Abstract
Rahnella aquatilis HX2, a biocontrol agent for grapevine crown gall caused by Agrobacterium vitis, produces an antibacterial substance that inhibits the growth of A. vitis in vitro. In this study, we show that MH15 and MH16, two Tn5-induced mutants of HX2, have lost their abilities to inhibit A. vitis and have reduced biocontrol activities; they grow in logarithmic phase at a rate similar to that of the wild type and have single Tn5 insertions. They are also impaired in producing pyrroloquinoline quinone (PQQ) or glucose dehydrogenase (GDH). Complementation of MH15 and MH16 with cosmid clones of CP465 and CP104 from an HX2 DNA library restored the antibiosis, biocontrol, and PQQ or GDH production phenotypes. A 6.7-kb BamHI fragment from CP465 that fully restored the MH15-affected phenotypes was cloned and sequenced. Sequence analysis of the mutated DNA region resulted in the identification of seven open reading frames (ORFs), six of which share significant homology with PQQ-synthesizing genes in other bacteria, designated pqqA through pqqF. Meanwhile, A 5.5-kb PstI fragment from CP104 fully complemented the MH16 mutant and contained a single ORF highly similar to that of genes coding for GDHs. An in-frame gdh deletion mutant has the same phenotypes as the Tn5 mutant of MH16. Complementation of both deletion and Tn5 gdh mutants restored the affected phenotypes to wild-type levels. Our results suggest that an antibacterial substance plays a role in biocontrol of A. vitis by HX2.
Crown gall of grapevine (Vitis vinifera), caused by Agrobacterium vitis (previously referred to as Agrobacterium tumefaciens biotype 3) (42), is an economically important disease that can cause extensive damage to grapevine health and quality in most viticultural regions of the world (6). Systemic survival and translocation of A. vitis within xylem tissue of both symptomatic and asymptomatic vines have been well documented (see reference 5 and references therein). Recurrent and severe attacks of the disease are responsible for serious economic losses in nurseries and vineyards. With no effective chemical control available, the disease can be managed only by using certain cultivation techniques, such as covering the flowering trunks of vines with soil in winter to reduce freeze injury and using A. vitis-free propagation material (6). However, A. vitis can persist in decaying grapevine debris in the soil (7) and can spread to uninfected plants, necessitating the development of alternative or complementary methods for disease management, such as biological control. Biological control of crown gall diseases has been utilized since Agrobacterium rhizogenes K84 (previously referred to as Agrobacterium radiobacter K84) successfully mitigated crown gall disease in many plant species (47). However, K84 could not prevent the infection of grapevines by A. vitis (6). Consequently, several bacterial strains were identified for suppression of grapevine crown gall, including nontumorigenic Agrobacterium strains, such as A. vitis E26 (35), A. vitis F2/5 (8), and A. vitis VAR03-1 (29), and two Pseudomonas strains (3, 31), as well as two strains of Rahnella aquatilis (3, 12). Although these bacteria have been shown to provide some level of control of grapevine crown gall, effective methods of biological control in fields are not clearly defined.
The gram-negative bacterium R. aquatilis is ubiquitous and is characterized by its beneficial traits in plant-bacterium interactions, including mineral phosphate solubilization (MPS), antimicrobial activity, nitrogen fixation, and plant disease suppression (9). We have examined the biological control of grapevine crown gall by nonpathogenic R. aquatilis HX2, which was isolated from vineyard soils in Beijing, China (12). Wild-type HX2 produces an antibacterial substance (ABS) that exhibits a broad spectrum of inhibition activity against bacterial phytopathogens, including Agrobacterium tumefaciens, A. rhizogenes, A. vitis, Pectobacterium carotovorum, Xanthomonas campestris, Xanthomonas oryzae, Pseudomonas syringae, and Clavibacter michiganensis, in vitro (11, 12). HX2 also significantly suppresses crown gall formation on sunflowers (Helianthus annuus L.) and grapes (Vitis vinifera cv. Muscat Hamburg) in the greenhouse (12). In field trials, 3-year average grape crown gall tumor inhibition by HX2 is 80.2% (12). Furthermore, the culture supernatant of HX2 is more efficient in controlling tumor formation than the cell suspension, and the crude extract of the bacteria containing ABS exhibited significant potency in controlling crown gall diseases (12).
Pyrroloquinoline quinone (PQQ), present in a wide variety of foods and possessing remarkable antioxidant properties (24, 34, 41), has attracted considerable interest. In gram-negative bacteria, PQQ functions as a noncovalently bound redox cofactor of several dehydrogenases, including methanol dehydrogenase, ethanol dehydrogenase and glucose dehydrogenase (EC 1.1.5.2) (GDH) (encoded by gdh) (18, 39, 49). GDH is one of the quinoproteins, which are enzymes with an o-quinone prosthetic group, and it has been found both in soluble fractions and in membrane fractions as membrane-bound GDH (mGDH) (see reference 39 and references therein). Both mGDH and soluble-fraction GDH use PQQ as a cofactor and are associated with the periplasmic oxidation of glucose to gluconic acid, although they have different substrate specificities (39). mGDH has been identified in a variety of bacteria, and its genes have been shown to be highly conserved (see reference 1 and references therein).
Some information on the biochemical functions of PQQ following formation of the GDH-PQQ holoenzyme has been documented for plant-bacterium interactions. There are reports that plant growth-promoting bacteria are able to solubilize inorganic and/or organic phosphates in soil, which is dependent on the GDH-PQQ holoenzyme activity (22, 36). In addition, transfected PQQ genes enhanced the phenotype of MPS in two rhizobacterial strains, Burkholderia cepacia IS-16 and a Pseudomonas sp. strain (43). Moreover, PQQ-linked GDH was recently shown to be required by Sinorhizobium meliloti for optimal nodulation efficiency and competitiveness on alfalfa roots (4). Several reports claim that the GDH-PQQ holoenzyme is involved in antimicrobial substance production in Pseudomonas fluorescens (17, 28, 46) and in Enterobacter intermedium (22), respectively. Additionally, it has been reported that PQQ plays a role beyond that of a cofactor of the PQQ-dependent dehydrogenase. PQQ promotes growth of both mammals and plants because of its antioxidant activity (14, 48) and is also found to be a chemotactic attractant for Escherichia coli (15).
Genes associated with PQQ production have been identified for various bacteria, including Acinetobacter calcoaceticus (21), E. intermedium 60-2G (32), Gluconobacter oxydans (26), Klebsiella pneumoniae (40), Methylobacterium extorquens AM1 (51), P. fluorescens B16 (14), and P. fluorescens CHA0 (46). The pqqABCDEF genes are conserved in these bacteria, but the accurate biochemical functions of the encoded proteins remain to be determined.
Previous study in this laboratory suggests that the ABS may contribute to the ability of R. aquatilis HX2 for biocontrol of crown gall diseases (11, 12). Recent experiments revealed that the ABS may be a thermostable and alkali-sensitive substance containing sugar(s) and an unknown 285-nm-absorbing substance. This substance exhibited a bactericidal effect against A. vitis both in vitro and in vivo, effects that were attributed mainly to inhibition of RNA and protein synthesis in A. vitis cells (11). However, the molecular mechanisms and the microbial determinants involved in biological control and antibacterial activity of strain HX2 are largely unknown. In this article, both PQQ and GDH produced by R. aquatilis strain HX2 are suggested to be necessary for biosynthesis of the ABS. We identified six previously known pqq genes and one gdh gene. Our data suggest that the ABS produced by R. aquatilis strain HX2 is involved in the biological control of crown gall diseases.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. aquatilis strains were grown at 28°C on potato dextrose agar (PDA) medium or potato dextrose broth (PDB). A. vitis strain K308 was cultured at 28°C on yeast extract broth (54) or yeast extract broth agar. Escherichia coli strains DH5α and S17-1(λ-pir) were grown at 37°C on Luria-Bertani (LB) medium. When required, antibiotics were added at the following concentrations in medium: for growth of Rahnella, 50 μg ml−1 kanamycin and 5 μg ml−1 gentamicin; for E. coli, 50 μg ml−1 kanamycin, 50 μg ml−1 ampicillin, and 20 μg ml−1 tetracycline. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added at 40 μg ml−1.
TABLE 1.
