. suis and the cervine, chipmunk, rabbit, and horse genotypes have been shown to cause disease in humans (13, 14, 36, 38). Of these, C. parvum and C. hominis cause over 95% of the reported cases of human cryptosporidiosis (52).
To reduce health risks posed by this parasite, the USEPA promulgated the Long Term 2 Enhanced Surface Water Treatment Rule (LT2). Under this rule, drinking water utilities that use surface water or groundwater under the direct influence of a surface water as their primary drinking water source are required to monitor their source waters for Cryptosporidium oocysts by using USEPA method 1622 or 1623 (43-45). These microscopy-based methods produce total counts of live and dead Cryptosporidium oocysts in water samples without distinguishing species or genotypes that can infect humans from those that cannot. Results are then used to determine the need for additional water treatment in order to reduce the public health burden of waterborne cryptosporidiosis. These methods can overestimate the concentration of oocysts infectious to humans in water supplies and health risks posed by waterborne exposures to this parasite.
Many genotyping methods have been used to identify anthroponotic and zoonotic Cryptosporidium species in water supplies (52). These include the use of PCR-restriction fragment length polymorphism, single-strand conformational polymorphism, PCR-randomly amplified polymorphic DNA (15, 54), and PCR-DNA sequencing (54). Despite being instrumental in illustrating the immense diversity of Cryptosporidium spp. present in the environment, these methods cannot distinguish viable oocysts from nonviable oocysts.
Alternative approaches that do distinguish viable from nonviable oocysts have been developed. For example, rodent models of infection are used to measure the infectivity of this parasite (3, 23, 29), but this approach is time-consuming, labor-intensive, and expensive. In addition, it can determine infectivity of only a select number of Cryptosporidium species. More-rapid alternative techniques that do not use animal models include cell culture (37, 39) and in vitro excystation assays (23). The cell culture assay, however, is also limited in its range. It can only be used to detect viable C. andersoni, C. hominis, C. parvum, C. meleagridis, and C. muris oocysts but not viable oocysts of other Cryptosporidium species (1, 8, 19). In vitro excystation assays can overestimate oocyst viability, particularly if oocysts have been treated with various disinfectants (6, 7, 16). Other techniques use vital dye staining approaches, like 4′,6-diamidino-2-phenylindole (DAPI), propidium iodide (PI), and SYTO-9. These approaches can also be unreliable and can overestimate the total number of viable oocysts in water samples, especially if the water was chemically disinfected (6, 7, 9, 21). Other more recent approaches utilize molecular biology-based techniques to assess oocyst viability. Fluorescence in situ hybridization (5, 10, 20, 26, 40, 47), nucleic acid sequence-based amplification (4), reverse transcriptase PCR (RT-PCR) (17, 22, 50), and an integrated cell culture/PCR assay (12, 25) all show promise in providing a rapid and relatively inexpensive viability assay for Cryptosporidium. However, it still remains to be determined if these methods can detect all Cryptosporidium species and genotypes found in the environment.
It has been demonstrated recently that propidium monoazide (PMA) or ethidium monoazide (EMA) treatment, in conjunction with molecular methods, can be used for selective detection and genotyping of viable bacteria (30, 34) and fungi (48). PMA and EMA are new photoactive vital dyes that can preferentially penetrate dead cells, or cells with damaged or permeabilized cell membranes, but not viable cells with intact cell membranes. Once inside the cell, these molecules then intercalate into the DNA and become covalently bound to DNA upon exposure to bright white light. This photoactivation process results in the formation of a stable DNA-PMA/EMA complex that renders the DNA inaccessible for PCR amplification (31, 33). However, Nocker and colleagues have reported that EMA can also penetrate live bacteria and suggested that PMA is more effective at discriminating live bacteria from dead bacteria (31). In this paper, we developed and evaluated the use of PMA to selectively block PCR amplification of DNA from dead Cryptosporidium oocysts. Following PMA treatment, the small-subunit (SSU) rRNA gene and hsp70 gene were PCR amplified and sequenced to determine the genotypes of these oocysts. This “CryptoPMA-PCR” (Cryptosporidium PMA-PCR) approach was then successfully applied to selectively detect viable oocysts in a mixed suspension of viable and dead oocysts that were spiked into various environmental matrices. This new CryptoPMA-PCR assay, which allows genotyping and viability determination, has a potential to substantially improve the data on waterborne exposures to Cryptosporidium and enhance the validity of human risk assessment.
MATERIALS AND METHODS
Cryptosporidium oocysts.
C. parvum oocysts (Iowa isolate, Harley Moon strain) were purchased from Waterborne, Inc., and propagated in C57BL/6 mice at the USEPA facility in Cincinnati, OH, as previously described (28). Oocysts were purified from the feces by sieving, step sucrose gradients, and cesium chloride purification (3). Purified oocysts were then suspended in reagent-grade water containing 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco, Gaithersburg, MD) and stored at 4°C. C. muris oocysts (RN66), originally obtained from U. Iseki (Osaka University, Medical School, Osaka, Japan), were propagated in CF-1 female mice, purified from feces by sieving and sucrose flotation, and stored at 4°C in 0.01% Tween 20 containing 100 U/ml penicillin and 100 μg/ml streptomycin as described by Miller and Schaefer (28). For all experiments, oocysts were used within 3 months of isolation, with the exception of intentionally aged oocysts that were kept at ambient temperatures (20 to 22°C) for 14 months in the presence of Tween 20 and antibiotics as described above.
Heat inactivation.
Two-milliliter Costar natural clear microcentrifuge tubes (Corning Inc., Corning, NY) with oocysts suspended in phosphate-buffered saline (PBS) were heat treated in a water bath at 70°C for 30 min. Oocyst infectivity was determined by cell culture as described below.
PMA treatment of oocysts.
PMA (Biotium, Inc., Hayward, CA) was received in lyophilized form and reconstituted with 20% dimethyl sulfoxide to a stock concentration of 20 mM and aliquoted into a volume of 5 μl and stored at 4°C for no longer than 2 months. The PMA stock solution was diluted with PBS to final concentrations ranging from 5 μM to 150 μM depending on the experiment. Oocysts were incubated with PMA for 5 to 30 min in a 2-ml Costar natural clear microcentrifuge tube (Corning Inc.) with vortexing for 10 s every 5 min. Samples were then exposed to a Cosmobeam 800-W halogen light source (Cosmolight, Rome, Italy) at a distance of 20 cm while horizontal in ice for photoinduced cross-linking of PMA to oocyst DNA. Light exposure time ranged between 1 and 5 min depending on the experiment. Exposures were in 30-s increments with 1 min rest in between exposures to avoid damaging the oocysts.
Flow sorting of C. parvum samples.
