Abstract
Members of the genus Bifidobacterium are gram-positive bacteria that commonly are found in the gastrointestinal tract (GIT) of mammals, including humans. Because of their perceived probiotic properties, they frequently are incorporated as functional ingredients in food products. From probiotic production to storage and GIT delivery, bifidobacteria encounter a plethora of stresses. To cope with these environmental challenges, they need to protect themselves through stress-induced adaptive responses. We have determined the response of B. breve UCC2003 to various stresses (heat, osmotic, and solvent) using transcriptome analysis, DNA-protein interactions, and GusA reporter fusions, and we combined these with results from an in silico analysis. The integration of these results allowed the formulation of a model for an interacting regulatory network for stress response in B. breve UCC2003 where HspR controls the SOS response and the ClgR regulon, which in turn regulates and is regulated by HrcA. This model of an interacting regulatory network is believed to represent the paradigm for stress adaptation in bifidobacteria.
Bifidobacteria are strict anaerobic bacteria with a typical forked (bifid) shape, and they represent one of the most common inhabitants of the mammalian and animal gastrointestinal tract (GIT) (42). From a phylogenetic perspective, the genus Bifidobacterium belongs to the subclass Actinobacteridae of the phylum Actinobacteria of high-GC-content, gram-positive bacteria (44). They are among the first colonizers of the sterile GIT of newborns, and they constitute the dominant genus in gut bacterial populations of healthy breastfed infants (28). In healthy adults, the proportion of Bifidobacterium species in the GIT remains relatively stable, about 3% (42). In the human GIT, their presence has been associated with various health-promoting or probiotic effects, e.g., they are thought to play an essential role in the development of the immune system (see reference 22 and references therein). This has led to the widespread use of various bifidobacterial strains as functional components of probiotic foods (22).
Commercially exploited bifidobacteria are exposed to a plethora of environmental stresses during large-scale production, product storage, and passage through the oral cavity, stomach, and small intestine. In particular, exposure to oxygen or other oxygen-derived radicals, organic acids, and bile, as well as osmotic, heat, and cold stress, have a major negative impact on bifidobacterial viability and, consequently, probiotic functionality. Bifidobacteria, like other bacteria, are capable of synthesizing a particular set of proteins protecting the cell from the deleterious effects caused by the accumulation of unfolded and/or misfolded proteins as a result of the stress conditions described above. Several of these protective proteins act as molecular chaperones, such as GroEL (Hsp60), DnaK (Hsp70), and ClpB (Hsp100) (19), or as proteases, such as Lon, ClpP, and ClpC (54), and they play key roles in several posttranslational events to prevent protein denaturation, aggregation, and/or misfolding. Traditionally, heat shock proteins (Hsps) have been classified according to their molecular mass into the principal families designated Hsp100, Hsp90, Hsp70, Hsp60, and small Hsp (for a review of high-GC-content bacteria, see reference 45). The analysis of the Bifidobacterium breve UCC2003 genome sequence has revealed a conserved set of chaperone-encoding genes represented by groEL, groES, dnaK, grpE, dnaJ1, and dnaJ2, as well as the Clp-ATPase-specifying genes clpB and clpC and the protease-encoding genes clpP1and clpP2 (45).
Although Hsps appear to be universally conserved among bacteria, the mechanisms underlying their regulated expression seem to be rather variable (45). B. breve UCC2003 encodes three heat stress-related transcriptional regulators, which, assuming that this strain is representative of other bifidobacteria, may explain the differential induction profiles observed for bifidobacterial Hsp-encoding genes (43, 46-50), and which also are found in other members of the Actinobacteria: (i) the HrcA (heat regulation at CIRCE) regulator, which binds to a DNA sequence referred to as controlling inverted repeat of chaperone expression (CIRCE) (35); (ii) the heat shock protein repressor (HspR), which binds to an HspR-associated inverted repeat (HAIR) consensus motif (8); and (iii) the Clp gene regulator (ClgR) (4), which binds to an imperfect inverted repeat.
The HrcA-encoding gene and HrcA recognition sequence, CIRCE (TTAGCACTC-N9-GAGTGCTAA), are highly conserved among eubacteria, e.g., Bacillus subtilis, where they regulate the transcription of the chaperone-encoding genes dnaK, groEL, grpE, dnaJ, and groES (35). In contrast, in Streptomyces spp., HrcA appears only to regulate the transcription of groELS, hrcA itself, and an adjacent DnaJ-encoding gene (18). In B. subtilis and possibly others, a negative regulatory feedback loop exists between HrcA and GroEL, since GroEL, when not sequestered by misfolded proteins, enhances the HrcA-directed repression of its gene targets (27).
In Streptomyces, as well as in other high-GC-content gram-positive bacteria, the regulation of dnaK, grpE, dnaJ, hspR itself, and clpB is mediated by HspR, which binds to the conserved HAIR sequence CTTGAGT-N7-ACTCAAG (8, 17, 49, 51). HspR-encoding genes are confined to members of the Actinomycetales order, in addition to Helicobacter, Campylobacter, Deinococcus, and related species (1, 34, 37). In Streptomyces and possibly others, a negative regulatory feedback loop between HspR and DnaK exists, similarly to what has been described for HrcA, where unsequestered DnaK forms a complex with HspR to dramatically increase the binding activity of the latter protein (6, 7).
The least conserved heat shock regulator, ClgR, differs somewhat between (currently sequenced) bifidobacteria and other high-GC-content gram-positive bacteria, as in the former group of bacteria the ClgR regulator possesses an N-terminal extension, the function of which presently is unknown. ClgR regulates the expression of protease-encoding genes such as clpP, clpC, and lon (4, 13, 48, 50). It also has been implicated in the response to DNA damage (12). Its deduced binding site, CGCT-N[4]-GCGNAC in Streptomyces, CGCT-N[4]-GCCNA in Bifidobacterium, and WNNWCGCYNANRGCGWWS in Corynebacterium, suggests that it is not particularly well conserved. Furthermore, the relevant binding sequence was not detected in the promoter region of bifidobacterial clpC, even though ClgR was shown to bind to this region (48). In Streptomyces and Corynebacterium species, it was shown that ClgR activity is controlled by degradation by the ClpCP protease (13). However, the proposed degradation signal, consisting of two adjacent alanine residues at the C terminus of ClgR, is present only in characterized Streptomyces species (3, 52). The degradation of ClgR by ClpCP thus provides a negative regulatory feedback loop that is analogous to what has been described for HrcA and HspR.
Additional heat shock-related regulators are known, e.g., RheA in Streptomyces (36), CtsR in many gram-positive bacteria (21), and stress-related sigma factors; however, these do not appear to be encoded by bifidobacterial genomes (see reference 45 and references therein).
We show here that various stress responses in B. breve UCC2003 are governed by a complex interactive network of regulators that defines a stimulon involved in bifidobacterial adaptation to different environmental insults.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Plasmids and strains used in this study are listed in Table 1. E. coli was grown in TY medium (33) in shaking flasks at 37°C. B. breve was grown at 37°C in MRS medium (Difco Laboratories, Detroit, MI) as standing cultures supplemented with 0.05% cysteine or on RCM agar plates containing 1.5% (wt/vol) agar under anaerobic conditions in a Modular Atmosphere Controlled System (Davidson & Hardy Ltd.). Where appropriate, ampicillin (Roche Diagnostics, East Sussex, United Kingdom) was used for Escherichia coli at 100 μg/ml, while tetracycline (Roche) was added at 12.5 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Name | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| B. breve UCC2003 | 25 | |
| E. coli | ||
| BL21(DE3) | F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE3 [lacI lacUV5-T7gene 1 ind1 sam7 nin5]) | Laboratory collection |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB lacIqZΔM15 Tn10 [TetR]) | Stratagene |
| Plasmids | ||
| pG-KJE8 | Cmr, dnaK-dnaJ-grpE and groES-groEL overexpression plasmid | Takara Bio |
| pQE30 | Apr, T5 promoter expression plasmid for N-terminal His tag fusions | Qiagen |
| pQE60 | Apr, T5 promoter expression plasmid for C-terminal His tag fusions | Qiagen |
| pQE60-hrcA | Apr, pQE60 carrying the hrcA gene of B. breve UCC2003 | This work |
| pQE30-clgR | Apr, pQE30 carrying the clgR gene of B. breve UCC2003 | This work |
| pQE30-clgR-T | Apr, pQE30 carrying the truncated clgR gene of B. breve UCC2003 | This work |
| pQE30-hspR | Apr, pQE30 carrying the hspR gene of B. breve UCC2003 | 49 |
| pNZ272 | Cmr, pSH71 derivative containing promoterless glucuronidase gene for promoter screening | 30 |
| pNZ272-PgroEL | pNZ272 derivative carrying the groEL promoter | This work |
| pNZ272-PrbsA1 | pNZ272 derivative carrying the rbsA1 promoter | This work |
Cmr, Tetr, and Apr indicate resistance to chloramphenicol, tetracycline, and ampicillin, respectively.