Bacterial strains and plasmidsa
| Strains/plasmids | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F−recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1Δ (argE-lacZYA)169φ80lazAΔM15 | 23 |
| S17-1(λ-pir) | thi pro hsdR hsdM+recA RP4-2 Tc::Mu-Km::Tn7 λ-pir | 16 |
| Rahnella aquatilis | ||
| HX2 | Apr, wild type, ABS+, biocontrol | This study |
| MH15 | Apr Kmr, HX2 derivative, containing a Tn5 insertion, ABS−, reduced biocontrol | This study |
| MH16 | Apr Kmr, HX2 derivative, containing a Tn5 insertion, ABS−, reduced biocontrol | This study |
| HX2Δgdh | Apr, HX2 derivative with 2.1-kb deletion in gdh gene, ABS−, reduced biocontrol | This study |
| CMH15(p465) | Apr Kmr Tcr, MH15 containing cosmid CP465, complemented strain, ABS+, biocontrol | This study |
| CMH15(pqq) | Apr Kmr Tcr, MH15 containing plasmid pCH15 with pqq genes, complemented strain, ABS+, biocontrol | This study |
| CMH16(p104) | Apr Kmr Tcr, MH16 containing cosmid CP104, complemented strain, ABS+, biocontrol | This study |
| CMH16(gdh) | Apr Gmr Kmr Tcr, MH16 containing plasmid pCH16 with gdh gene, complemented strain, ABS+, biocontrol | This study |
| CHX2Δgdh | Apr, Gmr; Tcr, HX2Δgdh containing plasmid pCH16 with the gdh gene, complemented strain, ABS+, biocontrol | This study |
| Agrobacterium vitis | ||
| K308 | Pathogen of grapevine crown gall, octopine type Ti plasmid | 44 |
| Plasmids | ||
| pUTkm1 | Apr Kmr, delivery plasmid for Tn5, R6K replicon | 25 |
| pGTN | Gmr Kmr, pBSL202 carrying 2-kb XbaI-SmaI fragment containing gfp-nptII fusion operon | 50 |
| pBluescript II SK+ | Apr, ColE1 origin, cloning vector | Stratagene |
| pRK415G | Gmr Tcr; broad-host-rang cloning vector, IncP1 replicon; polylinker of pUC19 | 30 |
| pRK600 | Cmr, ColE1 oriV; RP4; tra+; RP4 oriT; helper plasmid in triparental matings | 19 |
| pLAFR-5 | Tcr, oriT cosmid; | 30 |
| pHSG299 | Kmr, ColE1 origin, cloning vector | TaKaRa |
| CP465 | Tcr, pLAFR-5 containing pqq genes, cosmid | This study |
| CP104 | Tcr, pLAFR-5 containing gdh gene, cosmid | This study |
| pMH16 | Gmr Tcr, pRK415G containing ∼9.0-kb PstI fragment with gdh from cosmid CP104 | This study |
| pCH16 | Gmr Tcr, pRK415G containing 3.2-kb fragment with gdh from plasmid pMH16 | This study |
| pCH15 | Gmr Tcr, pRK415G containing ∼8.0-kb BamHI fragment including pqq genes from cosmid CP465 | This study |
| pHSG16L | Kmr, pHSG299 containing 1.1-kb upstream flanking sequences of gdh | This study |
| pHSG16R | Kmr, pHSG299 containing 1.0-kb downstream flanking sequences of gdh | This study |
| pHSGΔgdh | Kmr, pHSG299 containing 2.2-kb EcoRI -PstI fragment with a 2.1-kb deletion in gdh | This study |
Apr, Cmr, Kmr, Gmr, and Tcr indicate resistance to ampicillin, chloromycetin, kanamycin, gentamicin, and tetracycline, respectively.
Genetic techniques.
Chromosomal DNA from strain HX2 was isolated using the method of Sambrook et al. (45). Isolation of plasmid DNA from E. coli was performed by the alkaline lysis method (45). Restriction enzyme digestions were performed as recommended by the suppliers (Takara, Japan). Gel electrophoresis was performed in 0.8% agarose gels. Construction of a genomic library of strain HX2 was performed as follows: a 20- to 30-kb DNA insert was prepared by partial digestion of the total genomic DNA with MobI and the fragments ligated into the BamHI and ScaI sites of pLAFR5. The ligated DNA was packaged into bacteriophage λ, as described by the manufacturer (Promega, Madison, WI) and then transfected into E. coli DH5α. All other basic molecular techniques were employed according to standard protocols (45). Pfu DNA polymerase (Promega) was used for PCR amplification of inserts for cloning, and Taq DNA polymerase (TaKaRa) was used for PCR amplification in test reactions (e.g., colony PCR). Primers are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | DNA sequencea |
|---|---|
| Primers for cloning Tn5-flanking sequences | |
| PI | 5′-AGATCTGATCAAGAGACAG-3′ |
| PO | 5′-ACTTGTGTATAAGAGTCAG-3′ |
| Primers for amplifying sequences to screen cosmid clones | |
| P1 | 5′-TGTCCGTATTGCTCGAACCC-3′ |
| P2 | 5′-CAGCACGAAGTTCAGCACC-3′ |
| P3 | 5′-CACGTCGTATCATTCTGCC-3′ |
| P4 | 5′-CTTCATAACGCAGTTT GTGG-3′ |
| Primers for amplifying sequences used in in-frame deletion mutagenesis | |
| P16L1(EcoRI) | 5′-ATGAATTCGTGGAACCTGACATCGC-3′ |
| P16L2(BamHI) | 5′-ATGGATCCAGATACCGCCGACAAG-3′ |
| P16R1(BamHI) | 5′-ATGGATCCTACCTGCGTGCATTTGA-3′ |
| P16R2(PstI) | 5′-TAACTGCAGTCTGACCCTGGAACCTC-3′ |
| Primers for amplification of gdh | |
| Gdh-up (PstI) | 5′-AATCTGCAGCATGGGCTACGCTTCTCTTGA-3′ |
| Gdh-down (EcoRI) | 5′-ATGAATTCGATTACGCCCGCATCTTTG-3′ |
Underlined bases are restriction enzyme cut sites.
Transposon mutagenesis and screening of ABS-deficient mutants.
Tn5 mutagenesis of strain HX2 was accomplished by conjugal mating with E. coli S17-1 (pUTkm1) (25). Briefly, HX2 was grown for 48 h at 28°C on PDA, and S17-1 was grown on LB medium amended with kanamycin. Growth from two culture plates of each strain was suspended in 1.0 ml of sterile distilled water (SDW), and suspensions of the bacteria were combined. Volumes of 500 μl (each) of the mixed suspension were spread on the surfaces of five MG/L plates (10) (100 μl per plate) and incubated at 28°C for 8 h. Resulting bacterial growth from the matings was scraped from the plates, suspended in SDW, and dilution plated on AB minimal medium (13) containing kanamycin. Transconjugants that grew within 48 to 60 h were recultured on the same medium and assayed for their ability to inhibit growth of A. vitis strain K308 in vitro as previously described (12).
To determine whether single Tn5 insertions occurred in the ABS-deficient mutants, Southern blot hybridizations were conducted using standard Southern blot methods (45). Total genomic DNA was isolated from HX2 and mutants and incubated for at least 10 h overnight at 37°C in the presence of KpnI, SalI, or PstI (which do not cut within the transposon Tn5) under conditions specified by the manufacturer (TaKaRa). A total of 20 μl from each digestion sample was loaded onto a 0.8% agarose gel, transferred onto a nylon membrane, and hybridized with the labeled probe DNA from the pTGN plasmid carrying mini-Tn5gfp-Km (kanamycin resistance gene) (50). Probe labeling and Southern analysis were conducted using the DIG High Primer DNA labeling and detection starter kit I (Roche Applied Science) as per the manufacturer's instructions.
Characterization of the Tn5-flanking sequences.
Genomic DNA of the Tn5 mutant was isolated, digested with KpnI, ligated to plasmid vector pBluescript II SK(+) according to standard protocols (45), and transformed into E. coli DH5α competent cells by heat shock. Transformants containing inserts that included Tn5 were selected on LB plates containing X-Gal, ampicillin, and kanamycin. The clones were characterized by restriction analysis (KpnI) and sequencing. The Tn5-flanking sequence was determined on an ABI model 370 DNA sequencer at Invitrogen Biotechnology Co., Ltd. (Shanghai, China), using primer PI or PO (Table 2) based on the I-end or O-end terminal region of the Tn5 transposon. Sequences were analyzed using the DNAMAN DNA analysis software package (DNAMAN version 5.22; Lynnon Biosoft, Montreal, Canada). Homologous sequences in the National Center for Biotechnology Information (NCBI) database were identified by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
Complementation of MH15 and MH16 by HX2 cosmid clone.
We designed two primer pairs flanking the Tn5 insertion in MH15 or MH16 that amplify a 402-bp region or an 825-bp region from the HX2 genome. These primer pairs (P1 and P2, as well as P3 and P4; Table 2) were used to screen the HX2 cosmid library by using the colony PCR technique. The PCR products from cosmids CP104 and CP465 were subsequently sequenced to verify the presence of the MH16 or MH15 sequences of interest. Cosmids CP104 and CP465 were introduced into MH16 and MH15 by triparental mating using the helper strain DH5α, containing the plasmid pRK600 (19). The complemented strains CMH15 (p465) and CMH16 (p104) were used to conduct complementation assays, including the gall inhibition assay.
Construction of gdh in-frame deletion mutant strain.
