Abstract
The major flagellin of Campylobacter jejuni strain 81-176, FlaA, has been shown to be glycosylated at 19 serine or threonine sites, and this glycosylation is required for flagellar filament formation. Some enzymatic components of the glycosylation machinery of C. jejuni 81-176 are localized to the poles of the cell in an FlhF-independent manner. Flagellin glycosylation could be detected in flagellar mutants at multiple levels of the regulatory hierarchy, indicating that glycosylation occurs independently of the flagellar regulon. Mutants were constructed in which each of the 19 serine or threonines that are glycosylated in FlaA was converted to an alanine. Eleven of the 19 mutants displayed no observable phenotype, but the remaining 8 mutants had two distinct phenotypes. Five mutants (mutations S417A, S436A, S440A, S457A, and T481A) were fully motile but defective in autoagglutination (AAG). Three other mutants (mutations S425A, S454A, and S460A) were reduced in motility and synthesized truncated flagellar filaments. The data implicate certain glycans in mediating filament-filament interactions resulting in AAG and other glycans appear to be critical for structural subunit-subunit interactions within the filament.
Flagellins from many polarly flagellated bacteria are glycosylated (reviewed in reference 22). The best-characterized examples are the flagellins from Campylobacter spp. that are decorated with as many as 19 O-linked glycans that can contribute ∼10% to the weight of flagellin (38). The genes encoding the enzymes for biosynthesis of the glycans found on Campylobacter flagellins and the respective glycosyltransferases are located adjacent to the flagellin structural genes in one of the more hypervariable regions of the Campylobacter genome (3, 16, 28, 37). Most strains appear to carry the genes for synthesis of two distinct nine-carbon sugars that decorate flagellin: pseudaminic acid (PseAc) and an acetamidino form of legionaminic acid (LegAm) (23). In contrast, Campylobacter jejuni strain 81-176 contains only the pathway for synthesis of PseAc (9) and derivatives of PseAc that include an acetylated form (PseAcOAc), an acetamidino form (PseAm), and a form of PseAm with a glutamic acid moiety attached (PseAmOGln) (25, 34, 38). The flagellins of C. jejuni strain NCTC 11168 have recently been shown to be glycosylated with PseAc and LegAm, as well as two novel derivatives of PseAc, a di-O-methylglyceric acid and a related acetamidino form (24). Thus, although all of the flagellar glycans appear to be based on either PseAc and/or LegAm, there are variations among strains that contribute to serospecificity and reflect the heterogeneity of the flagellin glycosylation loci (23, 24).
The function of the glycosyl modifications to flagellar structure and to the biology of campylobacters is not fully understood. Although most polarly flagellated bacteria appear to glycosylate flagellin, mutation of the genes involved in glycosylation does not generally result in loss of motility (22). However, flagella from C. jejuni, Campylobacter coli, and Helicobacter pylori, all members of the epsilon division of Proteobacteria, are unable to assemble a filament in the absence of a functional glycosylation system (7, 33). Also, changes in the glycans on campylobacter flagellins have been shown to affect autoagglutination (AAG) and microcolony formation on intestinal epithelial cells in vitro (5, 9). Thus, a mutant of C. jejuni 81-176 that was unable to synthesize PseAm assembled a flagellar filament, but the sites on the flagellin subunits that were normally glycosylated with PseAm were instead glycosylated with PseAc. This mutant was reduced in AAG, adherence, and invasion of INT407 cells and was also attenuated in a ferret diarrheal disease model (9). C. coli VC167 has both PseAc and LegAm pathways. Mutants that were defective in either pathway could still assemble flagellar filaments composed of subunits that were modified with the alternate sugar, but these mutants showed defects in AAG (7). A VC167 double mutant, defective in both PseAc and LegAm synthesis, was nonflagellated (7). Collectively, these data suggest that some glycosylation is required for either secretion of flagellin or for interactions between subunits within the filament.