A fluorescence-activated cell sorter (FACS VantageSE; Beckton Dickinson, Palo Alto, CA) equipped with CloneCyt software was used for enumerating oocysts (49). Oocysts were gated by forward and side scatter (FSC and SSC), with FSC and SSC measured linearly with a gain of 1, SSC set at 399 V, and FSC threshold set at 32 V, reflecting their size and shape. This gate was used for live and heat-killed oocysts. Between 1 × 104 and 1 × 106 sorted oocysts were collected in sterile 1.5-ml microcentrifuge tubes containing 0.5 ml of sterile PBS.
Infectious focus detection and in vitro cell culture of Cryptosporidium oocysts.
Human ileocecal adenocarcinoma cells (HCT-8) were purchased from the American Type Culture Collection (ATCC CCL 244; Manasas, VA) and maintained as previously described (11). Briefly, HCT-8 cells were grown as monolayers in 75-cm2 cell culture flasks in maintenance media consisting of RPMI 1640 medium (Gibco, Gaithersburg, MD), 5% fetal bovine serum (HyClone, Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco) at 37°C with 5% CO2. For infectivity experiments, cells were grown on 25-mm coverslips in six-well Costar tissue culture plates (Corning, Inc.) to 80 to 85% confluence. Cells were then washed three times with PBS, and fresh maintenance medium was added prior to the addition of oocysts.
Live or heat-killed C. parvum oocysts were pretreated with 5% sodium hypochlorite (Sigma-Aldrich, St. Louis, MO) for 5 min on ice, followed by three washes with PBS prior to inoculation. A total of 1 × 106 C. parvum oocysts was then added to the coverslips as previously published (39). At 24 and 72 h postinoculation, coverslips containing the infected cells were washed three times in ice-cold PBS and fixed with 4% paraformaldehyde for 10 min. Cells were then permeabilized with 0.25% Triton X-100 (Sigma) for 10 min, followed by a blocking step using fresh 1% bovine serum albumin in PBS. Vicia villosa lectin (Vector Laboratories, Burlingame, CA) was used to detect parasites as previously described (18).
Genomic DNA extraction, PCR amplification, and sequencing.
Genomic DNA from 1 × 104 to 1 × 105 PMA-treated and untreated live and heat-killed oocysts was isolated using the MasterPure DNA purification kit in accordance with the manufacturer's protocol (Epicentre Biotechnologies, Madison, WI). Briefly, oocysts resuspended in tissue and cell lysis solution were subjected to five freeze-thaw cycles followed by an overnight proteinase K digestion at 55°C. Samples were then treated with RNase and protein precipitation reagent. After the removal of cell debris by centrifugation, genomic DNA was precipitated with isopropanol, followed by two 75% ethanol wash steps, and then resuspended in 35 μl of nuclease-free water (Invitrogen, Grand Island, NY).
Primers that amplified a 346-bp length of the C. parvum hsp70 gene, as described by Di Giovanni and LeChevallier (12), were used to determine presence or absence of amplifiable DNA following PMA treatment. Sequences were as follows: forward primer, 5′-TCC TCT GCC GTA CAG GAT CTC TTA-3′; reverse primer, 5′-TGC TGC TCT TAC CAG TAC TCT TAT CA-3′. PCR conditions consisted of an initial denaturation at 95°C for 10 min followed by 30 cycles at 95°C for 30 s, 60°C for 1 min, 72°C for 2 min, with a final extension step at 72°C for 10 min.
An 834-bp portion of the C. parvum SSU rRNA gene or an 824-bp portion of the C. muris SSU rRNA gene was also amplified to determine the presence or absence of amplifiable DNA following PMA treatment, using the following previously published sequences: forward primer, 5′-GGA AGG GTT GTA TTT ATT AGA TAA AG-3′; reverse primer, 5′-AAG GAG TAA GGA ACA ACC TCC A-3′ (53). PCR conditions for the SSU rRNA gene reactions were as follows: an initial denaturation step at 95°C for 4 min, followed by 30 cycles at 94°C for 45 s, 58°C for 45 s, and 72°C for 1 min, with a final extension step at 72°C for 10 min. All 40-μl PCR mixtures contained 2.5 U AmpliTaq Gold, 4 mM MgCl2, 1× PCR buffer 2 (Applied Biosystems, Foster City, CA), 1 μM of each primer, and 2 μl of genomic DNA. One microliter of the PCR product was visualized using the DNA 1000 kit for the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA).
For sequence analysis, PCR fragments were purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA) and sent to the Cincinnati Children's Hospital Medical Center DNA Core facility for sequencing. The Lasergene 7 (DNASTAR, Madison, WI) software package was used to analyze DNA sequences.
Analysis of Cryptosporidium oocysts by immunofluorescence.
Oocysts were stained using a Crypt-a-Glo fluorescent antibody staining kit in accordance with the manufacturer's instructions (Waterborne, New Orleans, LA). Microscopic examinations were performed with a Zeiss Axiophot2 epifluorescence microscope (Carl Zeiss, Thornwood, NY) equipped with bright-field, differential interference contrast and epifluorescence optics. The oocysts stained with fluorescein isothiocyanate (FITC)-conjugated antibody were observed using an FITC filter set, while oocysts with PMA-labeled DNA were observed using a rhodamine/Texas Red filter set. Photomicrographs were taken using AxioVision (Carl Zeiss) and analyzed with Adobe Photoshop (Adobe, San Jose, CA).
Detection of live and dead oocysts in a mixed sample.
FACS-sorted oocysts of live or heat-killed C. parvum or C. muris were mixed at defined ratios of 0:100 (0:10,000 oocysts), 1:99 (100:9,900 oocysts), 10:90 (1,000:9,000 oocysts), 50:50 (5,000:5,000 oocysts), 90:10 (9,000:1,000 oocysts), 99:1 (9,900:100 oocysts), or 100:0 (10,000:0 oocysts), respectively treated with PMA, and then exposed to an 800-W halogen light. Total genomic DNA from the oocyst mixture was then extracted and analyzed using a single round of PCR amplification of the SSU rRNA gene and the hsp70 gene, followed by sequencing, as described previously (53).
Detection of viable oocysts in environmental samples.
Twenty-liter water samples were collected in carboys from the Ohio River. Water turbidity was 17 nephelometric turbidity units. Samples were filtered in the laboratory by using a portable gear pump and Filta-Max compressed foam filters (IDEXX Laboratories, Westbrook, ME). Filters were eluted into 500-ml conical centrifuge tubes by use of a Filta-Max xpress automatic pressure elution station in accordance with the manufacturer's instructions and method 1623 (in 2007, USEPA accepted the use of Filta-Max xpress as a sample-processing option for methods 1622 and 1623) (44). The eluate was centrifuged at 3,000 × g for 15 min. The supernatant was then carefully aspirated and discarded. The centrifuged pellet (2-ml volume) was resuspended in 50 ml PBS and divided into five 10-ml aliquots containing 0.4 ml packed pellet each. These aliquots were dispensed into 12-ml flat-sided Leighton tubes.
