Abstract
A conjugative plasmid from the catheter-associated urinary tract infection strain Escherichia coli MS2027 was sequenced and annotated. This 42,644-bp plasmid, designated pMAS2027, contains 58 putative genes and is most closely related to plasmids belonging to incompatibility group X (IncX1). Plasmid pMAS2027 encodes two important virulence factors: type 3 fimbriae and a type IV secretion (T4S) system. Type 3 fimbriae, recently found to be functionally expressed in E. coli, played an important role in biofilm formation. Biofilm formation by E. coli MS2027 was specifically due to expression of type 3 fimbriae and not the T4S system. The T4S system, however, accounted for the conjugative ability of pMAS2027 and enabled a non-biofilm-forming strain to grow as part of a mixed biofilm following acquisition of this plasmid. Thus, the importance of conjugation as a mechanism to spread biofilm determinants was demonstrated. Conjugation may represent an important mechanism by which type 3 fimbria genes are transferred among the Enterobacteriaceae that cause device-related infections in nosocomial settings.
Bacterial biofilms are complex communities of bacterial cells living in close association with a surface (17). Bacterial cells in these protected environments are often resistant to multiple factors, including antimicrobials, changes in the pH, oxygen radicals, and host immune defenses (19, 38). Biofilm formation is a property of many bacterial species, and a range of molecular mechanisms that facilitate this process have been described (2, 3, 11, 14, 16, 29, 33, 34). Often, the ability to form a biofilm is dependent on the production of adhesins on the bacterial cell surface. In Escherichia coli, biofilm formation is enhanced by the production of certain types of fimbriae (e.g., type 1 fimbriae, type 3 fimbriae, F1C, F9, curli, and conjugative pili) (14, 23, 25, 29, 33, 39, 46), cell surface adhesins (e.g., autotransporter proteins such as antigen 43, AidA, TibA, EhaA, and UpaG) (21, 34, 35, 40, 43), and flagella (22, 45).
The close proximity of bacterial cells in biofilms creates an environment conducive for the exchange of genetic material. Indeed, plasmid-mediated conjugation in monospecific and mixed E. coli biofilms has been demonstrated (6, 18, 24, 31). The F plasmid represents the best-characterized conjugative system for biofilm formation by E. coli. The F pilus mediates adhesion to abiotic surfaces and stabilizes the biofilm structure through cell-cell interactions (16, 30). Many other conjugative plasmids also contribute directly to biofilm formation upon derepression of the conjugative function (16).
One example of a conjugative system employed by gram-negative Enterobacteriaceae is the type 4 secretion (T4S) system. The T4S system is a multisubunit structure that spans the cell envelope and contains a secretion channel often linked to a pilus or other surface filament or protein (8). The Agrobacterium tumefaciens VirB-VirD4 system is the archetypical T4S system and is encoded by 11 genes in the virB operon and one gene (virD4) in the virD operon (7, 8). Genes with strong homology to genes in the virB operon have also been identified on other conjugative plasmids. For example, the pilX1 to pilX11 genes on the E. coli R6K IncX plasmid and the virB1 to virB11 genes are highly conserved at the nucleotide level (28).
We recently described identification and characterization of the mrk genes encoding type 3 fimbriae in a uropathogenic strain of E. coli isolated from a patient with a nosocomial catheter-associated urinary tract infection (CAUTI) (29). The mrk genes were located on a conjugative plasmid (pMAS2027) and were strongly associated with biofilm formation. In this study we determined the entire sequence of plasmid pMAS2027 and revealed the presence of conjugative transfer genes homologous to the pilX1 to pilX11 genes of E. coli R6K (in addition to the mrk genes). We show here that biofilm formation is driven primarily by type 3 fimbriae and that the T4S apparatus is unable to mediate biofilm growth in the absence of the mrk genes. Finally, we demonstrate that conjugative transfer of pMAS2027 within a mixed biofilm confers biofilm formation properties on recipient cells due to acquisition of the type 3 fimbria-encoding mrk genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are described in Table 1. Cells were routinely grown at 37°C using solid or liquid Luria-Bertani (LB) medium supplemented with appropriate antibiotics, unless otherwise stated. M9 minimal medium supplemented with 0.2% glucose and synthetic urine were prepared as previously described (26, 32).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5-α | Commercial laboratory E. coli cloning strain | Promega, United States |
| MS427 | K-12 MG1655flu | 21 |
| MS661 | MS427recA::tet | 21 |
| MS2027 | E. coli CAUTI isolate | 29 |
| MS2091 | MS2027 attB::bla-PA1/04/03-gfpmut3b*- T0;Gfp | This study |
| MS2103 | MS2027(pMAS2027pilX::kan) | This study |
| MS2181 | MS2027(pMAS2027mrk::cam) | This study |
| MS2183 | MS2027(pMAS2027mrk::cam pilX::kan) | This study |
| MS2199 | MS661 harboring pAR163 | This study |
| MS2362 | MS2027(pMAS2027orf27::cam) | This study |
| MS2517 | MS2027(pMAS2027mrk::gfp-kan) | This study |
| MS2518 | MS2027(pMAS2027pilX::gfp-kan) | This study |
| MS2519 | MS2027(pMAS2027orf27::gfp-kan) | This study |
| Plasmids | ||
| pAR163 | dsRED2.T3-containing plasmid | 34 |
| pCO13 | Kanamycin resistance gene inserted into HindIII-digested pKEN2 | This study |
| pKD3 | Deletion mutant template plasmid (cam) | 12 |
| pKD4 | Deletion mutant template plasmid (kan) | 12 |
| pKEN2 | gfp-containing plasmid | 10 |
| pMAS2027 | mrk-containing conjugative plasmid isolated from MS2027 | This study |
| pTW10 | pGEM-T Easy with modified kan cassette without HindIII | 43 |
DNA manipulations and genetic techniques.