DNA techniques and transformation.
Molecular cloning techniques were performed essentially as described previously (33). Restriction enzymes and T4 DNA ligase were obtained from Roche and used according to their instructions. PCRs were performed using Taq PCR mastermix (Qiagen GmBH, Hilden, Germany), and high-fidelity PCR was performed with KOD polymerase (Novagen, Darmstadt, Germany). Synthetic oligonucleotides were synthesized by MWG Biotech AG (Ebersberg, Germany) and are described in Table S1 in the supplemental material. PCR products were purified using the High-Pure PCR product purification kit (Roche). Plasmid DNA was introduced into E. coli and B. breve by electrotransformation as previously described (25). Analytical-grade chemicals were obtained from Merck (Darmstad, Germany) or BDH (Poole, United Kingdom).
DNA microarray and quantitative reverse transcription-PCR (qRT-PCR) experimental procedures.
DNA microarrays containing oligonucleotide primers representing each of the 1,864 annotated genes of the genome of B. breve UCC2003 were obtained from Agilent Technologies (Palo Alto, CA).
An overnight culture of B. breve was used to inoculate (1% inoculum) 350 ml of MRS broth. Cells were incubated at 37°C until an optical density at 600 nm (OD600) of 0.5 was reached. The culture then was divided into seven different cultures. Six of these were exposed to one of the following stress conditions: growth at 42, 44, 47, or 50°C for 1 h, exposure to 0.5 M NaCl for 1 h, or exposure to 8% ethanol for 1 h. The remaining culture acted as a control (growth at 37°C for 1 h). Following these treatments, cells were harvested by centrifugation at 8,000 × g for 1 min at room temperature and immediately frozen in liquid nitrogen prior to RNA isolation. Methods for cell disruption, RNA isolation, RNA quality control, cDNA synthesis, and indirect labeling were performed as described previously (40). Labeled cDNA was hybridized using the Agilent Gene Expression hybridization kit (part number 5188-5242) as described in the Agilent Two-Color Microarray-Based Gene Expression Analysis (v4.0) manual (publication number G4140-90050). Following hybridization, microarrays were washed as described in the manual and scanned using Agilent's DNA microarray scanner G2565A. The scanning results were converted to data files with Agilent's Feature Extraction software (version 9.5). DNA microarray data were processed as previously described (16, 40). Differential expression tests were performed with the Cyber-T implementation of a variant of the t test (23). A gene was considered differentially expressed between a test condition and a control when an expression ratio of >3 or <0.33 relative to the result for the control was obtained with a corresponding P value that was <0.001. Final data presented are the averages from at least two independent array experiments. The original DNA microarray data are available at http://bioinfo.ucc.ie/heatshock/. The qRT-PCR amplifications were performed on an Roche Lightcycler 480 using SYBR green I dye assay chemistry. A 20-μl PCR assay for each gene of interest consisted of 10 μl of 2× RT-PCR master mix (Roche), 1 μl 1/3,000 SYBR green dye (Biogene), 3.75 μl of H2O, 0.625 μl (2.5 μmol) of forward and 0.625 μl (2.5 μmol) of reverse primers, and 4 μl (20 ng) of diluted cDNA template. All qRT-PCRs were run in quadruplicate; two biological replicates, independent from the microarray samples, were taken for both induced and uninduced cultures, resulting in eight measurements per gene for each environmental condition. No amplification was observed for the qRT-PCR controls (no reverse transcriptase and no template). Cycling conditions used for all amplifications were one cycle of 95°C for 10 min and 45 cycles of 95°C for 15 s, 55°C for 15 s, and 68°C for 20 s. From the qRT-PCR data, an average cycle threshold (CT) value was calculated from the duplicate reactions. Averaged CT values then were normalized (to adjust for various amounts of cDNA for each reaction) relative to values for the control gene, rnpA, which was selected because its average gene expression did not change in any of the experiments. The average ratio of differences (n-fold) from the biological duplicates of selected genes from uninduced (control) and induced (test) samples was determined as previously described (29).
Cloning of promoter fragments and construction of overexpression vectors.
Oligonucleotides used to amplify the described promoter regions and the hrcA and clgR genes from B. breve UCC2003 are listed in Table S1 in the supplemental material. The DNA fragment encompassing hrcA was cut with BfuAI and BglII and was inserted in pQE60 digested with NcoI and BglII enzymes. The resulting plasmid, designated pQE60-hrcA, was introduced in E. coli BL21(DE3)(pG-KJE8). The DNA fragments containing clgR and a 286-bp 5′ truncation of clgR were cut with BamHI and HindIII and inserted in pQE30 digested with the same, resulting in plasmids pQE30-clgR and pQE30-clgR-T, respectively, following introduction in and correct identification from E. coli XL1-Blue. The DNA fragments encompassing the groEL and rbsA1 promoter were cut with PstI/BamHI and BamHI/ScaI, respectively, and inserted into promoter probe vector pNZ272 cut with same to generate pNZ272-PgroEL and pNZ272-PrbsA1, respectively, following introduction in and correct identification from E. coli XL1-Blue and subsequent transfer to B. breve UCC2003. The sequence fidelity of the inserts in these recombinant plasmids was checked by sequence analysis (MWG, Ebersberg, Germany).
Expression and purification of H6-ClgR and H6-ClgR-T.
The B. breve UCC2003-derived, His-tagged ClgR and ClgR-T proteins were purified according to the QIAexpressionist 03/2001 protocol with the following modifications. An overnight culture of E. coli XL1-Blue [pQE30-ClgR(-T)] was diluted 1:50 into fresh TY medium supplemented with 100 μg/ml of ampicillin and grown at 37°C with vigorous shaking. At an OD600 of 0.6, the expression of the recombinant protein was induced by the addition of 1 mM isopropyl thio-ß-d-galactoside (IPTG). Growth was continued for 4 h, and cells from 200 ml of culture were collected by centrifugation (10 min, 8,000 × g, 4°C) in an Avanti J-20 XP centrifuge (Beckmann Coulter, Mijdrecht, The Netherlands). The pellet was washed with 50 ml of buffer A (50 mM NaHPO4, 300 mM NaCl, 10 mM imidazole, 3.5% glycerol, 1 mM ß-mercaptoethanol, pH 8.0) and stored at −80°C for future use. The pellet was resuspended in 10 ml of buffer A, and cells were disrupted by sonication. Cellular debris was removed by centrifugation (30 min, 20,000 × g, 4°C), and the supernatant fraction was used to purify the desired protein as described above. Purified protein (1 ml) was dialyzed against 20 mM Tris-HCl (pH 8) buffer containing 10% glycerol and 100 mM KCl to remove excess salts. Purified protein was examined for protein content and purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and for (the absence of) DNA by the ethidium bromide staining of agarose gels. Protein concentrations were quantified using the RC/DC protein determination kit (Bio-Rad Laboratories, Richmond, CA.).
Expression and purification of H6-HrcA.