For raw wastewater samples, 50 ml of raw wastewater was divided into five 10-ml aliquots, each containing 58 mg of solids and transferred into flat-sided 12-ml Leighton tubes.
A total of 1 × 105 live or heat-killed C. parvum oocysts was then spiked into each tube containing resuspended Ohio River packed pellet or raw wastewater. Oocysts were separated from the sample matrix by using a Dynabeads immunomagnetic separation (IMS) kit, which employs magnetic beads covalently coupled to anti-Cryptosporidium antibody (Invitrogen Dynal AS, Oslo, Norway) in accordance with the manufacturer's guidelines and method 1622. After separation from the sample matrix, the oocyst-bead complexes were resuspended in 200 μl PBS and treated with PMA as described above.
RESULTS
Microscopic analysis of live and dead C. parvum oocysts treated with PMA.
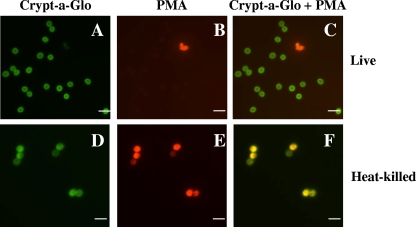
Live or heat-killed oocysts, which were incubated with 25 μM PMA for 10 min, were stained with FITC-conjugated anti-Cryptosporidium antibody (Crypt-a-Glo) and analyzed using fluorescence microscopy (Fig. 1). Following PMA treatment, live oocysts remained impermeable to PMA and did not stain red (Fig. 1B), whereas heat-killed oocysts stained bright red (Fig. 1E). Figure 1C and F are overlaid images of Crypt-a-Glo (Fig. 1A and D)- and PMA (Fig. 1B and E)-stained oocysts. Dead oocysts (Fig. 1F) that are both Crypt-a-Glo and PMA positive appear yellow. Further analysis revealed that less than 10% of oocysts which were not treated with heat were positive for PMA, while greater than 95% of heat-killed oocysts were PMA positive. The observed PMA staining of some untreated oocysts seen in Fig. 1B was likely due to the natural attrition of oocysts that occurs over time and/or oocysts killed during the purification process. To confirm that PMA-negative and PMA-positive oocysts were indeed live or dead, respectively, we performed a cell culture infectivity analysis of these oocysts by using HCT-8 cells. PMA-negative oocysts infected HCT-8 cells easily, as indicated by the presence of multiple infectious foci at 24 h postinfection, which increased in number at 72 h postinfection. In contrast, no infectious foci were detected at both time points when cells were inoculated with heat-killed oocysts that previously stained positive for PMA (data not shown).
FIG. 1.
Microscopic analyses distinguishing live from dead C. parvum oocysts using PMA. Live (A to C) or heat-killed (D to F) (70°C, 20 min) C. parvum oocysts treated with PMA and stained with Crypt-a-Glo. Crypt-a-Glo-labeled oocysts (green) are shown in panels A and D; the same cysts stained with PMA (red) are shown in panels B and E; and panels C and F represent overlaid images with Crypt-a-Glo and PMA (yellow) staining. Bar = 10 μm.
CryptoPMA-PCR assay to detect live but not dead Cryptosporidium oocysts.
To determine optimal PMA concentrations for live/dead oocyst discrimination using PCR, oocysts were stained with PMA concentrations from 0.5 to 150 μM. A total of 1 × 104 live or heat-killed oocysts was incubated with PMA for 10 min, followed by light exposure for 2 min. This was followed by DNA extraction and PCR amplification of the SSU rRNA gene (Fig. 2A). Our results revealed that PMA concentrations as high as 150 μM had no noticeable inhibitory effects on the detection of live oocysts by PCR, as indicated by the presence of a PCR product of the expected size with band intensity similar to that of PCR products detected from live untreated oocysts (Fig. 2A, lanes 2 and 3 to 6). In contrast, PMA treatment as low as 0.5 μM resulted in a marked reduction in detectable PCR products compared to heat-killed oocysts which were not treated with PMA (Fig. 2A, lane 7 and 8). More remarkable is the complete loss of amplicons from dead oocysts incubated with 5, 50, or 150 μM PMA (Fig. 2A, lanes 9 to 11).
FIG. 2.
Effects of different PMA concentrations on detecting live and heat-killed C. parvum oocysts. Live or heat-killed C. parvum oocysts were incubated with 0.5, 5, 50, or 150 μM PMA for 10 min followed by 2 min of light exposure. This was followed by the extraction of genomic DNA and PCR amplification with primers targeting the SSU rRNA gene (A) or the hsp70 gene (B). PCR products were analyzed using an Agilent Bioanalyzer 2100. M, marker; −, no PMA control; Δ, oocysts treated at 70°C for 30 min; C, PCR control that lacks a template.
To determine if this CryptoPMA-PCR assay can also work on another Cryptosporidium gene locus used for genotyping, the hsp70 gene was also tested. As shown in Fig. 2B, the hsp70 gene was easily amplified from live untreated C. parvum oocysts and was also detected with no marked inhibition of PCR amplification in live C. parvum oocysts treated with PMA (Fig. 2B, lanes 2 to 6). PCR products were also detected from untreated dead oocysts, albeit with slightly reduced band intensity (Fig. 2B, lane 7). In contrast, treatment with as little as 0.5 μM PMA (Fig. 2B, lane 8) resulted in an almost complete loss of the hsp70 PCR product. Treatment with higher PMA concentrations up to 150 μM resulted in undetectable levels of PCR products (Fig. 2B, lanes 9 to 11). These results demonstrate that the SSU rRNA gene and the hsp70 gene, two gene loci commonly used for genotyping Cryptosporidium spp. (15), are useful targets for this CryptoPMA-PCR detection assay. More importantly, pretreatment of Cryptosporidium oocysts with PMA resulted in the selective PCR amplification of the SSU rRNA gene or the hsp70 gene from live oocysts and the concomitant inhibition of PCR amplification of these genes from dead oocysts.
Optimizing PMA incubation times and photoactivation conditions for the CryptoPMA-PCR assay.
