Abstract
The protein Clp from Xanthomonas axonopodis pv. citri regulates pathogenesis and is a member of the CRP (cyclic AMP receptor protein) superfamily. We show that unlike the DNA-binding activity of other members of this family, the DNA-binding activity of Clp is allosterically inhibited by its effector and that cyclic di-GMP serves as that effector at physiological concentrations.
Xanthomonas is a genus of bacteria whose members cause disease in a wide variety of commercially important plants throughout the world. The transcription factor Clp (for CRP [cyclic AMP receptor protein]-like protein) regulates the expression of approximately 300 genes involved in pathogenesis in xanthomonads (5). Clp is a homologue (45% sequence identity) of the model transcription factor CRP of Escherichia coli. The six amino acids in CRP that contact cAMP (10) are largely conserved in Xanthomonas axonopodis pv. citri Clp, which suggested that Clp activity might be regulated by some nucleotide.
The paradigm for the effector response in the CRP family is activation of DNA binding upon effector binding. However, previous studies using X. campestris pv. campestris Clp have demonstrated that the protein is competent to bind target DNA with high affinity in the absence of any effector (8). This suggests that effector binding by Clp might cause inhibition, which would represent the first reported case of negative allosteric regulation of a CRP superfamily member. Neither adenylate cyclase nor cAMP has been found experimentally in X. campestris pv. campestris (2), and BLAST searches for cya homologues in the sequenced Xanthomonas genomes have also been negative. We therefore hypothesized that Clp is allosterically inhibited by a nucleotide other than cAMP.
The signaling network controlling virulence in the xanthomonads is largely encoded in the rpf cluster (for regulation of pathogenicity factors), including RpfG, a phosphodiesterase that degrades the signaling molecule cyclic di-GMP (c-di-GMP) (9). In X. campestris pv. campestris, deletion of either rpfG or clp significantly decreases expression of virulence-associated functions (5). The deletion of ravR, which encodes another c-di-GMP-specific phosphodiesterase in X. campestris, has effects very similar to those of deletion of rpfG, and an rpfG ravR mutant is even more severely affected (4). Collectively, these results suggest that both c-di-GMP and Clp have roles in virulence regulation and that these roles are linked in some way. Based on physiology, we hypothesized that c-di-GMP was the most obvious candidate as a Clp inhibitor.
In order to test our hypothesis in vitro, Clp (Xac0483) was PCR amplified from X. axonopodis pv. citri 306 genomic DNA using Pfu Turbo polymerase (Stratagene) with the addition of a C-terminal six-His tag and cloned into the SacI and SmaI sites of pEXT20. His-tagged X. axonopodis pv. citri Clp protein was overexpressed and purified as described previously for CRP (12). Protein purity was >90%, based on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). In vitro DNA-binding assays were carried out using a fluorescence polarization method with a Beacon 2000 fluorescence polarization detector (Invitrogen Corp., Carlsbad, CA). The method measures rotational speed of the DNA probe, which decreases upon binding to a transcription factor. The consensus Clp target DNA sequence appears to be the same as the consensus CRP target DNA sequence (3), so for our DNA binding assays we used a 26-bp DNA probe containing a Texas Red-labeled CCpmelR sequence (a synthetic consensus CRP binding site), as described in our report on work with CRP (12), at a concentration of 10 nM. Specificity of DNA binding is ensured by the inclusion of 6.4 μM salmon sperm DNA.