Analysis of the DNA sequence flanking the Tn5 insertion in MH16 revealed that a putative gene may have been disrupted by the transposon, i.e., a GDH homolog that we designate gdh. Therefore, an in-frame deletion mutant of the putative gene was generated as follows. Two fragments flanking the gdh gene were amplified by PCR. A 1,173-bp fragment was created by primers P16L1 and P16L2, which were designed based on the upstream flanking sequences of the gdh open reading frame (ORF) from HX2 and included the EcoRI and BamHI sites (Table 2); the other, 1,084-bp fragment was created by primers P16R1 and P16R2, designed based on the downstream flanking sequences of the gdh ORF from HX2 and containing the BamHI and PstI sites (Table 2). The standard PCR was 5 min at 94°C and 35 cycles of 40 s at 94°C, 40 s at 65°C, 1 min at 72°C, and 10 min at 72°C. After being digested with appropriate restriction enzymes, the two fragments were inserted into pHSG299 to create pHSG16L and pHSG16R. The EcoRI-BamHI fragment from pHSG16L was inserted into pHSG16R, resulting in pHSGΔgdh, from which a 2.2-kb EcoRI-PstI fragment was excised, including a gdh gene with a 2,100-bp deletion. The suicide plasmid pHSGΔgdh was used to introduce the shortened gdh locus into the chromosome of HX2 by electroporation. The primers Gdh-up and Gdh-down, containing PstI and EcoRI restriction sites, respectively (Table 2), were used to confirm a double-crossover event. Mutants were tested for their abilities to inhibit the growth of K308 in vitro, to provide biocontrol of grapevine crown gall, and to produce GDH.
Cloning of gdh and complementation.
Based on the flanking sequences of the gdh ORF from HX2, primers Gdh-up and Gdh-down were used to amplify the gdh gene from HX2 genomic DNA (Table 2). A 3.2-kb PCR product was amplified, digested with PstI and EcoRI, and ligated to the broad-host-range vector pRK415G (30) to make the complemented CMH16(gdh). The ligated DNA was transformed into DH5α-competent cells by heat shock and then introduced into MH16 by triparental mating, as described above. The positive colonies were selected on LB plates containing kanamycin, tetracycline, and X-Gal. The consensus clone in the recombinant plasmid was verified by restriction digestion and sequencing. The complemented MH16 clones were screened for abilities to inhibit the growth of K308 in vitro, to provide biocontrol of grapevine crown gall, and to produce GDH.
Bacterial growth assays.
Bacterial growth and pH change of the medium were tested in PDB medium. The bacteria were initially suspended to 1.0 × 108 CFU ml−1 in SDW and diluted 10-fold in broth. Cultures were incubated on a shaker at 28°C, and growth was determined by counting bacterial CFU on PDA medium via serial-dilution plating methods at 2-h intervals over 44 h. The assay was repeated three times.
Detection of PQQ.
PQQ was isolated and assayed as previously described (52) with slight modifications. Briefly, bacteria were grown at 28°C in 100 ml AB minimal medium (13) for 48 h, and 1 volume of cell culture was diluted with 9 volumes of methanol. Precipitated material was removed by centrifugation (30 min at 8,000 × g), and the methanol was evaporated. The sample was acidified with HCl to pH 2.0 and loaded onto the Sep-Pak C18 (Agilent Technologies) cartridge. The cartridge was washed with 10 ml of methanol, 10 ml of water, and then 10 ml of 2 mM HCl. PQQ was eluted with 70% methanol. To identify the PQQ peak, 200 μl of the sample was mixed with 100 μl of 0.2 M Na2B4O7 buffer and adjusted to pH 8.0 with HCl and 90 μl 0.5% (vol/vol) acetone. Reverse-phase high-performance liquid chromatography (RP-HPLC) was performed using a Shimadzu LC-6A HPLC system with a fluorescence detector on Zorbax SB- C18 columns (4.6 by 250 mm, 5 μm; Agilent Technologies) that were eluted with 27% (vol/vol) methanol-0.4% (vol/vol) H3PO4 at a flow rate of 0.8 ml min−1. The excitation wavelength of the fluorescence detector was set at 360 nm, and the light emitted was at 480 nm.
GDH assays.
GDH was partly isolated, and its activity was measured according to the method of Liucija et al. (37). Bacteria were incubated in 100 ml of AB minimal medium at 28°C for 18 h. Cells were collected by centrifugation (5,000 × g, 30 min), washed with 0.9% NaCl, and suspended in Tris-HCl buffer (pH 8.0). The collected cells were disrupted by ultrasound sonication, and the fractions were acquired by centrifugation (8,000 × g, 30 min) and suspended in 50 mM potassium phosphate buffer (pH 7.0, containing 1.3% Triton X-100 and 0.5 M NaCl). A crude enzyme sample of GDH was prepared by stirring the suspension for 2 h at room temperature and then removing the insoluble material. Activities were determined by measuring the rate of discoloration of 2,6-dichloroindophenol (DCPIP) at 600 nm in a mixture containing 50 mM potassium phosphate buffer, pH 7.3, and 20 mM glucose. One activity unit (U) corresponds with the amount of enzyme converting 1 μmol of glucose or DCPIP per min under the specified assay conditions. Enzyme units per mg protein are compared.
Greenhouse assays for biocontrol of crown gall.
Gall inhibition assays were performed on grapevine shoots of potted plants (Vitis vinifera cv. Muscat Hamburg) according to a previously described method (12). In brief, the suspension of K308 (ca. 2 × 108 CFU/ml) was mixed with an equal volume of HX2 or its derivative suspension (ca. 2 × 108 CFU/ml). A 10-μl drop of bacterial mixture was placed in a 0.8-cm longitudinal incision. The inoculation site was wrapped with Parafilm. Galls were excised and weighed 42 days after inoculation. SDW was applied as a negative control, and strain K308 mixed with SDW was applied as a positive control. The effectiveness index (EI) was calculated by the following formula: EI (%) = [(C − T)/C] × 100, where C is the average fresh weight of the crown gall tumor of the positive control group and T is the average weight of the crown gall tumor of the treated group. The assay was performed with three repeats of each treatment and 10 inoculation sites for each repeat.
Sequence analysis of cosmids CP104 and CP465.
An approximately 6.7-kb region of clone CP465 that carries pqq and a 5.5-kb fragment of clone CP104 that carries gdh were sequenced by a strategy used previously in our laboratory (56). Cosmid DNA was digested with BamHI for CP465 or with PstI for CP104 and subcloned into the plasmid vector pRK415G. Subclones were selected based on colony PCR (using the primer pair P1 and P2 or P3 and P4; Table 2) and insert size and then sequenced. The orders of the 6.7-kb BamHI fragment within CP465 and the 5.5-kb PstI fragment within CP104 were determined by identifying overlapping sequences. DNA sequence homology searches were performed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST/). An analysis of DNA and deduced protein sequences was carried out using the DNAMAN software program. Analysis of putative promoter regions was carried out using the Neural Network Promoter Prediction tool (http://www.fruitfly.org/seq_tools/promoter.html).
Statistical analysis.
An analysis of variance was performed on the data using the SAS software program (version 8.2; SAS, Inc., Cary, NC). The mean square error values were tested with a t test at a Ρ value of <0.05.
Nucleotide sequence accession numbers.
The nucleotide sequences of CP465 and CP104 have been deposited in GenBank under accession numbers FJ868974 and EF090904, respectively.
RESULTS
Isolation and characterization of ABS-deficient mutants.
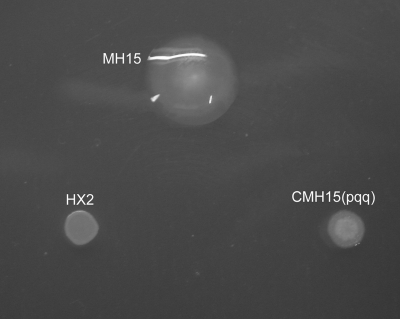
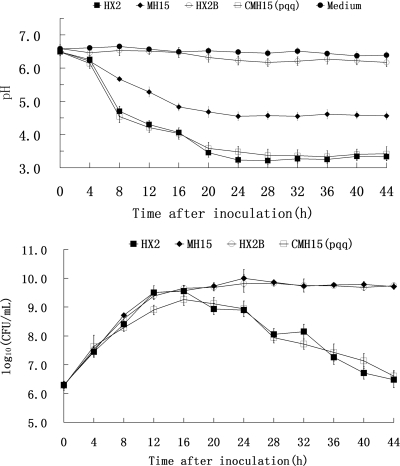
A total of 3,640 Tn5-induced kanamycin-resistant mutants of HX2 were tested for ABS production. Two mutants, MH15 and MH16, which no longer inhibited the growth of A. vitis K308 on PDA plates (Table 3), and five other mutants exhibiting reduced growth inhibition were obtained. The MH15 and MH16 mutants were further analyzed. Southern blotting indicated that single Tn5 insertions were present in both MH15 and MH16, indicated by a single hybridizing band upon digestion with KpnI, SalI, or PstI (data not shown). There were no differences in partial physiological and biochemical characteristics tested between HX2 and MH15 or MH16 according to previous methods (27). All showed positive reactions for catalase, citrate utilization, Voges-Proskauer, and d-glucose oxidation, and they exhibited negative reaction for oxidase or methyl red. Colony morphology of the MH15 and MH16 mutants on PDA plates was clearly different from that of the wild type (Fig. 1). MH15 and MH16 grew continuously during 44 h of cultivation in PDB liquid medium, whereas wild-type growth declined 24 h after inoculation (Fig. 2). However, MH15 and MH16, compared to the wild type, exhibited reduced biological control of grapevine crown gall (Table 3; Fig. 3); they had EIs of 82.1 and 74.4, respectively, whereas HX2 had an EI of 96.3.