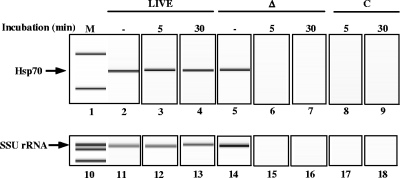
To optimize PMA incubation time, C. parvum oocysts were incubated on ice for 5 min or 30 min with 150 μM PMA followed by a 2-min exposure to bright light, a photoactivation step required to covalently bind PMA to DNA (Fig. 3) (31, 33). Incubation of live oocysts with PMA for 5 min (Fig. 3, lanes 3 and 12) or 30 min (Fig. 3, lanes 4 and 13) followed by bright light exposure did not have an appreciable inhibitory effect on amplifying the SSU rRNA gene or the hsp70 gene. Conversely, the PMA molecule readily penetrated and intercalated into DNA of heat-killed oocysts, as seen by fluorescence microscopy (Fig. 1E), resulting in the loss of detectable SSU rRNA gene or hsp70 PCR amplicons at 5 min (Fig. 3, lanes 6 and 15) or 30 min (Fig. 3, lanes 7 and 16) incubation times. These results revealed that incubation of oocysts for at least 5 min allowed sufficient time for PMA to cross several complex barriers (oocyst wall, sporozoite cell and nuclear membranes) and intercalate within the DNA structure to prevent amplification of genomic DNA from dead oocysts. Incubation of live oocysts with PMA for up to 30 min did not prevent amplification of their DNA nor did it result in any observable reduction in PCR products detected compared with live untreated oocysts (Fig. 3, lanes 2 to 4 and 11 to 13).
FIG. 3.
Effects of PMA incubation time on detecting live and heat-killed C. parvum oocysts. Live or heat-killed C. parvum oocysts were incubated with 150 μM PMA for either 5 or 30 min and then exposed to light for 2 min. Genomic DNA was purified as described in Materials and Methods. Amplification of a portion of the hsp70 gene (lanes 2 to 9) or the SSU rRNA gene (lanes 11 to 18) was performed using PCR. Gel electrophoresis analysis was performed using an Agilent Bioanalyzer 2100. M, marker; −, no PMA control; Δ, oocysts treated at 70°C; C, PCR control that lacks a template.
To minimize excessive heating of the oocysts, heat-induced oocyst excystation, or light-induced DNA damage during the PMA photoactivation step, light exposure times between 1 min and 5 min were evaluated. Figure 4 illustrates that a 1-min exposure to bright light (800 W) was sufficient to photoactivate PMA and prevent amplification of both target genes from heat-killed oocysts (Fig. 4, lanes 9 and 23), with no observable reduction of PCR products detected from live oocysts (Fig. 4, lanes 3 and 17). Exposure times up to 4 min also had no detectable inhibitory effects on amplifying the SSU rRNA gene or the hsp70 gene in live-oocyst samples; however, a 5-min light exposure showed a slight reduction in detectable SSU rRNA gene but not in the hsp70 amplicons from live oocysts (Fig. 4, lane 7 versus lane 21). Based on these experiments, oocysts were incubated with 150 μM PMA for 5 min, followed by a 2-min light exposure time in all the experiments described below.
FIG. 4.
Effects of the duration of exposure to light after PMA treatment on detecting live and heat-killed C. parvum oocysts. Live or heat-killed C. parvum oocysts were incubated with 150 μM PMA for 5 min and exposed to light for 1, 2, 3, 4, or 5 min. Genomic DNA was purified, and the hsp70 gene (lanes 2 to 14) or SSU rRNA gene (lanes 16 to 28) was amplified by PCR. Gel electrophoresis analysis was performed using an Agilent Bioanalyzer 2100. M, marker; −, no PMA control; Δ, oocysts treated at 70°C; C, PCR control that lacks a template.
Molecular genotyping of live Cryptosporidium oocysts in a mixture of live and dead oocysts of different species.
To determine if this CryptoPMA-PCR assay can be applied for the detection and genotyping of live Cryptosporidium oocysts in a mixed population containing different Cryptosporidium species, live or heat-killed C. muris oocysts were mixed with live or heat-killed C. parvum at defined ratios (Table 1). These samples were then treated with PMA followed by PCRs amplifying the SSU rRNA gene region. Detected amplicons were then sequenced to identify the species. Results in Table 1 demonstrate that in two of three experiments performed, live C. parvum oocysts were detected when mixed with dead C. muris at a 10:90 C. parvum-to-C. muris ratio. Additional experiments also revealed that only C. parvum DNA was detected in all three experiments when live C. parvum and dead C. muris oocysts were mixed at a 50:50 ratio. No C. muris SSU rRNA gene sequences were detected when dead C. muris oocysts were analyzed. Similarly, when ratios of 50:50 and 10:90 live C. muris-to-heat-killed C. parvum oocysts were analyzed, only the C. muris SSU rRNA gene sequence was detected in all six experiments performed. The ability of this CryptoPMA-PCR assay to detect 100 live C. muris oocysts in the presence of 9,900 heat-killed C. parvum oocysts (1:99 ratio) in one of three experiments performed is also notable.
TABLE 1.
Molecular genotyping of C. muris (Cm) and C. parvum (Cp) in a mixture of live and heat-killed oocysts using CryptoPMA-PCRa
| Oocyst |
Ratio | Oocyst detected in exptb: |
Livec | Deadc | |||
|---|---|---|---|---|---|---|---|
| Live | Heat-killed | 1 | 2 | 3 | |||
| Cp | Cm | 0:100 | − | − | − | 0/3 | 0/3 |
| Cp | Cm | 1:99 | − | − | − | 0/3 | 0/3 |
| Cp | Cm | 10:90 | Cp | − | Cp | 2/3 | 0/3 |
| Cp | Cm | 50:50 | Cp | Cp | Cp | 3/3 | 0/3 |
| Cp | Cm | 100:0 | Cp | Cp | Cp | 3/3 | 0/3 |
| Cm | Cp | 0:100 | − | − | − | 0/3 | 0/3 |
| Cm | Cp | 1:99 | Cm | − | − | 1/3 | 0/3 |
| Cm | Cp | 10:90 | Cm | Cm | Cm | 3/3 | 0/3 |
| Cm | Cp | 50:50 | Cm | Cm | Cm | 3/3 | 0/3 |
| Cm | Cp | 100:0 | Cm | Cm | Cm | 3/3 | 0/3 |
The SSU rRNA gene and the hsp70 gene were amplified and sequenced.
−, no PCR products detected.
Number of experiments in which PCR products from live or dead oocysts were detected/total number of experiments.
Application of the CryptoPMA-PCR assay for the detection of live oocysts in environmental matrices.
