Abstract
The 6.10-Mb genome sequence of the aerobic chitin-digesting gliding bacterium Flavobacterium johnsoniae (phylum Bacteroidetes) is presented. F. johnsoniae is a model organism for studies of bacteroidete gliding motility, gene regulation, and biochemistry. The mechanism of F. johnsoniae gliding is novel, and genome analysis confirms that it does not involve well-studied motility organelles, such as flagella or type IV pili. The motility machinery is composed of Gld proteins in the cell envelope that are thought to comprise the “motor” and SprB, which is thought to function as a cell surface adhesin that is propelled by the motor. Analysis of the genome identified genes related to sprB that may encode alternative adhesins used for movement over different surfaces. Comparative genome analysis revealed that some of the gld and spr genes are found in nongliding bacteroidetes and may encode components of a novel protein secretion system. F. johnsoniae digests proteins, and 125 predicted peptidases were identified. F. johnsoniae also digests numerous polysaccharides, and 138 glycoside hydrolases, 9 polysaccharide lyases, and 17 carbohydrate esterases were predicted. The unexpected ability of F. johnsoniae to digest hemicelluloses, such as xylans, mannans, and xyloglucans, was predicted based on the genome analysis and confirmed experimentally. Numerous predicted cell surface proteins related to Bacteroides thetaiotaomicron SusC and SusD, which are likely involved in binding of oligosaccharides and transport across the outer membrane, were also identified. Genes required for synthesis of the novel outer membrane flexirubin pigments were identified by a combination of genome analysis and genetic experiments. Genes predicted to encode components of a multienzyme nonribosomal peptide synthetase were identified, as were novel aspects of gene regulation. The availability of techniques for genetic manipulation allows rapid exploration of the features identified for the polysaccharide-digesting gliding bacteroidete F. johnsoniae.
Flavobacterium johnsoniae (formerly Cytophaga johnsonae) is a member of the large and diverse phylum of gram-negative bacteria known as the Bacteroidetes. Members of this group of organisms have a number of unique characteristics that distinguish them from other bacteria. Some have novel cell surface machinery to utilize polysaccharides (85, 95, 96). Rapid gliding motility over surfaces is also common among these bacteria (59), as are unusual outer membrane sulfonolipids (29) and flexirubin pigments (78). Bacteroidete gene expression and regulation also have novel aspects (10, 11, 20, 39, 92). The many unusual features of these common but understudied bacteria provide numerous avenues for further exploration, which can be greatly aided by analysis of genome sequences.
F. johnsoniae digests many polysaccharides and proteins, but it is best known for its ability to rapidly digest insoluble chitin (87). Chitin is one of the most abundant biopolymers on earth (63). F. johnsoniae and other members of the Bacteroidetes phylum are thought to play important roles in the turnover of this compound in many environments (47). F. johnsoniae has become a model system for the study of bacteroidete gliding motility biochemistry and molecular biology (20, 27-29, 59, 72). This paper highlights novel features of the F. johnsoniae genome, with particular emphasis on genes and proteins likely to be involved in polysaccharide utilization, gliding motility, and the novel biochemistry of this organism.
MATERIALS AND METHODS
Sequencing of the F. johnsoniae genome.
The random shotgun method was used to sequence the genome of F. johnsoniae UW101 (ATCC 17061). Large-insert (40-kb), medium-insert (8-kb), and small-insert (3-kb) random libraries were partially sequenced, and sequences were assembled with parallel phrap (High Performance Software, LLC). Possible misassemblies were corrected with Dupfinisher (30) or by analysis of transposon insertions in bridge clones. Gaps between contigs were closed by editing, custom primer walking, or PCR amplification.
Annotation.
Gene predictions were obtained using Glimmer (23), and tRNAs were identified using tRNAScan-SE (53). Basic analyses of the gene predictions were performed by comparing coding sequences with the PFam, BLOCKS, and Prodom databases. Protein localizations were predicted with PSORTb (26), and lipoproteins were identified using LipoP (42). A team of annotators added gene definitions and functional classes using BLAST results and information from the Pfam (http://pfam.janelia.org/index.html) (86), BLOCKS (33), Prodom (84), and SMART (82) databases. Metabolic pathways were constructed using MetaCyc as a reference data set (17). Genes encoding candidate glycoside hydrolases, polysaccharide lyases, and carbohydrate esterases were detected with routines used for updates of the Carbohydrate Active Enzyme database (16) at http://www.cazy.org. Because sequence-based families of carbohydrate-active enzymes contain enzymes with various substrate specificities, functional annotation was guided by the distance between the protein model and biochemically characterized enzymes. As a result, members of a particular family do not necessarily have the same predicted function. Information regarding predicted peptidases of F. johnsoniae was obtained from the MEROPS peptidase database (76) at http://merops.sanger.ac.uk/. Putative susC-like and susD-like genes were identified as previously described (97) using an iterative amino acid BLAST search that initially used the starch-binding SusC and SusD sequences from Bacteroides thetaiotaomicron. Detailed information about the genome properties and genome annotation can be obtained from the JGI Integrated Microbial Genomes website (54) at http://img.jgi.doe.gov/pub/main.cgi.
Utilization of carbohydrates.
To asses growth on different carbohydrates, F. johnsoniae was cultured in SD minimal medium (18) containing individual substrates as sole carbon sources at a concentration of 5 mg/ml, except for rhamnogalacturonan I, which was used at a concentration of 10 mg/ml. Monosaccharides and disaccharides were sterilized by filtration (pore size, 0.22 μm), and polysaccharides were sterilized by autoclaving them in distilled water as 2× stocks. Carbohydrates (75 μl of each stock) were arrayed in quadruplicate in a 96-well microtiter plate. F. johnsoniae cells were cultured overnight in CYE medium, and 10 ml was collected by centrifugation, washed once in 2× SD medium that did not contain any carbohydrate, suspended in 10 ml of 2× concentrated SD medium, and diluted 100-fold in 2× SD medium. Seventy-five microliters of the resulting cell suspension was added to each well of the 96-well plate containing the carbohydrate stocks. Plates were incubated at 22°C, and the growth in each well was measured by determining the absorbance at 600 nm at 5-min intervals for 88 h.
Analysis of genes involved in flexirubin synthesis.
A 450-bp internal fragment of Fjoh_1102, a homolog of Pseudomonas aurantiaca darB, was amplified from chromosomal DNA using primer 834 (5′-GCTAGGGATCCACAAGCCGTTATTACGCTGTTGAC-3′) and primer 833 (5′-GCTAGCTGCAGAAATGCACCGGCACCGTCAGATAA-3′), which were designed with engineered BamHI and PstI restriction sites, respectively. The product was inserted into pCR2.1 using an original TA cloning kit according to the manufacturer's instructions (Invitrogen) to generate pRR07. pRR07 was digested with BamHI and PstI, and the internal fragment of Fjoh_1102 was introduced into the suicide vector pLYL03 (50) that had been digested with the same enzymes to generate pRR08. pRR08 was introduced into F. johnsoniae by triparental conjugation, and erythromycin-resistant colonies were obtained. Disruption of Fjoh_1102 was confirmed by PCR using primer 838 (5′-CCTTCTAATCCTTTAGATCGCGGGCA-3′), which is 1,012 bp upstream of the Fjoh_1102 translation start site, and primer 737 (5′-AGGCACCCCAGGCTTTACACT-3′), which is specific for the suicide vector pLYL03.
A library of wild-type genomic fragments in cosmid pCP22 (37) was constructed to identify additional genes involved in flexirubin synthesis. Chromosomal DNA was partially digested with EcoRI, and fragments were ligated into pCP22, packaged in lambda phage particles (MaxPlax; Epicentre Technologies, Madison, WI), and introduced into Escherichia coli DH5αMCR. Cosmid DNA from approximately 10,000 colonies was transferred to the flexirubin-negative mutant F. johnsoniae UW102-154 by triparental conjugation essentially as previously described (37), except that transconjugants were plated on CYE medium with 100 μg/ml erythromycin. Colonies were screened for pigmentation after 2 days of incubation at 30°C. A pigmented flexirubin-positive colony was isolated, and plasmid pMM340, which carries a 12.8-kbp region spanning Fjoh_1078 to Fjoh_1089, was obtained.
Colonies were tested for the presence of flexirubin pigments by exposing them to 50 μl 10 N KOH, which resulted in a change from yellow to red if flexirubin pigments were present, followed by neutralization with 42 μl 12 N HCl, which resulted in a return to yellow pigmentation.
Nucleotide sequence accession number.
The genome sequence of F. johnsoniae has been deposited in the GenBank database under accession number CP000685.
RESULTS AND DISCUSSION
General genome features.
