Abstract
Actinomycetes can be symbionts in diverse organisms, including both plants and animals. Some actinomycetes benefit their host by producing small molecule secondary metabolites; the resulting symbioses are often developmentally complex. Actinomycetes associated with three cone snails were studied. Cone snails are venomous tropical marine gastropods which have been extensively examined because of their production of peptide-based neurological toxins, but no microbiological studies have been reported on these organisms. A microhabitat approach was used in which dissected tissue from each snail was treated as an individual sample in order to explore bacteria in the tissues separately. Our results revealed a diverse, novel, and highly culturable cone snail-associated actinomycete community, with some isolates showing promising bioactivity in a neurological assay. This suggests that cone snails may represent an underexplored reservoir of novel actinomycetes of potential interest for drug discovery.
Interest in natural products and drug discovery has been a major driving force for the study of microbial communities associated with marine invertebrates. Sponges, which have provided more bioactive metabolites than any other marine invertebrate group (see reference 4 and previous references in that series) have been the main focus of these investigations, yielding numerous reports of associated bacteria and complex microbial communities (17, 43). Other examples of marine invertebrate-associated microbes explored for their involvement in natural products include bryozoans (9, 41), ascidians (37), and shipworms (46). Thus far, there are literature reports of diverse bacterial taxa involved in natural product biosynthesis in marine animals (9, 10). Notably less well studied are the symbiotic actinomycetes, for which the biology of host-actinomycete associations is just beginning to be explored in a methodical way. Actinomycetes are known to be important symbionts in a number of biological systems, such as plants, insects, and marine invertebrates, contributing as nitrogen fixers in plants (39) or utilizing their chemical arsenal for defense purposes (8, 22, 38).
An initial investigation of a cone snail yielded a surprisingly high number of actinomycetes (data not shown), prompting this follow-up study on additional samples. Cone snail mollusks belong to the genus Conus, which contains about 500 closely related species (11). These mollusks are well known for their complex neurologically active venoms that they use to immobilize their prey, including fish, worms, and other mollusks. The venom of cone snails has been extensively studied, but to our knowledge no study of cone snail microbial communities has been reported. Cone snails are part of the larger superfamily Conoidea, comprising ∼20,000 species, making it an extremely diverse group (25, 34). Given the initial observation of cultivable actinomycetes from cone snails, this large group seemed like a potentially excellent source of new bacterial natural products and new models of actinomycete symbiosis.
The goal of this study was to examine the actinobacteria community associated with tropical marine snails of the genus Conus, using a microhabitat approach by which individual organs are treated as independent samples, and to assess the bioactivities of the isolates obtained by using a neurological assay. Although similar approaches have previously been used in microbial ecology (3, 42), this is a novel approach in drug discovery for the identification of bioactive bacteria. We report here the association between three cone snails, Conus pulicarius, Conus rolani, and Conus tribblei, and their actinobacteria as well as the bioactivities of some of these actinobacteria.
MATERIALS AND METHODS
Collection and processing of snails.
Cone snails were collected in Cebu, Philippines, by professional collectors with appropriate permits. Typically, the water temperature in Cebu ranges between 26°C and 29°C. The three different snails, C. pulicarius, C. tribblei, and C. rolani, were identified according to their shell patterns (Fig. 1). Snails were kept in seawater overnight before being processed. Snails were dissected under sterile conditions, and each tissue (body, hepatopancreas, and venom duct) was rinsed three times with sterile seawater. Body and hepatopancreas were divided in two, with one half used for cultivation and the other half transferred into RNAlater (Ambion) and kept frozen until DNA was extracted for molecular analysis.
FIG. 1.
Radial cladogram showing the evolutionary relationships between C. rolani, C. pulicarius, and C. tribblei (12, 31; N. Puillandre, M. Watkins, and B. M. Olivera, unpublished data). The tree was generated using mitochondrial 16S sequences from the Olivera database. This figure shows that snails used in this study occupy distinct branches of the Conus radiation.
Cultivation of actinobacteria.