As expected from the previous studies, we found that in the absence of any effector, purified His-tagged X. axonopodis pv. citri Clp bound target DNA with an affinity of approximately 60 nM, which is similar to the DNA-binding affinity of CRP for this binding site in the presence of cAMP (Fig. 1). Furthermore, we found that c-di-GMP inhibits X. axonopodis pv. citri Clp DNA binding with an apparent affinity for Clp of approximately 1 μM (Fig. 2). This is within the measured range of physiological c-di-GMP levels in proteobacterial species (7, 11) and is also similar to the affinity for c-di-GMP in other known c-di-GMP sensors (6). This result provides a critical missing link in the virulence signaling pathway of xanthomonads by establishing a direct interaction between c-di-GMP and Clp. Other nucleotides (cAMP, cGMP, GMP, ATP, ADP, AMP, and pGpG) had no effect on DNA affinity of X. axonopodis pv. citri Clp at concentrations up to 1 mM (data not shown). Cyclic di-GMP and pGpG were obtained from BioLog Life Science Institute (Germany). Other nucleotides were obtained from Sigma-Aldrich. We also found that the DNA affinities of E. coli CRP and Pseudomonas aeruginosa Vfr are unaffected by up to 40 μM c-di-GMP in both the presence and absence of cAMP (data not shown). Finally, as a first step in analyzing the binding site for c-di-GMP, we asked if cAMP could compete for binding, since most of the cAMP-contacting residues are conserved in Clp. As shown in Fig. 2, addition of a very high level of cAMP to a Clp sample that had been inhibited by c-di-GMP reversed that inhibition. Thus, while cAMP is not necessary in vitro, or indeed relevant physiologically, for activation of Clp, it can reverse inhibition by c-di-GMP, presumably by stabilizing the active form of Clp. While suggestive, this effect does not prove that the two molecules bind the same region of Clp, and identification of the exact c-di-GMP binding site will require structural analysis.
FIG. 1.
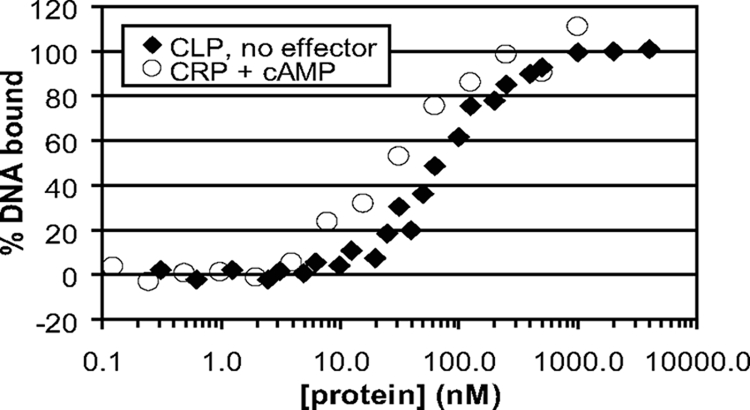
Extent of DNA binding by CRP in the presence of 1 mM cAMP compared to that of C-terminally His-tagged X. axonopodis Clp in the absence of any effector, as determined by fluorescence anisotropy using 10 nM CCpmelR DNA probe. Data points for Clp are compiled from two independent experiments.
FIG. 2.
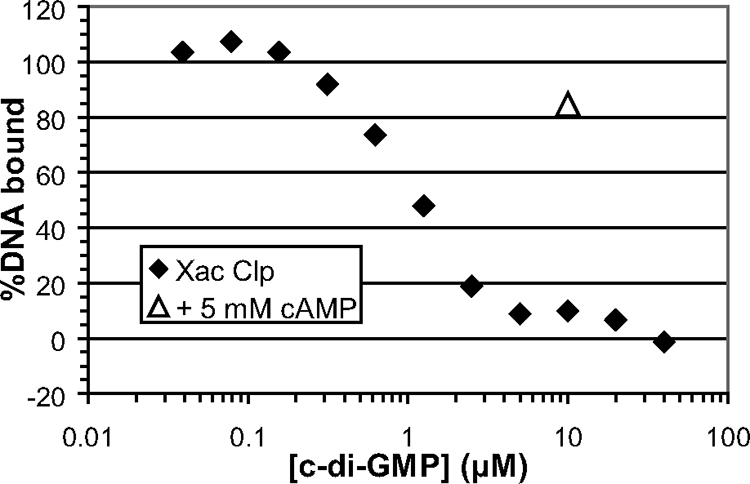
c-di-GMP inhibition of DNA binding by C-terminally His-tagged X. axonopodis Clp as measured by fluorescence anisotropy, using 200 nM protein. A single point shows the restoration of DNA binding upon addition of 5 mM cAMP to the 10 μM c-di-GMP-inhibited Clp sample.