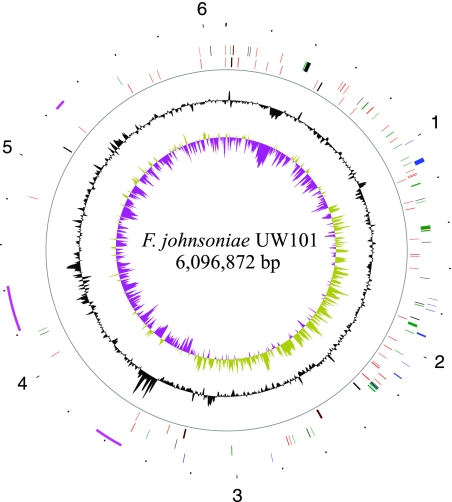
The F. johnsoniae genome consists of a single circular 6,096,872-bp chromosome with a G+C content of 34.11% (Fig. 1). It is one of the largest sequenced bacteroidete genomes and is more than twice as large as the genome of the other sequenced member of the genus Flavobacterium, the fish pathogen Flavobacterium psychrophilum (24). Six rRNA operons were identified, and 5,056 protein-encoding genes were predicted. The GC skew allowed prediction of the site of the origin of replication near nucleotide 3322321. Alignment of conserved genomic sequences of F. johnsoniae and F. psychrophilum with MAUVE revealed a 4.17-Mbp region between 5.43 Mbp and 3.5 Mbp that contained most of the conserved sequences (see Fig. S1 in the supplemental material). Not surprisingly, this region harbors most of the genes involved in central energy-generating metabolism (glycolysis, tricarboxylic acid cycle, oxidative phosphorylation) and the core information-processing genes involved in transcription and translation (Fig. 1). The remaining 1.93 Mbp of the genome harbors several transposon islands and a large number of genes encoding hypothetical proteins of unknown function. This region is also enriched for genes predicted to be involved in polysaccharide utilization, and 45% of the predicted glycoside hydrolases and 67% of the polysaccharide lyases are encoded by genes in this region.
FIG. 1.
Map of the F. johnsoniae genome. The innermost circle (circle 1) shows the GC skew [(G − C)/(G + C)] (yellow-green, values more than 1; purple, values less than 1). Circle 2 shows the G+C content. Circle 3 indicates the backbone ring. Circle 4 shows rRNA (black) and tRNA (red). Circle 5 shows core protein-encoding genes, including genes encoding ribosomal proteins (black), aminoacyl tRNA synthetases (red), enzymes involved in glycolysis, the tricarboxylic acid cycle, oxidative phosphorylation, and synthesis of electron transport chain menaquinones (green), and the RNA polymerase core (teal). Circle 6 shows motility genes (blue). Circle 7 shows transposon islands (pink). The outermost circle (circle 8) shows a ruler, and the numbers indicate Mbp of sequence.
Polysaccharide utilization.
F. johnsoniae utilizes a variety of polysaccharides as nutrients. Analysis of the genome identified 138 predicted glycoside hydrolases and nine predicted polysaccharide lyases. F. johnsoniae was originally described as a chitinolytic bacterium (87), and analysis of the genome sequence identified genes encoding possible chitinolytic enzymes. These enzymes include five chitinases that cut the long chitin polymers and five β-N-acetylglucosaminidases that release N-acetylglucosamine and/or chitobiose from the oligomers (Table 1). The predicted chitinases are diverse and include enzymes that are related to chitinases of bacteria (Fjoh_4175, Fjoh_4555, Fjoh_4757), animals (Fjoh_4560), and plants (Fjoh_2608). Other predicted enzymes that may be involved in chitin utilization include an N-acetylglucosamine kinase (Fjoh_4589) related to mouse NagK (34), an N-acetylglucosamine-6-phosphate deacetylase (Fjoh_3974) related to Vibrio cholerae NagA (98), and four glucosamine-6-phosphate isomerases/deaminases (Fjoh_2029, Fjoh_2381, Fjoh_4812, Fjoh_4557) related to E. coli NagB (79). NagK, NagA, and NagB are predicted to function in sequence to convert N-acetylglucosamine into fructose-6-phosphate for entry into the Embden-Meyerhof-Parnas pathway.
TABLE 1.
Predicted F. johnsoniae glycohydrolases involved in chitin digestion
| Gene | Predicted functiona | Homologsb | Predicted localizationc | Molecular mass (kDa)d | Enzymatic and other domainse |
|---|---|---|---|---|---|
| Fjoh_0674 | Candidate β-N-acetylglucosaminidase | P. gingivalis NahA (38% identity over 774 amino acids [52]) | Periplasmic | 87.6 | GH20 |
| Fjoh_2039 | Candidate β-N-acetylglucosaminidase | P. gingivalis NahA (40% identity over 610 amino acids [52]) | Periplasmic | 87.3 | GH20 |
| Fjoh_2118 | Candidate β-glycosidase, related to N-acetylglucosaminidases | Pseudoalteromonas piscicida Cht60 (30% identity over 389 amino acids [91]) | Cytoplasmic | 59.9 | GH3 |
| Fjoh_2608 | Distantly related to plant chitinases | Solanum tuberosum (potato) CHTB2 endochitinase 2 (24% identity over 231 amino acids [12]) | Unknown | 90.8 | GH19 |
| Fjoh_4175 | Candidate chitinase | Bacillus circulans ChiA1 (26% identity over 317 amino acids [94]) | Unknown | 57.8 | GH18-CBM6-D5 |
| Fjoh_4555 | Candidate chitinase | B. circulans ChiA1 (33% identity over 513 amino acids [94]), B. circulans ChiD (37% identity over 353 amino acids [93]) | Outer membrane or extracellular | 168.9 | GH18-GH18 |
| Fjoh_4556 | Candidate β-N-acetylglucosaminidase | P. gingivalis NahA (36% identity over 447 amino acids [52]) | Periplasmic | 77.3 | GH20 |
| Fjoh_4560 | Candidate chitinase | Bos taurus (bovine) ChiA (25% identity over 244 amino acids [89]) | Lipoprotein | 38.1 | GH18 |
| Fjoh_4757 | Candidate chitinase | B. circulans ChiA1 (37% identity over 298 amino acids [94]) | Unknown, not cytoplasmic | 41.2 | GH18 |
| Fjoh_4808 | Candidate β-N-acetylglucosaminidase | P. gingivalis NahA (32% identity over 633 amino acids [52]) | Periplasmic | 94.8 | GH20 |
Predicted functions were assigned by routines used for updating the Carbohydrate Active Enzymes database (http://www.cazy.org/) using the following criteria: typically, 70% or greater amino acid identity with a protein domain with a biochemically determined function at the time of analysis resulted in “candidate” status; 30% to 70% amino acid identity with a protein domain with a known function resulted in “related to” status; and less than 30% amino acid identity with a protein domain with a known function resulted in “distantly related to” status. Because the threshold of similarity that correlates with a change of substrate specificity is variable from one glycoside hydrolase family to another, the criteria were tightened or loosened appropriately for several families. All analyses were conducted domain by domain to avoid problems arising from the modular structure of many of the proteins.
Homologs were identified by a BlastP search with the Swiss-Prot database. Fjoh_4555 has two catalytic domains, so a homolog for each domain is listed. ChiA1 is similar to the N-terminal GH18 domain, and ChiD is similar to the C-terminal GH18 domain. The numbers in brackets are reference numbers.
Localization was predicted using the default settings of PSORTb (26). Predicted lipoproteins were identified using LipoP (42).
Predicted molecular mass of the primary product of translation, including any predicted signal peptide.
CBM6, family 6 CBM, as assigned by CAZY; D5, carboxy-terminal domain of R. marinus xylanases predicted to be involved in attachment to the cell surface (43); GH, glycoside hydrolase as assigned by CAZY (the numbers indicate families).
In addition to genes predicted to be involved in chitin utilization, F. johnsoniae also has genes predicted to encode numerous glycohydrolases, polysaccharide lyases, and esterases that are likely involved in the digestion of other polysaccharides (see Tables S1, S2, and S3 in the supplemental material). It appears to have an arsenal of enzymes for digestion of plant cell wall polysaccharides, which may explain the prevalence of this organism in soil and rhizosphere habitats (71, 87). F. johnsoniae is known to digest pectin, and glycohydrolases, lyases, and esterases likely to be involved in this process were identified (see Tables S1, S2, and S3 and Fig. S2 in the supplemental material). Many genes predicted to encode glycohydrolases and esterases that could be involved in utilization of plant cell wall hemicelluloses, such as xylans (β-1,4-linked polymers of xylose often substituted with acetyl, arabinofuranoside, and glucuronosyl residues), mannans (heteropolysaccharides containing β-1,4-linked mannose residues), and xyloglucans (β-1,4-linked polymers of glucose substituted with xylose and other sugars), were also identified (see Tables S1 and S3 and Fig. S2 in the supplemental material). Candidate xylanases, β-xylosidases, arabinofuranosidases, glucuronidases, and carbohydrate esterases involved in xylan digestion, candidate β-mannanases and β-mannosidases involved in mannan digestion, and candidate β-glucosidases and α-glycosidases and an endoglucanase that could be involved in xyloglucan digestion were identified. Digestion of hemicelluloses has not been reported previously for F. johnsoniae. We tested growth of F. johnsoniae on a battery of carbohydrates and determined that it grows on many hemicellulosic substrates (Table 2). The highest growth rates were observed with three polysaccharides: glucomannan, polygalacturonate, and laminarin. F. johnsoniae also utilized the major monosaccharides in these glycans (mannose, galacturonic acid, and glucose, respectively), but the growth rates were lower. A similar phenomenon has been observed for B. thetaiotaomicron, which grows more rapidly on α-glucans, such as pullulan and dextran, than on monomeric glucose (49). F. johnsoniae does not utilize crystalline cellulose as a carbon and energy source, but its genome contains genes that encode a predicted endoglucanase (Fjoh_4946) and six β-glucosidases (Fjoh_1567, Fjoh_3392, Fjoh_3521, Fjoh_3861, Fjoh_4857, Fjoh_4963) (see Table S1 in the supplemental material). These enzymes may be involved in utilization of glucose-containing hemicelluloses, such as xyloglucans, and the β-1,3-glucan laminarin, or they may allow partial digestion of cellulose. The β-glucosidases also likely account for the ability of F. johnsoniae to utilize cellobiose and cellohexaose (Table 2) (21). F. johnsoniae is also known to utilize starch (21, 87) and dextran (40), and genes encoding candidate enzymes involved in the utilization of these polysaccharides were identified (Fig. 2; see Fig. S2 and Table S1 in the supplemental material). Carbohydrate-binding modules (CBMs) are present in 11 of the 138 glycohydrolases and in seven additional proteins that do not have obvious catalytic domains (Fjoh_0913, Fjoh_1470, Fjoh_1765, Fjoh_2035, Fjoh_2869, Fjoh_3324, Fjoh_4174). Most of the potential polysaccharide utilization proteins are predicted to be extracytoplasmic, and 11 of them have carboxy-terminal domains that are similar to the “D5” domains of Rhodothermus marinus cell-associated xylanolytic enzymes (43). The R. marinus D5 domains have been postulated to be involved in attachment to the cell surface. One cluster of four genes (Fjoh_4174 to Fjoh_4177) has a high density of CBMs and D5 domains. The proteins encoded by these genes account for 6 of the 21 CBMs identified and 4 of the 11 D5 domains. The function of this cluster is not known, but it encodes a candidate chitinase, two possible β-1,3-glucanases, and a protein that lacks an obvious catalytic domain but contains two CBMs and a D5 domain.