Cone snail tissue extract was obtained by grinding ca. 5 mm3 of tissue in 2 ml of sterile artificial seawater using a sterile mortar and pestle. Three different medium types were used: marine agar 2216 (Difco), ISP2 (Difco), and R2A agar (Difco). ISP2 and R2A are media designed for selective growth of actinomycetes and were supplemented with a final concentration of 10 μg of nalidixic acid ml−1, 10 μg of cycloheximide ml−1, 25 μg of nystatin ml−1, and 2% NaCl. Serial dilutions of the extracts were plated on the different media, and cultures were incubated at 30°C for up to 4 weeks. One bacterial colony of each morphotype was selected and subcultured until pure.
Analysis of snail-associated actinobacteria.
DNA from preserved snail organs was extracted using the Puregene tissue kit (Qiagen) and quantified using a Nanodrop (Thermo Scientific). Bacterial isolates were grown in 50 ml of their medium of isolation and incubated for a week prior to DNA extraction using an UltraClean microbial DNA isolation kit (MoBio). The 16S rRNA genes were amplified using 100 ng/μl of DNA with PrimeStar polymerase (Takara) in a DNA Engine thermocycler (Bio-Rad). Universal bacterial 16S primers 8-27f/1492r (33, 44) were applied to pure cultures and to DNA from whole tissues. To amplify actinomycete sequences from whole tissues, specific primers 243F (19) and AB1165R (28) were applied to the universal primer PCR product in a nested PCR strategy. Cycling conditions for the universal primers were a hot start at 98°C for 5 min, 30 cycles of 98°C for 30 s, 48°C for 10 s, and 72°C for 45 s, and a final extension step at 72°C for 5 min. The PCR product was then gel purified on a 1% (wt/vol) agarose gel, and bands of approximately 1,500 bp were excised and recovered using a QIAQuick gel extraction kit (Qiagen) and eluted in a 20-μl volume. PCR amplifications using actinomycete-specific primers 243f/AB1165R (28) were carried out under the following cycling conditions: a hot start at 98°C for 5 min, 30 cycles of 98°C for 30 s, 55°C for 10 s, and 72°C for 45 s, and a final extension step at 72°C for 5 min. The PCR product was then gel purified on a 1% (wt/vol) agarose gel, extracted using the QIAQuick gel extraction kit (Qiagen), and eluted in 20 μl of water before being used for cloning. Purified PCR products were cloned with a TOPO-XL cloning kit (Invitrogen) according to the manufacturer's instructions. A total of 1,017 sequences were obtained, 960 from clone library sequencing (480 from universal primer-generated libraries and 480 from actinobacteria primer-generated libraries) and 57 from isolates.
Sequencing and phylogenetic analysis.
Sequencing was performed using an ABI 3730 automated sequencer (PE Applied Biosystems, Foster City, CA) with M13 forward and reverse primers. Partial sequences were compared to those in the GenBank database with the Basic Local Alignment Search Tool algorithm to identify known closely related sequences. Sequences were examined for the formation of chimeras using the program CHECK_CHIMERA (6). Sequences were manually compiled and aligned using Phydit software (5). Evolutionary trees were generated using the neighbor-joining (36), Fitch-Margoliash (14), and maximum parsimony (24) algorithms in the PHYLIP package (13). Evolutionary distance matrices for the neighbor-joining and Fitch-Margoliash methods were generated as described by Jukes and Cantor (21). The robustness of inferred tree topologies was evaluated after 1,000 bootstrap resamplings of the neighbor-joining data, and only values of >50% are shown.
Rarefaction and statistical analysis.
The assemblages of 16S rRNA gene sequences in C. pulicarius and C. tribblei libraries were analyzed by rarefaction analysis using EcoSim (15) to assess the extent to which the diversity of microbial communities was represented by the library. A 99% and 97% clone-to-clone maximum identity cutoff was used for the generation of the rarefaction curves. The phylogenetic tree branches statistical analysis was done with Unifrac (http://bmf2.colorado.edu/unifrac/index.psp) (27). Using the Unifrac significance algorithm, the reported P value was ≤0.06, which is in the suggestive range, while a P value if >0.1 would not be significant.