Plasmid DNA was isolated using QIAprep Spin Miniprep or Midiprep kits (Qiagen, Australia). Restriction endonucleases were used according to the manufacturer's specifications (New England Biolabs). PCR was performed using Taq polymerase according to the manufacturer's instructions (New England Biolabs). DNA sequencing was performed by the Australian Genome Research Facility. E. coli strain MS2027 was genetically marked by insertion of gfpmut3b* into the chromosomal attachment site of bacteriophage λ (attB) as previously described (34), which resulted in E. coli MS2091. Mutation of pMAS2027 was performed by performing λ-red-mediated homologous recombination as previously described (12), except that the gfp-kan mutants were constructed using pCO13 as the template DNA. Primers used in this study are described in Table 2. All deletion mutants were confirmed by PCR and subsequent DNA sequencing.
TABLE 2.
Primers used in this study
| Primer | Descriptiona | Sequence (5′-3′) |
|---|---|---|
| 1293 | 50-bp overhang mrk knockout forward primer | TCTTCTCTCTGCAGCAATGGCAACCGCGTTTTTTGGCATGACTGCTGCCCGTGTAGGCTGGAGCTGCTTCG |
| 1294 | 50-bp overhang mrk knockout reverse primer | GGTGTGAGCGGGATAGTTGTCTGAGTCACAGGCAGTTTCCTCTTCACCAGCATATGAATATCCTCCTTAG |
| 1295 | Screening forward primer (mrk) | GGCAGCATAACCGAACAAAT |
| 1296 | Screening reverse primer (mrk) | TAAATTTTCTGCGGCAAACC |
| 1297 | 50-bp overhang pilX knockout forward primer | GTGGAATATTTATGTTATCTACTTCTACTTTTCTTGCGCTTGCCATGCAATGTGTAGGCTGGAGCTGCTTCG |
| 1298 | 50-bp overhang pilX knockout reverse primer | TTTAATACTCATGATAATATCAATATTACTCAGTACACGTCTTAAAATGACATATGAATATCCTCCTTAG |
| 1299 | Screening forward primer (pilX) | CCGCAACGCAATGTACTCTA |
| 1300 | Screening reverse primer (pilX) | CGTTTTTCCGTTCAGGAAGT |
| 1301 | 50-bp overhang Cmr tag forward primer | AACGGGAAAACGAAAACAACAGATGAAAAGACACATACATTCAATACAGCGTGTAGGCTGGAGCTGCTTCG |
| 1302 | 50-bp overhang Cmr tag reverse primer | TTGCACTGGAGTTGATAATGGCCTCAATGCACAAATTGAGGTAAATTTATGTGTAGGCTGGAGCTGCTTCG |
| 1303 | Screening forward primer (tagging) | AGCAGTCCTGCAACCTAAGC |
| 1304 | Screening reverse primer (tagging) | TAATGGGCGTCCTAAACTGG |
| 1305 | GFP-kan mrk knockout forward primer | TCTTCTCTCTGCAGCAATGGCAACCGCGTTTTTTGGCATGACTGCTGCCCTAGGGCGAATTGACATTGTG |
| 1306 | GFP-kan mrk knockout reverse primer | GGTGTGAGCGGGATAGTTGTCTGAGTCACAGGCAGTTTCCTCTTCACCAGTGGACCAGTTGGTGATTTTG |
| 1307 | GFP-kan pilX knockout forward primer | GTGGAATATTTATGTTATCTACTTCTACTTTTCTTGCGCTTGCCATGCAATTAGGGCGAATTGACATTGTG |
| 1308 | GFP-kan pilX knockout reverse primer | TTTAATACTCATGATAATATCAATATTACTCAGTACACGTCTTAAAATGATGGACCAGTTGGTGATTTTG |
| 1309 | GFP-kan tagging forward primer | AACGGGAAAACGAAAACAACAGATGAAAAGACACATACATTCAATACAGCTAGGGCGAATTGACATTGTG |
| 1310 | GFP-kan tagging reverse primer | TTGCACTGGAGTTGATAATGGCCTCAATGCACAAATTGAGGTAAATTTATTGGACCAGTTGGTGATTTTG |
| pCO_F | pCO13 internal screening forward primer (kan region) | CGAAAGATCCCAACGAAAAG |
| pCO13-R2 | pCO13 internal screening reverse primer (GFP region) | TCGATAGATTGTCGCACCTG |
| pCO13-F3 | pCO13 external screening forward primer (GFP region) | CAGCAACACCTTCTTCACGA |
| pCO13-R3 | pCO13 external screening reverse primer (kan region) | TCACCTTCACCCTCTCCACT |
GFP, green fluorescent protein.
Sequencing and annotation of pMAS2027.
Both strands of plasmid pMAS2027 were sequenced by employing a primer walking strategy. Primers were designed so that they read progressively outward from the mrkA and mrkF genes, until a complete, overlapping sequence was obtained. The DNA sequence was assembled using Contig Express (Invitrogen), and annotation was performed by using Vector NTi (Invitrogen) and Artemis (version 10) (5). BLASTn and BLASTp searches were performed using the National Center for Biotechnology Information website (1).