The B. breve UCC2003-derived H6-HrcA protein was purified as follows. An overnight culture of E. coli BL21(DE3)(pQE60-HrcA)(pG-KJE8) supplemented with chloramphenicol and ampicillin was grown until an OD600 of 0.6 was reached, after which the expression of the recombinant protein was induced by the addition of 1 mM IPTG, while the expression of GroEL and GroES (from plasmid pG-KJE8) was induced by the inclusion of 10 ng/ml tetracycline. Growth was continued for 16 h at 18°C, and purification was performed as described above.
Expression and purification of H6-HspR.
The B. breve UCC2003-derived H6-HspR protein was purified as described previously (49), with the following modifications: for all refolding buffers 200 mM KCl was added, which was shown to increase the concentration of soluble H6-HspR protein.
DNA band shift assays.
DNA fragments encompassing various promoter regions were prepared by PCR using primers described in Table S1 in the supplemental material. DNA band shift assays were performed as described previously (41), with the following modification. The PCR products were generated using IRD-800-labeled fluorescent probes. Protein and probe were mixed on ice and subsequently incubated for 20 min at 37°C. Samples were loaded onto a 6% nondenaturing polyacrylamide gel prepared with 1× TAE (40 mM Tris-acetate [pH 8.0], 2 mM EDTA) and run in a 0.5× to 2.0× gradient of TAE at 100 V for 60 min in a mini-protean electrophoresis system (Bio-Rad Laboratories). Following electrophoresis, the presence and mobility position of the fluorescent PCR products in the gel were detected using Odyssey (LI-COR Biosciences UK Ltd., Cambridge, United Kingdom).
Promoter predictions and motif searches.
DNA sequences encompassing 400 bp of the upstream intergenic regions of the genes that were overexpressed under various stress conditions used, as detected by the DNA microarray experiments, were collected from the genome sequence of B. breve UCC2003. This data set was used as the input for the MEME software tool (2) to search for overrepresented nucleotide sequences. A graphical representation of the identified motif was obtained using WebLOGO software (9). Additionally, motif searches were done with MotifLocator (39) using a stringent cutoff of 0.95 and with HMMer (11).
RESULTS
Global analysis of B. breve UCC2003 stress transcriptomes using functional category enrichment.
To identify genes whose expression changes upon exposure to various levels or different types of environmental stress known to cause protein denaturing and/or misfolding, the transcriptional profile of B. breve UCC2003 growing at 37°C was compared to transcriptional profiles obtained from this strain when it was grown at 42, 44, 47, or 50°C for 1 h, exposed to 0.5 M NaCl for 1 h, or exposed to 8% ethanol for 1 h.
Exposure to severe heat stress (47 or 50°C), 0.5 M NaCl, or 8% ethanol had a pronounced negative effect on the growth rate of B. breve UCC2003. Moderate heat stress, i.e., exposure at 44 or 42°C, was shown to have either a minor or no effect on B. breve UCC2003 growth, respectively (see Fig. S1 in the supplemental material). The compiled analyzed results of the transcriptome comparisons between stressed and unstressed cultures are presented in Table 2 and show that the highest number of genes was transcriptionally affected when the culture was exposed to a 47°C heat shock. The strongest transcriptional induction of a gene (more than 400-fold; clgR) occurs following a 50°C heat shock.
TABLE 2.
Functional category enrichment analysis of transcriptomics dataa
| Functional category | No. of genes responding to stress condition and regulation status |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 42°C |
44°C |
47°C |
50°C |
Ethanol |
Osmotic |
|||||||
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | |
| Cell cycle control, cell division, and chromosome partitioning | 2 | 2 | 2 | |||||||||
| Cell wall/membrane/envelope biogenesis | 1 | 1 | 10 | 2 | 11 | 1 | 6 | 2 | 6 | |||
| Defense mechanisms | 1 | 5 | 1 | 6b | 2 | 1 | 1 | |||||
| Intracellular trafficking, secretion, and vesicular transport | 2 | 1 | 1 | |||||||||
| Posttranslational modification, protein turnover, and chaperones | 3b | 4b | 4 | 9b | 5 | 8 | 9 | 11b | 5 | 1 | ||
| Signal transduction mechanisms | 1 | 3 | 2 | 2 | 1 | |||||||
| Replication, recombination, and repair | 2 | 3 | 13 | 12 | 24 | 12 | 6 | 4 | 10 | 2 | ||
| RNA processing and modification | ||||||||||||
| Transcription | 2 | 3 | 18 | 6 | 19 | 8 | 2 | 4 | 12 | 4 | ||
| Translation, ribosomal structure, and biogenesis | 7 | 8 | 8 | 21 | 2 | 4 | 1 | |||||
| Amino acid transport and metabolism | 2 | 2 | 9 | 16 | 8 | 24 | 5 | 1 | 11 | 5 | ||
| Carbohydrate transport and metabolism | 4 | 1 | 32b | 36 | 25 | 41 | 5 | 27b | 7 | 3 | ||
| Coenzyme transport and metabolism | 1 | 3 | 4 | 7 | 1 | 1 | 1 | 3 | ||||
| Energy production and conversion | 1 | 4b | 3 | 6 | 13 | 1 | 19b | 3 | 1 | 3 | 5 | |
| Inorganic ion transport and metabolism | 1 | 3 | 7 | 7 | 4 | 7 | 3 | 2 | 8 | 1 | ||
| Lipid transport and metabolism | 1 | 1 | 1 | 1 | 2 | 1 | ||||||
| Nucleotide transport and metabolism | 3 | 10 | 2 | 17b | 1 | 2 | 4 | |||||
| Secondary metabolite biosynthesis, transport, and catabolism | 1 | 2 | 1 | 3 | 4 | 4 | 1 | |||||
| General function prediction only | 4 | 25 | 31 | 27 | 27 | 8 | 11 | 23 | 9 | |||
| Function unknown | 2 | 4 | 12 | 32 | 136b | 82 | 217b | 89 | 27 | 31 | 133b | 35 |
| Total no. of genes regulatedc | 5 | 11 | 17 | 92 | 267 | 266 | 346 | 324 | 82 | 99 | 238 | 93 |
Genes were considered regulated when the fold change ratio was ≤3 or P < 0.0001.
Significantly enriched categories. P < 0.01 using a Fisher Exact test, right-tailed values.
Some genes are present in multiple functional categories.
To facilitate the interpretation of the obtained results, the genes exhibiting a significantly altered level of transcription were grouped on the basis of the predicted functions of the encoded proteins (38), and the significance of the enrichment of their functional categories was calculated with a Fisher Exact test (15, 26). Many of the affected genes had either no known function or were present in the category carbohydrate transport and metabolism. Nevertheless, and as expected, genes belonging to the protein fate functional category were clearly the most strongly upregulated under the stress conditions used, both by their significance (Table 2) and by the level (n-fold) of induction (Table 3).
TABLE 3.