FISH.
Samples for fluorescence in situ hybridization (FISH) were fixed in 4% paraformaldehyde in artificial seawater and then stored in 70% ethanol prior to embedding and sectioning as described previously (47). Sections were hybridized with fluorescein-labeled eubacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) (1) and a Cy-3 HG69a actinobacteria probe (5′-TATAGTTACCACCGCCGT-3′) (35). Nonsense fluorescein-labeled EUB338 (5′-CTCCTACGGGAGGCAGC-3′) (1) was used as a negative control. Images were captured by laser scanning confocal microscopy using the FV-300 XY system (Olympus). Final images were merged and artificially colored using FV10-ASW software (Olympus).
Extraction of cultivated isolates.
Purified actinomycetes were grown in 50 ml of ISP2 or R2A broth for 7 days at 30°C. For each isolate, the culture was centrifuged and the supernatant was incubated for 45 min with HP20SS resin. The resin was then washed three times with Milli-Q water and eluted with methanol. The methanol was dried, and the resulting extract was partitioned three times between ethyl acetate and water. The organic fraction was transferred to a clean vial and dried using a rotovap. The dried extract was resuspended in methanol and passed through a C18 column before being dried by rotary evaporation.
DRG assay.
Dorsal root ganglia (DRG) cells from cervical and lumbar regions were obtained from mice and used in an assay with bacterial culture extracts. DRG cells were suspended in medium with additives and loaded with Fura-2 AM (Molecular Probes), a fluorescent dye used to measure intracellular calcium levels. Experiments were performed at room temperature (20 to 25°C) in a 24-well plate format using fluorescence microscopy. Individual cells were treated as single samples, so that the individual responses of diverse neuron subtypes from the DRG could be examined. After baseline measurements, the cells were treated with KCl solution and then washed. After return to baseline, bacterial extracts, fractions, or pure compounds were applied. This solution was then later replaced with KCl solution. The use of KCl permitted observation of direct, modulatory, as well as inhibitory and excitatory effects of the extracts. Full experimental details can be found in the supplemental material section.
RESULTS AND DISCUSSION
Three cone snails, C. pulicarius, C. rolani, and C. tribblei, were dissected freshly after collection, and three tissue parts, body, hepatopancreas, and venom duct, were used to conduct an analysis of the snail-associated actinomycete community. A total of 229 morphologically distinct bacteria were isolated from C. rolani (n = 121), C. pulicarius (n = 121), and C. tribblei (n = 105). Morphologically, 96 actinomycetes were identified: 47 from C. rolani, 28 from C. pulicarius, and 21 from C. tribblei. At the end of the isolation and purification process, 57 unique actinomycete strains remained viable (see Table S1 in the supplemental material). Most isolates were recovered from the hepatopancreas (n = 27) and the body (n = 18), while the venom duct yielded 11 isolates. Phylogenetic analysis of the isolates using the 16S rRNA gene showed that 16 different genera (Fig. 2; see also Fig. S2 in the supplemental material) were present, representing 11 families. Four isolates had 97% or less maximum identity to their closest relative in GenBank. The snails had in common four actinomycete genera: Streptomyces, Microbacterium, Gordonia, and Brevibacterium.
FIG. 2.
Neighbor-joining phylogenetic tree of 16S rRNA gene sequences from uncultivated and cultivated actinobacteria. Dots indicate cultivated isolates, while capital letters indicate sequences obtained from the body of C. pulicarius (CPB), C. rolani (CRB), and C. tribblei (CTB) and hepatopancreas of C. tribblei (CTHP) and C. pulicarius (CPHP). Colors indicate source snails: black (C. tribblei), blue (C. pulicarius), and red (C. rolani). Reference strains from GenBank are marked by an asterisk. f and p indicate branches that were also found using the Fitch-Margoliash or maximum parsimony methods, respectively. The numbers at the nodes are percentages indicating the levels of bootstrap support, based on a neighbor-joining analysis of 1,000 resampled data sets. Only values of >50% are shown. Bar, 0.01 substitutions per nucleotide position. All actinobacteria identified fall into the actinomycete group.