TABLE 2.
Growth of F. johnsoniae on various carbohydrates
| Substratea | Substrate type | Relative growth rateb |
|---|---|---|
| d-Glucose | Monosaccharide | 1.0 (0.2) |
| d-Fructose | Monosaccharide | 0.9 (0.1) |
| d-Galactose | Monosaccharide | 0.7 (0.1) |
| d-Mannose | Monosaccharide | ± |
| d-Xylose | Monosaccharide | 1.2 (0.2) |
| d-Galacturonic acid | Monosaccharide | 0.5 (0.1) |
| d-Glucosamine | Monosaccharide | 0.6 (0.1) |
| N-Acetylglucosamine | Monosaccharide | ± |
| Cellobiose | Disaccharide | 1.0 (0.1) |
| Cellohexaose | Hexasaccharide | ± |
| Pullulan | Linear water-soluble polymer of glucose with α-1,4 and α-1,6 glycosidic bonds | 0.6 (0.1) |
| Laminarin (brown algae) | Linear polymer of glucose with β-1,3 and β-1,6 glycosidic bonds | 1.3 (0.1) |
| Amylopectin (potato) | Insoluble component of plant starch; branched polymer of glucose with α-1,4 and α-1,6 glycosidic bonds | + |
| Dextran (from L. mesenteroides) | Branched polymer of glucose with primarily α-1,6 and α-1,4 glycosidic bonds | 0.8 (0.1) |
| Polygalacturonate (citrus peel) | Pectic polysaccharide | 1.5 (0.1) |
| Arabinoxylan (wheat) | Hemicellulosic polysaccharide | + |
| β-Glucan (barley) | Hemicellulosic polysaccharide | + |
| Methyl glucuronyl xylan | Hemicellulosic polysaccharide | ± |
| Xylan (oat spelt, water-soluble fraction) | Hemicellulosic polysaccharide | 0.7 (0.1) |
| Xyloglucan (tamarind) | Hemicellulosic polysaccharide | 0.8 (0.1) |
| Galactomannan (carob) | Hemicellulosic polysaccharide | 1.2 (0.1) |
| Glucomannan (konjac) | Hemicellulosic polysaccharide | 1.9 (0.4) |
All carbohydrates were used at a concentration of 5 mg ml−1 in SD minimal medium, except for rhamnogalacturonan I, which was used at a concentration of 10 mg ml−1. The following carbohydrates did not support growth: d-arabinose, l-fucose, ribose, d-glucuronic acid, N-acetylgalactosamine, l-rhamnose, sucrose, lactose, carboxymethyl cellulose, α-mannan (Saccharomyces cerevisiae), α-arabinan (sugar beet), arabinogalactan (larch), inulin (chicory), β-1,4-galactan (potato and lupin), λ-carrageenan (seaweed), levan (Erwinia herbicola), rhamnogalacturonan I (potato pectin), RNA, DNA (salmon sperm), and xanthan gum. The sources of some of the polysaccharides are indicated in parentheses.
Relative growth rates were determined by dividing the doubling time (in min) in SD medium plus glucose by the doubling time in SD plus the test substrate. Thus, a relative growth rate greater than 1 for a substrate indicates that growth on the substrate was more rapid than growth on glucose. Each substrate was tested in quadruplicate, and standard deviations are indicated in parentheses. ±, cells grew too slowly to calculate the relative growth rate, but the optical density at 600 nm was at least 0.1 after 88 h of incubation; +, cells grew on the substrate, but the growth rate could not be determined accurately due to the high turbidity of the initial growth medium containing the insoluble polysaccharide.
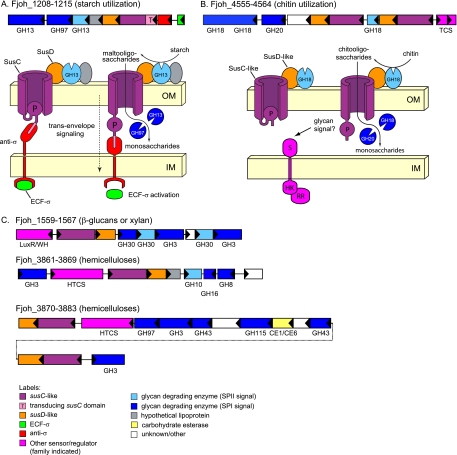
FIG. 2.
Representative PULs of F. johnsoniae. (A) Putative starch utilization PUL with components that are similar to those of the prototypic starch utilization system (Sus) of B. thetaiotaomicron. Like the prototypic Sus system, this system includes two GH13 α-amylases, one of which has a lipidation signal similar to the endo-acting surface enzyme SusG (light blue), and a single GH97 α-glucosidase. Notably, this system lacks a homolog of the inner-membrane-spanning maltose sensor, SusR, and instead is linked to an extracytoplasmic function sigma (ECF-σ)-anti-σ transcriptional regulator pair (green and red, respectively). These regulatory elements function by coupling to a specialized N-terminal “transducing domain” (pink) attached to the SusC-like transporter and together comprise a “transenvelope signaling” pathway spanning both bacterial membranes. (B) Putative chitin utilization PUL. In contrast to the system shown in panel A, this system includes three glycoside hydrolases predicted to target the β-1,4-N-acetylglucosamine linkages found in chitin (GH18 and GH20 enzymes). One of these enzymes has a secretion signal predicted to position it on the cell surface or in the extracellular space (light blue). Unlike other PULs in other bacteroidetes delineated so far, the chitin utilization PUL is associated with a classic two-component regulatory system (dark pink). Other functional labels for each system's schematic diagram are indicated at the bottom. (C) Representative PULs with predicted roles in hemicellulose utilization (see Fig. S2 in the supplemental material for a more complete list). OM, outer membrane; IM, inner membrane; TCS, two-component regulatory system; HTCS, hybrid two-component regulatory system.
Other predicted proteins that may be involved in polysaccharide utilization include proteins with similarity to the B. thetaiotaomicron outer membrane starch utilization proteins SusC and SusD. B. thetaiotaomicron is an anaerobic inhabitant of the human large intestine and is a distant relative of F. johnsoniae. The B. thetaiotaomicron polysaccharide-degrading enzymes are primarily cell associated (6, 81). The outer membrane lipoprotein SusD and the outer membrane protein SusC are involved in binding starch on the cell surface and in transport of oligomers across the outer membrane. Homologs of SusC and SusD are common in the phylum Bacteroidetes (31). Analysis of the F. johnsoniae genome revealed the presence of 44 susC-like genes and 42 susD-like genes (see Table S4 in the supplemental material). Each of the susD-like genes was located immediately downstream of a susC-like gene. In many cases the genes adjacent to the susC-like and susD-like genes are predicted to be involved in polysaccharide utilization (Fig. 2; see Fig. S2 in the supplemental material). In Bacteroides species such regions have been referred to as polysaccharide utilization loci (PULs) (13). The SusC-like and SusD-like proteins are likely involved in binding and uptake of the polysaccharide substrates that are attacked by the products of the neighboring genes. The SusC-like proteins are part of a larger family of TonB receptor-like proteins. In addition to the SusC-like proteins, 50 other TonB receptor-like proteins were identified using the genome information. Some of these proteins are predicted to function in Fe uptake, but the roles of the others are not known.
The SusC-like and SusD-like proteins that have been studied are involved in the utilization of soluble polysaccharides (56, 77). Analysis of the F. johnsoniae genome suggests that some SusC-like and SusD-like proteins may also function in utilization of insoluble polysaccharides, such as chitin and hemicelluloses. The region from Fjoh_4555 to Fjoh_4564 is likely to be involved in chitin utilization (Table 1 and Fig. 2). It contains genes encoding several candidate chitinases (Fjoh_4555, Fjoh_4560), a candidate 1,4-β-N-acetylglucosaminidase (Fjoh_4556), a glucosamine-6-phosphate isomerase/deaminase (Fjoh_4557), two SusC-like proteins (Fjoh_4559, Fjoh_4562), and two SusD-like proteins (Fjoh_4558, Fjoh_4561). The 1,4-β-N-acetylglucosaminidases thought to be involved in chitin digestion are all predicted to be periplasmic or cytoplasmic enzymes (Table 1), suggesting that chitin oligomers are transported across the outer membrane before digestion, as expected if SusC-like and SusD-like proteins are involved in chitin utilization. This suggests a model for chitin utilization in which cell surface proteins bind chitin and perform the initial digestion to form soluble oligomers that are transported into the periplasm for further digestion by 1,4-β-N-acetylglucosaminidases. This model may explain why intimate contact with insoluble chitin is needed for efficient utilization (61, 88). This strategy may also be used by F. johnsoniae for digestion of hemicelluloses, and it may be used by other bacteroidetes that digest insoluble substrates. For example, Cytophaga hutchinsonii has SusC-like and SusD-like proteins that may be involved in utilization of insoluble cellulose (95).