Regulated gene members of the putative stress regulatory networka
| Locus | Product | Description | Level (n-fold) of induction under stress condition: |
Putative regulator/motif (position) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 42°Cb | 42°Cc | 44°Cb | 44°C | 47°C | 47°C | 50°Cb | 50°Cc | EtOHb | EtOHc | osmb | osmc | ||||
| Bbr_0076 | Hsp20 | Chaperone | 19.4 | ND | 26.7 | ND | 51.8 | ND | 119.3 | ND | 1.8 | ND | 5.4 | ND | |
| Bbr_0124 | DnaK | Chaperone protein | 3.5 | 1.4 | 7.0 | 4.4 | 10.2 | 40.7 | 29.3 | 59.8 | 3.6 | 14.3 | 16.7 | 38.8 | HspR, GAAGTTGAGTGATGTAGGCTCAAGTTTT (−15) and AAACCTGAGCCTACATCGCTCAACTTTT (−74) |
| Bbr_0125 | GrpE | GrpE protein | 3.2 | 1.4 | 6.7 | 6.0 | 7.8 | 36.5 | 22.8 | 37.0 | 3.6 | 18.7 | 13.1 | 22.2 | |
| Bbr_0126 | DnaJ1 | Chaperone protein | 3.6 | 2.8 | 8.2 | 8.7 | 10.2 | 31.1 | 24.9 | 48.2 | 1.6 | 10.5 | 16.2 | 7.4 | |
| Bbr_0127 | HspR | Regulator | 2.6 | ND | 6.1 | ND | 8.8 | ND | 23.8 | ND | 1.9 | ND | 15.4 | ND | |
| Bbr_1046 | Nfo | Endonuclease IV | 1.1 | 1.0 | 1.7 | 1.0 | 8.9 | 3.2 | 18.1 | 1.6 | 5.0 | 5.6 | 1.9 | 1.7 | HspR, ATGCTTAAGTCTCCTTCGGTCAGATGGTe |
| Bbr_1171 | HrdB | Principal sigma factor | 1.1 | 1.0 | 1.7 | 1.0 | 5.8 | 4.6 | 13.1 | 8.9 | 1.7 | 2.5 | 1.3 | 1.5 | HspR, TCCGTGGAGGGTGGTTTACTCACATAGGe |
| Intergenic region 1182-1181 | ND | ND | 19.4 | 16.8 | 15.3 | 6.7 | |||||||||
| Bbr_1182 | 1.0 | 1.0 | 1.7 | 1.8 | 9.1 | 36.6 | 66.9 | 37.9 | 3.6 | 38.5 | 1.2 | 14.7 | |||
| Bbr_1183 | ClgR | Regulator | 1.6 | 1.1 | 3.6 | 2.0 | 57.2 | 31.8 | 416.3 | 40.5 | 7.1 | 31.0 | 3.3 | 13.4 | HspR, AAACTTGAGTCTATTCACAGCAAGTTTT |
| Bbr_1781 | ClpB | Chaperone protein | 4.2 | 1.6 | 12.2 | 7.7 | 27.3 | 34.5 | 94.6 | 84.1 | 4.4 | 12.0 | 8.0 | 21.0 | HspR, AAACTTGAGTCACTTACACTCAACTTTT (−54) |
| Bbr_1004 | HrcA | Regulator | 2.0 | 1.0 | 3.0 | 2.6 | 11.2 | 17.7 | 3.5 | 13.2 | 5.7 | 13.0 | 6.9 | 1.3 | HrcA, TTAGCACTCGAAACCAATGAGTGCTAA (−23); ClgR, CTTCGCCGCAAACGAAC (−49) |
| Bbr_1005 | DnaJ2 | Chaperone protein | 1.4 | 1.9 | 2.4 | 2.9 | 2.0 | 7.6 | −4.4 | −5.8 | 3.8 | 7.3 | 1.5 | 1.0 | |
| Bbr_1364 | GroEL | Chaperone protein | 2.1 | 2.0 | 3.6 | 5.2 | 3.8 | 3.8 | 1.1 | 3.0 | 3.8 | 5.6 | 1.1 | 1.2 | HrcA, TTGGCACTCGGCAAGGGCGAGTGCTAA (−22)d and TTAGCACTCGGAGATCGAGAGTGATAA (+14)d |
| PgroEL-gusA | 2.5 | 10.2 | 1.0 | 1.0 | 14.7 | 2.2 | |||||||||
| Bbr_1668 | GroES | Chaperone protein | 1.5 | 1.6 | 2.3 | 6.1 | 3.3 | 2.7 | 3.3 | 2.3 | 3.3 | 5.7 | 1.8 | 1.2 | HrcA, TTAGCACTCGAGTTGGCAGAGTGCTAA (−35) |
| Bbr_0794 | ClpP1 | Protease | 1.0 | 1.6 | 1.2 | 4.0 | 1.0 | 2.1 | −1.3 | 2.7 | 3.4 | 12.2 | −2.3 | −3.6 | ClgR, CTACGCTGGCCGCCAAA (−61) |
| Bbr_0795 | ClpP2 | Protease | −1.1 | 1.5 | 1.2 | 3.2 | −1.4 | 2.4 | −3.5 | 2.3 | 3.3 | 10.2 | −1.5 | 1.0 | |
| Bbr_1356 | ClpC | Protease | −1.1 | 1.2 | 1.2 | 3.5 | −1.2 | 1.4 | −2.7 | −1.2 | 2.4 | 8.9 | −1.6 | −1.2 | ClgR, CTCCTCTCACGGCGCAA (−60) |
| Bbr_0216 | Modification methylase | −1.3 | −1.2 | 1.3 | 1.0 | 5.3 | 3.9 | 3.1 | 3.0 | −1.1 | 2.2 | 1.9 | 1.7 | LexA, TCGAACAAATGTTCTA | |
| Bbr_0496 | MutY | DNA glycosylase | 1.1 | 1.3 | 2.3 | 4.0 | 2.5 | 11.6 | −1.7 | 6.9 | 3.9 | 5.8 | 1.7 | 2.2 | LexA, TCGAACATCTTCTCGA |
| Bbr_0585 | ImpB | DNA repair | 1.3 | 1.3 | 4.3 | 4.0 | 33.0 | 24.5 | 18.3 | 17.1 | 1.9 | 3.8 | 4.7 | 5.0 | LexA, TCGAACAAATGTTCGA |
| Bbr_1013 | RuvA | Holliday junction DNA helicase | 1.1 | 1.0 | 2.0 | 1.4 | 5.7 | 2.5 | 6.7 | 1.8 | 2.6 | 1.6 | 1.5 | −1.1 | LexA, TCGAACAGTTGTTCGA |
| Bbr_1014 | RuvB | Holliday junction DNA helicase | 1.0 | 1.1 | 1.9 | 1.6 | 2.2 | 1.6 | −2.4 | 1.5 | 2.4 | 1.1 | 1.0 | 1.5 | |
| Bbr_1180 | RecX | Recombination regulator | 1.2 | 1.2 | 2.9 | 2.4 | 17.7 | 13.9 | 10.9 | 3.5 | 3.5 | 2.8 | 2.6 | 3.2 | |
| Bbr_1181 | RecA | Recombinase | 1.4 | 1.4 | 3.3 | 3.3 | 10.2 | 19.2 | 12.0 | 18.5 | 4.3 | 16.7 | 2.4 | 3.5 | LexA, TCGAACAAATGTTCGC |
| Bbr_1271 | LexA | Regulator | 1.1 | 1.1 | 1.9 | 1.4 | 2.8 | 4.8 | 7.0 | 4.6 | 3.4 | 2.7 | 6.8 | 4.5 | LexA, TCGAACAGATGTTTGA and TCGAACAAGTGTTCTA; LexA, TCGAACATCTGTTCTT and TGGCACAAATGTTCGA |
Underlining indicates that the locus is the first member of the operon. The respective stresses are the following: 42 to 50°C, 1 h of exposure to the temperature given; EtOH, 1 h of exposure to 8% ethanol; osm, 1 h of exposure to 0.5 M NaCl.
Microarray upregulation (n-fold). Significantly regulated genes are shown in boldface.
qRT-PCR results. ND, not determined.
Transcriptional start site predicted.
These motifs do not bind H6-HspR.
Identification of the regulatory elements governing stress response.