Flagellar biogenesis in C. jejuni is a complex process that is highly controlled by the alternate sigma factors σ28 and σ54, a two-component regulatory system composed of the sensor kinase FlgS and the σ54-response regulator FlgR, and the flagellar export apparatus (15, 39). Both flgR and flgS genes undergo slip strand mismatch repair in C. jejuni strain 81-176, resulting in an on/off-phase variation of flagellar expression (13, 14). The major flagellin gene, flaA, and some other late flagellar genes are regulated by σ28; the genes encoding the minor flagellin, flaB, and the hook and rod structures are regulated by σ54. Here, we examine several aspects of glycosylation to flagellar function in C. jejuni 81-176. We demonstrate that some components of the flagellar glycosylation machinery are localized to the poles of the cell, but independently of the signal recognition particle-like flagellar protein, FlhF, and that flagellin glycosylation occurs independently of the flagellar regulon. We also show that the glycans on some amino acids appear to play a structural role in subunit interactions in the filament, while others affect interactions with adjacent filaments that result in AAG.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni strain 81-176 has been described previously (2). Escherichia coli strains DH5α and XL1-Blue were the hosts for routine cloning experiments. The C. jejuni 81-176 mutants used in this study are shown in Table 1. All mutants except fliP and flhF have been described previously (7, 9). Both fliP and flhF insertions were constructed in an E. coli host using an in vitro Tn5-based transposition system (Epicentre, Madison, WI) with a Campylobacter chloramphenicol resistance (Cmr) cassette as previously described (8-10). The insertion point was mapped by sequence analysis with primers mapping within the Cmr cassette, and selected clones were used to electroporate C. jejuni 81-176 to Cmr. The insertion point in the flhF gene was at bp 705 within the 1,455-bp gene; the insertion into fliP was at bp 308 of the 734-bp open reading frame. C. jejuni strains were grown on Mueller-Hinton (MH) agar supplemented with kanamycin (50 μg/ml) and/or chloramphenicol (15 μg/ml) as needed at 37°C under microaerobic conditions. E. coli strains were grown on Luria agar supplemented with kanamycin (50 μg/ml), chloramphenicol (20 μg/ml), and ampicillin (62.5 μg/ml), as needed.
TABLE 1.
81-176 mutants used in this study
| Gene(s) | NCTC 11168 no. | CJJ81176 no. | Function | Source or reference |
|---|---|---|---|---|
| flgR | 1024 | 1043 | σ54 transcriptional activator | 7 |
| flgS | 0793 | 0814 | Histidine kinase of FlgR | 7 |
| flgE | 0043 | 0025 | Hook protein | 6 |
| fliR | 1179 | 1194 | Export component | 7 |
| fliP | 0820 | 0837 | Export component | This work |
| flaA flaB | 1339/1338 | 1339/1338 | Major and minor flagellins | 4, 7 |
| flhF | 0064 | 0102 | Signal recognition particle; export component | This work |
| flhA | 0882 | 0890 | Export component | 7 |
| flhB | 0335 | 0357 | Export component; substrate specificity switch | 6 |
| pseA | 1316 | 1333 | Synthesis of PseAm | 39 |
| pseB | 1293 | 1310 | Dehydratase that converts UDP-GlcNAc to UDP-2-acetamidino-2,6-dideoxy-β-l-arabino-hexos-4-ulose; first enzyme in PseAc synthesis | 9 |
| pseC | 1294 | 1311 | Converts the product of PseB to UDP-4-amino-4,6-dideoxy-β-l-AltNAc; second enzyme in PseAc synthesis | 9 |
| pseD | 1333 | 1336 | PseAm transferase | 9 |
| pseE | 1337 | 1337 | PseAc transferase | 9 |
GFP-tagged protein fusions.