Proteases.
Casein, gelatin, and other proteins are digested by F. johnsoniae and can serve as sole C, N, and energy sources (19, 21, 87). The MEROPS peptidase database recognized 124 predicted F. johnsoniae peptidases (see Table S5 in the supplemental material) and another 35 “nonpeptidase homolog” proteins that exhibit sequence similarity to peptidases but appear to lack residues critical for activity. The number of predicted peptidases is greater than that for any other member of the Bacteroidetes with a completed genome sequence. In addition to peptidases identified by MEROPS, F. johnsoniae has one additional predicted peptidase (Fjoh_0798), a homolog of the F. psychrophilum protease Fpp1 (83) that is not currently included in the MEROPS database. Ten of the predicted peptidase-encoding genes listed in Table S5 in the supplemental material are located near susC-like genes (see Fig. S2 in the supplemental material; data not shown). For eight of these genes, genes encoding obvious polysaccharide utilization proteins have not been found nearby, raising the possibility that these susC-like genes may function in protein utilization rather than in polysaccharide utilization. Proteases of the pathogenic bacteroidetes Porphyromonas gingivalis and F. psychrophilum have been suggested to play important roles in virulence (24, 69). F. johnsoniae has homologs of several of the extracellular proteases of these organisms, but it lacks homologs of the P. gingivalis gingipain proteases, which have been most clearly associated with virulence.
Protein export and secretion.
As mentioned above, extracellular and cell surface proteins appear to be involved in polymer digestion by F. johnsonaie. Export of proteins across the cytoplasmic membrane is apparently mediated by SecA, SecE, SecY, SecDF, SecG, YidC, and YajC of the Sec system and TatA and TatC of the twin-arginine transport system. Several systems are available to mediate secretion of proteins across the outer membrane. Components of an apparent type II secretion system (GspD, GspE, GspF, GspG, and GspJ) are present. In addition, multiple copies of type IV secretion system genes encoding proteins related to VirB4 and VirD4 are also present and are associated with apparent conjugative transposons. These proteins may be involved in translocation of DNA and/or proteins. Possible ATP-binding cassette transporters that may transport specific proteins (type I transport) were also identified. Finally, F. johnsoniae has predicted components of a proposed bacteroidete-specific protein secretion system (K. Sato, M. Naito, H. Yukitake, H. Hirakawa, M. Shoji, M. J. McBride, R. G. Rhodes, and K. Nakayama, submitted for publication). This system, which is required for cell movement, is discussed below. The F. johnsoniae genome is predicted to encode 423 lipoproteins (8.37% of all proteins), as well as machinery for their processing (Lgt, LspA) and translocation (LolA).
Central metabolism and biosynthetic capabilities.
F. johnsoniae carries out aerobic respiration of glucose, and genome analysis indicated the presence of a complete Embden-Meyerhof-Parnas pathway and a tricarboxylic acid cycle. Genes encoding each of the NADH dehydrogenase subunits, cytochrome c, cytochrome c oxidase, and components of ATP synthase were also present. F. johnsoniae uses menaquinones instead of ubiquinones as respiratory electron transport chain components, and genes encoding the enzymes for menaquinone synthesis were identified. F. johnsoniae does not have genes encoding the components of the bc1 complex (also known as complex III) that functions as a quinol:cytochrome c oxidoreductase in many aerobes. Instead, F. johnsoniae has genes encoding components of an alternative complex III that is thought to function as a menaquinol:cytochrome c oxidoreductase. The components of this complex were recently recognized in the distantly related bacteroidete R. marinus and in the photosynthetic bacterium Chloroflexus aurantiacus (70, 99). Involvement of menaquinones and replacement of the bc1 complex with the novel menaquinol oxidoreductase appear to be universal among aerobic bacteroidetes for which genome sequence data are available, including F. johnsoniae, F. psychrophilum, Gramella forsetii, C. hutchinsonii, and R. marinus. Some apparent metabolic abilities of F. johnsoniae were not anticipated. For example, unlike most other bacteroidetes, this species has a cluster of genes (Fjoh_3902 to Fjoh_3913) predicted to encode a nickel-dependent uptake hydrogenase complex and accessory proteins for maturation of the enzyme. The role of this hydrogenase complex in the energy metabolism of this aerobic respiratory heterotrophic bacterium is not yet clear. F. johnsoniae grows on minimal media with glucose as a sole carbon and energy source and thus has the ability to synthesize all of its organic components from this substrate. As expected, analysis of the genome sequence revealed genes encoding biosynthetic enzymes needed to synthesize amino acids, nucleotides, fatty acids, heme, and many vitamins and coenzymes.
Novel outer membrane lipids and secondary metabolites.
Many gliding bacteroidetes contain novel components in their outer membranes. These components include sulfonolipids, which may give the membrane increased fluidity and be important for gliding (1), and flexirubin pigments (3). Flexirubin pigments are found in many bacteroidetes, including F. johnsoniae, F. psychrophilum, and C. hutchinsonii, for which complete genome sequences are available. Flexirubin pigments typically consist of an ω-phenyloctaenic acid chromophore esterified with resorcinol carrying two hydrocarbon chains (Fig. 3). Variations in the length of the polyenic acid, variations in the R group hydrocarbon chains, and substitutions on the phenyl rings result in production of a variety of different flexirubin pigments. A single strain of F. johnsoniae was reported to produce 25 different flexirubin pigments (2). Flexirubins are yellow at neutral pH but undergo a reversible switch to red under alkaline conditions. This allows simple identification of flexirubin-negative mutants.
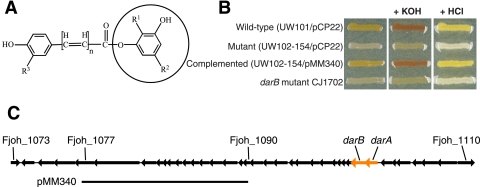
FIG. 3.
Identification of a cluster of genes involved in synthesis of flexirubin. (A) Structure of F. johnsoniae flexirubin with the dialkyl resorcinol moiety circled. “n” indicates that the length of the polyene units varies from 6 to 8. R1 and R2 indicate alkyl chains that are various lengths and have various structures, and R3 indicates either H or Cl. (B) Analysis of wild-type, mutant, and complemented strains for flexirubin pigment. Cells were grown on CYE agar, and the same samples were photographed before treatment (unlabeled), after exposure to 50 μl of 10 N KOH (+ KOH), and after exposure to KOH followed by exposure to 42 μl 12 N HCl (+ HCl). Flexirubin-positive cells were yellow at neutral pH and red under alkaline conditions. (C) Gene cluster linked to flexirubin synthesis. The 37-kbp region of DNA spanning Fjoh_1073 to Fjoh_1110 is shown. The predicted dialkyl resorcinol biosynthesis genes darA and darB are orange. pMM340, which complements the flexirubin mutant UW102-154, carries a 12.8-kbp region spanning Fjoh_1078 to Fjoh_1089.
A combination of genome analysis and genetic experiments resulted in identification of a cluster of genes involved in flexirubin synthesis. Comparative genome analysis identified two genes, Fjoh_1103 and Fjoh_1102, with likely roles in flexirubin synthesis. Homologs of these genes were identified in the genomes of the three flexirubin-producing bacteroidetes, F. johnsoniae, F. psychrophilum, and C. hutchinsonii, but not in the flexirubin nonproducers Bacteroides fragilis, B. thetaiotaomicron, Bacteroides vulgatus, G. forsetii, Parabacteroides distasonis, and P. gingivalis. Fjoh_1103 and Fjoh_1102 are similar to P. aurantiaca darA, and darB, respectively, which are involved in biosynthesis of the antifungal compound 2-hexyl-5-propyl-alkylresorcinol (68). Similar 2,5-dialkylresorcinol compounds are likely intermediates in F. johnsoniae flexirubin biosynthesis. An insertion mutation was constructed in F. johnsoniae darB (Fjoh_1102), and the resulting strain, CJ1702, lacked flexirubin pigments. Cells of the mutant were cream colored, whereas wild-type cells were yellow at neutral pH and red under alkaline conditions (Fig. 3B). The conversion of yellow wild-type cells to red cells by exposure to KOH was reversed by addition of HCl, as previously reported for flexirubin pigments (3). F. johnsoniae darA and darB are part of a large cluster of genes transcribed in the same direction and organized in what appear to be several operons (Fig. 3C). Genome comparisons revealed that in addition to darA and darB, homologs of seven other genes in this cluster (Fjoh_1080, Fjoh_1084, Fjoh_1095, Fjoh_1097, Fjoh_1098, Fjoh_1100, Fjoh_1108) are present in the flexirubin-positive bacteroidetes mentioned above but not in bacteroidetes that are flexirubin negative. Most of these genes encode hypothetical proteins of unknown function, but Fjoh_1095 encodes a predicted component of an ABC-2-type transporter that could be involved in localization of flexirubin pigments. Many other genes in this region are predicted to encode enzymes involved in lipid synthesis, such as a (3-oxoacyl)-acyl carrier protein synthase (Fjoh_1087), beta-ketoacyl synthases (Fjoh_1088, Fjoh_1093, Fjoh_1106), and a beta-hydroxyacyl-(acyl carrier protein) dehydratase (Fjoh_1081), and some of these enzymes could have roles in flexirubin synthesis. darA, darB, and perhaps other genes in this region may also be involved in the synthesis of the mammalian cell growth-promoting dialkylresorcinol resorcinin, which is produced by strains of F. johnsoniae (38). Cosmid complementation of the spontaneous flexirubin-negative mutant F. johnsoniae UW102-154 (19, 37) confirmed that genes in this region other than darA and darB are involved in flexirubin synthesis, since the complementing plasmid, pMM340, carried a region spanning Fjoh_1078 to Fjoh_1089 (Fig. 3B and 3C). Further analysis of this region will likely elucidate the steps involved in the biosynthesis of flexirubins and of other bacteroidete dialkylresorcinols. Similar approaches could identify genes involved in sulfonolipid synthesis.