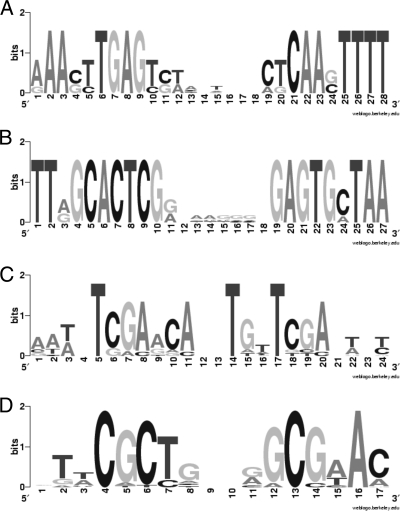
An in silico sequence analysis was performed to determine whether the genes upregulated by stress treatment in B. breve UCC2003 contain conserved regulatory sequences in their promoter regions. Data sets of sequences containing 400 bp upstream of the putative translation start sites of the upregulated genes per stress condition (see Table S1 in the supplemental material) were generated. We assumed that these genes were under the direct control of one or more stress response regulators, which elicit this control through binding to specific sequences in their target promoter regions. In this way, data sets of 5 (in the case of 42°C heat shock) to 400 (in the case of 50°C heat shock) sequences were examined for the occurrence of common elements with a length between 10 and 30 bp using the MEME algorithm (2). Highly conserved inverted repeat sequences indeed were present upstream of the translational start of many of the analyzed promoter regions. The first common motif detected was found in the promoter regions of the genes encoding DnaK, ClpB, ClgR, Nfo, and HrdB (Table 3 and Fig. 1A). The motif described is very similar to the known HAIR motif that previously had been described for B. breve UCC2003 for the promoter region of clpB, which is bound by HspR (49). For those promoters where the transcriptional start has been determined experimentally, this motif is present on or upstream of the −35 and −10 sequences (Table 3). The second motif detected was similar to the known CIRCE motif (Fig. 1B) and was found to be present in the promoter regions upstream of the genes encoding GroEL, GroES, and HrcA, the latter being the regulatory protein that is known to bind the CIRCE motif (35, 47). For characterized promoter regions, the CIRCE motif is overlapping the −35 or −10 sequence (Table 3). The third detected motif represents a presumptive SOS box that is involved in the SOS response (14), as it is present in the promoter regions of genes whose homologs are known to be involved in DNA damage response and repair, such as those predicted to specify LexA, RuvA, RecA, and MutY (Table 3).
FIG. 1.
Deduced stress regulatory binding sites described in Table 3 depicted in WebLOGO format based on the comparative sequence analysis of actual target sequences in the B. breve UCC2003 genome. (A) HAIR; (B) CIRCE; (C) SOS box; and (D) ClgR binding site.
The inspection of the upstream regions of the 82 identified ethanol stress-induced genes (see Table S2 in the supplemental material) with an HMM model of the ClgR binding site, generated from known ClgR binding sites (4, 12, 50), revealed the presence of putative binding sites in the promoter regions of the genes encoding ClpP1, ClpC, and HrcA. The ClgR motif detected in the promoter region of ClpC deviates somewhat from the previously identified ClgR recognition sites, explaining why it had not been detected before (48). An analysis of the location of the putative ClgR-binding box in the upstream region of each gene revealed a preference of the center of the first (upstream) heptamer of the motif for positions −60/61 or for −50 relative to the transcriptional start site (Table 3).
Binding of HspR, ClgR, and HrcA to promoter regions containing predicted operators.
To examine whether HspR, ClgR, and HrcA interact directly with the promoter regions of their suspected target genes, EMSAs were performed with purified, N-terminally His-tagged HspR (H6-HspR) and ClgR (H6-ClgR) or C-terminally His-tagged HrcA (H6-HrcA). H6-ClgR and H6-HrcA were overproduced and isolated under native conditions, while HspR had to be isolated under denaturing conditions and refolded to acquire this protein in a soluble form (results not shown). To obtain soluble H6-HrcA, it was necessary to produce this protein in conjunction with E. coli GroEL and GroES, encoded on pG-KJE8 (see Materials and Methods), because refolding procedures following the purification of H6-HrcA under denaturing conditions had yielded soluble but inactive protein. Obtaining soluble and active HrcA homologs from other microorganisms previously has been described to be troublesome, as the protein easily forms aggregates and/or inclusion bodies (see reference 31 and references therein).
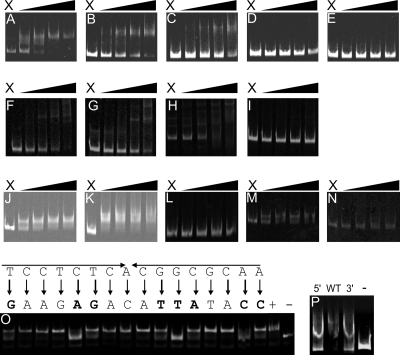
Gel shift retardation assays showed that H6-HspR indeed is capable of binding the dnaK, clpB, and clgR promoter regions. In contrast, H6-HspR was unable to retard the mobility of the nfo and hrdB promoter regions (Fig. 2A), although in these cases a HAIR motif was detected using in silico methods. Upon closer inspection it was noted that the motifs capable of interacting with HspR all are part of a larger inverted repeat structure consisting of an A-rich and T-rich extension at the 3′ and 5′ end of the HAIR motif, respectively (Table 3). The mutation of either of these extensions was shown to result in a decreased binding efficiency of the H6-HspR protein (Fig. 2b), suggesting that the extension is a valid part of the bifidobacterial HAIR consensus sequence.
FIG. 2.
(A to E) EMSAs of H6-HspR interaction with DNA fragments encompassing the promoter regions of dnaK (A), clpB (B), clgR (C), nfo (D), and hrdB (E). DNA fragments were obtained by PCR using IRD-800-labeled primers. Lane X contained the probe without added protein. The remaining lanes contained probe samples incubated with increasing concentrations of H6-HspR (concentrations ranged from 50 to 400 nM). For each successive lane from left to right, the concentration of H6-HspR was doubled. (F to I) EMSAs of H6-HrcA interactions with promoter regions of hrcA (F), groEL (G), groES (H), and hspR (I) (negative control). DNA fragments were obtained by PCR using IRD-800-labeled primers. Lane X contained the probe without added protein. The remaining lanes contained probe samples incubated with increasing concentrations of H6-HrcA (concentrations ranged from 50 to 400 nM). For each successive lane from left to right, the concentration of H6-HrcA was doubled. (J to N) EMSAs of H6-ClgR-T interactions with promoter regions of clpC (J), clpP (K), clgR (L), and hrcA (M). Panel N represents an EMSA in which full-length ClgR was used in combination with the labeled promoter region of clpC. DNA fragments were obtained by PCR using IRD-800-labeled primers. Lane X contained the probe without added protein. The remaining lanes contained probe samples incubated with increasing concentrations of H6-ClgR-T or H6-ClgR (concentrations ranged from 15 to 120 nM). For each successive lane from left to right, the concentration of H6-ClgR-T or H6-ClgR was doubled. (O) EMSAs of H6-ClgR-T with a concentration of 60 nM on mutated promoter regions of the ClgR motif of PclpC. Original sequences and mutations are shown above the image. +, original promoter sequence; −, promoter region of hsp20. (P) EMSAs of H6-HspR with a concentration of 175 nM on mutated promoter regions of the HAIR motif of PclpB. N, mutation of the AAAA 3′ extension to GGGG; WT, original promoter sequence; and C, mutation of the TTTT 5′ extension to CCCC.
Gel shift retardation assays also showed that H6-HrcA is capable of the retardation of the promoter regions upstream of the groES and hrcA genes, as well as that of the intergenic region located between cspA and groEL (Fig. 2F and G). In all cases a clear CIRCE motif is present in these regions, as detected using in silico methods (Table 3).
EMSAs show direct interaction of N-terminally truncated ClgR with the B. breve ClgR motif.
H6-ClgR is unable to form complexes with its target sequences, the promoters of clpP1 (not shown) and clpC (Fig. 2N), in the absence of cofactor(s), confirming previously obtained results (48, 50). It also had been noted previously that bifidobacterial ClgR homologs possess an N-terminal extension, in contrast to ClgR homologs encoded by other high-GC, gram-positive bacteria (48). The in silico analysis, using IUpred (10), of the amino acid sequence of ClgR shows that this N-terminal extension is an intrinsically unstructured region, prompting the possibility that this region prevents proper folding and consequently precludes the binding activity of ClgR. To investigate the involvement of the N-terminal extension of ClgR in interfering with its DNA binding activity, we generated a 94-residue N-terminally truncated and His-tagged version of ClgR (H6-ClgR-T) and performed EMSAs with the purified H6-ClgR-T.
H6-ClgR-T indeed was shown to form complexes with fluorescently labeled DNA fragments containing 250 bp of the upstream region of the clpC and clpP1 genes (Fig. 2J and K). Also, the third DNA fragment containing a predicted ClgR binding site, the promoter region upstream of the hrcA gene, displayed the ability to bind to H6-ClgR-T (Fig. 2M). Although in the cases of clpP1 and hrcA a clear ClgR motif was detected using in silico methods, the motif in the clpC promoter region appeared to deviate from the consensus ClgR-binding motif. Nevertheless, this apparent degenerate motif did not appear to affect H6-ClgR-T binding, as equal amounts of this protein retarded all tested promoters similarly. No binding was observed for the promoter region of the ClgR-encoding gene (Fig. 2L), unlike what has been shown for ClgR in Streptomyces lividans.