In conclusion, Clp represents a novel type of c-di-GMP binding motif with no similarity to known c-di-GMP binding proteins from other organisms (i.e., PilZ proteins, diguanylate cyclases, PelD, FleQ, and LapD) but with strong similarity to the cNMP-binding protein family. Furthermore, c-di-GMP inhibition of Clp provides a missing link in the virulence signaling pathways of xanthomonads: Because of the similarity of the rpf and clp genes in X. campestris and X. axonopodis (1), we expect that the in vivo results with X. campestris described above apply to X. axonopodis and that our in vitro Clp results with X. axonopodis will be applicable to X. campestris. While allosteric inhibition is not unprecedented among transcription factors in general, Clp represents the first characterized CRP superfamily member that is allosterically inhibited upon effector binding. Clp also appears to be the closest characterized homologue of CRP that responds to a physiological effector other than cAMP. For these reasons, comparison of Clp and the well-characterized CRP provides a fascinating case study in the evolution of effector response and specificity in a global regulatory protein.
Acknowledgments
This work was supported by the College of Agricultural and Life Sciences at UW-Madison and by NIH GM53228 (to G.P.R).
We thank Robert Kerby, Caroline Harwood, Patricia Kiley, Katrina Forest, and Richard Gourse for critical reading of the manuscript, Hwan Youn for helpful discussions, and Chuck Farah for a gift of X. axonopodis pv. citri genomic DNA.
Footnotes
Published ahead of print on 24 July 2009.
REFERENCES
- 1.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 2.de Crecy-Lagard, V., P. Glaser, P. Lejeune, O. Sismeiro, C. E. Barber, M. J. Daniels, and A. Danchin. 1990. A Xanthomonas campestris pv. campestris protein similar to catabolite activation factor is involved in regulation of phytopathogenicity. J. Bacteriol. 172:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong, Q., and R. H. Ebright. 1992. DNA binding specificity and sequence of Xanthomonas campestris catabolite gene activator protein-like protein. J. Bacteriol. 174:5457-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He, Y. W., C. Boon, L. Zhou, and L. H. Zhang. 2009. Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol. Microbiol. 71:1464-1476. [DOI] [PubMed] [Google Scholar]
- 5.He, Y. W., A. Y. Ng, M. Xu, K. Lin, L. H. Wang, Y. H. Dong, and L. H. Zhang. 2007. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64:281-292. [DOI] [PubMed] [Google Scholar]
- 6.Hengge, R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263-273. [DOI] [PubMed] [Google Scholar]
- 7.Hickman, J. W., and C. S. Harwood. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao, Y. M., H. Y. Liao, M. C. Lee, T. C. Yang, and Y. H. Tseng. 2005. Clp upregulates transcription of engA gene encoding a virulence factor in Xanthomonas campestris by direct binding to the upstream tandem Clp sites. FEBS Lett. 579:3525-3533. [DOI] [PubMed] [Google Scholar]
- 9.Ryan, R. P., Y. Fouhy, J. F. Lucey, L. C. Crossman, S. Spiro, Y. W. He, L. H. Zhang, S. Heeb, M. Camara, P. Williams, and J. M. Dow. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 103:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Weber, I. T., and T. A. Steitz. 1987. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J. Mol. Biol. 198:311-326. [DOI] [PubMed] [Google Scholar]
- 11.Weinhouse, H., S. Sapir, D. Amikam, Y. Shilo, G. Volman, P. Ohana, and M. Benziman. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416:207-211. [DOI] [PubMed] [Google Scholar]
- 12.Youn, H., R. L. Kerby, M. Conrad, and G. P. Roberts. 2006. Study of highly constitutively active mutants suggests how cAMP activates cAMP receptor protein. J. Biol. Chem. 281:1119-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]