Gene fusions were done using plasmids based on the pRY107 (Kmr) and pRY111 (Cmr) Campylobacter shuttle plasmids (40). Expression forms of these plasmids, in which a σ28 promoter of flaA was cloned between the XbaI-BamHI sites of these vectors, have been described previously (21). The green fluorescent protein (GFP) gene from pZSGreen (Clontech) was cloned as a BamHI-EcoRI fragment into each plasmid, generating pCPE107/28/GFP (Kmr) and pCPE111/28/GFP (Cmr). The pseC, pseD, and pseE genes of strain 81-176 were PCR amplified with primers that contained BamHI sites, and the resulting amplicons were cloned into the shuttle plasmid pCPE107/28/GFP (Kmr) or pCPE111/28/GFP (Cmr) such that the glycosylation genes were fused to the 5′ end of the gene encoding GFP. The primers used for cloning of pseC were GAAGGGATCCATGATTACTTATTCTCATCAAAATATTG and GAAGGGATCCTCCACAATATCCCTTTTTAACTTTTTC, the primers for pseD were GAAGGGATCCATGAAATTTAATTTAAATCAAAAAGAGC and GAAGGGATCCTTTGTTTGCATTTTTTATCCTTCTTAGG, and the primers for pseE were GAAGGGATCCGATGCAAACAAATGAAATTTTTAAAAAAAATTTAG and GAAGGGATCCGATTAAGCTTCTTTTTTCTAGCTCATCC. The flhF gene was also PCR amplified using the primers GAAGGGATCCATGGGACAACTTATACATACTTTTACCGTTG and GAAGGGATCCTTCATTATTTTTTCCTTTGTTAAACCCTTC. BamHI sites are underlined, and the translational start sites are shown in bold. Each plasmid was transformed into DH5α containing the conjugative plasmid RK212.2 (11) and then mobilized into C. jejuni 81-176 or into the C. jejuni flhF::aph3 strain with selection on the appropriate antibiotics. Localization of GFP-tagged proteins was determined by examination in a Nikon Eclipse E400 fluorescence microscope.
Immunoblotting.
Proteins were separated on 8.25% or 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and immunodetected with rabbit polyclonal antisera to either flagellin (9), FlgR, FlgS, or FspA (29) at the indicated dilutions. Blots were detected with goat anti-rabbit antibodies conjugated to alkaline phosphatase (Caltag, Burlingame, CA) followed by detection with NBT/BCIP (nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate) (Promega, Madison, WI) or with goat anti-rabbit antibodies conjugated to horseradish peroxidase followed by detection with Supersignal West Pico detection kits (Pierce, Rockland, IL). Chemiluminescence was detected using a Kodak Image Station 2000R (Rochester, NY).
Site-directed mutagenesis.
Site-directed mutagenesis was done as described by Galkin et al. (4) using QuikChange kits from Stratagene (La Jolla, CA) as recommended by the supplier. An 81-176 mutant in which most of flaA and flaB were replaced with a cat cassette, PG2112, has been described previously (4). The wild-type σ28-regulated flaA gene and an aph3 gene were inserted into astA on plasmid pCPE2644. This plasmid was used previously as a template for site-directed mutagenesis to isolate straight flagellar mutants for structural studies (4) and was used in this study to mutate each glycosylation site in flaA. Primers are listed in Table 2. Plasmids that had been subjected to mutagenesis were confirmed by DNA sequence analysis of the complete flagellin gene to confirm that only the desired mutation had occurred. DNA sequence analysis was done with dye terminator chemistry on an Applied Biosystems model 3100 DNA sequencer. Each site-directed mutant was electroporated into PG2112 with selection for Cmr and Kmr. Transformants were screened for loss of expression of arylsulfatase by growth on a chromogenic substrate, X-S (Sigma) (41). All mutants were compared to the motile strain PG2645, which was PG2112 electroporated with pCPE2644 expressing the wild-type flaA gene.
TABLE 2.