TABLE 3.
Inhibition effects of Rahnella aquatilis wild-type strain HX2 and its mutants and complementary strains on growth of Agrobacterium vitis strain K308 and tumor formation on grapevine (Vitis vinifera cv. Muscat Hamburg) seedlings
| Strain | Inhibition zone diam (mm)a | EI (%)b |
|---|---|---|
| HX2 | 24.7 b | 96.3 a |
| HX2Bc | 22.9 b | 95.2 a |
| MH15 | 0.00 c | 82.1 b |
| MH16 | 0.00 c | 74.4 b |
| HX2Δgdh | 0.00 c | 76.8 b |
| CMH15(p465) | 23.6 b | 95.7 a |
| CMH15(pqq) | 25.1 b | 98.4 a |
| CMH16(p104) | 22.7 b | 96.4 a |
| CMH16(gdh) | 24.2 b | 96.7 a |
| CHX2Δgdh | 25.2 b | 95.4 a |
| MH15 + PQQd | 32.8 a | 97.3 a |
HX2 and its mutants were spot inoculated onto PDA medium and incubated at 28°C for 24 h. Production of ABS was assessed by overlaying the plates with a suspension of A. vitis strain K308 as the indicator, as described by Chen et al. (12). Data with the same letters in the same column are not significantly different (P < 0.05).
EI was calculated by the formula EI (%) = [(C − T)/C] × 100, where C is the average fresh weight of the crown gall tumor of the positive control group (strain K308 mixed with SDW) and T is the average weight of the crown gall tumor of the treated group. Galls were excised and weighed 42 days after inoculation. Data are the means of three replicates. Data with the same letters in the same column are not significantly different (P < 0.05).
HX2 grown in PDA or PDB with additional phosphate-buffered saline (pH 6.8).
PDA or PDB supplemented with PQQ (0.1 μM).
FIG. 1.
Colony morphology of Rahnella aquatilis HX2 and its Tn5 mutant MH15 and complemented mutant CMH15(pqq) on PDA plate. Colony morphology of the MH16 mutant and the complemented mutant gave results similar to those observed with the MH15 mutant (data not shown).
FIG. 2.
Growth and changes in the pH of culture filtrates of the wild type and Tn5 mutant MH15 of Rahnella aquatilis HX2. Cells were grown in PDB at 28°C. CFU of bacterial cultures were determined by plating serial dilutions of the culture on LB agar plates. The pH of the culture filtrate was determined at 4-h intervals. HX2B, HX2 grown in PDB with additional phosphate-buffered saline (pH 6.8); Medium, noninoculated PDB medium. The means for three experiments are presented, and the vertical bars represent the standard errors. Treatment with the MH16 mutant gave results similar to those observed with the MH15 mutant (data not shown).
FIG. 3.
Biological control phenotypes of wild-type Rahnella aquatilis HX2 and its Tn5 mutants MH15 and MH16, deletion mutant HX2Δgdh, and complemented mutants CMH15(p465), CMH15(pqq), CMH16(p104), CMH16(gdh), and CHX2Δgdh on grapevine (Vitis vinifera cv. Muscat Hamburg) shoots. Shoots were inoculated by placing 10-μl drops of bacterial strains (108 CFU ml−1), alone or in equal-number mixtures, at sites in which wounds of 1.0-cm longitudinal incisions were made with a sterile scalpel. Tumors were readily apparent 7 weeks after inoculation. The treatments were SDW alone (a), Agrobacterium vitis K308 alone (b), HX2 plus K308 (c), MH16 plus K308 (d), CMH16(p104) plus K308 (e), CMH16(gdh) plus K308 (f), HX2Δgdh plus K308 (g), CHX2Δgdh plus K308 (h), MH15 plus K308 (i), CMH15(p465) plus K308 (j),CMH15(pqq) plus K308 (k), and MH15 plus PQQ (l).
Effect of glucose on ABS biosynthesis.
When the wild-type HX2 strain was grown on potato agar (PA) rather than PA supplemented with 2% glucose (PDA), no bacterial inhibition activity was observed. Other carbon sources were tested for their effect on bacterial inhibition. Fructose, mannitol, and lactose were unable to substitute for glucose in causing HX2 to produce a high level of antibacterial activity. When the wild type was grown on PA supplemented with these carbon sources, weaker bacterial inhibition was observed than on PDA (data not shown). Additionally, when the wild-type strain was grown on PA supplemented with gluconate or sucrose, bacterial inhibition was not observed.
pqqE and gdh are associated with ABS production and biocontrol.
Tn5-containing clones of MH15 and MH16 were produced by shotgun cloning. A 0.6-kb Tn5-flanking DNA fragment from MH15 and a 1.7-kb fragment from MH16 were sequenced by PCR with the P2 primer (Table 2), which was designed to amplify the I end of the Tn5 sequences. A homology search of the GenBank database revealed that the MH15 fragment showed 92.0% identity to a PQQ-biosynthesizing protein gene, pqqE, from R. aquatilis (33), and the MH16 fragment showed 84.0% identity to a GDH gene from Serratia marcescens (GenBank accession no. AF441442); therefore, it was suggested that Tn5 insertions in MH15 and MH16 occurred in pqqE, a homologue of a PQQ-biosynthesizing protein gene, and gdh, a GDH gene. To verify the accuracy of transposon insertions in the sequence, PCR amplification was performed using primers designed from the sequences flanking the pqqE or gdh ORF. The size of the PCR product from the MH15 or MH16 mutant was increased by insertion of a transposon, indicating that the transposon was inserted in the pqqE or gdh gene of the HX2 genome (data not shown).
Detection of PQQ.
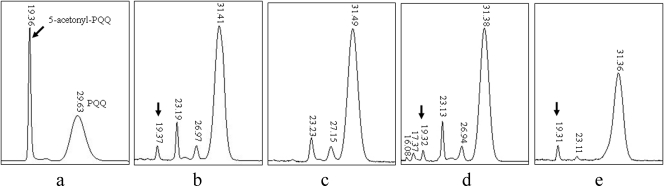
Based on the fact that the mutations in a pqq gene abolished antibiosis activity of wild-type HX2, we examined whether the strain produces PQQ by analyzing culture supernatants using RP-HPLC. PQQ was detected as 5-acetonyl-PQQ by comparison with the elution times of synthetic PQQ and 5-acetonyl-PQQ from the RP-HPLC chromatograms (Fig. 4). The data revealed that wild-type HX2 produces PQQ in vitro. The MH15 mutant defective in bacterial growth inhibition did not produce PQQ, and the constructed plasmid pCH15, which carries all of the pqq genes, conferred PQQ production in the mutants. The MH16 mutant was not impaired in PQQ production.
FIG. 4.
HPLC detection of PQQ (5-acetonyl-PQQ) synthesized by wild-type Rahnella aquatilis HX2, its Tn5 mutants MH15 and MH16, and the complemented mutant CMH15(pqq). (a) Synthetic PQQ; (b) HX2; (c) MH15 (pqqE::Tn5); (d) CMH15(pqq); (e) MH16 (gdh::Tn5). Arrows indicate 5-acetonyl-PQQ.
GDH levels.
The MH16 mutant grown in minimal glucose broth had reduced levels of GDH (Table 4). In this mutant, GDH levels were only about 10% of the levels found in the wild-type strain. In MH15, GDH levels were not significantly different from those observed in the wild type. Enzyme activities were also measured in HX2 and in the MH15 and MH16 mutants grown on PDA to confirm these results under conditions of the ABS biosynthesis. In MH16 mutants, GDH activities were reduced to levels observed in minimal glucose broth. HX2 and MH15 had a level of GDH of 0.81 U mg−1 protein and 0.74 U mg−1 protein, respectively, whereas MH16 had a level of GDH of 0.07 U mg−1 protein.
TABLE 4.