Strains of F. johnsoniae are known to produce several secondary metabolites in addition to resorcinin, including monobactam and quinoline antibiotics (25, 44, 45), and numerous other secondary metabolites are produced by related bacteria (66). Several clusters of genes likely to be involved in secondary metabolite production were identified in the F. johnsoniae genome. Most striking was a cluster of genes in the 70.7-kbp region spanning Fjoh_2083 to Fjoh_2104 that appear to encode components of a multienzyme nonribosomal peptide synthetase assembly line with a novel predicted product having an unknown function (see Fig. S3 in the supplemental material). F. johnsoniae also appears to have the capacity to produce an aerobactin-like siderophore. Genes located between Fjoh_3170 and Fjoh_3179 are predicted to be involved in the regulation, synthesis, and export of this compound.
Signal transduction and regulation of gene expression.
F. johnsoniae genes encode a variety of proteins that are predicted to regulate gene expression in response to external or internal stimuli. The sigma factors include an RpoD (σ70) homolog, an RpoN (σ54) homolog, and 27 sigma factors belonging to the ECF subfamily. F. johnsoniae RpoD is similar in size (32.7 kDa) and sequence to other bacteroidete RpoD proteins and is much smaller than E. coli σ70. Like the other bacteroidete RpoD proteins, F. johnsoniae RpoD lacks regions found in most nonbacteroidete RpoD proteins, such as N-terminal region 1.1 and the segment between regions 1.2 and 2.1 (92). The novel structure of bacteroidete RpoD sigma factors may account for some of the unusual features of bacteroidete housekeeping promoters (10, 20). For example, the −33/−7 consensus promoter sequence of F. johnsoniae and other members of the Bacteroidetes phylum that have been studied (TTG/TANNTTTG) differs from the −35/−10 consensus promoter sequences of other well-studied bacteria (20). A search of the F. johnsoniae genome with the promoter consensus sequence (with spacing between the two motifs set at the optimal value, 19 bp) revealed 109 exact matches in intergenic regions on the coding strand within 300 bp upstream of a start codon, whereas a similar search of the E. coli genome with the same consensus sequence revealed less than 10 sequences (20). Not surprisingly many of the putative F. johnsoniae promoters identified were associated with housekeeping genes that are likely to be highly expressed. Promoters with slight variations in the consensus sequence would have been missed by this analysis, so it is likely that many more RpoD-dependent promoters are present in the F. johnsoniae genome.
The functions of F. johnsoniae RpoN (σ54) are not known, but σ54-like proteins are required for transcription of genes involved in a wide variety of processes in other bacteria. In these other bacteria σ54-RNA polymerase holoenzyme (σ54-holoenzyme) binds to promoter sequences to form a closed complex, but it cannot proceed further in transcription initiation in the absence of an activator. Activators of σ54-holoenzyme generally bind to sites located upstream of the promoter and contact σ54-holoenzyme through DNA looping to stimulate transcription. Binding sites for σ54-dependent activators are referred to as bacterial enhancers because some of them can function when they are moved several kilobases from their native locations.
Activators of σ54-holoenzyme usually consist of an N-terminal regulatory domain, a central domain required for ATP hydrolysis and transcriptional activation, and a C-terminal DNA-binding domain. F. johnsoniae has six predicted σ54-dependent activators (Fjoh_0470, Fjoh_0638, Fjoh_1332, Fjoh_1848, Fjoh_1977, and Fjoh_5047). Five of these activators have N-terminal response regulator receiver domains, whereas the remaining activator (Fjoh_1848) contains an N-terminal PAS domain. The stimuli controlling these activators are not known, but Fjoh_0470, Fjoh_0638, and Fjoh_1332 are adjacent to genes that likely encode the presumptive cognate sensor kinases.
Fjoh_5047 is unique among the F. johnsoniae σ54-dependent activators in that it lacks a DNA-binding domain. When expressed in E. coli, Fjoh_5047 activated transcription from a σ54-dependent lacZ reporter gene (data not shown), indicating that it is a functional activator. Examples of σ54-dependent activators lacking DNA-binding domains are found in other members of the Bacteroidetes (11). A problem of specificity of gene activation arises for these unusual activators. In the few bacteria outside the Bacteroidetes phylum that are known to have such activators, specificity is conferred by one of two mechanisms. Some of these bacteria have only a single σ54-dependent activator and thus avoid the specificity problem entirely, and specificity is conferred by the sigma factor (67). In other cases multiple σ54-dependent activators are present, but these activators interact with different versions of σ54 (74, 75). F. johnsoniae has multiple σ54-dependent activators and a single σ54, so it is unclear how specificity could be conferred without a DNA-binding domain. Fjoh_5047 may indiscriminately activate transcription of all of the genes in the RpoN regulon. Alternatively, Fjoh_5047 may gain specificity by interacting with an unidentified DNA-binding protein, or it may engage σ54-holoenzyme in solution to alter the promoter specificity of the enzyme.
The genes regulated by RpoN and its activators are not known, but analysis of genes located near the genes encoding σ54-dependent activators and identification of sequences that match the σ54 consensus sequence suggest likely candidates (see Table S6 in the supplemental material). Two apparent operons (Fjoh_0471, Fjoh_0472, and Fjoh_0473; and Fjoh_1331, Fjoh_1330, and Fjoh_1329) are particularly good candidates for σ54-dependent regulation because they are located adjacent to genes encoding σ54-dependent activators and they have sequences that match the σ54 promoter consensus sequence. In addition, multiple regions of dyad symmetry that may function as binding sites for the σ54-dependent activators are located within 63 bp upstream of these putative promoters. Both of these operons are predicted to encode ABC-type antibacterial peptide transport systems, suggesting roles for σ54 in resistance to such compounds.
The presence of large numbers of ECF sigma factors is a common property of members of the phylum Bacteroidetes (46). ECF sigma factors in other bacteria regulate expression of genes with extracellular functions. This regulation often involves transmembrane FecR-like anti-sigma factors that sense extracytoplasmic stimuli, and 12 of the 27 F. johnsoniae genes encoding ECF sigma factors are adjacent to genes predicted to encode such anti-sigma factors. Eight of the ECF sigma factors are encoded by genes that are adjacent to susC-like and susD-like genes (Fig. 2; see Fig. S2 in the supplemental material), and each of these genes is accompanied by a fecR-like anti-sigma factor gene. The eight SusC-like proteins encoded by these regions have an extra N-terminal signaling domain that is thought to interact with the relevant anti-sigma factor during signaling (Fig. 2) (48). Expression of nearby genes involved in macromolecule utilization may be regulated in response to the substrates via interaction of a SusC-like protein with components of the corresponding ECF sigma factor-anti-sigma factor system. The other susC-like and susD-like gene-containing PULs lack ECF sigma factor genes but contain genes encoding other types of regulatory proteins (Fig. 2; see Fig. S2 in the supplemental material). These proteins include members of two-component regulatory systems, LuxR-winged helix regulators, LacI-type repressors, XRE-like repressors, and AraC family regulators.
The diversity of regulatory mechanisms extends beyond genes involved in utilization of polysaccharides. F. johnsoniae genes are predicted to encode 107 transcription regulatory proteins. Several regulatory systems found in F. johnsoniae were unexpected. F. johnsoniae has genes encoding components of the rsb global stress response system, which is not widely distributed among members of the phylum Bacteroidetes. This system involves control of an alternative sigma factor by a complex anti-sigma factor-anti-sigma factor agonist interaction that is modulated by phosphorylation (55). F. johnsoniae genes are predicted to encode 30 cyclic-nucleotide-binding proteins, and this organism probably uses cyclic AMP to regulate gene expression and enzymatic activities. In contrast, it does not appear to use the common bacterial signaling molecule cyclic di-GMP because, as is the case for most bacteroidetes, genes encoding GGDEF and EAL domain proteins have not been identified.