The snail-associated actinomycete community was then examined using a non-culture-based approach that entailed PCR amplification and cloning of 16S rRNA genes obtained from cone snail tissues. An initial amplification of 16S rRNA genes using the bacterial universal primer pair 8-27F and 1492r yielded only a limited number of actinomycete sequences, while the majority of the clone libraries (480 clones total) were dominated by clones closely related to common marine invertebrate-associated bacteria, such as Vibrio, Photobacterium, and Staphylococcus species. By using a set of specific actinobacteria primers, 243F (19) and AB1165R (27), in a nested PCR approach, the number of actinobacteria sequences in the clone libraries (480 clones total) increased substantially and accounted for 97% of the clone library bacterial sequences.
All of the actinobacterial sequences belonged to the Actinomycetales order. Relatively complete data were obtained for the body and the hepatopancreas, while more limited data were available for the venom ducts. Thus, hepatopancreas (C. pulicarius and C. tribblei) and body (C. pulicarius, C. tribblei, and C. rolani) analyses are presented here. One 96-well plate of clones was analyzed for each tissue type, resulting in 103 unique actinomycete sequences. Rarefaction analysis using a 97% and 99% identity threshold level (see Fig. S1 in the supplemental material) showed that the libraries were appropriately sampled at the 97% level. At the 99% level more clones would need to be sequenced to provide a better bacterial diversity representation.
The molecular analysis of the 16S rRNA gene actinobacteria clone libraries from the body of C. pulicarius, C. rolani, and C. tribblei and the hepatopancreas from C. pulicarius and C. tribblei showed representatives of 10 actinomycete families (Fig. 2; see also Fig. S2 in the supplemental material). The body clone libraries contain members of the Streptomycetaceae (C. tribblei), Sporichthyaceae (C. rolani), Microbacteriaceae (C. pulicarius), Nocardioidaceae (C. pulicarius), Pseudonocardiaceae (C. rolani), Dietziaceae (C. pulicarius, C. rolani, and C. tribblei), Propionibacteriaceae (C. tribblei), and Corynebacteriaceae (C. pulicarius and C. tribblei). The hepatopancreas libraries of C. tribblei and C. pulicarius contained members of Streptomycetaceae (C. tribblei), Micrococcaceae (C. tribblei), Intrasporangiaceae (C. tribblei), Nocardioidaceae (C. pulicarius and C. tribblei), Dietziaceae (C. tribblei), and Corynebacteriaceae (C. pulicarius). No clone sequences were identical to those of isolated actinomycetes, although there were representatives of many groups.
Although the purpose of this study was to use the microhabitat approach to obtain bioactive actinomycetes for drug discovery, a few other features were evident in the uncultivated analysis. It was observed that sequences of clones from given snails and tissues appeared to cluster according to their snail of origin. A statistical analysis of these clusters using Unifrac showed that the observed clustering was not random and was statistically suggestive (P ≤ 0.06). However, the sample size was not large enough to provide statistically significant results (P ≤ 0.05).
During the microbiological and molecular analyses of the snails that yielded over a thousand 16S rRNA gene sequences, only actinomycetes were observed and no other actinobacteria. Since the specific primers used were aimed at the actinobacteria group, this indicates that the samples are strongly dominated by the actinomycetes group.
The reasons for the association between cone snails and actinomycetes remain unclear. From the clone library phylogenetic analysis, it was observed that each snail and each tissue contain members of similar subfamilies. However, at the species level, differences are evident. These differences are statistically suggestive of true differences between the groups. It is possible that the bacterial groups common to all snails represent a core of symbionts, performing similar tasks required by all host snails, while other bacterial groups specific to their host snail, i.e., Streptomycetaceae for C. tribblei and Pseudonocardiaceae for C. rolani, have been selected to fit the biology of this host. In order to test this hypothesis, more snails should be sampled, including multiple replicates of the same species.