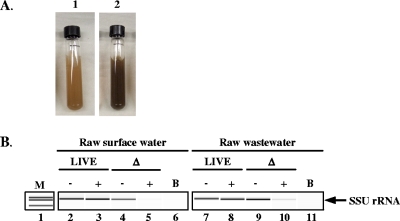
To determine if the CryptoPMA-PCR assay is suitable for the detection and genotyping of live oocysts in complex matrices, samples of untreated water from the Ohio River and raw wastewater samples from a municipal sewage treatment plant were each spiked with live or dead C. parvum oocysts. These matrices were chosen because they contain debris and dissolved compounds (Fig. 5A), which may not be completely separated from oocysts by the IMS procedure and can physically block entry of PMA into oocysts, sequester PMA away from the oocysts and/or mask light exposure during the photoactivation step. There was nearly a complete loss of the SSU rRNA gene PCR product from heat-killed PMA-treated oocysts spiked into the raw surface water (Fig. 5B, lane 5). Similarly, a marked reduction in the amount of amplicons was observed when analyzing the PMA-treated heat-killed oocysts, which were spiked into the raw wastewater (Fig. 5B, lane 10). The presence of a faint band in the raw wastewater sample may be a result of the presence of excess free DNA in the sample binding PMA and/or masking effects of debris present in the matrix preventing the complete removal and/or inhibition of PCR amplification of spiked dead oocysts.
FIG. 5.
Detection of C. parvum oocysts in Ohio River and raw wastewater samples, using CryptoPMA-PCR. (A) Environmental samples spiked with oocysts prior to immunomagnetic separation, PMA treatment, and PCR. Resuspended 0.4-ml pellet from raw surface water concentrate (panel 1) and raw wastewater (panel 2). (B) SSU rRNA gene PCR amplicons from live and dead oocysts spiked in raw surface water or raw wastewater. Amplification of a portion of the SSU rRNA gene was performed using PCR (lanes 2 to 11). Gel electrophoresis analysis was performed using an Agilent Bioanalyzer 2100. M, marker; −, no PMA; +, PMA added; Δ, oocysts treated at 70°C; B, blank sample that did not receive oocyst spike. Data represent the results of one of two independent experiments performed.
Using CryptoPMA-PCR assay to determine viability of Cryptosporidium oocysts stored at ambient temperatures.
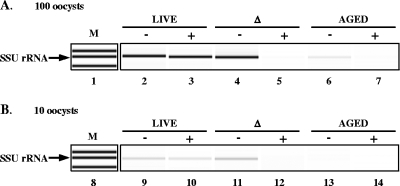
Previous studies by Widmer and others (50) have shown that long-term storage of oocysts at ambient temperatures resulted in the loss of oocyst infectivity due to natural oocyst attrition as a consequence of time and temperature. To determine if the CryptoPMA-PCR assay can be used to discriminate viable Cryptosporidium oocysts from oocysts that lost viability as a result of long-term storage at room temperature, 1 × 104 live, heat-killed, or aged (stored at 20 to 22°C for 14 months) oocysts were analyzed as described above. PCR amplification of the SSU rRNA gene from DNA equivalent to 100 live or heat-killed oocysts in the absence of PMA pretreatment resulted in a distinct band (Fig. 6A, lanes 2 and 4). No difference in band intensity was observed with the addition of PMA in live oocysts, while no PCR product was observed in heat-killed oocysts treated with PMA (Fig. 6A, lanes 3 and 5).
FIG. 6.
Differentiation of live C. parvum oocysts from heat-killed oocysts (Δ) and oocysts inactivated by long-term storage at room temperature (aged) using CryptoPMA-PCR. Live (lanes 2, 3, 9, and 10), heat-killed (lanes 4, 5, 11, and 12), and aged (lanes 6, 7, 13, and 14) C. parvum oocysts were incubated with 150 μM PMA for 5 min and then exposed to light for 2 min (lanes 3, 5, 7, 10, 12, and 14). Genomic DNA was purified as described in Materials and Methods. SSU rRNA gene PCR amplicons from DNA equivalent to 100 (A) or 10 (B) live, heat-killed, or aged oocysts were visualized using an Agilent Bioanalyzer 2100. M, marker; −, no PMA; +, PMA added; Δ, treated at 70°C.
Furthermore, when the SSU rRNA gene region was amplified from DNA equivalent to 10 oocysts, amplicons were still detected both in live-oocyst samples with or without PMA and in heat-killed oocysts in the absence of PMA (Fig. 6B, lanes 9 to 11). When the SSU rRNA gene was PCR amplified from 100 aged oocysts, a distinct, albeit faint, band was detected in the absence of PMA, which was completely abolished upon the addition of PMA (Fig. 6A, lanes 6 and 7, respectively). No amplicons were detected from 10 PMA-treated heat-killed oocysts (Fig. 6B, lane 12) or 10 aged oocysts in the presence or absence of PMA (Fig. 6B, lanes 13 and 14, respectively).
These results demonstrate that the CryptoPMA-PCR assay is effective at preventing the amplification of dead oocysts in a sample that has been kept at ambient temperatures for 14 months, conditions that oocysts can be exposed to in the natural environment. Moreover, these results also demonstrate that the single round of PCR used for the CryptoPMA-PCR is able to detect DNA equivalent to 10 live oocysts.
DISCUSSION
Current methods for determining viability employ cell culture or rodent models of infection (37). However, cell culture-based and animal bioassay approaches have inherent limitations. For example, it is not known if all species and genotypes of Cryptosporidium can grow in cell culture (2). Little is known about the susceptibility of rodents to infection with many species and genotypes of this parasite that can contaminate water supplies (42). Vital dyes like PI, DAPI, and SYTO-9 have been used as rapid alternatives to animal bioassays or cell culture assays in determining viability of oocysts, but results from these approaches can be variable and offer limited genotyping capabilities (6, 7, 9, 21, 29). PMA and EMA represent a new generation of vital dyes that contain a photoreactive azide group that can covalently bind to nucleic acids following photolysis. And unlike PI, which can diffuse in and out of oocysts (6, 7), photoactivated PMA or EMA forms a stable complex with nucleic acids that does not diffuse out of oocysts (31). Studies have shown, however, that EMA can overestimate the amount of viable bacteria (31). Thus, this study used PMA as a pre-PCR treatment for the CryptoPMA-PCR assay. An advantage of the CryptoPMA-PCR assay is that it is a rapid approach that does not involve cell culture. In addition, this approach allows for downstream genotyping following PMA treatment.