GDH activities in crude extracts of Rahnella aquatilis wild-type strain HX2 and its mutants after growth in ABM mediuma
| Strain | Protein content (mg ml−1) | Activity (U ml−1) | Activity (U mg−1 protein) |
|---|---|---|---|
| HX2 | 15.4 a | 11.4 a | 0.74 a |
| MH15 | 15.1 a | 10.8 a | 0.72 a |
| MH16 | 14.8 a | 1.21 b | 0.08 b |
| HX2Δgdh | 14.4 a | 1.42 b | 0.10 b |
| CMH16(p104) | 16.7 a | 12.8 a | 0.76 a |
| CMH16(gdh) | 16.1 a | 11.8 a | 0.73 a |
| CHX2Δgdh | 16.3 a | 13.6 a | 0.83 a |
Bacterial cells in overnight cultures were collected by centrifugation and disrupted by ultrasound sonication. The fractions were acquired by centrifugation and suspended in 50 mM potassium phosphate buffer (pH 7.0; containing 1.3% Triton X-100 and 0.5 M NaCl). Crude enzyme samples were prepared by stirring the suspension for 2 h at room temperature and then removing the insoluble material. Activities were determined by measuring the rate of discoloration of DCPIP at 600 nm in a mixture containing 50 mM potassium phosphate buffer (pH 7.3) and 20 mM glucose. One activity unit corresponds to the amount of enzyme converting 1 μmol of glucose or DCPIP per min under the specified assay conditions. Each value represents the average of triplicate determinations of the activities. Data with the same letters in the same column are not significantly different (P < 0.05).
Phenotypes generated by mutation of the gdh in-frame deletion.
The gdh in-frame deletion mutant HX2Δgdh was determined to have the same phenotype as the original Tn5 mutant of MH16. HX2▵gdh lacked the ability to inhibit growth of A. vitis K308 on PDA and was impaired in biological control and production of GDH to the same level as MH16. HX2Δgdh did not show any inhibition zone on PDA plates seeded with strain K308 (Table 3). Strains of HX2Δgdh with an EI of 76.8 showed reduced biological control activity on grapevines compared to the wild-type HX2 strain (EI = 93.6) (Table 3). An average of 0.08 U mg−1 protein enzyme activity was detected for both HX2Δgdh and MH16 in GDH assays, but an average of 0.74 U mg−1 protein was detected for wild-type HX2 (Table 4). Colony morphology of the HX2Δgdh mutant was identical to that of MH16 on PDA plates, both of which were clearly different from wild-type morphology (Fig. 1). Compared to growth in PDB, HX2Δgdh and MH16 both grew continuously during 44 h of cultivation, while that of the wild type declined (Fig. 2).
Complementation of MH15 and MH16.
MH15 was first complemented by transforming it with cosmid CP465 via triparental mating. The abilities of A. vitis K308 to inhibit growth in vitro and to suppress gall development on grape plants are fully restored in the complemented MH15 strain of CMH15(p465). The MH15 mutant did not produce an inhibition zone on PDA plates seeded with strain K308, whereas CMH15(p465) exhibited an inhibition zone at about the same level as HX2, with a diameter of about 25 mm (Table 3). Similarly, CMH15(p465) showed the same level of biological control as HX2 on grapevines (Table 3; Fig. 3). Colony morphology of the CMH15(p465) strain was identical to that of the wild type, HX2, on PDA plates (Fig. 1). As with the growth in PDB, the MH15 mutants grew continuously during 44 h of cultivation, while the wild type and CMH15(p465) both declined beginning 24 h after inoculation.
To further verify that pqqE was involved in antibiosis, biological control, and PQQ production, a 6.7-kb BamHI fragment from cosmid CP465 was subcloned into the expression vector pRK415G and transferred to the mutant MH15 by triparental mating. Again, the complemented MH15 strain, CMH15(pqq), was fully restored in its growth dynamics in PDB and in its abilities to inhibit growth of A. vitis K308 in vitro (Table 3), to suppress gall development on grape plants (Table 3; Fig. 3), and to produce PQQ (Fig. 4).
To confirm that PQQ is involved in ABS production, synthetic PQQ was applied to complement the MH15 mutant. Concentrations of synthetic PQQ ranging from 0 to 1.0 μM were used. When the MH15 strain was grown on PDA supplemented with 0.1 μM PQQ, an obvious bacterial inhibition zone was observed with the same level as that of the wild type (Table 3).
The MH16 mutant was first complemented with cosmid CP104. Growth inhibition of A. vitis K308 in vitro, suppression of gall development on grape plants, production of GDH, and growth dynamics in PDB were fully restored in the complemented MH16 strain of CMH16(p104). MH16 presented no inhibition zone on PDA seeded with strain K308, whereas CMH16(p104) presented an inhibition zone at about the same level as that of HX2, with a diameter of about 25 mm (Table 3). Similarly, CMH16(p104) exhibited the same level of biocontrol as HX2 on grapevines (Table 3; Fig. 3). Colony morphology of the CMH16(p104) strain was identical to that of the wild type, HX2, on PDA (Fig. 1). About 0.08 U mg−1 protein enzyme activity was detected for MH16 in GDH assays, but about 0.8 U mg−1 protein was detected for CMH16(p104) and for the wild type, HX2 (Table 4). The growth of CMH16(p104) in PDB, as well as that of the wild type, declined at 24 h after inoculation, while MH16 grew continuously (Fig. 2).
To determine that gdh was associated with antibiosis, biological control, and GDH production, the gene was cloned into the expression vector pRK415G and transferred to the MH16 and HX2Δgdh mutants by triparental mating. The complemented strains, CMH16(gdh) and CHX2Δgdh, were both fully restored in their abilities to inhibit growth of A. vitis K308 on plates (Table 3), suppress gall development on grape plants (Table 3; Fig. 3), and produce GDH (Table 4).
Analysis of gdh flanking sequence and pqqE flanking sequence.
A 5.5-kb gdh-including fragment of CP104 contained only one ORF, the homologue of the GDH gene (gdh). The putative gdh sequence encodes a predicted 799-amino-acid protein with a molecular mass of 86.4 kDa and had the highest sequence identity to the GDH quinoprotein from Serratia proteamaculans 568 (81.5%; GenBank accession no. ABV42200) (Fig. 5). The predicted structure of the deduced GDH quinoprotein is highly similar to that of the E. coli quinoprotein GDH, which consists of five N-terminal transmembrane helices and a catalytic C-terminal domain facing the periplasm (55). Regions similar to the tryptophan-docking motifs of the “propeller fold,” the common structure of PQQ-dependent dehydrogenases (1), were also found in the deduced GDH (Fig. 5). As revealed by the X-ray structures of several identified GDHs, the PQQ molecule is covered by a Trp residue and a His residue, which correspond to Trp-404 and His-262 in the structure of E. coli GDH (1). A Trp residue (Trp-408) and a His residue (His-262) were also found in the corresponding region of the deduced GDH (Fig. 5). In addition, the deduced GDH shares the conserved Asp residue (Asp-470) present in all studied PQQ-dependent dehydrogenases, which is believed to initiate the catalytic reaction and corresponds to Asp-466 in E. coli GDH. Furthermore, the deduced GDH shares the conserved Asp (Asp-358), Asn (Asn-359), and Thr (Thr-428) residues of other GDHs (Asp-353, Asn-354, and Thr-424 in E. coli; Fig. 5), which likely interact with the Ca2+ ion in the active site (1).
FIG. 5.
Alignment of deduced gdh amino acid sequence with sequences of diverse members of the GDH family. Proteins whose sequences have similarities with the amino acid sequence deduced from gdh include QGDH_S.pro (81.5%) from Serratia proteamaculans 568 (GenBank accession number ABV42200), QGDH_E.tas (75.7%) from Erwinia tasmaniensis Et1/99 (accession number YP_001908309), QGDH_K.pne (71.6%) from Klebsiella pneumoniae 342 (accession number YP_002240396), QGDH_B.xen (71.2%) from Burkholderia xenovorans LB400 (accession number YP_555448), and QGDH_E.col (70.3%) from Escherichia coli (accession number P15877). Regions of transmembrane helices determined for GDH (55) and predicted for QGDH-HX2 are underlined. Functionally important residues located in the active sites binding to Ca2+ and PQQ are numbered and denoted with “•,” and the predicted so-called “propeller fold” region is indicated by “□.” The solid arrow indicates the Tn5 insertion site.