Results obtained from cultivation compare favorably to those from other marine invertebrates, such as sponges, which are often considered microbial fermenters (18). For example, Zhang et al. (47) identified 63 different actinomycete morphotypes isolated from five different sponges, with Streptomyces accounting for 90% of the morphotypes. In this study, of the 96 actinomycete morphotypes identified and 57 actinomycetes purified, Streptomyces accounted for ca. 38% of the morphotypes.
The analysis of cone snail-associated actinomycete communities from clone libraries and cultivation yielded a total of 12 actinomycete families. This diversity rivals and/or surpasses that seen in other marine invertebrates, such as sponges (45). The actinomycetes obtained are novel and, based on 16S rRNA gene identity level, include three new Streptomyces species.
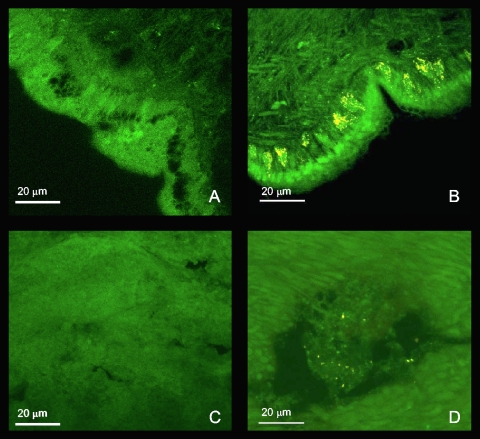
FISH analysis of C. pulicarius.
FISH analysis using the actinobacteria probe HGC69a and a scrambled probe as a control revealed that living actinobacteria are present in both the hepatopancreas and the body tissues (Fig. 3). In the hepatopancreas, actinobacteria are present in moderate numbers with no clear distribution pattern. In the body, actinobacteria formed dense clusters near the body wall, inside large oval cells, while other bacteria could be seen in other parts of the tissue. These large oval cells open toward the outside and are similar in shape and position to mucocytes that produce the mucus lubricating the head and foot of the mollusk (Y. Kantor, personal communication). This analysis confirmed that actinobacteria are associated with the snail and are actively dividing. Snails have been known to use mucus to cover their shells (20), and mucus has been reported to contain bioactive compounds (23), indicating one of many potential roles for these bacteria. Moreover, since actinobacterial sequences in the samples were derived solely from actinomycetales, it is probable that these identified bacteria fall within this group.
FIG. 3.
Epifluorescence micrograph section of Conus pulicarius tissue visualized by FISH. (A) Body region hybridized with fluorescein-labeled non-EUB338. (B) Body region hybridized with fluorescein-labeled EUB338 and Cy3-labeled HGC69a. (C) Hepatopancreas region hybridized with fluorescein-labeled non-EUB338. (D) Hepatopancreas region hybridized with fluorescein-labeled EUB338 and Cy3-labeled HGC69a.
Dorsal root ganglion assay.
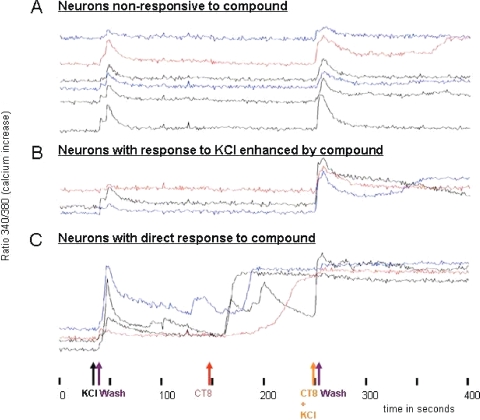
This neurological assay consists of mouse neurons from DRG (26, 29). Since the DRG contains many types of neurons, responses on many different channel and receptor types can be observed in real time by microscopy. In this particular assay, freshly obtained DRG cells were treated with a fluorophore that binds to cellular calcium. Using fluorescence microscopy, the calcium response of DRG cells to chemicals and extracts is followed in real time. The protocol is optimized to observe effects that are immediate and are highly biased toward direct interactions with ion channels or with receptors that are closely coupled to ion channels. Using this DRG assay, we tested a total of 87 extracts from cone snail isolates (actinomycetes and nonactinomycetes), which yielded four leads, CT8 (Fig. 4), CP32, CP41, and CR60. Although the active compounds remain to be elucidated, results from the DRG assay suggest that the compounds act directly on channels or receptors. The data support either a decrease in K+ channel activation, and/or an increase in Na+ channel activation. The leads are potent molecules on which structural elucidations are ongoing and for which individual active high-performance liquid chromatography fractions containing small bioactive molecules have been obtained. In addition, six of the isolates exhibited antifungal activity during isolation. Together, these results indicate that the cone snail microhabitats are suitable sources for drug discovery studies.