To confirm the position of the ClgR binding site in the promoter region of clpC, every base in the putative ClgR motif TCCTCTCACGGCGCAA (Fig. 2E) was mutated (A⇂C and G⇂T), and EMSAs were performed with these mutated fragments using H6-ClgR-T. The mutation of the individual bases shown in boldface in Fig. 2E had the most profound effect on ClgR-DNA complex formation, indicating that these bases are important for interaction with the protein.
Alignments of the ClgR motifs of clpC, clpP1, and hrcA, combined with the EMSA experiments described in this report, suggest that the ClgR binding site is TNCGCTNNNGGCGNAA instead of CGCT-N[4]-GCCNA.
Heat shock-responsive genes show distinct induction patterns.
It had been reported previously that the induction of heat shock-responsive genes in B. breve UCC2003 follows a specific induction pattern (see reference 45 and references therein). Microarray analysis, supported by qRT-PCR data, enabled us to investigate this in more detail. The experimentally determined heat stress-regulated genes indeed follow a distinct induction profile. The strong induction of candidate HspR-regulated genes occurs mainly at severe heat shock, but overexpression already is observed at lower temperatures, even at 42°C, as well as under ethanol and osmotic stress conditions (Table 3). Genes regulated by HrcA are induced mainly at 42 and 44°C, as well as under ethanol stress. Although the transcription of hrcA itself is induced when B. breve UCC2003 cells are subjected to severe heat shock and osmotic shock, its target genes are not (Table 3). The transcription of groEL is very high, representing one of the most transcribed genes in most microarray and qRT-PCR experiments, even under seemingly unstressed conditions, and at 44°C, 47°C, and under ethanol stress it becomes the most transcribed gene as determined by the microarray experiments (see Table S3 in the supplemental material). In contrast to previous observations, our data show that candidate ClgR-regulated genes are induced only following ethanol stress (3- to 4-fold in the microarray results and 10- to 12-fold in the qRT-PCR experiments). No induction was seen at 42 or 44°C in the microarray results, and only a relatively minor induction (three- to fourfold) was seen in the qRT-PCR experiments at these temperatures, while a three- to fourfold downregulation was observed at severe heat shock. However, it should be noted that the transcription of clpC and clpP already occurs under nonstressed conditions, with clpP ranking as the 63rd most expressed gene compared to other genes represented on the microarray (see Table S3 in the supplemental material). The clear induction of DNA damage-related genes occurs when cells are exposed to 47 or 50°C, as well as following ethanol stress (Table 3). Under all stresses tested, except for ethanol stress, the small Hsp encoded by gene hsp20 was strongly upregulated; however, no binding motifs were observed in the promoter region, and no binding was observed for the regulators used in this study.
Putative HspR-based control of SOS response.
Following severe heat stress, the transcription of genes containing an SOS box was upregulated, showing that the SOS response is induced by heat stress, a feat also observed in other bacteria (14). Because the upregulation of this regulon always coincided with the upregulation of the HspR regulon and the proximity of the recA and recX genes to the strongest upregulated HspR regulon member, clgR, we decided to investigate whether HspR is able to activate the SOS response by inducing the expression of RecA via the increased transcription of the putative clgR-Bbr_1182-recA-recX operon. qRT-PCR experiments were performed on the entire intergenic region between Bbr_1182 and recA, and a clear induction was shown for this fragment, showing that clgR-Bbr_1182-recA-recX probably exists as a single transcriptional unit in addition to the recA-recX transcriptional unit. The level of upregulation (n-fold) and CT of this fragment were similar to those of clgR and Bbr_1182 (Table 3).
The groEL gene is transcribed from its own promoter.
The large difference between the expression of groEL and cspA (see Table S3 in the supplemental material) suggests that groEL is transcribed from its own promoter, in contrast to what was suggested previously (47). The intergenic region between cspA and groEL was cloned into promoter probe vector pNZ272 to generate plasmid pNZ272-PgroEL and to drive the expression of the promoterless β-glucuronidase-encoding gene gusA, which is present on this plasmid. High levels of β-glucuronidase were observed (600 Miller units) at 37°C, which clearly demonstrates that this region contains an active promoter. However, no further increase of GusA activity was measured at increased temperatures (results not shown). Subsequent experiments with a constitutive promoter (PrbsA1) showed that GusA activity is severely impaired when bifidobacterial cells are exposed to the heat shock conditions employed here (results not shown). Therefore, qRT-PCR experiments were performed to monitor the transcription of the gusA gene on pNZ272-PgroEL, which showed a clear expression and heat shock-dependent transcriptional induction, proving that the intergenic region upstream of groEL indeed contains a strong promoter that is bound and presumed to be regulated by HrcA (Table 3).
DISCUSSION
In this paper, we investigated the moderate and severe heat shock stimulons of B. breve as well as the response to osmotic shock and solvent stress. Moderate heat stress has only a limited effect on the transcriptome and growth rate (at 42 and 44°C only 5 and 17 genes, respectively, are induced), while severe heat shock, ethanol stress, and osmotic stress were shown to have a profound effect on the transcriptome while also severely affecting growth (267 and 347 genes upregulated, respectively), consistently with the finding that the maximum growth temperature of B. breve UCC2003 is 43°C (48).
Because of the apparent pleiotropic effects of the tested conditions, the up- and downregulated genes were grouped in functional categories, which then were analyzed for significant enrichment. The severely impaired growth under the applied severe heat shock and ethanol stress conditions was reflected in the large negative regulatory effect on genes belonging to the carbohydrate transport and metabolism, energy production and conversion, and nucleotide transport and metabolism functional categories, whereas the upregulation of a large number of genes also occurred (Table 2). Although only a fraction of these genes are known to be involved in response to protein misfolding or DNA damage conditions (Table 3), genes presumed to encode metabolic functions also are upregulated. It is possible that these are aspecific effects caused by the denaturation of repressor proteins, resulting in the derepression of a large variety of operons. Combined with the overexpression of the main sigma factor (hrdB) (Table 3) under heat shock, this will result in a higher transcription level of genes and operons that are under negative control. Nevertheless, and as expected, genes belonging to the protein fate functional category clearly were the most strongly upregulated under the stress conditions applied.
Previously it had been shown that the induction of heat shock-responsive genes in B. breve UCC2003 appears to be subject to a specific induction pattern (see reference 45 and references therein): (i) the analyzed HspR-regulated gene clpB was induced 15- to 20-fold under severe heat shock, while only a ∼2-fold induction was observed for moderate heat shock (49); (ii) the dnaK-grpE-dnaJ1-hspR operon was shown previously to exhibit an approximately twofold induction in transcription following exposure to moderate heat stress, while severe heat shock was not tested (51). Our microarray analysis indeed confirmed the strong induction of candidate HspR-regulated transcriptional units (represented by dnaK, grpE, dnaJ1, hspR, clgR, Bbr_1182, recA, recX, and clpB) at severe heat shock. However, overexpression already is observed at lower temperatures and also with ethanol stress and osmotic stress (Table 3). Although previous data suggested that clgR is not under the transcriptional control of HspR (45), our microarray results, qRT-PCR, in silico analysis, and EMSA experiments clearly show that clgR is the most highly induced candidate HspR-regulated gene.
Although the presence of a so-called HAIR motif was detected in several promoters (Table 3), the binding of H6-HspR was shown only for the promoters of the dnaK operon, the clgR operon, and the clpB gene. No binding was observed for the promoters of nfo and hrdB. The closer inspection of the functional binding sites revealed the presence of a larger inverted repeat structure around the HAIR motif (Table 3). The mutation of either the 3′ or 5′ extension of the HAIR motif resulted in reduced binding at the used H6-HspR concentration. Based on these results and on alignments of the binding sites, we suggest that the bifidobacterial HAIR motif has the consensus sequence AAAsTTGAGysw-N[5]-CTCAAsTTTT, which is 8 bp longer than the HAIR motif TTGAGy-N[7]-ACTCAA from other bacteria (6).