Plasmid mobilization study.
Plasmid mobility was monitored by using filter paper bacterial conjugation as previously described (44). An overnight culture of the donor strain was concentrated 10-fold and left to stand at 37°C to allow growth of the sex pili. The donor and recipient (MS661) were then mixed at a ratio of 1:10 and then incubated on filter paper for 3 to 4 h. The filter paper mixture was then resuspended in LB medium, and dilutions were plated on LB agar containing tetracycline and kanamycin to select for transconjugants. Plates were incubated overnight at 37°C, and the plasmid conjugation efficiency was calculated by determining the ratio of transconjugant colonies to donor colonies.
Biofilm study.
Biofilm formation on polyvinyl chloride (PVC) surfaces was monitored by using 96-well microtiter plates (Falcon) essentially as previously described (33). Briefly, cells were grown for 24 h in urine or M9 minimal medium (containing 0.2% glucose) at either 28°C or 37°C (statically or with shaking), washed to remove unbound cells, and stained with crystal violet. Quantification of bound cells was performed by addition of acetone-ethanol (20:80) and measurement of the dissolved crystal violet using the optical density at 595 nm. Flow chamber biofilm experiments were performed as previously described (21). Briefly, biofilms were allowed to form on glass surfaces in a multichannel flow system that permitted online monitoring of community structures. Flow cells were inoculated with overnight cultures that were standardized using the optical density at 600 nm and were prepared in M9 medium. Biofilm development was monitored by confocal scanning laser microscopy at 16 to 24 h after inoculation. All experiments were performed in triplicate. In the flow chamber experiments with mixed biofilms, the donor and recipient strains were incubated for 6 h before they were subjected to the medium flow. Biofilm development was monitored by confocal scanning laser microscopy at 16 and 40 h after inoculation. All experiments were performed in triplicate. Both single-strain biofilm and mixed-biofilm flow chamber experiments were done using M9 minimal medium supplemented with 100 μg/μl ampicillin.
SEM.
Scanning electron microscopy (SEM) was performed essentially as previously described (29). Cells were grown as described above for the biofilm study on PVC surfaces, except that the experiment was performed using a 12-well microtiter plate (Griener bio-one) with a polystyrene disk placed at the bottom. The disk was fixed with 3% glutaraldehyde in 0.1 M cacodylate buffer and postfixed with 1% osmium tetroxide in 0.1 M cacodylate buffer. The sample was then infiltrated with glycerol and frozen in liquid nitrogen. The sample was freeze substituted in 100% ethanol containing a molecular sieve and incubated at −80°C for 10 h; then the temperature was increased from −80°C to −20°C over a 10-h period, and the sample was critical point dried. The sample was then mounted on carbon tabs and sputter coated with platinum at 15 mA for 120 s.
Statistical analysis.
Differences in plasmid conjugation efficiency between pMAS2027mrk::kan and pMAS2027orf24::cam were determined by using a t test (with two samples assuming unequal variances). Differences in biofilm formation were analyzed using the analysis of variance single-factor test, and values were compared with values obtained for E. coli MS2027 (wild type) or E. coli MS2362 (MS2027::cam) (Minitab 15 statistical software).
Nucleotide sequence accession number.
The pMAS2027 plasmid sequence has been deposited in the GenBank database under accession number FJ666132.
RESULTS
Complete nucleotide sequence of pMAS2027.
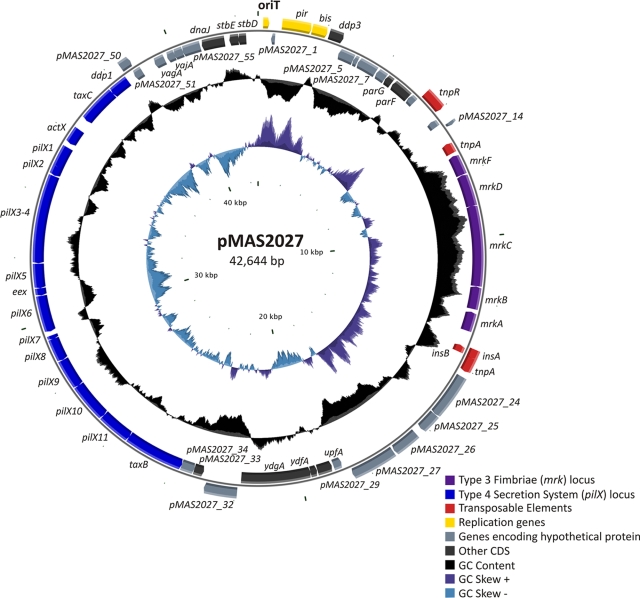
Plasmid pMAS2027 was determined to be a 42,644-bp circular plasmid with a G+C content of 43.43% (Fig. 1). Bioinformatic analysis predicted the presence of 58 open reading frames (ORFs), which represent 88.83% of the total plasmid DNA. Plasmid pMAS2027 is most closely related to the IncX group of plasmids, a narrow-host-range family of conjugative plasmids commonly found in the family Enterobacteriaceae (13). The backbone structure of pMAS2027 contains genes associated with plasmid stability and partitioning, as well as a 14,628-kb region comprising genes predicted to encode a T4S system (Fig. 1). This region includes the pilX1 to pilX11 genes analogous to the genes described for R6K (28) and most likely involved in conjugative DNA transfer. pMAS2027 also contains the mrkABCDF genes (encoding type 3 fimbriae). The mrk genes are located on a 5,536-bp segment and have an overall G+C content of 56.6%. Genes predicted to encode proteins associated with transposition are located both upstream (tnpR and tnpA) and downstream (insA, insB, and tnpA) of the mrk cluster, suggesting that the mrk genes are located on a mobile genetic element. A summary of each ORF of pMAS2027, including the predicted function of each encoded protein, is shown in Table 1S in the supplemental material. No genes were predicted to encode proteins associated with antibiotic resistance. The remainder of our study focused on the relative contributions of the mrk and pilX1 to pilX11 genes to biofilm formation and conjugative transfer.