FIG. 4.
Dorsal root ganglion assay of CT8 extract. The initial KCl injection and first wash provided the control. Once the cells return to their original state, a series of injections follow, starting with injection of the CT8 extract. In panel C, it can be seen that some cells respond with an influx of calcium directly upon injection of the extract, while others attenuate the response until a second KCl injection (B and C). Other samples show no response to the compound (A).
Significance.
The need for new bioactive compounds has led to the pursuit of actinomycetes from novel habitats, including remote tropical forests on land (2) and coral reefs or benthic sediments at sea (32). It has been increasingly common to examine animals (7, 16, 30) or plants (40) for symbiotic actinomycetes and to study the biology of these relationships. The microhabitat approach can bridge both drug discovery and microbial ecology studies. By looking at individual organs and tissue types as independent samples, it is possible to simultaneously explore the natural product components while studying details of the host-symbiont relationship.
In this study, a relatively large number of novel actinomycetes were obtained by using the microhabitat approach. From a drug discovery point of view, the novel actinomycete strains are attractive, as they are likely to contain new compounds. Additionally, the remarkable accessibility to the snail-associated actinomycete community is quite unique and will help circumvent the supply issue which so often plagues natural products. If this diversity truly scales with the 20,000 species in the Conoidean superfamily, this diversity is potentially quite large. At least some of these actinomycetes are actively living and dividing within the snail, as evidenced by FISH.
Cone snails have shown to be an unexpectedly rich and diverse reservoir for novel actinomycetes. The promising bioactivity from snail-associated actinomycetes and important rate of cultivation make cone snails animals of interest for drug discovery, while their biology may make them a good model for studying symbiosis.
Supplementary Material
Acknowledgments
We thank the government of the Philippines and the people of Cebu for opportunities to work on these specimens. All field work was performed with appropriate regional and national permits (to G. Concepcion and B. Olivera), including prior informed consent permission from local communities. Yuri I. Kantor (Severtzov Institute of Ecology and Evolution of the Russian Academy of Sciences, Moscow, Russia) provided key information regarding snail anatomy. Mary Anne Astilla and Jasmin Jao (University of the Phillippines Marine Science Institute) helped in processing of samples. We thank Margo Haygood for her comments.
This work was supported by a Synergy Grant from the University of Utah, NIH GM071425, and NIH GM48677. NIH ICBG U01TW008163 supported the analysis of samples.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balick, M. J., E. Elisabethsky, and S. A. Laird. 1996. Medicinal resources of the tropical forest: biodiversity and its importance to human health. Columbia University Press, New York, NY.
- 3.Birkby, K. M., and T. F. Preece. 1987. The ligules of green leaves of cock's-foot grass, Dactylis glomerata L., are a micro-habitat for bacteria and fungi. J. Appl. Microbiol. 63:505. [Google Scholar]
- 4.Blunt, J. W., B. R. Copp, W. P. Hu, M. H. Munro, P. T. Northcote, and M. R. Prinsep. 2009. Marine natural products. Nat. Prod. Rep. 26:170-244. [DOI] [PubMed] [Google Scholar]
- 5.Chun, J. 1995. Computer-assisted classification and identification of actinomycetes. Ph.D. thesis. University of Maryland, College Park.