F. johnsoniae has several regulatory systems that may respond to light. The genes encoding these systems include genes encoding two phytochromes (Fjoh_3820 and Fjoh_3936) and genes encoding proteins related to the cyanobacterial circadian rhythm regulatory proteins KaiB (Fjoh_ 4009 and Fjoh_4010) and KaiC (Fjoh_4008). The functions of these genes in F. johnsoniae are not known, but their presence suggests the possibility that this heterotrophic bacterium may respond to light and regulate gene expression in a circadian manner. The two phytochromes are similar to other bacterial phytochromes and contain PAS, GAF, phytochrome, and histidine kinase domains. In addition to the putative phytochromes, F. johnsoniae genes are predicted to encode 13 other PAS domain-containing proteins of unknown function and four GAF domain-containing proteins of unknown function.
Gliding motility.
F. johnsoniae cells cannot swim in liquid, but they attach to and move along surfaces at speeds of up to 5 μm/s in a process known as gliding motility. Gliding motility is characteristic of many members of the phylum Bacteroidetes (58). In some bacteria belonging to other phylogenetic branches, flagella and type IV pili allow cells to move over surfaces (32, 57). Electron microscopic analyses have failed to identify these organelles on cells of F. johnsoniae, and analysis of the genome also failed to identify genes encoding critical components of flagella and type IV pili, suggesting that F. johnsoniae gliding motility is achieved by another mechanism.
Fourteen genes (gldA, gldB, gldD, gldF, gldG, gldH, gldI, gldJ, gldK, gldL, gldM, gldN, sprA, and sprB) involved in F. johnsoniae gliding motility have been identified (4, 14, 15, 35-37, 60, 61, 64, 65). These genes are scattered across 2.24 Mb of DNA (Fig. 1) and are organized in 11 operons. The proteins encoded by these genes all localize or are predicted to localize to the cell envelope, and six of them are lipoproteins. Many of the motility proteins are novel and lack homologs outside the Bacteroidetes. Some or all of the Gld proteins are thought to be components of the “motor” that propels the cell (41). SprB is a large cell surface protein that is thought to function as an adhesin and appears to be propelled along the cell surface by the Gld motor (64). Disruption of gld genes results in a complete loss of motility, but disruption of sprB causes only a partial defect in motility (64). Cells of sprB mutants fail to move on agar, but they exhibit some motility on glass surfaces. Analysis of the F. johnsoniae genome suggests an explanation for this finding, because multiple paralogs of sprB are present. Cells with mutations in sprB and in one of its paralogs (Fjoh_0808) have much more severe motility defects than cells with mutations in either gene alone (73). Such “synthetic” motility defects suggest that SprB and the other SprB-like proteins are semiredundant mobile cell surface components of the motility machinery that allow attachment to and movement over the many different types of surfaces that cells encounter.
F. johnsoniae genes encode numerous proteins predicted to be involved in the biosynthesis and export of exopolysaccharides. Many of these genes are located in a 76.5-kbp region of DNA spanning Fjoh_0290 to Fjoh_0361, which contains closely spaced genes that are all transcribed in the same direction. Recent results indicate that some of these genes have roles in motility. Strains with mutations in several of these genes have synthetic motility defects in an sprB mutant background (73). Specific exopolysaccharides may be required for efficient movement over some surfaces. SprB and some of the SprB-like proteins have lectin-like domains predicted to interact with carbohydrates. The polysaccharides may coat the substratum and allow productive contact with SprB-like cell surface adhesins, thus facilitating cell movement over diverse surfaces.
The genomes of other bacteroidetes known to exhibit gliding motility, such as F. psychrophilum and C. hutchinsonii, have homologs of each of the 14 F. johnsoniae motility genes described above (see Table S7 in the supplemental material). Not surprisingly, the amino acid sequences of the F. johnsoniae and F. psychrophilum motility proteins are similar, with identities ranging from 32% to 87%. C. hutchinsonii is not closely related to the two flavobacteria, and the levels of amino acid identity with the F. johnsoniae motility proteins are lower, ranging from 22% to 50%. In spite of the divergence in amino acid sequence, the organization of several gene clusters has been highly conserved. For example, gldF and gldG, which have been demonstrated to constitute an operon in F. johnsoniae (35), are also likely to be cotranscribed in F. psychrophilum and in C. hutchinsonii based on genome sequence analyses. GldF and GldG localize in the cytoplasmic membrane of F. johnsoniae and interact with GldA (35). Together, these proteins are thought to constitute an ATP-binding cassette transporter that is required for gliding. C. hutchinsonii gldA appears to be cotranscribed with gldF and gldG, which is consistent with the hypothesis that the products of these genes function together and form a complex. In F. johnsoniae, gldK, gldL, gldM, and gldN are clustered together on the genome and form two adjacent operons, and gldK is transcribed separately from the other three genes (14). A similar arrangement is found in F. psychrophilum and C. hutchinsonii. This may suggest that the protein products of these genes function together as part of a complex and that coordinated expression may be important for assembly of the complex in the cell envelope.
Nonmotile bacteroidetes, such as P. gingivalis, B. fragilis, B. thetaiotaomicron, B. vulgatus, P. distasonis, and Salinibacter ruber, have homologs of some, but not all, of the F. johnsoniae motility genes (see Table S7 in the supplemental material). The SprA homolog in P. gingivalis, Sov, is required for secretion of gingipain proteases (80). Recent results suggest that homologs of GldK, GldL, GldM, and GldN are also required for P. gingivalis gingipain secretion and may be components of a novel protein secretion system (Sato et al., submitted). Conservation of many of the genes encoding these proteins in B. fragilis and P. distasonis suggests that similar protein secretion systems may function in many gliding and nongliding bacteroidetes. The F. johnsoniae secretion system may function in translocation of the motility protein SprB to the cell surface (Sato et al., submitted). The presence of a protein translocation machine at the heart of a motility apparatus is not unique, because the bacterial flagellum is built around a type III secretion system that is involved in secretion of flagellins and some other flagellar proteins (5). P. gingivalis lacks homologs of gldD, gldF, gldG, and gldJ. The proteins encoded by these genes may have functions that are essential for motility but not for protein secretion. The presence of homologs of each of the motility genes in the genome of the relatively unstudied marine bacteroidete G. forsetii (9) suggests that it may have the ability to glide, a property that has not been experimentally observed yet.
Most motile bacteria control their motility to move in a favorable direction. This typically involves a complex signal transduction system related to the E. coli chemotaxis system. F. johnsoniae has homologs of E. coli cheY (Fjoh_3353), cheB (Fjoh_3351), and cheR (Fjoh_3352). These genes are clustered together on the genome, supporting the idea that their products may function together. F. johnsoniae CheB is unusual in that it lacks the N-terminal response regulatory domain that is found in most CheB proteins. Genes encoding other expected components of a bacterial chemotaxis system, such as homologs of methyl-accepting chemotaxis proteins (MCPs), and of CheA, CheW, and CheZ, are apparently not present. The absence of obvious MCPs is surprising since CheB (methylesterase) and CheR (methyltransferase) are expected to modify MCPs. The region surrounding F. johnsoniae cheB, cheR, and cheY contains several genes whose products are likely to be involved in signal transduction. These products include an apparent histidine kinase with a PAS domain (Fjoh_3360), a protein that contains a histidine kinase domain and a response regulatory domain (Fjoh_3355), and a large protein (Fjoh_3354) that contains a CHASE3 domain, a GAF domain, a histidine kinase domain, and a response regulatory domain. PAS, CHASE3, and GAF domains are commonly found in sensory or signal transduction proteins (7, 90, 100) and could have roles in F. johnsoniae tactic responses. However, the absence of central components of bacterial chemotaxis systems, such as CheA and MCPs, and the fact that the closely related gliding bacterium F. psychrophilum lacks cheB, cheR, and other obvious chemotaxis genes suggest that the genes and proteins involved in control of the bacteroidete motility machinery may be novel. Alternatively, cells of F. johnsoniae and F. psychrophilum may lack chemotaxis entirely, as has been suggested for several other motile bacteria that appear to lack critical chemotaxis genes (8, 22, 62). Negative chemotaxis of F. johnsoniae in response to H2O2 and several other chemicals has been reported based on observations of swarming patterns of groups of cells (51), but an analysis of the behavior of individual cells in response to potential chemoeffectors has not been conducted yet.
Numerous aspects of the physiology and molecular biology of F. johnsoniae were revealed in this initial analysis of its genome sequence. Many of these aspects, such as the unusual gliding motility machinery, protein secretion system, and polysaccharide utilization strategy, are shared by other members of the phylum Bacteroidetes. The genome sequence data and the availability of techniques to genetically manipulate F. johnsoniae should result in rapid progress toward a better understanding of the many novel features of these common but understudied bacteria.
Supplementary Material
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy's Office of Science, Biological and Environmental Research Program and at the University of California Lawrence Livermore National Laboratory under contract W-7405-Eng-48, at Lawrence Berkeley National Laboratory under contract DE-AC02-05CH11231, and at Los Alamos National Laboratory under contract DE-AC02-06NA25396. M.J.M. and R.G.R. were supported by National Science Foundation grant MCB-0641366. W.W. and J.X. were supported by a One Hundred Talents grant from the Chinese Academy of Sciences.