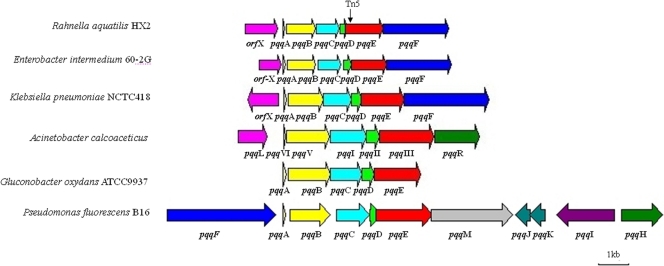
The 6.7-kb fragment of CP465 that includes pqqE contained seven putative ORFs, and six of these ORFs were similar to those of other ppq genes (14, 32), i.e., pqqABCDEF (Table 5; Fig. 6). A putative intact ORF (orfX) with homology to a membrane dipeptidase was located upstream of the proposed pqqA gene. The putative 72-bp pqqA gene encoded a peptide of 23 amino acids, with a molecular mass of 2.8 kDa. The presence of glutamic acid and tyrosine residues at positions 15 and 19 is similar to sequences of other deduced PqqA proteins. The 912-bp pqqB gene encodes a 303-amino-acid peptide with a molecular mass of 33.0 kDa. The 768-bp pqqC gene encoded a predicted 255-amino-acid peptide of approximately 29.5 kDa. The 258-bp pqqD gene encoded a predicted 85-amino-acid peptide of approximately 9.8 kDa. The 1,191-bp pqqE gene encodes a predicted 396-amino-acid protein with a molecular mass of 44.3 kDa. The Tn5 insertion in pqqE occurred near the conserved Radical_SAM domain toward the 5′ end of the gene (Fig. 6). The 2,094-bp pqqF gene encoded a predicted 76.0-kDa, 697-amino-acid peptide. Nucleotide sequence analyses suggest that R. aquatilis pqqABCDEF might be regulated as an operon in which the pqqD and pqqE genes were fused. The intergenic regions within the putative operon were short and have a low promoter probability (promoter prediction score below 0.2), whereas the region upstream of the pqqA protein gene is likely a promoter (promoter prediction score of 0.85). The deduced amino acid sequences of the R. aquatilis pqq genes had 30 to 90% identity with those corresponding to previously identified pqq genes from E. intermedium, K. pneumoniae, and R. aquatilis (Table 5). A sequence comparison revealed that all of the proteins deduced from the CP465 sequence had corresponding homologues in E. intermedium 60-2G (32), K. pneumoniae NCTC418 (40), and P. fluorescens B16 (14), except for that deduced from the membrane dipeptidase-like gene (orfX), which had no homologues in P. fluorescens B16. Sequence comparisons showed that ORFs in CP465 were homologues of genes located on the circular chromosomes of K. pneumoniae (GenBank accession no. CP000647). The relative order of pqqA to pqqF was identical for all four bacterial species, except that pqqF in P. fluorescens B16 existed in a different location (Fig. 6).
TABLE 5.
Comparison of genes in the 6.7-kb region of cosmid CP465 from Rahnella aquatilis HX2 with genes of Enterobacter intermedium 60-2G,a Klebsiella pneumoniae NCTC418,b and Rahnella aquatilis ISL19c
| HX2 ORF | Size of deduced product (aa) | Homologue |
|||
|---|---|---|---|---|---|
| Gene | Putative product | Size (aa) | Identity (%) | ||
| ORF1 | 345 | orfX | Dipeptidase-like protein | >239d | 28.2 |
| orfX | Membrane dipeptidase | >271e | 43.2 | ||
| ORF2 | 23 | pqqA | PqqA involved in pyrroloquinoline biosynthesis | 23 | 86.9 |
| pqqA | PqqA involved in pyrroloquinoline biosynthesis | 23 | 86.9 | ||
| ORF3 | 303 | pqqB | PqqB involved in pyrroloquinoline biosynthesis | 307 | 71.8 |
| pqqB | PqqB involved in pyrroloquinoline biosynthesis | 308 | 74.2 | ||
| ORF4 | 255 | pqqC | PqqC involved in pyrroloquinoline biosynthesis | 251 | 82.5 |
| pqqC | PqqC involved in pyrroloquinoline biosynthesis | 251 | 82.9 | ||
| ORF5 | 85 | pqqD | PqqD involved in pyrroloquinoline biosynthesis | 92 | 58.7 |
| pqqD | PqqD involved in pyrroloquinoline biosynthesis | 92 | 60.9 | ||
| pqqD | Coenzyme PQQ synthesis protein D | 85 | 98.8 | ||
| ORF6 | 396 | pqqE | PqqE involved in pyrroloquinoline biosynthesis | 374 | 72.5 |
| pqqE | PqqE involved in pyrroloquinoline biosynthesis; coenzyme PQQ synthesis protein E | 380 | 78.7 | ||
| pqqE | 396 | 96.2 | |||
| ORF7 | 697 | pqqF | PqqF involved in pyrroloquinoline biosynthesis | 693 | 30.7 |
| pqqF | PqqF involved in pyrroloquinoline biosynthesis | 761 | 32.5 | ||
FIG. 6.
Comparis on of ORF organization within the 6.7-kb region in CP465 that includes pqqE to corresponding regions in Enterobacter intermedium 60-2G, Klebsiella pneumoniae NCTC418, Acinetobacter calcoaceticus, Gluconobacter oxydans ATCC 9937, and Pseudomonas fluorescens B16. The length of each arrow represents the relative ORF size and indicates the direction of transcription. The solid triangles represent the Tn5 insertion sites. The same colors represent homologous encoded proteins. The information and sizes of the genes are depicted based on nucleotide sequence data from GenBank. Genes of the following strains were used: E. intermedium 60-2G (GenBank accession no. AY216683), K. pneumoniae NCTC418 (accession no. X58778), A. calcoaceticus (accession no. accession no. X06452), G. oxydans ATCC 9937 (accession no. AJ277117), and P. fluorescens B16 (accession no. AY780887).
DISCUSSION
Two ABS-deficient mutants of HX2, MH15 and MH16, suppressed crown gall in a greenhouse setting but were inferior to the parental strain. Our data indicate that the logarithmic growth rate of the two mutants is similar to that of the wild-type strain, HX2. Furthermore, a previous study in this laboratory showed that the epiphytic fitness of strain HX2 on the explant tissues of grapevine was not compromised by the Tn5 insertion. Differences in suppression of crown gall by HX2 and MH15 or MH16 are therefore likely due to differences in antibiosis rather than in bacterial growth or attachment to grapevine tissues. A cloned DNA fragment from the HX2 genome containing either previously identified pqq genes or the gdh gene fully complemented the affected phenotypes of corresponding mutant genes. Resultant data implied that PQQ and GDH, individually or in combination, contributed to ABS production and biological control by HX2.
Colony morphologies of both the pqq and gdh mutants on PDA plates were clearly different from those of the wild type (Fig. 1), a similar situation to that found for pqq mutants of E. intermedium 60-2G (22). The growth decline of the wild type, HX2, and the complemented mutants was correlated with a decrease in the pH of the medium (Fig. 2). These results indicate that acidification of the medium caused by the wild type and the complemented mutants might be involved in antibacterial activity.
In gram-negative bacteria, acidification during growth on glucose is due to the accumulation of gluconate, resulting from the oxidation of glucose (53). This suggests that the behavior of the ABS-deficient mutants could be due to a deficiency in PQQ-linked GDH, which catalyzes the oxidation of glucose. The results presented in Table 4 reveal that GDH levels were significantly reduced in the gdh mutants of MH16 and HX2Δgdh. Interestingly, in the pqq mutant MH15, although GDH levels were not significantly different from those in the wild-type strains, PQQ levels were significantly reduced (Fig. 4). Taken together, these data suggest that both GDH and PQQ, with the formation of the GDH-PQQ holoenzyme, played a role in acidification during growth on glucose, which may be essential for bacterial growth inhibition by HX2.
Using Tn5 tagging, six previously known pqq genes and one gdh gene from the R. aquatilis genome were identified for the first time. The deduced PqqA, -B, -C, -D, -E, and -F proteins all revealed extensive sequence similarity to those from other bacteria described previously (14, 32). Furthermore, our data lend further support to the notion that the pqqABCDEF genes are conserved in gram-negative bacteria (14). It was reported that the pqqF gene appears to be absent in R. aquatilis, since the nucleotide sequence downstream of pqqE showed no similarity to pqqF, and the physical organization of the pqq genes of R. aquatilis may be similar to that of A. calcoaceticus (33). However, our data on the nucleotide sequence of the pqq locus of R. aquatilis HX2 showed that the organization of the R. aquatilis pqq genes appears to be same as that for E. intermedium and K. pneumoniae (Fig. 6). This conclusion can be strengthened by our sequence data that indicate that these ORFs are similar (greater than 58% identity) to the pqq genes of K. pneumoniae and E. intermedium, except for the pqqF gene. The biochemical functions of the PqqA, -B, -C, and -F proteins in other PQQ-synthesizing bacteria have been well documented; however, functions of the deduced encoded proteins in R. aquatilis remain to be determined. Recently the pqq operon in P. fluorescens B16 was found to contain 11 pqq genes: pqqA, pqqAB, pqqAC, pqqAD, pqqAE, pqqAF, pqqAH, pqqAI, pqqAJ, pqqAK, and pqqAM (14). The functions of PqqA, -B, -C, -D, -E, and -F remain highly conserved to those of other reported pqq genes. PqqH was suggested to be a transcriptional activator of pqq gene expression, while the functions of PqqI, -J, -K, and -M remain unknown (14). It is also currently unknown whether there exist genes similar to pqqH, pqqI, pqqJ, pqqK, and pqqM in R. aquatilis.