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currie, C. R., M. Poulsen, J. Mendenhall, J. J. Boomsma, and J. Billen. 2006. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science 311:81-83. [DOI] [PubMed] [Google Scholar]
- 8.Currie, C. R., J. A. Scott, R. C. Summerbell, and D. Malloch. 1999. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature 398:701-704. [Google Scholar]
- 9.Davidson, S. K., S. W. Allen, G. E. Lim, C. M. Anderson, and M. G. Haygood. 2001. Evidence for the biosynthesis of bryostatins by the bacterial symbiont “Candidatus Endobugula sertula” of the bryozoan Bugula neritina. Appl. Environ. Microbiol. 67:4531-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rosa, S., A. Milone, A. Kujumgiev, K. Stefanov, I. Nechev, and S. Popov. 2000. Metabolites from a marine bacterium Pseudomonas/Alteromonas, associated with the sponge Dysidea fragilis. Comp. Biochem. Physiol. B 126:391-396. [DOI] [PubMed] [Google Scholar]
- 11.Duda, T. F., W. Korn, A. J. Kohn, and S. R. Palumbi. 2001. Origins of diverse feeding ecologies within Conus, a genus of venomous marine gastropods. Biol. J. Linnean Soc. 73:391-409. [Google Scholar]
- 12.Espiritu, D. J. D., M. Watkins, V. Dia-Monje, G. E. Cartier, L. J. Cruz, and B. M. Olivera. 2001. Venomous cone snails: molecular phylogeny and the generation of toxin diversity. Toxicon 39:1899-1916. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1993. PHYLIP (Phylogenetic Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 14.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees: a method based on mutation distances as estimated from cytochrome c sequences is of general applicability. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 15.Gotelli, N. J., and G. L. Entsminger. 2006. EcoSim: null models software for ecology, version 7. Acquired Intelligence, Inc., and Kesey-Bear, Jericho, VT. http://garyentsminger.com/ecosim.htm.
- 16.Haeder, S., R. Wirth, H. Herz, and D. Spiteller. 2009. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 106:4742-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentschel, U., K. M. Usher, and M. W. Taylor. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167-177. [DOI] [PubMed] [Google Scholar]
- 19.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes, S. P., C. J. Sturgess, A. Cherill, and M. S. Davies. 2001. Shell wiping in Clliostoma zizyphinum: the use of pedal mucus as aprovendering agent and its contribution to daily energetic requirements. Mar. Ecol. Prog. Ser. 212:171-181. [Google Scholar]
- 21.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 22.Kaltenpoth, M., W. Göttler, G. Herzner, and E. Strohm. 2005. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15:475-479. [DOI] [PubMed] [Google Scholar]
- 23.Kelley, W. P., A. M. Wolters, J. T. Sack, R. A. Jockusch, J. C. Jurchen, E. R. Williams, J. V. Sweedler, and W. F. Gilly. 2003. Characterization of a novel gastropod toxin (6-bromo-2-mercaptotryptamine) that inhibits shaker K channel activity. J. Biol. Chem. 278:34934-34942. [DOI] [PubMed] [Google Scholar]
- 24.Kluge, A. G., and F. S. Farris. 1969. Quantitative phyletics and the evolution of annurans. Syst. Zool. 18:1-32. [Google Scholar]
- 25.Kohn, A. J. 1990. Tempo and mode of evolution in Conidae. Malacologia 32:55-67. [Google Scholar]
- 26.Light, A. R., R. W. Hughen, J. Zhang, J. Rainier, Z. Liu, and J. Lee. 2008. Dorsal root ganglion neurons innervating skeletal muscle respond to physiological combinations of protons, ATP, and lactate mediated by ASIC, P2X, and TRPV1. J. Neurophysiol. 100:1184-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac: an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludemann, H., and R. Conrad. 2000. Molecular retrieval of large 16S rRNA gene fragments from an Italian rice paddy soil affiliated with the class Actinobacteria. Syst. Appl. Microbiol. 23:582-584. [DOI] [PubMed] [Google Scholar]
- 29.Martin, D. J., D. McClelland, M. B. Herd, K. G. Sutton, M. D. Hall, K. Lee, R. D. Pinnock, and R. H. Scott. 2002. Gabapentin-mediated inhibition of voltage-activated Ca2+ channel currents in cultured sensory neurones is dependent on culture conditions and channel subunit expression. Neuropharmacology 42:353-366. [DOI] [PubMed] [Google Scholar]
- 30.Mueller, U. G., D. Dash, C. Rabeling, and A. Rodrigues. 2008. Coevolution between attine ants and actinomycete bacteria: a reevaluation. Evolution 62:2894-2912. [DOI] [PubMed] [Google Scholar]
- 31.Olivera, B. M., M. Quik, M. Vincler, and J. M. McIntosh. 2008. Subtype-selective conopeptides targeted to nicotinic receptors: concerted discovery and biomedical applications. Channels 2:143-152. [DOI] [PubMed] [Google Scholar]
- 32.Proksch, P., R. A. Edrada, and R. Ebel. 2002. Drugs from the seas: current status and microbiological implications. Appl. Environ. Microbiol. 59:125-134. [DOI] [PubMed] [Google Scholar]
- 33.Reysenbach, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rockel, D., W. Korn, and A. J. Kohn. 1995. Manual of the living Conidae, vol. I, Indo-Pacific. Christa Hemmen Verlag, Wiesbaden, Germany.