FIG. 1.
Circular diagram of plasmid pMAS2027. The outer circle indicates (to scale) the genetic organization of ORFs within the plasmid. The direction of transcription of each ORF is indicated. The middle circle indicates the G+C content, and the inner circle indicates the GC skew. Genes are color coded as indicated. The image was constructed using cgview (37). CDS, coding sequences.
Bioinformatic analysis of the conjugative transfer locus of plasmid pMAS2027.
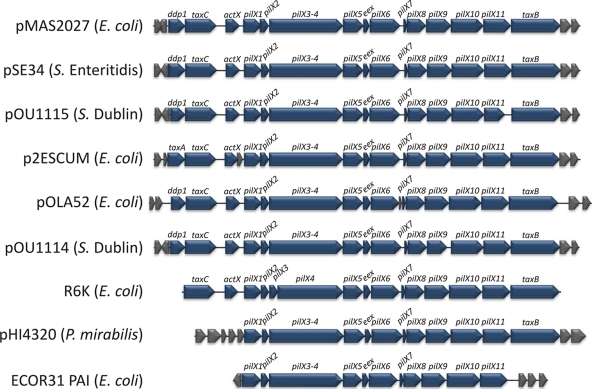
The pilX1 to pilX11 genes of pMAS2027 are 43% identical at the nucleotide sequence level to the virB1 to virB11 genes of the archetypical transferred DNA transfer system of the plant pathogen A. tumefaciens (42). These genes have also been identified on conjugative plasmids isolated from other Enterobacteriaceae, including Salmonella enterica serovar Enteritidis, S. enterica serovar Dublin, Proteus mirabilis, and other E. coli strains (Fig. 2). The pilX1 to pilX11 regions of three plasmids (pSE34 from Salmonella serovar Enteritidis, pOU1115 from Salmonella serovar Dublin, and p2ESCUM from E. coli) share the strongest overall sequence identity with the pilX1 to pilX11 genes of pMAS2027 (Table 3). The sequences of two other plasmids (pOLA52 from E. coli and pOU1114 from Salmonella serovar Dublin) were also highly conserved in this region, except for the eex and pilX6 genes. The nucleotide sequences of the pilX1 to pilX11 genes of pMAS2027, R6K (an archetypical E. coli IncX plasmid), and pHI4320 (P. mirabilis) were more divergent (Table 3). Despite this, the genetic organization of the pilX1 to pilX11 (and adjacent) genes on pMAS2027 and a number of other conjugative plasmids is highly conserved (Fig. 2).
FIG. 2.
Physical map comparing the genes encoding the T4S system (and adjacent genes) in plasmid pMAS2027 to the corresponding regions in plasmids pSE34 (Salmonella serovar Enteritidis; EU219533), pOU1115 (Salmonella serovar Dublin; DQ115388), p2ESCUM (E. coli; CU928149), pOLA52 (E. coli; EU370913), pOU1114 (Salmonella serovar Dublin; DQ115387), R6K (E. coli; AJ006342), and pHI4320 (P. mirabilis; AM942760) and the pathogenicity island (PAI) in E. coli ECOR31 (AY233333). The genes encoding the T4S system are blue and include ddp1 (taxA), taxC, actX, pilX1 to pilX11, and taxB. Adjacent genes (gray) encode hypothetical proteins with unknown functions. No sequence information is available for genes outside the T4S system gene cluster in E. coli R6K. The direction of transcription of each gene is indicated.
TABLE 3.