The deduced GDH showed significant identity with mGDH from a variety of bacteria (Fig. 5) and contained all five conserved membrane-spanning α-helices in the N-terminal region, which ensure strong anchorage of the protein in the inner membrane (55). The remaining C-terminal portion is assumed to have a catalytic domain including PQQ and Ca2+ binding sites (1). In addition, our data suggest that the cloned GDH was a functional membrane-bound PQQ-linked GDH.
The results clearly show that gdh disruption in strain HX2 is partially responsible for its abolished antibacterial phenotype and impaired biocontrol phenotype. To determine that the affected phenotypes of HX2 were not due to other, unknown mutations that may be present in this strain, we constructed a second gdh mutant, HX2Δgdh, via reverse genetics. Since the phenotype of HX2Δgdh is identical to that of MH16, it is evident that the phenotype is due to the gdh mutations in these strains. Moreover, analysis of the genome of K. pneumoniae (GenBank accession no. CP000647), a close relative of R. aquatilis, indicates that there are no genes downstream of gdh (gcd; KPN_00132)in the same orientation. Thus, insertions within the gdh gene would not be expected to have polar effects on the transcription of downstream genes (http://img.jgi.doe.gov). These data strongly suggest that GDH is a primary determinant of the production of the ABS, which is an important factor in HX2 biocontrol of A. vitis-induced crown gall of grapevine.
It has been demonstrated that PQQ, with the formation of the GDH-PQQ holoenzyme, is significantly involved in the MPS by bacteria, which is essential for plant growth promotion by plant growth-promoting bacteria (22, 36, 43). Additionally, PQQ is suggested to be involved in production of antibiotics by several bacterial biocontrol agents of plant disease and thereby contributes to disease suppression (22, 28, 46) as well as acting as a growth promotion factor of several plants (14). Bernardelli et al. (4) observed that PQQ-linked GDH is necessary for optimal nodulation efficiency and competitiveness on alfalfa roots for S. meliloti. The present study provides evidence that both PQQ and GDH are necessary for beneficial functions of R. aquatilis strain HX2, including the production of antibacterial activity and the biological control of crown gall disease.
Conversion of glucose to gluconolactone and then gluconic acid by gram-negative bacteria is facilitated by PQQ-dependent periplasmic GDH, which is a constitutive member of the direct oxidative pathway of glucose catabolism in several organisms including enteric bacteria (1, 57). The resultant data suggest that the glucose metabolism related to GDH-PQQ holoenzyme activity is involved in biosynthesis of the ABS in HX2 and that gluconolactone and/or its metabolic intermediates might function as a precursor in the ABS synthesis pathway or as a sensor for external glucose concentration that might regulate ABS biosynthesis in HX2. The identity of the antibacterial substance(s) produced by HX2 and the effect of glucose on its production require further investigation.
Although a role for antibiosis in biological control was demonstrated in this study, the ABS was not the sole factor involved in suppression of grapevine crown gall by R. aquatilis HX2. The ABS-deficient mutants were generally less effective than the wild-type strain, HX2, in suppressing crown gall, but they significantly reduced the incidence of crown gall in the greenhouse compared with results for the water-treated control. Therefore, other mechanisms, such as competitive exclusion and/or competition for nutrients, are also likely to contribute to the suppression of crown gall by HX2, as in the case of Azospirillum brasilens protecting tomato seedlings from infection by Pseudomonas syringae pv. Tomato (2).
Acknowledgments
We are grateful to Deqin Ma (Institute of Microbiology, The Chinese Academy of Sciences, Beijing, China) for kindly providing bacterial strains. We thank Liqun Zhang, Zaifeng Fan, and Xili Liu (Department of Plant Pathology, China Agricultural University, Beijing, China) for their assistance with the work.
This research was supported by the National Key Technology R&D Program (project no. 2007BAC15B05) and Gongyixing Hangye Keji (grant no. 200803033).
Footnotes
Published ahead of print on 4 September 2009.
REFERENCES
- 1.Anthony, C. 2004. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 428:2-9. [DOI] [PubMed] [Google Scholar]
- 2.Bashan, Y., and Luz E. de-Bashan. 2002. Protection of tomato seedlings against infection by Pseudomonas syringae pv. tomato by using the plant growth-promoting bacterium Azospirillum brasilense. Appl. Environ. Microbiol. 68:2637-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, C. R., G. A. Dickie, and J. W. Y. F. Chan. 1995. Variable response of bacteria isolated from grapevine xylem to control grape crown gall disease in planta. Am. J. Enol. Vitic. 46:499-508. [Google Scholar]
- 4.Bernardelli, C. E., M. F. Luna, M. L. Galar, and J. L. Boiardi. 2008. Symbiotic phenotype of a membrane-bound glucose dehydrogenase mutant of Sinorhizobium meliloti. Plant Soil 313:217-225. [Google Scholar]
- 5.Burr, T. J., C. Bazzi, S. Süle, and L. Otten. 1998. Crown gall of grape: biology of Agrobacterium vitis and the development of disease control strategies. Plant Dis. 82:1288-1297. [DOI] [PubMed] [Google Scholar]
- 6.Burr, T. J., and L. Otten. 1999. Crown gall of grape: biology and disease management. Annu. Rev. Phytopathol. 37:53-80. [DOI] [PubMed] [Google Scholar]
- 7.Burr, T. J., C. L. Reid, E. Tagliati, C. Bazzi, and S. Süle. 1997. Biological control of grape crown gall by strain F2/5 is not associated with agrocin production or competition for attachment sites on grape cells. Phytopathology 87:706-711. [DOI] [PubMed] [Google Scholar]
- 8.Burr, T. J., C. L. Reid, M. Yoshimura, E. A. Momol, and C. Bazzi. 1995. Survival and tumorigenicity of Agrobacterium vitis in living and decaying grape roots and canes in soil. Plant Dis. 79:677-682. [Google Scholar]
- 9.Calvo, J., V. Calvente, M. E. de Orellano, D. Benuzzi, and M. I. Sanz de Tosetti. 2007. Biological control of postharvest spoilage caused by Penicillium expansum and Botrytis cinerea in apple by using the bacterium Rahnella aquatilis. Int. J. Food Microbiol. 113:251-257. [DOI] [PubMed] [Google Scholar]
- 10.Cangelosi, G. A., E. A. Best, G. Martinetti, and E. W. Nester. 1991. Genetic analysis of Agrobacterium. Methods Enzymol. 204:384-397. [DOI] [PubMed] [Google Scholar]
- 11.Chen, F., J. Y. Li, Y. B. Guo, J. H. Wang, and H. M. Wang. 2009. Biological control of grapevine crown gall: purification and partial characterisation of an antibacterial substance produced by Rahnella aquatilis strain HX2. Eur. J. Plant Pathol. DOI 10.1007/s10658-009-9429-z. [DOI]
- 12.Chen, F., Y. B. Guo, J. H. Wang, J. Y. Li, and H. M. Wang. 2007. Biological control of grape crown gall by Rahnella aquatilis HX2. Plant Dis. 91:957-963. [DOI] [PubMed] [Google Scholar]
- 13.Chilton, M. D., C. C. Thomas, S. K. Farrand, A. J. Bendich, M. P. Gordon, and E. W. Nester. 1974. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc. Natl. Acad. Sci. USA 71:3672-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, O., J. Kim, J.-G. Kim, Y. Jeong, J. S. Moon, C. S. Park, and I. Hwang. 2008. Pyrroloquinoline quinone is a plant growth promotion factor produced by Pseudomonas fluorescens B16. Plant Physiol. 146:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Jonge, R., M. Joost Teixeira de Mattos, J. B. Stock, and O. M. Neijssel. 1996. Pyrroloquinoline quinone, a chemotactic attractant for Escherichia coli. J. Bacteriol. 178:1224-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotype in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:387-405. [DOI] [PubMed] [Google Scholar]
- 17.de Werra, P., M. Péchy-Taur, C. Keel, and M. Maurhofer. 17 April 2009. Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. doi: 10.1128/AEM.00295-09. [DOI] [PMC free article] [PubMed]
- 18.Duine, J. A., R. A. van der Meer, and B. W. Groen. 1990. The cofactor pyrroloquinoline quinone. Annu. Rev. Nutr. 10:297-318. [DOI] [PubMed] [Google Scholar]