Previously, it had been shown that the induction of ClgR-regulated genes (encoding the ClpC and ClpP1P2 proteases) occurred only when a moderate heat shock was applied (48, 50). ClpC and ClpP1P2 are required for the proteolysis of proteins when they are irreversibly denatured or damaged, for instance, when protein aggregates are formed; therefore, the induction of these proteins under moderate heat shock conditions was somewhat surprising, as under the conditions used in this work moderate heat shock caused only a relatively minor growth defect. In the current study, the transcriptional induction of clpC and clpP1P2 was detected only when the cells had been subjected to solvent stress (Table 3), whereas no induction was observed for moderate heat stress in the microarray experiments and only a very moderate induction in the qRT-PCR experiments, while a three- to fourfold downregulation was observed at severe heat shock. The differences observed between this study and previous studies (48, 50) may be caused by the difference in culturing methods. In this study, B. breve was grown under anaerobic conditions in a Modular Atmosphere Controlled System (Davidson & Hardy Ltd.), while in the previous studies B. breve UCC2003 was grown in anaerobic jars. Very recently it was shown by Maisonneuve et al. (24) that in E. coli, the amount of protein aggregates is dependent on how many reactive oxygen species the cell is exposed to. The B. breve cells were more exposed to oxygen, and therefore to more reactive oxygen species, in the anaerobic jars, and this might have increased the levels of protein aggregates, thereby facilitating the induction of ClpC and ClpP1P2 expression. In contrast, the expression of clpC and clpP already occurs under nonstressed conditions. Perhaps the growth conditions used in this study already cause the high transcription of clpC and clpP, thereby masking induction effects by ClgR. Further comparative studies will need to be performed to resolve these differences.
The ClgR-binding motif was detected in the promoter regions of the clpC, clpP1, and hrcA genes (Table 3). This motif was present at positions −49 or at a 10.5 bp decrease (or one DNA helical turn) at −60/61 relative to the transcriptional start. It is tempting to speculate that the activating effect of ClgR is a helix face-dependent phenomenon, either by interacting with RNA polymerase or by bending the DNA, thereby allowing the alpha subunit of RNA polymerase to interact with UP sequences, highly rich in AT, to enhance transcription. The bending of DNA has been suggested to play a role in the activity of C. glutamicum ClgR by Russo et al. (32).
In the absence of a cofactor, the full-length his-tagged form of ClgR, H6-ClgR, is unable to form complexes with its target sequences, the promoters of clpP1 (not shown) and clpC (Fig. 2N), thus confirming previously obtained results (48, 50). We suspect that the N-terminal extension, which is absent in ClgR homologs encoded by other high-GC gram-positive bacteria (48), interferes with the ability of ClgR to reach its active state in the absence of chaperones such as GroEL, which has been implicated as (one of) the ClgR cofactor(s) (48).
Alignments of the ClgR motifs of clpC, clpP, and hrcA, combined with the EMSA experiments described in this report, suggest that the ClgR binding site is TNCGCTNNNGGCGNAA instead of CGCT-N[4]-GCCNA, larger than the motif found in Streptomyces lividans, GTTCGC-N[5]-GCG (4), but with similarities to the motif WNNWCGCYNANRGCGWWS proposed for Corynebacterium species (12).
One of the strongest induced genes, even under moderate stresses, is the small Hsp encoding gene hsp20 (Table 3). The analysis of its promoter region shows that binding sites for the above-described regulators are not present. Regulation might occur via an alternative sigma factor, as suggested by Ventura et al. (46). In addition, the in silico analysis of the upstream region of this gene using WebSIDD (5) does reveal the presence of a so-called stress-induced DNA duplex destabilization (SIDD) site at position −71 or −98 relative to the transcriptional start sites described by Ventura et al. (46). SIDD sites in the promoter region of genes are assumed to be involved in the increased expression of these genes in response to environmental changes that are known to transiently alter superhelicity, such as the heat stress or osmotic stress employed in this study (see references 20 and 53 and references therein).
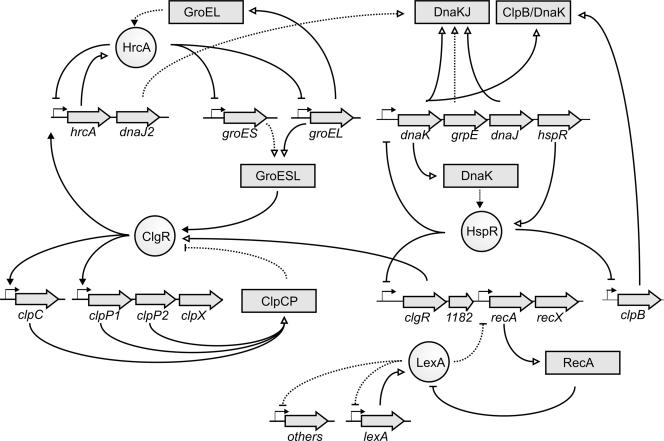
Reconstruction of a model for the bifidobacterial stress gene regulatory network.
A model for the stress regulatory network of B. breve UCC2003 can be constructed using the current knowledge obtained from the research described in this work and other data available in the literature. Although simplified, this model shows the complexity of stress gene regulation in B. breve UCC2003 (Fig. 3). Connections between the various regulons occur at different levels, which is indicative of complex interactions. The high level of the conservation of the regulons and binding sites suggest that this model can be used for all characterized Bifidobacterium species.
FIG. 3.
Schematic representation of the stress gene regulatory network of B. breve UCC 2003. Dotted lines indicate the predicted interaction, and closed lines indicate a proven interaction. A dash at the end of a line indicates repression, while a triangle at the end of a line indicates activation.
The regulation of the HrcA regulon depends on the activity of the HrcA regulator, where binding very likely is dependent on the availability of unsequestered GroEL, similarly to what has been described in the literature for other HrcA proteins (27). Misfolded protein would sequester GroELS and relieve the repression of the HrcA regulon. DnaJ2, a member of the HrcA regulon, possibly could interact with DnaK of the HspR regulon, forming a DnaKJ2 complex and thereby relieving the binding of HspR to its target promoters, since its activity likely is dependent on unsequestered DnaK (6, 7). Additionally, misfolded protein also would sequester DnaK by the formation of the DnaKJ complex. In turn, the derepression of the clgR-Bbr_1182-recA-recX operon would occur, sensitizing the SOS response through the overproduction of RecA and, if certain conditions are met, the activation of the ClgR regulon by the overproduced ClgR, represented by clpC, clpP1P2, and hrcA, thereby making a full circle to HrcA. In addition, the binding activity of ClgR appears to be dependent on GroEL and/or GroES (48), and it is possible that GroEL and/or GroES needs to be present in large quantities in an unsequestered form, perhaps when GroEL and/or GroES no longer is able to refold proteins when protein aggregates are formed under severe stress conditions. The induction of ClpC and ClpP would ensure the complete degradation of these large protein aggregates or facilitate entrance for the other chaperones by breaking up the large complexes into smaller fragments.
Supplementary Material
Acknowledgments
Aldert Zomer acknowledges financial support through an EMBARK fellowship from the Irish Research Council for Science Engineering and Technology. Douwe van Sinderen was supported by the Science Foundation Ireland Alimentary Pharmabiotic Centre located at University College Cork. Marco Ventura was financially supported by the Italian Award for Outstanding Young Researcher scheme Incentivazione Alla Mobilita di Studiosi Stranieri e Italiani Residenti All'estero and by a Marie Curie reintegration grant (MERG-CT-2005-03080). Matilde Fernandez was supported by a postdoctoral fellowship from the Spanish Ministry of Education and Culture (MEC), Madrid, Spain.