Comparison of the E. coli plasmid pMAS2027 T4S system protein and nucleotide sequences with the corresponding sequences from other T4S system conjugative plasmids
| T4S system protein | % Amino acid identity (% nucleotide sequence conservation) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| pSE34 | pOU1115 | p2ESCUM | pOLA52 | pOU1114 | R6K | pHI4320 | ECOR31 pathogenicity island | pTib | |
| Ddp1 | 97 (98) | 87 (88) | 85 (89) | 88 (88) | 88 (88) | NAc | NA | NA | NA |
| TaxC | 97 (98) | 97 (97) | 97 (97) | 98 (98) | 97 (98) | 77 (71) | NA | NA | NA |
| ActX | 99 (100) | 99 (100) | 29 (48) | 99 (99) | 99 (100) | 25 (47) | NA | NA | NA |
| PilX1/VirB1 | 93 (93) | 94 (94) | 93 (93) | 94 (94) | 93 (93) | 62 (64) | 44 (53) | 41 (47) | 22 (42) |
| PilX2/VirB2 | 89 (90) | 89 (89) | 91 (93) | 89 (89) | 89 (90) | 76 (68) | 29 (42) | 20 (42) | 13 (45) |
| PilX3- PilX4/VirB3- VirB4a | 99 (99) | 100 (99) | 99 (99) | 100 (99) | 99 (99) | 80 (74) | 63 (63) | 42 (53) | 24 (47) |
| PilX5/VirB5 | 100 (100) | 98 (98) | 98 (98) | 93 (95) | 100 (100) | 63 (67) | 48 (60) | 27 (49) | 11 (46) |
| Eex | 97 (100) | 97 (100) | 97 (100) | 35 (45) | 49 (56) | 28 (46) | 43 (57) | 33 (48) | NA |
| PilX6/VirB6 | 99 (100) | 99 (99) | 98 (99) | 68 (69) | 81 (79) | 58 (61) | 54 (58) | 30 (46) | 13 (39) |
| PilX7/VirB7 | 100 (100) | 100 (100) | 100 (100) | 100 (100) | 100 (100) | 72 (77) | 40 (61) | 33 (51) | 18 (36) |
| PilX8/VirB8 | 99 (99) | 99 (100) | 99 (100) | 99 (100) | 99 (99) | 75 (72) | 45 (54) | 33 (46) | 17 (43) |
| PilX9/VirB9 | 98 (98) | 99 (100) | 98 (99) | 100 (100) | 77 (79) | 80 (74) | 54 (60) | 39 (54) | 23 (48) |
| PilX10/VirB10 | 99 (99) | 100 (100) | 100 (98) | 100 (100) | 100 (100) | 63 (65) | 50 (59) | 42 (55) | 23 (44) |
| PilX11/VirB11 | 98 (98) | 72 (74) | 98 (97) | 99 (100) | 100 (100) | 73 (71) | 59 (65) | 43 (53) | 26 (47) |
| TaxB | 99 (99) | 99 (99) | 98 (96) | 99 (100) | 99 (99) | 78 (70) | 72 (67) | NA | NA |
| Hyp | 93 (95) | 96 (98) | 96 (98) | 100 (100) | 96 (98) | NA | 44 (57) | NA | NA |
| Hyp | 77 (79) | 85 (86) | 92 (90) | 100 (100) | 85 (86) | NA | NA | NA | NA |
pilX3 and pilX4 and virB3 and virB4 in R5K and pTi are separate genes, but in all other plasmids they are fused in one ORF.
pTi contains the virB operon, which was compared to the pilX operon of pMAS2027.
NA, not applicable.
The pilX locus promotes conjugative transfer of plasmid pMAS2027.
To demonstrate the function of the pilX1 to pilX11 genes, the entire segment was deleted by λ-red-mediated homologous recombination and replaced by insertion of a kanamycin resistance gene (to generate plasmid pMAS2027pilX::kan). Plasmid pMAS2027pilX::kan could not be mobilized into a recipient E. coli strain, confirming the function of the pilX1 to pilX11 genes in conjugative transfer. To examine whether type 3 fimbriae (encoded by the mrkABCDF genes on pMAS2027) also contribute to the efficiency of conjugative transfer, we constructed two additional plasmids: pMAS2027mrk::kan and pMAS2027orf27::cam. Plasmid pMAS2027orf27::cam contained a gene encoding resistance to chloramphenicol inserted into orf27 (which encodes a hypothetical protein of unknown function) and thus served as a control. The frequency of conjugative transfer, calculated by determining the number of transconjugants per donor, was 0.0064 for pMAS2027orf27::cam, compared to 0.032 for pMAS2027mrk::kan (P < 0.05). Thus, the lack of type 3 fimbriae resulted in a fivefold increase in conjugative plasmid transfer.
Type 3 fimbriae, but not T4S pili, contribute to biofilm formation.
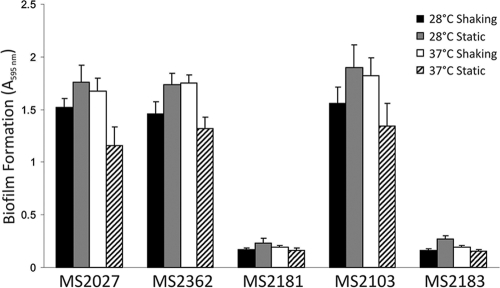
The contributions of type 3 fimbriae and T4S pili to biofilm growth were examined by using MS2027 harboring either pMAS2027mrk::kan, pMAS2027pilX::kan, or pMAS2027orf27::cam and performing two dynamic biofilm assays. First, we used a PVC microtiter plate assay and showed that type 3 fimbriae, but not T4S pili, promote biofilm formation after growth in M9 minimal medium and in urine (Fig. 3). Next, we tested the ability of the same strains to promote biofilm formation in a continuous-flow chamber. Consistent with the results of our microtiter plate assay, MS2072 harboring pMAS2027orf27::cam and MS2072 harboring pMAS2027pilX::kan produced equivalent, strong biofilms that were approximately 20 μm deep, while MS2072 harboring pMAS2027mrk::kan was unable to form a biofilm (Fig. 4). An additional plasmid construct with both the mrk and pilX genes deleted (pMAS2027mrk::kan pilX::cam) was also examined and did not mediate biofilm growth. Thus, although plasmid pMAS2027 encodes the capacity to produce conjugative T4S pili, these pili did not contribute to biofilm formation in these experiments.
FIG. 3.