Primers used for site-directed mutations
| Mutation | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| S206A | GATTTTAAATTTGATAATGTTGTGATTGCAACTTCAGTTGGAACAGGACTTGGAGC | GCTCCAAGTCCTGTTCCAACTGAAGTTGCAATCACAACATTATCAAATTTAAAATC |
| S342A | GATGGTAGAGATATCAATATAGCTGGAACCAATCTTAGTGCTATAGG | CCTATAGCACTAAGATTGGTTCCAGCTATATTGATATCTCTACCATC |
| S347A | GAGATATCAATATAAGTGGAACCAATCTTGCTGCTATAGGTATGGGTACAACAG | CTGTTGTACCCATACCTATAGCAGCAAGATTGGTTCCACTTATATTGATATCTC |
| T393A | CTTATAAAGGTGGTGGAAAATTTGTTTTTGCTCAAAATGTAAGTTCAATTTCTGCATTTATGAGTGC | GCACTCATAAATGCAGAAATTGAACTTACATTTTGCGTAAAAACAAATTTTCCACCACCTTTATAAG |
| S397A | GGAAAATTTGTTTTTACTCAAAATGTAGCTTCAATTTCTGCATTTATGAGTGC | GTGCACTCATAAATGCAGAAATTGAAGCTACATTTTGAGTAAAAACAAATTTTCCC |
| S400A | ACTCAAAATGTAAGTTCAATTGCTGCATTTATGAGTGCACAAGGTTC | GAACCTTGTGCACTCATAAATGCAGCAATTGAACTTACATTTTGAGT |
| S404A | GTAAGTTCAATTTCTGCATTTATGGCTGCACAAGGTTCAGGATTTTCTAG | CTAGAAAATCCTGAACCTTGTGCAGCCATAAATGCAGAAATTGAACTTAC |
| S408A | CAATTTCTGCATTTATGAGTGCACAAGGTGCAGGATTTTCTAGAGGTTCAGGATTTTCTG | CAGAAAATCCTGAACCTCTAGAAAATCCTGCACCTTGTGCACTCATAAATGCAGAAATTG |
| S417A | GGATTTTCTAGAGGTTCAGGATTTGCTGTGGGTAGTGGTAAAAATTTATCTGTTGGATTGAG | CTCAATCCAACAGATAAATTTTTACCACTACCCACAGCAAATCCTGAACCTCTAGAAAATCCTGAACC |
| S425A | CTGTGGGTAGTGGTAAAAATTTAGCTGTTGGATTGAGTCAAGGAATACAAATTATTTCAAGTGCGGCTTC | GAAGCCGCACTTGAAATAATTTGTATTCCTTGACTCAATCCAACCGATAAATTTTTACCACTACCCACAG |
| S429A | GTGGGTAGTGGTAAAAATTTATCTGTTGGATTGGCTCAAGGAATACAAATTATTTCAAGTGCGGCTTCAATG | CATTGAAGCCGCACTTGAAATAATTTGTATTCCTTGAGCCAATCCAACAGATAAATTTTTACCACTACCCAC |
| S436A | TCTGTTGGATTGAGTCAAGGAATACAAATTATTGCAAGTGCGGCTTCAATGAGCAATACTTATGTTGTTTC | GAAACAACATAAGTATTGCTCATTGAAGCCGCACTTGCAATAATTTGTATTCCTTGACTCAATCCAACAGA |
| S440A | GGAATACAAATTATTTTCAAGTGCGGCTGCAATGAGCAATACTTATGTTGTTTC | GAAACAACATAAGTATTGCTCATTGCAGCCGCACTTGAAAATAATTTGTATTCC |
| S448A | GCTTCAATGAGCAATACTTATGTTGTTGCAGCAGGTTCAGGATTTTCTTCTG | CAGAAGAAAATCCTGAACCTGCTGCAACAACATAAGTATTGCTCATTGAAGC |
| S451A | GAGCAATACTTATGTTGTTTCAGCAGGTGCAGGATTTTCTTCTGGCTCAGGAAATTCTCAATTTGC | GCAAATTGAGAATTTCCTGAGCCAGAAGAAAATCCTGCACCTGCTGAAACAACATAAGTATTGCTC |
| S454A | CTTATGTTGTTTCAGCAGGTTCAGGATTTGCTTCTGGCTCAGGAAATTCTCAATTTGCAGCCCTTAAAAC | GTTTTAAGGGCTGCAAATTGAGAATTTCCTGAGCCAGCAGAAAATCCTGAACCTGCTGAAACAACATAAG |
| S457A | GTTGTTTCAGCAGGTTCAGGATTTTCTTCTGGCGCAGGAAATTCTCAATTTGCAGCCCTTAAAACTACTGC | GCAGTAGTTTTAAGGGCTGCAAATTGAGAATTTCCTGCGCCAGAAGAAAATCCTGAACCTGCTGAAACAAC |
| S460A | GGTTCAGGATTTTCTTCTGGCTCAGGAAATGCTCAATTTGCAGCCCTTAAAAC | GTTTTAAGGGCTGCAAATTGAGCATTTCCTGAGCCAGAAGAAAATCCTGAACC |