This proof-of-concept study is the first to demonstrate that heat-killed oocysts and oocysts that were inactivated by long-term storage at room temperature were permeable to PMA, whereas live oocysts were impermeable to it. Since PMA can cross multiple parasite membranes to intercalate and covalently bind to Cryptosporidium genomic DNA, it can be used as a pre-PCR treatment that renders dead oocysts undetectable by PCR analysis. For example, in the absence of a PMA labeling step, the SSU rRNA gene and the hsp70 gene from live or heat-killed Cryptosporidium oocysts or Cryptosporidium oocysts inactivated by long-term storage were both amplified, whereas these genes were detected only from live Cryptosporidium oocysts when the PMA labeling step was performed. As illustrated in Fig. 6 and Table 1, this method was able to detect relatively low numbers of live oocysts (10 to 100 oocysts) in a pure preparation as well as in a mixture with at least 9,900 dead oocysts. Conversely, these target genes were not detected from oocysts exposed to room temperature for 14 months or heat-killed oocysts at doses up to 10,000 oocysts, further demonstrating the efficiency of PMA to prevent amplification of DNA from dead Cryptosporidium oocysts. This study builds upon previous findings by Nocker and others (32) on the use of PMA to selectively detect and genotype viable bacteria and extends the application of PMA treatment to a more complex parasite. In this study, a single round of PCR was effective at detecting DNA from as few as 10 live oocysts (Fig. 6). We anticipate that combining the PMA staining step with a more sensitive genotyping approach, such as nested PCR, will enable the detection of a single live oocyst in environmental samples (15, 53). However, great care should be taken with nested PCR, since this approach can also increase the risk of false-positive results, particularly when environmental samples are analyzed. Additional experiments are also warranted to further evaluate the utility of PMA for the differentiation of live and dead oocysts by the use of other molecular biology-based detection methods not described here.
The addition of PMA was effective at preventing the amplification of DNA from oocysts that were killed by exposure to heat or long-term storage at room temperature while suspended in distilled water (Fig. 6). However, a faint band was detected in oocyst-free sewage samples that were spiked with heat-killed oocysts. This result could be due to the presence of high levels of indigenous extracellular DNA in the wastewater matrix, which can sequester free PMA molecules away from oocysts, thereby reducing the effectiveness of PMA in inhibiting amplification of DNA from dead oocysts. Alternatively, photoactivation of PMA could have been prevented through a “masking” effect by debris and contaminants that may have carried over during the IMS step. It is also possible that the beads themselves can interfere with the photoactivation process. However, PCR amplification of the SSU rRNA gene from live oocysts treated with PMA and photoactivated in the presence of at least 105 magnetic beads had no effects on the levels of PCR amplicons detected (data not shown). A recent study by Varma and others (46) also reported similar findings when using PMA to detect bacteria in wastewater, indicating the difficulty in using PMA on wastewater matrices. To minimize the effects of indigenous DNA, suspended solids, and dead microorganisms on PMA staining of dead Cryptosporidium oocysts in environmental samples, adding excess amounts of PMA as well as increasing light exposure should be considered potential strategies. Nevertheless, this study demonstrated that the CryptoPMA-PCR assay was an improvement to the specificity of current PCR techniques used to detect live Cryptosporidium oocysts seeded in turbid waters, like raw surface water samples (Fig. 5B).
The mode of action of PMA is still not completely understood. Previous studies by Nocker and colleagues (31) demonstrated that once PMA intercalates into and becomes covalently bound to DNA of bacteria, the PMA-DNA complex becomes insoluble and is subsequently removed during DNA extraction procedures. These scientists also hypothesized that bound PMA may affect Taq polymerase-based PCR amplification of the target DNA (32). Studies using EMA to differentiate live Listeria monocytogenes bacteria from dead L. monocytogenes bacteria have suggested that the duration of light exposure of DNA in the presence of EMA can inhibit PCR amplification of target genes (41). Whether PMA has effects similar to those of EMA remains to be determined. Despite our limited knowledge of its mode of action, PMA treatment is very effective at preventing the amplification of the hsp70 and SSU rRNA genes from heat-killed Cryptosporidium oocysts. Interestingly, during the optimization of the photoactivation process, we observed that exposure to light for at least 5 min resulted in a marked decrease in the signal intensity of the SSU rRNA gene amplicon, but not in that of the hsp70 gene. This may be attributed to light-induced DNA strand breaks leading to decreased signal intensity for the SSU rRNA gene amplicon. These results were also seen in previous studies done on other eukaryotic cells (35). The SSU rRNA gene amplicon is longer than the hsp70 amplicons by nearly 500 bp. The relatively small size of the hsp70 amplicon (346 bp) may explain why it might have a lower probability of light-induced strand breaks. Since 2 min of light exposure did not result in any appreciable decrease in signal intensity for either SSU rRNA gene or hsp70 amplicons from live Cryptosporidium oocysts, we chose this duration of light exposure for all further experiments performed in this study. Nonetheless, great care should also be taken when optimizing this approach to detect other microorganisms.
Here, we have developed the CryptoPMA-PCR technique, which is very effective at specifically detecting and genotyping live oocysts seeded in distilled water samples and also has a potential to specifically detect viable oocysts in environmental water samples. Unlike RT-PCR, which is labor-intensive and requires advanced technical skills to manipulate mRNA (a very labile molecule that easily degrades), the CryptoPMA-PCR assay uses DNA as its template, which makes this technique more versatile and user friendly. The CryptoPMA-PCR can be modified to use other types of molecular genotyping techniques, such as nested PCR, single-strand conformational polymorphisms, or multilocus sequence typing. The use of quantitative real-time PCR with PMA pretreatment may also enable quantitation of viable pathogenic Cryptosporidium oocysts in environmental samples.
Future research efforts should also focus on determining sensitivity and versatility of PMA to distinguish live oocysts from dead oocysts that have been treated with different methods of disinfection (e.g., UV, chlorine, ozone, and natural sunlight). Previous studies have demonstrated that real-time PCR plus PMA treatment could not be used to differentiate UV-inactivated bacteria from viable ones, suggesting that PMA pretreatment may also be limited to preventing PCR amplification of DNA from oocysts that were treated with heat or chemicals (e.g., chlorine, ozone, aldehydes, and alcohols) that make the oocyst wall permeable for PMA (33). More importantly, standardization and interlaboratory performance comparison of this assay, using different water matrices spiked with low numbers of oocyst typically found in environmental samples, must be performed in order to evaluate the practical applicability of this approach for water quality monitoring.
This CryptoPMA-PCR approach is a novel tool that combines genotyping and viability determination. After further refinement and validation, this method may be used in source-tracking studies or for generating improved data on waterborne exposures to Cryptosporidium and assessing health risks posed by this parasite.
Acknowledgments
This research was supported, in part, by an appointment to the Postgraduate Research Participation Program administered by the Oak Ridge Institute for Science and Education, for C.C.B., through an interagency agreement between the U.S. DOE and the U.S. EPA. The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed the research described here. It has been subjected to the Agency's administrative review and approved for publication.
We thank Nick Dugan and Daniel Williams for providing raw surface water samples, Eric Rhodes for providing raw wastewater samples, and Rich Haugland and Stacy Pfaller for critical review of the manuscript.
C.C.B. and E.N.V. conceived and designed the experiments. C.C.B., S.M.G., M.W.W., E.A.V., and A.I.E. performed the experiments. C.C.B. and E.N.V. analyzed the data. C.C.B., E.N.V., S.M.G., M.W.W., E.A.V., and A.I.E. contributed reagents/materials/analysis tools. C.C.B. and E.N.V. wrote the paper.