Biofilm formation by E. coli MS2027 and derivatives. Strains were grown in urine for 16 h in PVC microtiter plates under various conditions, as indicated. Biofilm formation was quantified by staining adhered cells with 0.1% crystal violet, resuspending them in ethanol-acetate (80:20), and measuring the absorbance at 595 nm. The results are the averages and standard deviations of three independent experiments. The data are the results for MS2027, MS2362 [MS2027(pMAS2027orf27::cam)], MS2181 [MS2027(pMAS2027mrk::cam)], MS2103 [MS2027(pMAS2027pilX::kan)], and MS2183 [MS2027(pMAS2027mrk::cam, pilX::kan)].
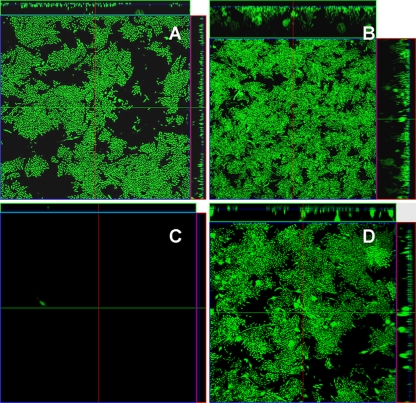
FIG. 4.
Flow chamber biofilm formation for (A) E. coli MS2091 (MS2027 attB::bla-PA1/04/03-gfpmut3b*-T0;Gfp), (B) MS2519 [MS2027(pMAS2027orf27::gfp-kan)], (C) MS2517 [MS2027(pMAS2027mrk::gfp-kan)], and (D) MS2518 [MS2027(pMAS2027pilX::gfp-kan)]. Biofilm development was monitored by confocal scanning laser microscopy 24 h after inoculation. The large micrographs show horizontal sections. To the right of and above each large image are images of the yz plane and the xz plane, respectively, obtained at the positions indicated by the lines.
Transfer of the mrk genes via conjugation in an evolving mixed biofilm.
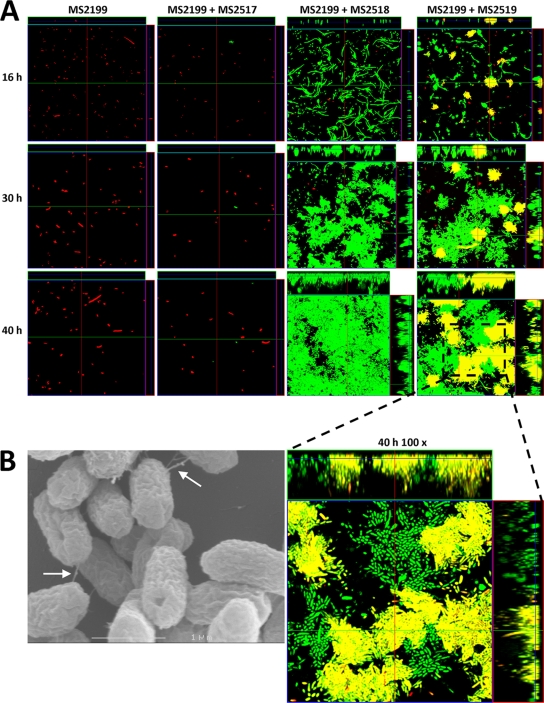
A further three deletion mutants were constructed by replacing the mrk, pilX, and orf27 genes on pMAS2027 with gfp and a kanamycin resistance gene, tagging pMAS2027 with gfp in the process. MS2027 cells harboring plasmid pMAS2027mrk::gfp-kan, pMAS2027pilX::gfp-kan, or pMAS2027orf27::gfp-kan were examined for the ability to form a mixed biofilm in the continuous-flow chamber system with E. coli K-12 strain MS2199, a non-biofilm-forming strain tagged with the rfp gene. Biofilm formation was assessed by performing scanning laser confocal microscopy at 16 h, 30 h, and 40 h after inoculation. E. coli MS2199 was unable to form a biofilm at all three time points when it was alone and when it was grown in a coculture with MS2027(pMAS2027mrk::gfp-kan) or MS2027(pMAS2027pilX::gfp-kan) (Fig. 5A). However, when E. coli MS2199 was grown in a coculture with MS2027(pMAS2027orf27::gfp-kan), a mixed biofilm consisting of MS2027(pMAS2027orf27::gfp-kan) cells (green) and transconjugant MS2199 cells (yellow) formed (Fig. 5A). Close examination of the mixed biofilm (magnification, ×100) revealed regions with a mixture of green and yellow cells, suggesting that there was conjugative transfer within the evolving biofilm. Taken together, the data demonstrate that conjugative transfer of pMAS2027orf27::gfp-kan from MS2027 to MS2199 enabled MS2199 transconjugant cells to form a strong biofilm due to production of type 3 fimbriae. No red-tagged MS2199 cells were observed in the mixed biofilm. When the mixed biofilm containing MS2199 and MS2027(pMAS2027orf27::gfp-kan) was viewed with an SEM, T4S pilus-like structures linking cells within the biofilm were observed (Fig. 5B). Thus, it is possible that the T4S pili encoded on pMAS2027 contribute to the structural composition of a mixed biofilm both directly and through their ability to transfer the type 3 fimbria-encoding mrk genes to recipient cells.
FIG. 5.