Footnotes
Published ahead of print on 4 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andersen, M. T., L. Brondsted, B. M. Pearson, F. Mulholland, M. Parker, C. Pin, J. M. Wells, and H. Ingmer. 2005. Diverse roles for HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151:905-915. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1995. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:21-29. [PubMed] [Google Scholar]
- 3.Bellier, A., M. Gominet, and P. Mazodier. 2006. Post-translational control of the Streptomyces lividans ClgR regulon by ClpP. Microbiology 152:1021-1027. [DOI] [PubMed] [Google Scholar]
- 4.Bellier, A., and P. Mazodier. 2004. ClgR, a novel regulator of clp and lon expression in Streptomyces. J. Bacteriol. 186:3238-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, C., and C. J. Benham. 2004. WebSIDD: server for predicting stress-induced duplex destabilized (SIDD) sites in superhelical DNA. Bioinformatics 20:1477-1479. [DOI] [PubMed] [Google Scholar]
- 6.Bucca, G., A. M. Brassington, G. Hotchkiss, V. Mersinias, and C. P. Smith. 2003. Negative feedback regulation of dnaK, clpB, and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol. Microbiol. 50:153-166. [DOI] [PubMed] [Google Scholar]
- 7.Bucca, G., A. M. Brassington, H. J. Schonfeld, and C. P. Smith. 2000. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol. Microbiol. 38:1093-1103. [DOI] [PubMed] [Google Scholar]
- 8.Bucca, G., Z. Hindle, and C. P. Smith. 1997. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J. Bacteriol. 179:5999-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dosztányi, Z., V. Csizmok, P. Tompa, and I. Simon. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433-3434. [DOI] [PubMed] [Google Scholar]
- 11.Eddy, S. R. 1996. Hidden Markov models. Curr. Opin. Struct. Biol. 6:361-365. [DOI] [PubMed] [Google Scholar]
- 12.Engels, S., C. Ludwig, J. E. Schweitzer, C. Mack, M. Bott, and S. Schaffer. 2005. The transcriptional activator ClgR controls transcription of genes involved in proteolysis and DNA repair in Corynebacterium glutamicum. Mol. Microbiol. 57:576-591. [DOI] [PubMed] [Google Scholar]
- 13.Engels, S., J. E. Schweitzer, C. Ludwig, M. Bott, and S. Schaffer. 2004. clpC and clpP1P2 gene expression in Corynebacterium glutamicum is controlled by a regulatory network involving the transcriptional regulators ClgR and HspR as well as the ECF sigma factor sigmaH. Mol. Microbiol. 52:285-302. [DOI] [PubMed] [Google Scholar]
- 14.Erill, I., S. Campoy, and J. Barbe. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol. Rev. 31:637-656. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, R. A. 1970. Statistical methods for research workers. Oliver and Boyd, Edinburgh, Scotland.
- 16.García de la Nava, G. J., S. A. F. T. van Hijum, and O. Trelles. 2003. PreP: gene expression data pre-processing. Bioinformatics 19:2328-2329. [DOI] [PubMed] [Google Scholar]
- 17.Grandvalet, C., V. de Crecy-Lagard, and P. Mazodier. 1999. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol. Microbiol. 31:521-532. [DOI] [PubMed] [Google Scholar]
- 18.Grandvalet, C., G. Rapoport, and P. Mazodier. 1998. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J. Bacteriol. 180:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 20.Hatfield, G. W., and C. J. Benham. 2002. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 36:175-203. [DOI] [PubMed] [Google Scholar]
- 21.Krüger, E., and M. Hecker. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 180:6681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leahy, S. C., D. G. Higgins, G. F. Fitzgerald, and D. van Sinderen. 2005. Getting better with bifidobacteria. J. Appl. Microbiol. 98:1303-1315. [DOI] [PubMed] [Google Scholar]
- 23.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 24.Maisonneuve, E., L. Fraysse, D. Moinier, and S. Dukan. 2008. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J. Bacteriol. 190:887-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazé, A., M. O'Connell-Motherway, G. F. Fitzgerald, J. Deutscher, and D. van Sinderen. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta, C. R., N. R. Patel, and A. A. Tsiatis. 1984. Exact significance testing to establish treatment equivalence with ordered categorical data. Biometrics 40:819-825. [PubMed] [Google Scholar]
- 27.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penders, J., C. Vink, C. Driessen, N. London, C. Thijs, and E. E. Stobberingh. 2005. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol. Lett. 243:141-147. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli beta-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roncarati, D., G. Spohn, N. Tango, A. Danielli, I. Delany, and V. Scarlato. 2007. Expression, purification and characterization of the membrane-associated HrcA repressor protein of Helicobacter pylori. Protein Expr. Purif. 51:267-275. [DOI] [PubMed] [Google Scholar]
- 32.Russo, S., J. E. Schweitzer, T. Polen, M. Bott, and E. Pohl. 2008. Crystal structure of caseinolytic protease gene regulator, a transcriptional activator in actinomycetes. J. Biol. Chem. 284:5208-5216. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Schmid, A. K., H. A. Howell, J. R. Battista, S. N. Peterson, and M. E. Lidstrom. 2005. HspR is a global negative regulator of heat shock gene expression in Deinococcus radiodurans. Mol. Microbiol. 55:1579-1590. [DOI] [PubMed] [Google Scholar]
- 35.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Servant, P., C. Grandvalet, and P. Mazodier. 2000. The RheA repressor is the thermosensor of the HSP18 heat shock response in Streptomyces albus. Proc. Natl. Acad. Sci. USA 97:3538-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spohn, G., and V. Scarlato. 1999. The autoregulatory HspR repressor protein governs chaperone gene transcription in Helicobacter pylori. Mol. Microbiol. 34:663-674. [DOI] [PubMed] [Google Scholar]
- 38.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thijs, G., Y. Moreau, F. De Smet, J. Mathys, M. Lescot, S. Rombauts, P. Rouze, B. De Moor, and K. Marchal. 2002. INCLUSive: integrated clustering, upstream sequence retrieval and motif sampling. Bioinformatics 18:331-332. [DOI] [PubMed] [Google Scholar]
- 40.van Hijum, S. A. F. T., J. A. De, R. J. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 42.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie van Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 43.Ventura, M., C. Canchaya, V. Bernini, C. A. Del, F. Dellaglio, E. Neviani, G. F. Fitzgerald, and D. van Sinderen. 2005. Genetic characterization of the Bifidobacterium breve UCC 2003 hrcA locus. Appl. Environ. Microbiol. 71:8998-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ventura, M., C. Canchaya, Z. Zhang, V. Bernini, G. F. Fitzgerald, and D. van Sinderen. 2006. How high G+C gram-positive bacteria and in particular bifidobacteria cope with heat stress: protein players and regulators. FEMS Microbiol. Rev. 30:734-759. [DOI] [PubMed] [Google Scholar]
- 46.Ventura, M., C. Canchaya, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2007. Molecular characterization of hsp20, encoding a small heat shock protein of bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:4695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ventura, M., C. Canchaya, R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 70:6197-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ventura, M., G. F. Fitzgerald, and D. van Sinderen. 2005. Genetic and transcriptional organization of the clpC locus in Bifidobacterium breve UCC 2003. Appl. Environ. Microbiol. 71:6282-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ventura, M., J. G. Kenny, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2005. The clpB gene of Bifidobacterium breve UCC 2003: transcriptional analysis and first insights into stress induction. Microbiology 151:2861-2872. [DOI] [PubMed] [Google Scholar]
- 50.Ventura, M., Z. Zhang, M. Cronin, C. Canchaya, J. G. Kenny, G. F. Fitzgerald, and D. van Sinderen. 2005. The ClgR protein regulates transcription of the clpP operon in Bifidobacterium breve UCC 2003. J. Bacteriol. 187:8411-8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ventura, M., R. Zink, G. F. Fitzgerald, and D. van Sinderen. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viala, J., and P. Mazodier. 2002. ClpP-dependent degradation of PopR allows tightly regulated expression of the clpP3 clpP4 operon in Streptomyces lividans. Mol. Microbiol. 44:633-643. [DOI] [PubMed] [Google Scholar]
- 53.Wang, H., and C. J. Benham. 2008. Superhelical destabilization in regulatory regions of stress response genes. PLoS Comput. Biol. 4:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.