Footnotes
Published ahead of print on 11 September 2009.
REFERENCES
- 1.Akiyoshi, D. E., J. Dilo, C. Pearson, S. Chapman, J. Tumwine, and S. Tzipori. 2003. Characterization of Cryptosporidium meleagridis of human origin passaged through different host species. Infect. Immun. 71:1828-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrowood, M. J. 2008. In vitro cultivation, p. 499-526. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press and IWA Publishing, Boca Raton, FL.
- 3.Arrowood, M. J., L. T. Xie, K. Rieger, and J. Dunn. 1996. Disinfection of Cryptosporidium parvum oocysts by pulsed light treatment evaluated in an in vitro cultivation model. J. Eukaryot. Microbiol. 43:88S. [DOI] [PubMed] [Google Scholar]
- 4.Baeumner, A. J., M. C. Humiston, R. A. Montagna, and R. A. Durst. 2001. Detection of viable oocysts of Cryptosporidium parvum following nucleic acid sequence based amplification. Anal. Chem. 73:1176-1180. [DOI] [PubMed] [Google Scholar]
- 5.Bednarska, M., A. Bajer, E. Sinski, A. S. Girouard, L. Tamang, and T. K. Graczyk. 2007. Fluorescent in situ hybridization as a tool to retrospectively identify Cryptosporidium parvum and Giardia lamblia in samples from terrestrial mammalian wildlife. Parasitol. Res. 100:455-460. [DOI] [PubMed] [Google Scholar]
- 6.Black, E. K., G. R. Finch, R. Taghi-Kilani, and M. Belosevic. 1996. Comparison of assays for Cryptosporidium parvum oocysts viability after chemical disinfection. FEMS Microbiol. Lett. 135:187-189. [DOI] [PubMed] [Google Scholar]
- 7.Bukhari, Z., M. M. Marshall, D. G. Korich, C. R. Fricker, H. V. Smith, J. Rosen, and J. L. Clancy. 2000. Comparison of Cryptosporidium parvum viability and infectivity assays following ozone treatment of oocysts. Appl. Environ. Microbiol. 66:2972-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buret, A. G., A. C. Chin, and K. G. Scott. 2003. Infection of human and bovine epithelial cells with Cryptosporidium andersoni induces apoptosis and disrupts tight junctional ZO-1: effects of epidermal growth factor. Int. J. Parasitol. 33:1363-1371. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, A. T., L. J. Robertson, and H. V. Smith. 1992. Viability of Cryptosporidium parvum oocysts: correlation of in vitro excystation with inclusion or exclusion of fluorogenic vital dyes. Appl. Environ. Microbiol. 58:3488-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, C. M., N. Altavilla, M. Krogh, C. M. Ferguson, D. A. Deere, and N. J. Ashbolt. 2005. Environmental inactivation of Cryptosporidium oocysts in catchment soils. J. Appl. Microbiol. 98:308-317. [DOI] [PubMed] [Google Scholar]
- 11.Di Giovanni, G. D., F. H. Hashemi, N. J. Shaw, F. A. Abrams, M. W. LeChevallier, and M. Abbaszadegan. 1999. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Appl. Environ. Microbiol. 65:3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Giovanni, G. D., and M. W. LeChevallier. 2005. Quantitative-PCR assessment of Cryptosporidium parvum cell culture infection. Appl. Environ. Microbiol. 71:1495-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fayer, R. 2008. General biology, p. 1-42. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press and IWA Publishing, Boca Raton, FL.
- 14.Fayer, R., M. Santin, and J. M. Trout. 2008. Cryptosporidium ryanae n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). Vet. Parasitol. 156:191-198. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, C., D. Deere, M. Sinclair, R. M. Chalmers, K. Elwin, S. Hadfield, L. Xiao, U. Ryan, R. Gasser, Y. A. El-Osta, and M. Stevens. 2006. Meeting report: application of genotyping methods to assess risks from Cryptosporidium in watersheds. Environ. Health Perspect. 114:430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finch, G. R., E. K. Black, L. Gyurek, and M. Belosevic. 1993. Ozone inactivation of Cryptosporidium parvum in demand-free phosphate buffer determined by in vitro excystation and animal infectivity. Appl. Environ. Microbiol. 59:4203-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallier-Soulier, S., and E. Guillot. 2003. An immunomagnetic separation-reverse transcription polymerase chain reaction (IMS-RT-PCR) test for sensitive and rapid detection of viable waterborne Cryptosporidium parvum. Environ. Microbiol. 5:592-598. [DOI] [PubMed] [Google Scholar]
- 18.Hashim, A., M. Clyne, G. Mulcahy, D. Akiyoshi, R. Chalmers, and B. Bourke. 2004. Host cell tropism underlies species restriction of human and bovine Cryptosporidium parvum genotypes. Infect. Immun. 72:6125-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hijjawi, N. S., B. P. Meloni, U. M. Ryan, M. E. Olson, and R. C. Thompson. 2002. Successful in vitro cultivation of Cryptosporidium andersoni: evidence for the existence of novel extracellular stages in the life cycle and implications for the classification of Cryptosporidium. Int. J. Parasitol. 32:1719-1726. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins, M., J. M. Trout, J. Higgins, M. Dorsch, D. Veal, and R. Fayer. 2003. Comparison of tests for viable and infectious Cryptosporidium parvum oocysts. Parasitol. Res. 89:1-5. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, M. B., L. J. Anguish, D. D. Bowman, M. J. Walker, and W. C. Ghiorse. 1997. Assessment of a dye permeability assay for determination of inactivation rates of Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 63:3844-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins, M. C., J. Trout, M. S. Abrahamsen, C. A. Lancto, J. Higgins, and R. Fayer. 2000. Estimating viability of Cryptosporidium parvum oocysts using reverse transcriptase-polymerase chain reaction (RT-PCR) directed at mRNA encoding amyloglucosidase. J. Microbiol. Methods 43:97-106. [DOI] [PubMed] [Google Scholar]
- 23.Korich, D. G., J. R. Mead, M. S. Madore, N. A. Sinclair, and C. R. Sterling. 1990. Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 56:1423-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer, M. H., B. L. Herwaldt, G. F. Craun, R. L. Calderon, and D. D. Juranek. 1996. Surveillance for waterborne-disease outbreaks—United States, 1993-1994. MMWR CDC Surveill. Summ. 45:1-33. [PubMed] [Google Scholar]