(A) Flow chamber biofilm formation for E. coli MS2199 (Rfp+) and mixed cultures of E. coli MS2199 (Rfp+) and MS2517 [MS2027(pMAS2027mrk::gfp-kan)] (Gfp+), MS2199 (Rfp+) and MS2518 [MS2027(pMAS2027pilX::gfp-kan)] (Gfp+), and MS2199 (Rfp+) and MS2519 [MS2027(pMAS2027orf27::gfp-kan)] (Gfp+). Magnification, ×40. Biofilm development was monitored by confocal scanning laser microscopy at 16 h, 30 h, and 40 h after inoculation. The micrographs show horizontal sections. To the right of and above each large panel are images of the yz plane and the xz plane, respectively, obtained at the positions indicated by the lines. The largest micrograph shows a higher magnification (×100) of a defined region of the MS2199-MS2519 mixed biofilm. (B) SEM micrograph of a mixed MS2199-MS2519 microtiter plate biofilm. Putative T4S pilus structures are indicated by arrows.
DISCUSSION
Biofilm formation by uropathogenic E. coli is mediated by a range of cell surface factors, including fimbriae, flagella, and adhesins. Often, the genes encoding these factors are located on mobile genetic elements, such as plasmids, transposons, and pathogenicity islands. Here we determined the complete nucleotide sequence of a conjugative plasmid isolated from a strain of uropathogenic E. coli that caused CAUTI and defined the properties of this plasmid that are associated with biofilm growth.
Plasmid pMAS2027 is most closely related to conjugative plasmids belonging to the IncX group (36), and its nucleotide sequence strongly suggested that it belonged to IncX1, a subset of this group (20). The genetic organization of pMAS2027 is similar to that previously described for several characterized virulence plasmids from S. enterica (9, 15, 41). Indeed, of the 58 ORFs identified on pMAS2027, 40 (69%) exhibit the strongest nucleotide sequence similarity to S. enterica genes. The major genetic load region of pMAS2027 comprised the mrk genes (encoding type 3 fimbriae), which appear to be located on a mobile genetic element. Recently, a large conjugative plasmid (pOLA52) isolated from swine manure that also contains both the mrkABCDF and pilX1 to pilX11 genes was described (27). The mrkABCDF genes of pOLA52 are flanked by transposon-like sequences, and the nucleotide sequences of these genes are >94% identical to the nucleotide sequences of the mrkABCDF genes of pMAS2027. A comparison of the pilX1 to pilX11 genes of the two plasmids also revealed a high degree of nucleotide sequence conservation, except for the eex and pilX6 genes. Although pOLA52 is approximately 10 kb larger than pMAS2027 and contains genes that impart multidrug resistance, the similarity between the backbone sequences of the two plasmids is striking considering that the plasmids were identified in strains isolated from two very different environments (i.e., swine manure and the urine of a patient with CAUTI).
The ability to produce type 3 fimbriae was an absolute requirement for biofilm growth of E. coli MS2027. No biofilm formation was observed with mutants that lacked the mrkABCDF genes but retained the ability to produce conjugative T4S pili. Thus, unlike the F pilus (16, 30), T4S pili do not mediate binding to abiotic surfaces and do not promote biofilm formation. Although the role of type 3 fimbriae in biofilm formation was consistent in previous studies performed with plasmid pOLA52 (4), we found that deletion of mrkABCDF resulted in a fivefold increase in the conjugation efficiency of pMAS2027. This finding is in contrast to the results of studies performed with pOLA52, where mutation of the mrkC gene (which abrogates production of type 3 fimbriae) caused a dramatic reduction in conjugation efficiency (4). It is possible that this discrepancy is due to differences in the makeup of other cell surface components that might interfere with this process between the E. coli strains harboring pMAS2027 and pOLA52.
Mixed-culture flow chamber assays were employed to examine the contribution of conjugative plasmid transfer to biofilm development. Maintenance of transconjugant MS2199(pMAS2027orf27::cam) cells within a biofilm was dependent on the production of type 3 fimbriae. Thus, the genetic load of pMAS2027 (mrkABCDF) defined its ability to spread laterally within the biofilm. Genes encoding type 3 fimbriae have been identified in many gram-negative uropathogens, including Klebsiella spp., E. coli, Enterobacter spp., P. mirabilis, Serratia spp., Yersinia spp., and Providentia spp. It is likely that the widespread occurrence of type 3 fimbria-encoding genes in these pathogens is associated with plasmid transfer within biofilms in the hospital setting.