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbanat, D. R., E. R. Leadbetter, W. Godchaux III, and A. Escher. 1986. Sulphonolipids are molecular determinants of gliding motility. Nature 324:367-369. [Google Scholar]
- 2.Achenbach, H., W. Kohl, and H. Reichenbach. 1979. Die Konstitutionen der Pigmente vom Flexirubin-typ aus Cytophaga johnsonae Cy j1. Chem. Ber. 112:1999-2011. [Google Scholar]
- 3.Achenbach, H., W. Kohl, W. Wachter, and H. Reichenbach. 1978. Investigations of the pigments from Cytophaga johnsonae Cy jl. New flexirubin-type pigments. Arch. Microbiol. 117:253-257. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal, S., D. W. Hunnicutt, and M. J. McBride. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 94:12139-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aizawa, S. I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, K. L., and A. A. Salyers. 1989. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J. Bacteriol. 171:3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravind, L., and C. P. Ponting. 1997. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22:458-459. [DOI] [PubMed] [Google Scholar]
- 8.Badger, J. H., T. R. Hoover, Y. V. Brun, R. M. Weiner, M. T. Laub, G. Alexandre, J. Mrázek, Q. Ren, I. T. Paulsen, K. E. Nelson, H. M. Khouri, D. Radune, J. Sosa, R. J. Dodson, S. A. Sullivan, M. J. Rosovitz, R. Madupu, L. M. Brinkac, A. S. Durkin, S. C. Daugherty, S. P. Kothari, M. G. Giglio, L. Zhou, D. H. Haft, J. D. Selengut, T. M. Davidsen, Q. Yang, N. Zafar, and N. L. Ward. 2006. Comparative genomic evidence for a close relationship between the dimorphic prosthecate bacteria Hyphomonas neptunium and Caulobacter crescentus. J. Bacteriol. 188:6841-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer, M., M. Kube, H. Teeling, M. Richter, T. Lombardot, E. Allers, C. A. Würdemann, C. Quast, H. Kuhl, F. Knaust, D. Woebken, K. Bischof, M. Mussmann, J. V. Choudhuri, F. Meyer, R. Reinhardt, R. I. Amann, and F. O. Glöckner. 2006. Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ. Microbiol. 8:2201-2213. [DOI] [PubMed] [Google Scholar]
- 10.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 11.Beck, L. L., T. G. Smith, and T. R. Hoover. 2007. Look, no hands! Unconventional transcriptional activators in bacteria. Trends Microbiol. 15:530-537. [DOI] [PubMed] [Google Scholar]
- 12.Beerhues, L., and E. Kombrink. 1994. Primary structure and expression of mRNAs encoding basic chitinase and 1,3-beta-glucanase in potato. Plant Mol. Biol. 24:353-367. [DOI] [PubMed] [Google Scholar]
- 13.Bjursell, M. K., E. C. Martens, and J. I. Gordon. 2006. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 281:36269-36279. [DOI] [PubMed] [Google Scholar]
- 14.Braun, T. F., M. K. Khubbar, D. A. Saffarini, and M. J. McBride. 2005. Flavobacterium johnsoniae gliding motility genes identified by mariner mutagenesis. J. Bacteriol. 187:6943-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun, T. F., and M. J. McBride. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J. Bacteriol. 187:2628-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caspi, R., H. Foerster, C. A. Fulcher, R. Hopkinson, J. Ingraham, P. Kaipa, M. Krummenacker, S. Paley, J. Pick, S. Y. Rhee, C. Tissier, P. Zhang, and P. D. Karp. 2006. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 34:D511-D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang, L. Y. E., and J. L. Pate. 1981. Nutritional requirements of Cytophaga johnsonae and some of its auxotrophic mutants. Curr. Microbiol. 5:235-240. [Google Scholar]
- 19.Chang, L. Y. E., J. L. Pate, and R. J. Betzig. 1984. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J. Bacteriol. 159:26-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, S., M. Bagdasarian, M. G. Kaufman, A. K. Bates, and E. D. Walker. 2007. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J. Bacteriol. 189:5108-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen, P. J. 1977. Synonomy of Flavobacterium pectinovorum Dorey with Cytophaga johnsonae Stanier. Int. J. Syst. Bacteriol. 27:122-132. [Google Scholar]
- 22.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 23.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duchaud, E., M. Boussaha, V. Loux, J. F. Bernardet, C. Michel, B. Kerouault, S. Mondot, P. Nicolas, R. Bossy, C. Caron, P. Bessières, J. F. Gibrat, S. Claverol, F. Dumetz, M. L. Hénaff, and A. Benmansour. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 25:763-769. [DOI] [PubMed] [Google Scholar]
- 25.Evans, J. R., E. J. Napier, and R. A. Fletton. 1978. G1499-2, a new quinoline compound isolated from the fermentation broth of Cytophaga johnsonii. J. Antibiot. 31:952-958. [DOI] [PubMed] [Google Scholar]
- 26.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. L. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 27.Gilmore, D. F., W. Godchaux III, and E. R. Leadbetter. 1989. Cysteine is not an obligatory intermediate in the biosynthesis of cysteate by Cytophaga johnsonae. Biochem. Biophys. Res. Commun. 160:535-539. [DOI] [PubMed] [Google Scholar]
- 28.Gilmore, D. F., W. Godchaux III, and E. R. Leadbetter. 1989. Regulation of sulfate assimilation in Cytophaga johnsonae. Arch. Microbiol. 152:387-392. [Google Scholar]
- 29.Godchaux, W., III, and E. R. Leadbetter. 1983. Unusual sulfonolipids are characteristic of the Cytophaga-Flexibacter group. J. Bacteriol. 153:1238-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han, C. S., and P. Chain. 2006. Finishing repetitive regions automatically with Dupfinisher, p. 142-147. In H. R. Arabnia and H. Valafar (ed.), Proceedings of the 2006 International Conference on Bioinformatics & Computational Biology. CSREA Press, Las Vegas, NV.
- 31.Hanley, S. A., J. Aduse-Opoku, and M. A. Curtis. 1999. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect. Immun. 67:1157-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harshey, R. M. 1994. Bees aren't the only ones: swarming in Gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 33.Henikoff, S., and J. G. Henikoff. 1994. Protein family classification based on searching a database of blocks. Genomics 19:97-107. [DOI] [PubMed] [Google Scholar]
- 34.Hinderlich, S., M. Berger, M. Schwarzkopf, K. Effertz, and W. Reutter. 2000. Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. Eur. J. Biochem. 267:3301-3308. [DOI] [PubMed] [Google Scholar]
- 35.Hunnicutt, D. W., M. J. Kempf, and M. J. McBride. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 184:2370-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunnicutt, D. W., and M. J. McBride. 2001. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes gldD and gldE. J. Bacteriol. 183:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunnicutt, D. W., and M. J. McBride. 2000. Cloning and characterization of the Flavobacterium johnsoniae gliding motility genes, gldB and gldC. J. Bacteriol. 182:911-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai, S., K. Fujioka, K. Furihata, R. Fudo, S. Yamanaka, and H. Seto. 1993. Studies on cell growth stimulating substances of low molecular weight. Part 3. Resorcinin, a mammalian cell growth stimulating substance produced by Cytophaga johnsonae. J. Antibiot. 46:1319-1322. [DOI] [PubMed] [Google Scholar]
- 39.Jackson, C. A., B. Hoffmann, N. Slakeski, S. Cleal, A. J. Hendtlass, and E. C. Reynolds. 2000. A consensus Porphyromonas gingivalis promoter sequence. FEMS Microbiol. Lett. 186:133-138. [DOI] [PubMed] [Google Scholar]
- 40.Janson, J. C. 1975. Studies on dextran-degrading enzymes. Isolation and identification of a dextranase-producing strain of Cytophaga johnsonii and studies on the formation of the surface-bound enzyme. J. Gen. Microbiol. 88:205-208. [Google Scholar]
- 41.Jarrell, K. F., and M. J. McBride. 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6:466-476. [DOI] [PubMed] [Google Scholar]
- 42.Juncker, A. S., H. Willenbrock, G. von Heijne, H. Nielsen, S. Brunak, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlsson, E. N., M. A. Hachem, S. Ramchuran, H. Costa, O. Holst, S. F. Svenningsen, and G. O. Hreggvidsson. 2004. The modular xylanase Xyn10A from Rhodothermus marinus is cell-attached, and its C-terminal domain has several putative homologues among cell-attached proteins within the phylum Bacteroidetes. FEMS Microbiol. Lett. 241:233-242. [DOI] [PubMed] [Google Scholar]
- 44.Kato, T., H. Hinoo, J. Shoji, K. Matsumoto, T. Tanimoto, T. Hattori, K. Hirooka, and E. Kondo. 1987. PB-5266 A, B and C, new monobactams. I. Taxonomy, fermentation and isolation. J. Antibiot. 55:135-138. [DOI] [PubMed] [Google Scholar]
- 45.Kato, T., H. Hinoo, Y. Terui, J. Nishikawa, Y. Nakagawa, Y. Ikenishi, and J. Shoji. 1987. PB-5266 A, B and C, new monobactams. II. Physico-chemical properties and chemical structures. J. Antibiot. 55:139-144. [DOI] [PubMed] [Google Scholar]
- 46.Kill, K., T. T. Binnewies, T. Sicheritz-Ponten, H. Willenbrock, P. F. Hallin, T. M. Wassenaar, and D. W. Ussery. 2005. Genome update: sigma factors in 240 bacterial genomes. Microbiology 151:3147-3150. [DOI] [PubMed] [Google Scholar]
- 47.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 48.Koebnik, R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343-347. [DOI] [PubMed] [Google Scholar]