- 19.Finan, T. M., B. Kunkel, G. F. de Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Goosen, N., H. P. A. Horsman, R. G. M. Huien, and P. V. D. Putte. 1989. Acinetobacter calcoaceticus genes involved in biosynthesis of the coenzyme pyrroloquinoline quinone: nucleotide sequence and expression in Escherichia coli K-12. J. Bacteriol. 171:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, S. H., C. H. Kim, J. H. Lee, J. Y. Park, S. M. Cho, S. K. Park, K. Y. Kim, H. B. Krishnan, and Y. C. Kim. 2008. Inactivation of pqq genes of Enterobacter intermedium 60-2G reduces antifungal activity and induction of systemic resistance. FEMS Microbiol. Lett. 282:140-146. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan, F. 1983., Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 24.He, K., H. Nukada, T. Urakami, and M. P. Murphy. 2003. Antioxidant and prooxidant properties of pyrroloquinoline quinone (PQQ): implications for its function in biological system. Biochem. Pharmacol. 65:67-74. [DOI] [PubMed] [Google Scholar]
- 25.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holscher, T., and H. Gorisch. 2006. Knockout and overexpression of pyrroloquinoline quinone biosynthetic genes in Gluconobacter oxydans 621H. J. Bacteriol. 188:7668-7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iimura, K., and A. Hosono. 1996. Biochemical characteristics of Enterobacter agglomerans and related strains found in buckwheat seeds. Int. J. Food Microbiol. 30:243-253. [DOI] [PubMed] [Google Scholar]
- 28.James, D. W., Jr., and N. I. Gutterson. 1986. Multiple antibiotics produced by Pseudomonas fluorescens HV37a and their differential regulation by glucose. Appl. Environ. Microbiol. 52:1183-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi, A., K. Inoue, and Y. Ichinose. 2008. Biological control of crown gall of grapevine, rose, and tomato by nonpathogenic Agrobacterium vitis strain VAR03-1. Phytopathology 98:1218-1225. [DOI] [PubMed] [Google Scholar]
- 30.Keen, N. T., S. Tamaki, and D. Kobayashi. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 31.Khmel, I. A., T. A. Sorokina, N. B. Lemanova, V. A. Lipasova, O. Z. Metlitski, T. V. Burdeinaya, and L. S. Chernin. 1998. Biological control of crown gall in grapevine and raspberry by two Pseudomonas spp. with a wide spectrum of antagonistic activity. Biocontrol Sci. Technol. 8:45-57. [Google Scholar]
- 32.Kim, C. H., S. H. Han, K. Y. Kim, B. H. Cho, Y. H. Kim, B. S. Koo, and Y. C. Kim. 2003. Cloning and expression of pyrroloquinoline quinone (PQQ) genes from a phosphate-solubilizing bacterium Enterobacter intermedium. Curr. Microbiol. 47:457-461. [DOI] [PubMed] [Google Scholar]
- 33.Kim, K. Y., D. Jordan, and H. B. Krishnan. 1998. Expression of genes from Rahnella aquatilis that are necessary for mineral phosphate solubilization in Escherichia coli. FEMS Microbiol. Lett. 159:121-127. [DOI] [PubMed] [Google Scholar]
- 34.Kumazawa, T., K. Sato, H. Seno, A. Ishii, and O. Suzuki. 1995. Levels of pyrroloquinoline quinone in various foods. Biochem. J. 307:331-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, Y. J., J. Y. Zhao, D. Q. Ma, and Y. B. Di. 1990. A biotype 3 strain of Agrobacterium radiobacter inhibits crown gall formation on grapevine. Acta Microbiol. Sin. 30:165-171. [Google Scholar]
- 36.Liu, S. T., L. Y. Lee, C. Y. Tai, C. H. Hung, Y. S. Chang, J. H. Wolfram, R. Rogers, and A. H. Goldstein. 1992. Cloning of an Erwinia herbicola gene necessary for gluconic acid production and enhanced mineral phosphate solubilization in Escherichia coli HB101. J. Bacteriol. 174:5814-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liucija, M., B. Irina, S. Rasa, R. Rolandas, B. Gediminas, M. Rita, and M. Rolandas. 1999. Purification and characterisation of PQQ-dependent glucose dehydrogenase from Erwinia sp. 34-1. Biotechnol. Lett. 21:187-192. [Google Scholar]
- 38.Reference deleted.
- 39.Matsushita, K., H. Toyama, M. Yamada, and O. Adachi. 2002. Quinoproteins: structure, function, and biotechnological applications. Appl. Microbiol. Biotechnol. 58:13-22. [DOI] [PubMed] [Google Scholar]
- 40.Meulenberg, J. J., E. Sellink, N. H. Riegman, and P. W. Postma. 1992. Nucleotide sequence and structure of the Klebsiella pneumoniae p operon. Mol. Gen. Genet. 232:284-294. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell, A. E., A. D. Jones, R. S. Mercer, and R. B. Rucker. 1999. Characterization of pyrroloquinoline quinone amino acid derivatives by electrospray ionization mass spectrometry and detection in human milk. Anal. Biochem. 269:317-325. [DOI] [PubMed] [Google Scholar]
- 42.Ophel, K., and A. Kerr. 1990. Agrobacterium vitis sp. nov. for strains of Agrobacteium biovar 3 from grapevines. Int. J. Syst. Bacteriol. 40:236-241. [Google Scholar]
- 43.Rodríguez, H., T. Gonzalez, and G. Selman. 2001. Expression of a mineral phosphate solubilizing gene from Erwinia herbicola in two rhizobacterial strains. J. Biotechnol. 84:155-161. [DOI] [PubMed] [Google Scholar]
- 44.Salomone, J. Y., P. Crouzet, P. De Ruffray, and L. Otten. 1996. Characterization and distribution of tartate utilization genes in the grapevine pathogen Agrobacterium vitis. Mol. Plant Microbe Int. 9:401-408. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 46.Schnider, U., C. Keel, C. Voisard, G. Defago, and D. Haas. 1995. Tn5-directed cloning of p genes from Pseudomonas fluorescens CHA0: mutational inactivation of the genes results in overproduction of the antibiotic pyoluteorin. Appl. Environ. Microbiol. 61:3856-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shim, J. S., S. K. Farrand, and A. Kerr. 1987. Biological control of crown gall: construction and testing of new biocontrol agents. Phyopathology 77:463-466. [Google Scholar]
- 48.Smidt, C. R., F. M. Steinberg, and R. B. Rucker. 1991. Physiological importance of pyrroloquinoline quinone. Proc. Soc. Exp. Biol. Med. 197:19-26. [DOI] [PubMed] [Google Scholar]
- 49.Stites, T. E., A. E. Mitchell, and R. B. Rucker. 2000. Physiological importance of quinoenzymes and the O-quinone family of cofactors. J. Nutr. 130:719-727. [DOI] [PubMed] [Google Scholar]
- 50.Tang, X., B. F. Lu, and S. Q. Pan. 1999. A bifunctional transposon mini-Tn5gfp-km which can be used to select for promoter fusions and report gene expression levels in Agrobacterium tumefaciens. FEMS Microbiol. Lett. 179:37-42. [DOI] [PubMed] [Google Scholar]
- 51.Toyama, H., L. Chistoserdova, and M. E. Lidstrom. 1997. Sequence analysis of pqq genes required for biosynthesis of pyrroloquinoline quinone in Methylobacterium extorquens AM1 and the purification of a biosynthetic intermediate. Microbiology 143:595-602. [DOI] [PubMed] [Google Scholar]
- 52.van der Meer, R. A., B. W. Groen, M. A. G. van Kleef, J. Frank, J. A. Jongejan, and J. A. Duine. 1990. Isolation, preparation, and assay of pyrroloquinoline quinone. Methods Enzymol. 188:260-283. [DOI] [PubMed] [Google Scholar]
- 53.Van Schie, B. J., O. H. de Mooy, J. D. Linton, J. P. van Dijken, and J. G. Keunen. 1987. PQQ-dependent production of gluconic acid by Acinetobacter, Agrobacterium and Rhizobium species. J. Gen. Microbiol. 133:867-875. [Google Scholar]
- 54.Vervliet, G., M. Holsters, H. Teuchy, M. van Montagu, and J. Schell. 1975. Characterization of different plaque forming and defective temperate phages in Agrobacterium strains. J. Gen. Virol. 26:33-48. [DOI] [PubMed] [Google Scholar]
- 55.Yamada, M., K. Sumi, K. Matsushita, O. Adachi, and Y. Yamada. 1993. Topological analysis of quinoprotein glucose dehydrogenase in Escherichia coli and its ubiquinone-binding site. J. Biol. Chem. 268:12812-12817. [PubMed] [Google Scholar]
- 56.Yang, Y. L., J. Y. Li, J. H. Wang, and H. M. Wang. 2008. Mutations affecting chemotaxis of Agrobacterium vitis strain E26 also alter attachment to grapevine roots and biocontrol of crown gall disease. Plant Pathol. doi: 10.1111/j.1365-3059.2008.02020.x. [DOI]
- 57.Zhao, C., and G. Wittstock. 2005. Scanning electrochemical microscopy for detection of biosensor and biochip surfaces with immobilized pyrroloquinoline quinine (PQQ)-dependent glucose dehydrogenase as enzyme label. Biosens. Bioelectron. 20:1277-1284. [DOI] [PubMed] [Google Scholar]