Supplementary Material
Acknowledgments
This work was supported by grant DP666852 from the Australian Research Council and by grant 455914 from the National Health and Medical Research Council.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Balestrino, D., J. A. J. Haagensen, C. Rich, and C. Forestier. 2005. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J. Bacteriol. 187:2870-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnhart, M. M., and M. R. Chapman. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burmolle, M., M. L. Bahl, L. B. Jensen, S. J. Sorensen, and L. H. Hansen. 2008. Type 3 fimbriae, encoded by the conjugative plasmid pOLA52, enhance biofilm formation and transfer frequencies in Enterobacteriaceae strains. Microbiology 154:187-195. [DOI] [PubMed] [Google Scholar]
- 5.Carver, T., M. Berriman, A. Tivey, C. Patel, U. Bohme, B. G. Barrell, J. Parkhill, and M. A. Rajandream. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, B. B., C. Sternberg, J. B. Andersen, L. Eberl, S. Moller, M. Givskov, and S. Molin. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, C. S., Y. Feng, A. C. Chien, S. N. Hu, C. H. Chu, and C. H. Chiu. 2008. Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics 92:339-343. [DOI] [PubMed] [Google Scholar]
- 10.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 11.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta, N., and V. M. Hughes. 1983. Plasmids of the same Inc groups in enterobacteria before and after the medical use of antibiotics. Nature 306:616-617. [DOI] [PubMed] [Google Scholar]
- 14.Di Martino, P., N. Cafferini, B. Joly, and A. Darfeuille-Michaud. 2003. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 154:9-16. [DOI] [PubMed] [Google Scholar]
- 15.Fierer, J., L. Eckmann, F. Fang, C. Pfeifer, B. B. Finlay, and D. Guiney. 1993. Expression of the Salmonella virulence plasmid gene spvB in cultured macrophages and nonphagocytic cells. Infect. Immun. 61:5231-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 17.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 18.Hausner, M., and S. Wuertz. 1999. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65:3710-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jefferson, K. K. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236:163-173. [DOI] [PubMed] [Google Scholar]
- 20.Jones, C. S., D. J. Osborne, and J. Stanley. 1993. Molecular comparison of the IncX plasmids allows division into IncX1 and IncX2 subgroups. J. Gen. Microbiol. 139:735-741. [DOI] [PubMed] [Google Scholar]
- 21.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 22.Lane, M. C., V. Lockatell, G. Monterosso, D. Lamphier, J. Weinert, J. R. Hebel, D. E. Johnson, and H. L. T. Mobley. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73:7644-7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasaro, M. A., N. Salinger, J. Zhang, Y. T. Wang, Z. T. Zhong, M. Goulian, and J. Zhu. 2009. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl. Environ. Microbiol. 75:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lebaron, P., P. Bauda, M. C. Lett., Y. Duval-Iflah, P. Simonet, E. Jacq, N. Frank, B. Roux, B. Baleux, G. Faurie, J. C. Hubert, P. Normand, D. Prieur, S. Schmitt, and J. C. Block. 1997. Recombinant plasmid mobilization between E. coli strains in seven sterile microcosms. Can. J. Microbiol. 43:534-540. [DOI] [PubMed] [Google Scholar]
- 25.Luo, H. L., K. Wan, and H. H. Wang. 2005. High-frequency conjugation system facilitates biofilm formation and pAMβ1 transmission by Lactococcus lactis. Appl. Environ. Microbiol. 71:2970-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martino, P. D., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443-449. [DOI] [PubMed] [Google Scholar]
- 27.Norman, A., L. H. Hansen, Q. X. She, and S. J. Sorensen. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59-74. [DOI] [PubMed] [Google Scholar]
- 28.Nunez, B., P. Avila, and F. de la Cruz. 1997. Genes involved in conjugative DNA processing of plasmid R6K. Mol. Microbiol. 24:1157-1168. [DOI] [PubMed] [Google Scholar]
- 29.Ong, C. L. Y., G. C. Ulett, A. N. Mabbett, S. A. Beatson, R. I. Webb, W. Monaghan, G. R. Nimmo, D. F. Looke, A. G. McEwan, and M. A. Schembri. 2008. Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation. J. Bacteriol. 190:1054-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 31.Reisner, A., B. M. Holler, S. Molin, and E. L. Zechner. 2006. Synergistic effects in mixed Escherichia coli biofilms: conjugative plasmid transfer drives biofilm expansion. J. Bacteriol. 188:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Schembri, M. A., and P. Klemm. 2001. Biofilm formation in a hydrodynamic environment by novel FimH variants and ramifications for virulence. Infect. Immun. 69:1322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stalker, D. M., and D. R. Helinski. 1985. DNA segments of the IncX plasmid R485 determining replication and incompatibility with plasmid R6K. Plasmid 14:245-254. [DOI] [PubMed] [Google Scholar]
- 37.Stothard, P., and D. S. Wishart. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537-539. [DOI] [PubMed] [Google Scholar]
- 38.Tenke, P., B. Kovacs, M. Jackel, and E. Nagy. 2006. The role of biofilm infection in urology. World J. Urol. 24:13-20. [DOI] [PubMed] [Google Scholar]
- 39.Ulett, G. C., A. N. Mabbett, K. C. Fung, R. I. Webb, and M. A. Schembri. 2007. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 153:2321-2331. [DOI] [PubMed] [Google Scholar]
- 40.Valle, J., A. N. Mabbett, G. C. Ulett, A. Toledo-Arana, K. Wecker, M. Totsika, M. A. Schembri, J. M. Ghigo, and C. Beloin. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J. Bacteriol. 190:4147-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valone, S. E., G. K. Chikami, and V. L. Miller. 1993. Stress induction of the virulence proteins (SpvA, -B, and -C) from native plasmid pSDL2 of Salmonella dublin. Infect. Immun. 61:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ward, J. E., D. E. Akiyoshi, D. Regier, A. Datta, M. P. Gordon, and E. W. Nester. 1988. Characterization of the Virb operon from an Agrobacterium tumefaciens Ti plasmid. J. Biol. Chem. 263:5804-5814. [PubMed] [Google Scholar]
- 43.Wells, T. J., O. Sherlock, L. Rivas, A. Mahajan, S. A. Beatson, M. Torpdahl, R. I. Webb, L. P. Allsopp, K. S. Gobius, D. L. Gally, and M. A. Schembri. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10:589-604. [DOI] [PubMed] [Google Scholar]
- 44.Williams, S. L., and J. F. Schildbach. 2006. Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Res. 34:426-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright, K. J., P. C. Seed, and S. J. Hultgren. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 73:7657-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.