- 49.Koropatkin, N. M., E. C. Martens, J. I. Gordon, and T. J. Smith. 2008. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16:1105-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, L.-Y., N. B. Shoemaker, and A. A. Salyers. 1995. Location and characterization of the transfer region of a Bacteroides conjugative transposon and regulation of the transfer genes. J. Bacteriol. 177:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, Z. X., and I. Fridovich. 1996. Negative chemotaxis in Cytophaga johnsonae. Can. J. Microbiol. 42:515-518. [DOI] [PubMed] [Google Scholar]
- 52.Lovatt, A., and I. S. Roberts. 1994. Cloning and expression in Escherichia coli of the nahA gene from Porphyromonas gingivalis indicates that beta-N-acetylhexosaminidase is an outer-membrane-associated lipoprotein. Microbiology 140:3399-3406. [DOI] [PubMed] [Google Scholar]
- 53.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34:D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marles-Wright, J., and R. J. Lewis. 2007. Stress responses of bacteria. Curr. Opin. Struct. Biol. 17:755-760. [DOI] [PubMed] [Google Scholar]
- 56.Martens, E. C., H. C. Chiang, and J. I. Gordon. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 58.McBride, M. J. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 59.McBride, M. J. 2004. Cytophaga-Flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 7:63-71. [DOI] [PubMed] [Google Scholar]
- 60.McBride, M. J., and T. F. Braun. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 186:2295-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McBride, M. J., T. F. Braun, and J. L. Brust. 2003. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 185:6648-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moran, M. A., A. Buchan, J. M. González, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. D. Selengut, and N. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 63.Muzzarelli, R. 1999. Native, industrial, and fossil chitins, p. 1-6. In P. Jolles and R. A. A. Muzzarelli (ed.), Chitin and chitinases. Birkhauser, Basel, Switzerland. [DOI] [PubMed]
- 64.Nelson, S. S., S. Bollampalli, and M. J. McBride. 2008. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 190:2851-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nelson, S. S., P. P. Glocka, S. Agarwal, D. P. Grimm, and M. J. McBride. 2007. Flavobacterium johnsoniae SprA is a cell surface protein involved in gliding motility. J. Bacteriol. 189:7145-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nett, M., and G. M. Konig. 2007. The chemistry of gliding bacteria. Nat. Prod. Rep. 24:1245-1261. [DOI] [PubMed] [Google Scholar]
- 67.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52:947-961. [DOI] [PubMed] [Google Scholar]
- 68.Nowak-Thompson, B., P. E. Hammer, D. S. Hill, J. Stafford, N. Torkewitz, T. D. Gaffney, S. T. Lam, I. Molnár, and J. M. Ligon. 2003. 2,5-Dialkylresorcinol biosynthesis in Pseudomonas aurantiaca: novel head-to-head condensation of two fatty acid-derived precursors. J. Bacteriol. 185:860-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pereira, M. M., P. N. Refojo, G. O. Hreggvidsson, S. Hjorleifsdottir, and M. Teixeira. 2007. The alternative complex III from Rhodothermus marinus—a prototype of a new family of quinol:electron acceptor oxidoreductases. FEBS Lett. 581:4831-4835. [DOI] [PubMed] [Google Scholar]
- 71.Peterson, S. B., A. K. Dunn, A. K. Klimowicz, and J. Handelsman. 2006. Peptidoglycan from Bacillus cereus mediates commensalism with rhizosphere bacteria from the Cytophaga-Flavobacterium group. Appl. Environ. Microbiol. 72:5421-5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pitta, T., W. Godchaux III, and E. R. Leadbetter. 1993. Protein content of peptidoglycan of liquid-grown cells differs from that of surface-grown, gliding Cytophaga johnsonae. Arch. Microbiol. 160:214-221. [Google Scholar]
- 73.Pochiraju, S., S. S. Nelson, J. Liu, S. Subramaniam, S. Bollampalli, and M. J. McBride. 2007. Abstr. 107th Gen. Meet. Am. Soc. Microbiol., abstr. I-064.
- 74.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2002. The four different σ54 factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 46:75-85. [DOI] [PubMed] [Google Scholar]
- 75.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2006. Transcriptional specificity of RpoN1 and RpoN2 involves differential recognition of the promoter sequences and specific interaction with the cognate activator proteins. J. Biol. Chem. 281:27205-27215. [DOI] [PubMed] [Google Scholar]
- 76.Rawlings, N. D., F. R. Morton, C. Y. Kok, J. Kong, and A. J. Barrett. 2008. MEROPS: the peptidase database. Nucleic Acids Res. 36:D320-D325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reeves, A. R., G. R. Wang, and A. A. Salyers. 1997. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J. Bacteriol. 179:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reichenbach, H., W. Kohl, and H. Achenbach. 1981. The flexirubin-type pigments, p. 101-108. In H. Reichenbach and O. B. Weeks (ed.), The Flavobacterium-Cytophaga group. Verlag Chemie, Weinheim, Germany.
- 79.Rogers, M. J., T. Ohgi, J. Plumbridge, and D. Söll. 1988. Nucleotide sequences of the Escherichia coli nagE and nagB genes: the structural genes for the N-acetylglucosamine transport protein of the bacterial phosphoenolpyruvate:sugar phosphotransferase system and for glucosamine-6-phosphate deaminase. Gene 62:197-207. [DOI] [PubMed] [Google Scholar]
- 80.Saiki, K., and K. Konishi. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol. Immunol. 51:483-491. [DOI] [PubMed] [Google Scholar]
- 81.Salyers, A. A., A. Reeves, and J. D'Elia. 1996. Solving the problem of how to eat something as big as yourself: diverse bacterial strategies for degrading polysaccharides. J. Ind. Microbiol. 17:470-476. [Google Scholar]
- 82.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Secades, P., B. Alvarez, and J. A. Guijarro. 2001. Purification and characterization of a psychrophilic, calcium-induced, growth-phase-dependent metalloprotease from the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 67:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Servant, F., C. Bru, S. Carrere, E. Courcelle, J. Gouzy, D. Peyruc, and D. Kahn. 2002. ProDom: automated clustering of homologous domains. Brief. Bioinform. 3:246-251. [DOI] [PubMed] [Google Scholar]
- 85.Shipman, J. A., J. E. Berleman, and A. A. Salyers. 2000. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 182:5365-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sonnhammer, E. L., S. R. Eddy, and R. Durbin. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405-420. [DOI] [PubMed] [Google Scholar]
- 87.Stanier, R. Y. 1947. Studies on non-fruiting myxobacteria. I. Cytophaga johnsonae, n. sp., a chitin-decomposing myxobacterium. J. Bacteriol. 53:297-315. [PMC free article] [PubMed] [Google Scholar]
- 88.Sundarraj, N., and J. V. Bhat. 1972. Breakdown of chitin by Cytophaga johnsonii. Arch. Mikrobiol. 85:159-167. [DOI] [PubMed] [Google Scholar]
- 89.Suzuki, M., M. Morimatsu, T. Yamashita, T. Iwanaga, and B. Syuto. 2001. A novel serum chitinase that is expressed in bovine liver. FEBS Lett. 506:127-130. [DOI] [PubMed] [Google Scholar]
- 90.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsujibo, H., K. Fujimoto, H. Tanno, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1994. Gene sequence, purification and characterization of N-acetyl-beta-glucosaminidase from a marine bacterium, Alteromonas sp. strain O-7. Gene. 146:111-115. [DOI] [PubMed] [Google Scholar]
- 92.Vingadassalom, D., A. Kolb, C. Mayer, T. Rybkine, E. Collatz, and I. Podglajen. 2005. An unusual primary sigma factor in the Bacteroidetes phylum. Mol. Microbiol. 56:888-902. [DOI] [PubMed] [Google Scholar]
- 93.Watanabe, T., W. Oyanagi, K. Suzuki, K. Ohnishi, and H. Tanaka. 1992. Structure of the gene encoding chitinase D of Bacillus circulans WL-12 and possible homology of the enzyme to other prokaryotic chitinases and class III plant chitinases. J. Bacteriol. 174:408-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Watanabe, T., K. Suzuki, W. Oyanagi, K. Ohnishi, and H. Tanaka. 1990. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J. Biol. Chem. 265:15659-15665. [PubMed] [Google Scholar]
- 95.Xie, G., D. C. Bruce, J. F. Challacombe, O. Chertkov, J. C. Detter, P. Gilna, C. S. Han, S. Lucas, M. Misra, G. L. Myers, P. Richardson, R. Tapia, N. Thayer, L. S. Thompson, T. S. Brettin, B. Henrissat, D. B. Wilson, and M. J. McBride. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 73:3536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 97.Xu, J., M. A. Mahowald, R. E. Ley, C. A. Lozupone, M. Hamady, E. C. Martens, B. Henrissat, P. M. Coutinho, P. Minx, P. Latreille, H. Cordum, A. Van Brunt, K. Kim, R. S. Fulton, L. A. Fulton, S. W. Clifton, R. K. Wilson, R. D. Knight, and J. I. Gordon. 2007. Evolution of symbiotic bacteria in the distal human intestine. PLOS Biol. 5:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamano, N., N. Oura, J. Wang, and S. Fujishima. 1997. Cloning and sequencing of the genes for N-acetylglucosamine use that construct divergent operons (nagE-nagAC) from Vibrio cholerae non-O1. Biosci. Biotechnol. Biochem. 61:1349-1353. [DOI] [PubMed] [Google Scholar]
- 99.Yanyushin, M. F., M. C. del Rosario, D. C. Brune, and R. E. Blankenship. 2005. New class of bacterial membrane oxidoreductases. Biochemistry 44:10037-10045. [DOI] [PubMed] [Google Scholar]
- 100.Zhulin, I. B., A. N. Nikolskaya, and M. Y. Galperin. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.