| T481A | GCTAATACAACTGATGAGACTGCAGGTGTAACCGCTCTTAAAGGTGCAATGGCGGTTATGGATATAGC | GCTAATACAACTGATGAGACTGCAGGTGTAACCGCTCTTAAAGGTGCAATGGCGGTTATGGATATAGC |
Motility testing.
Motility was determined by stabbing the center of a semisolid motility agar plate (MH broth plus 0.4% agar) with 1 μl of bacterial culture at an optical density at 600 nm (OD600) of 1.0. The zones of motility were measured after incubation for 48 h at 37°C under microaerobic conditions. The motility zone measurements were determined between four and seven times for mutants that showed motility defects. Mutants that showed motility defects in soft agar were also examined by phase microscopy.
TEM.
Bacteria were examined by transmission electron microscopy (TEM) following negative staining with uranyl acetate. Images were printed and filament lengths were measured.
AAG.
AAG was done as described previously (9, 26). Basically, bacteria were suspended to an OD600 of 1.00 in phosphate-buffered saline (PBS). Two milliliters of each suspension was incubated at room temperature for 24 h. The top 1 ml was removed from each tube, and the OD600 was determined. A reduction in OD600 indicated that AAG had occurred.
Secretion of flagellin to the supernatant.
Supernatants of selected site-directed mutants were collected and precipitated with trichloroacetic acid as described by Poly et al. (29).
Generation of anti-FlgR and anti-FlgS antibodies.
The flgR and flgS genes were PCR amplified from 81-176 using high-fidelity KOD Hi/Fi DNA polymerase (Novagen). The primers used for flgR were GAAGGGATCCGATGAATTTAGTCATAGTAGAAGATG and TAATGGATCCTTACTTATCCTTTATTTGATATTTTTTAATTTTTTC. The primers used for flgS were GAAGGGATCCGATGAATGAAAGTATTTTAAAAAGTTTAGACTC and TAATGGATCCTCACACCAAAGGTAAGGTAAAATAAAAATTC. These primers introduced BamHI sites at either end of the amplicon (underlined); start and stop codons are shown in bold. The PCR products were ligated with BamHI-digested pET-19b (Novagen) and cloned into E. coli DH5α. Candidate clones were sequenced to confirm the proper orientation in the vector. Appropriate clones were transferred into E. coli BL21(DE3), and proteins were induced with IPTG (isopropyl-β-d-thiogalactopyranoside) as recommended by the supplier (Invitrogen). Recombinant proteins were purified on Ni-nitrilotriacetic acid agarose (Qiagen). Rabbit polyclonal antibodies were generated against recombinant FlgR and FlgS by Harlan Biosciences (Madison, WI).
Statistical analyses.
All statistical analyses were done by t test using Graphpad Prism software.
RESULTS
Localization of the glycosylation machinery in C. jejuni 81-176.