- 35.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K. H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, E. W., J. T. Nelson, D. A. Rasko, S. Sudek, J. A. Eisen, M. G. Haygood, and J. Ravel. 2005. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102:7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott, J. J., D.-C. Oh, M. C. Yuceer, K. D. Klepzig, J. Clardy, and C. R. Currie. 2008. Bacterial protection of beetle-fungus mutualism. Science 322:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sellstedt, A., K. Huss-Danell, and A.-S. Ahlqvist. 1986. Nitrogen fixation and biomass production in symbioses between Alnus incana and Frankia strains with different hydrogen metabolism. Physiol. Plant. 66:99-107. [Google Scholar]
- 40.Strobel, G., and B. Daisy. 2003. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 67:491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudek, S., N. B. Lopanik, L. E. Waggoner, M. Hildebrand, C. Anderson, H. Liu, A. Patel, D. H. Sherman, and M. G. Haygood. 2007. Identification of the putative bryostatin polyketide synthase gene cluster from “Candidatus Endobugula sertula,” the uncultivated microbial symbiont of the marine bryozoan Bugula neritina. J. Nat. Prod. 70:67-74. [DOI] [PubMed] [Google Scholar]
- 42.Völksch, B., M. Ullrich, and W. Fritsche. 1992. Identification and population dynamics of bacteria in leaf spots of soybean. Microb. Ecol. 24:305-311. [DOI] [PubMed] [Google Scholar]
- 43.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xin, Y., J. Huang, M. Deng, and W. Zhang. 2008. Culture-independent nested PCR method reveals high diversity of actinobacteria associated with the marine sponges Hymeniacidon perleve and Spongia sp. Antonie van Leeuwenhoek 94:533-542. [DOI] [PubMed] [Google Scholar]
- 46.Yang, J. C., R. Madupu, A. S. Durkin, N. A. Ekborg, C. S. Pedamallu, J. B. Hostetler, D. Radune, B. S. Toms, B. Henrissat, P. M. Coutinho, S. Schwarz, L. Field, A. E. Trindade-Silva, C. A. Soares, S. Elshahawi, A. Hanora, E. W. Schmidt, M. G. Haygood, J. Posfai, J. Benner, C. Madinger, J. Nove, B. Anton, K. Chaudhary, J. Foster, A. Holman, S. Kumar, P. A. Lessard, Y. A. Luyten, B. Slatko, N. Wood, B. Wu, M. Teplitski, J. D. Mougous, N. Ward, J. A. Eisen, J. H. Badger, and D. L. Distel. 2009. The complete genome of Teredinibacter turnerae T7901: an intracellular endosymbiont of marine wood-boring bivalves (shipworms). PLoS ONE 4:e6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, H., W. Zhang, Y. Jin, M. Jin, and X. Yu. 2008. A comparative study on the phylogenetic diversity of culturable actinobacteria isolated from five marine sponge species. Antonie van Leeuwenhoek 93:241-248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.