- 25.LeChevallier, M. W., G. D. Di Giovanni, J. L. Clancy, Z. Bukhari, S. Bukhari, J. S. Rosen, J. Sobrinho, and M. M. Frey. 2003. Comparison of method 1623 and cell culture-PCR for detection of Cryptosporidium spp. in source waters. Appl. Environ. Microbiol. 69:971-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemos, V., T. K. Graczyk, M. Alves, M. L. Lobo, M. C. Sousa, F. Antunes, and O. Matos. 2005. Identification and determination of the viability of Giardia lamblia cysts and Cryptosporidium parvum and Cryptosporidium hominis oocysts in human fecal and water supply samples by fluorescent in situ hybridization (FISH) and monoclonal antibodies. Parasitol. Res. 98:48-53. [DOI] [PubMed] [Google Scholar]
- 27.Mac Kenzie, W. R., N. J. Hoxie, M. E. Proctor, M. S. Gradus, K. A. Blair, D. E. Peterson, J. J. Kazmierczak, D. G. Addiss, K. R. Fox, J. B. Rose, and J. P. Davis. 1994. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N. Engl. J. Med. 331:161-167. [DOI] [PubMed] [Google Scholar]
- 28.Miller, T. A., and F. W. Schaefer III. 2006. Characterization of a single dose methylprednisolone acetate immune suppression model using Cryptosporidium muris and Cryptosporidium parvum. Vet. Parasitol. 141:66-83. [DOI] [PubMed] [Google Scholar]
- 29.Neumann, N. F., L. L. Gyurek, G. R. Finch, and M. Belosevic. 2000. Intact Cryptosporidium parvum oocysts isolated after in vitro excystation are infectious to neonatal mice. FEMS Microbiol. Lett. 183:331-336. [DOI] [PubMed] [Google Scholar]
- 30.Nocker, A., and A. K. Camper. 2006. Selective removal of DNA from dead cells of mixed bacterial communities by use of ethidium monoazide. Appl. Environ. Microbiol. 72:1997-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nocker, A., C. Y. Cheung, and A. K. Camper. 2006. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67:310-320. [DOI] [PubMed] [Google Scholar]
- 32.Nocker, A., P. Sossa-Fernandez, M. D. Burr, and A. K. Camper. 2007. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 73:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nocker, A., K. E. Sossa, and A. K. Camper. 2007. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 70:252-260. [DOI] [PubMed] [Google Scholar]
- 34.Pan, Y., and F. Breidt, Jr. 2007. Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl. Environ. Microbiol. 73:8028-8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pflaum, M., S. Boiteux, and B. Epe. 1994. Visible light generates oxidative DNA base modifications in high excess of strand breaks in mammalian cells. Carcinogenesis 15:297-300. [DOI] [PubMed] [Google Scholar]
- 36.Robinson, G., K. Elwin, and R. M. Chalmers. 2008. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg. Infect. Dis. 14:1800-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rochelle, P. A., M. M. Marshall, J. R. Mead, A. M. Johnson, D. G. Korich, J. S. Rosen, and R. De Leon. 2002. Comparison of in vitro cell culture and a mouse assay for measuring infectivity of Cryptosporidium parvum. Appl. Environ. Microbiol. 68:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan, U. M., M. Power, and L. Xiao. 2008. Cryptosporidium fayeri n. sp. (Apicomplexa: Cryptosporidiidae) from the red kangaroo (Macropus rufus). J. Eukaryot. Microbiol. 55:22-26. [DOI] [PubMed] [Google Scholar]
- 39.Slifko, T. R., D. Friedman, J. B. Rose, and W. Jakubowski. 1997. An in vitro method for detecting infectious Cryptosporidium oocysts with cell culture. Appl. Environ. Microbiol. 63:3669-3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, J. J., T. S. Gunasekera, C. R. Barardi, D. Veal, and G. Vesey. 2004. Determination of Cryptosporidium parvum oocyst viability by fluorescence in situ hybridization using a ribosomal RNA-directed probe. J. Appl. Microbiol. 96:409-417. [DOI] [PubMed] [Google Scholar]
- 41.Soejima, T., K. Iida, T. Qin, H. Taniai, M. Seki, A. Takade, and S. Yoshida. 2007. Photoactivated ethidium monoazide directly cleaves bacterial DNA and is applied to PCR for discrimination of live and dead bacteria. Microbiol. Immunol. 51:763-775. [DOI] [PubMed] [Google Scholar]
- 42.Tzipori, S., and G. Widmer. 2008. Animal models, p. 485-498. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press and IWA Publishing, Boca Raton, FL.
- 43.USEPA. 2005. Method 1622: Cryptosporidium in water by filtration/IMS/FA. 815-R-05-001. Office of Water, U.S. EPA, Washington, DC.
- 44.USEPA. 2005. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. 815-R-05-002. Office of Water, EPA, Washington, DC.
- 45.USEPA. 2006. National primary drinking water regulations: long term 2 enhanced surface water treatment rule. 40 CFR Parts 9, 141, and 142. Fed. Regist. 71:654-786. [PubMed] [Google Scholar]
- 46.Varma, M., R. Field, M. Stinson, B. Rukovets, L. Wymer, and R. Haugland. 2009. Quantitative real-time PCR analysis of total and propidium monoazide-resistant fecal indicator bacteria in wastewater. Water Res. doi: 10.1016/j.watres.2009.05.031. [DOI] [PubMed]
- 47.Vesey, G., N. Ashbolt, E. J. Fricker, D. Deere, K. L. Williams, D. A. Veal, and M. Dorsch. 1998. The use of a ribosomal RNA targeted oligonucleotide probe for fluorescent labelling of viable Cryptosporidium parvum oocysts. J. Appl. Microbiol. 85:429-440. [DOI] [PubMed] [Google Scholar]
- 48.Vesper, S., C. McKinstry, C. Hartmann, M. Neace, S. Yoder, and A. Vesper. 2008. Quantifying fungal viability in air and water samples using quantitative PCR after treatment with propidium monoazide (PMA). J. Microbiol. Methods 72:180-184. [DOI] [PubMed] [Google Scholar]
- 49.Ware, M. W., L. Wymer, H. D. Lindquist, and F. W. Schaefer III. 2003. Evaluation of an alternative IMS dissociation procedure for use with method 1622: detection of Cryptosporidium in water. J. Microbiol. Methods 55:575-583. [DOI] [PubMed] [Google Scholar]
- 50.Widmer, G., E. A. Orbacz, and S. Tzipori. 1999. β-Tubulin mRNA as a marker of Cryptosporidium parvum oocyst viability. Appl. Environ. Microbiol. 65:1584-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao, L., and U. M. Ryan. 2008. Molecular epidemiology, p. 119-171. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press and IWA Publishing, Boca Raton, FL.
- 53.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, W., P. Chen, E. N. Villegas, R. B. Landy, C. Kanetsky, V. Cama, T. Dearen, C. L. Schultz, K. G. Orndorff, G. J. Prelewicz, M. H. Brown, K. R. Young, and L. Xiao. 2008. Cryptosporidium source tracking in the Potomac River watershed. Appl. Environ. Microbiol. 74:6495-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]