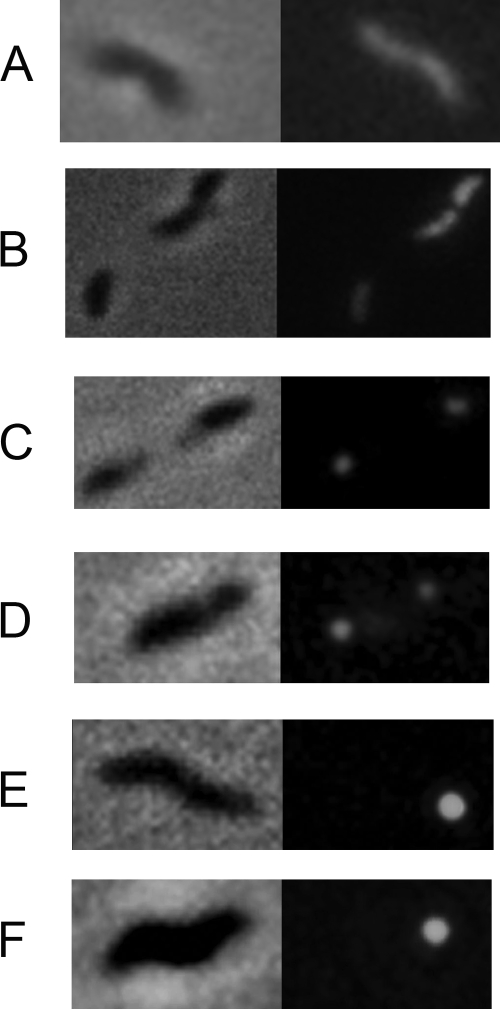
The genes encoding PseC, which is the enzyme involved in the second step of PseAc synthesis, PseE, the putative PseAc transferase, and PseD, the putative PseAm transferase, were fused to GFP on a Campylobacter shuttle vector (21). As a control, the flhF gene, which encodes a signal recognition-like flagellar protein that is itself localized to the poles of some bacteria (27), was also fused to the GFP gene. The resulting plasmids were transferred conjugatively into C. jejuni, and the cells were examined by fluorescence microscopy. C. jejuni 81-176 with the vector expressing GFP alone showed generalized fluorescence throughout the cell (Fig. 1A), as did 81-176 expressing the PseD-GFP fusion (Fig. 1B). In contrast, 81-176 expressing the PseE fusion (Fig. 1C) and the PseC fusion (not shown) displayed polar fluorescence, as did 81-176 expressing the FlhF-GFP fusion (Fig. 1E). The FlhF-GFP fusion localized to the poles in an flhF mutant (Fig. 1F). Interestingly, the FlhF-GFP fusion was able to fully restore motility to the flhF mutant (data not shown). PseE-GFP was also localized to the poles in the flhF mutant, as shown in Fig. 1D, as well as mutants in the export apparatus (fliR fliP) and in the flaA flaB double mutant (results not shown).
FIG. 1.
Localization of PseD, PseE, and PseC fusions to GFP. The pseE, pseD, and pseC genes were cloned into the shuttle plasmid pCPE107/28/GFP, and the flhF gene was cloned into pCPE111/28/GFP as C-terminal GFP fusions. The resulting vectors were transferred conjugatively from E. coli into wild-type C. jejuni 81-176 or the isogenic flhF mutant, as previously described (11). The pattern for PseC-GFP was the same as for PseE-GFP (not shown). Phase images are on the left, and fluorescent images are on the right. A, pCPE107/28/GFP in 81-176; B, PseD-GFP fusion in 81-176; C, PseE-GFP fusion in 81-176; D, PseE-GFP fusion in 81-176 fihF; E, FIhF-GFP fusion in 81-176; F, FIhF-GFP fusion in 81-176 fihF.
Flagellin glycosylation can occur in the absence of a functional flagellar export system.
The predicted molecular mass of 81-176 flagellin is 59.5 kDa, but glycosylated flagellin has an apparent molecular mass of 65.5 kDa on SDS-PAGE gels (7). In the nonmotile pseB mutant of 81-176, flagellin of an apparent molecular mass of 59.5 kDa was observed in whole-cell lysates, and mass spectrometry analysis of this flagellin confirmed the lack of glycosylation (7). Figure 2A and B show that flagellin of an apparent molecular mass of 59.7 kDa also accumulated in a pseC mutant, and when the mutant was complemented in trans (designated as “pseC/c”), the molecular mass of flagellin was restored to that of the wild type. The expression and apparent molecular mass of flagellin in a series of mutants at different levels of the flagellar hierarchy (Table 1) were examined by immunoblotting of whole-cell preparations. Flagellin could be detected in flgR, flgS, and flgE mutants, all of which are nonmotile and do not assemble a filament, that had the same apparent molecular mass as wild-type flagellin (Fig. 2B). Mutants in flhF, flhA, and flhB also accumulated flagellin of the same apparent molecular mass as the wild type (Fig. 2C). Similarly, flagellin from the fliP and fliR early flagellar mutants, which lack the flagellar export machinery, and the fliG mutant, which encodes a component of the flagellar motor/switch (Fig. 2D), also ran at an apparent molecular mass consistent with complete glycosylation.
FIG. 2.

Flagellin glycosylation in flagellar mutants. Whole cells of wild-type 81-176 (WT) and mutants with mutations in various flagellar genes were resuspended in SDS-PAGE solubilization buffer and electrophoresed on 8.25% SDS-PAGE gels. Following transfer to nitrocellulose membranes, flagellin was immunodetected using rabbit polyclonal antibody generated against purified flagellin from 81-176 (9) at a final dilution of 1:50,000 followed by chemiluminescent detection. fla, PG2112 (the 81-176 flaA flaB mutant); fla/c, PG2112 complemented in trans; pseC/c, 81-176 pseC::cat complemented in trans (9). Comparison of flagellins from WT, pseC, and pseC/c to flagellin from: A, PG2112; B, mutants in the middle of the flagellar hierarchy; C and D, mutants early in the flagellar hierarchy.
Effect of site-directed mutations that removed individual glycosylation sites on flagellin on motility and filament biogenesis.
A mutant in 81-176, PG2112, in which most of the flaA and flaB genes were replaced with a cat cassette, has been described previously (4, 6). The wild-type flaA gene and its σ28 promoter were inserted into the astA gene of PG2112 to generate plasmid pCPE2644. When plasmid pCPE2644 was electroporated into PG2112, the resulting strain, PG2645, produced a full-length flagellar filament and was fully motile (4) (Fig. 3). Plasmid pCPE2644 was used as the substrate in a series of site-directed mutagenesis experiments in which each of the 19 serine or threonine sites which have been shown to be glycosylated in FlaA (38) was changed to an alanine. The position of each serine or threonine residue in FlaA is shown in Fig. 4. Each mutated allele was electroporated into PG2112 with selection for Kmr and Cmr and screened on X-S agar to confirm insertion into astA (41). The motility of each mutant was determined using semisolid motility medium, and the flagellar structure was examined by TEM. Most mutants (16/19) were equally motile compared to PG2645 or the wild type and produced a filament that was indistinguishable from that of the wild type or PG2645; representative data for the S404A mutant are shown in Fig. 3. Three mutants (mutations S425A, S454A, and S460A) displayed significant but varied reductions in motility compared to either 81-176 or PG2645 (P < 0.001), although the S425A mutant showed the greatest reduction in motility (Fig. 3B). Quantitative measurement of flagellar filament length in these mutants confirmed that all three produced filaments that were statistically different in length from PG2645 (P < 0.001) (Fig. 3C).
FIG. 3.
Effect of site-directed mutations of specific glycosylation sites on flagella. (A) TEM of representative filaments. Strain PG2112 is the flaA flaB mutant that is nonmotile and does not produce a flagellar filament, and PG2645 is PG2112 complemented with the wild-type flaA gene (4). The S404A mutant is representative of mutants that showed no motility defects. (B) Zones of motility on motility agar. (C) Flagellar filament length of mutants that showed reduced motility compared to PG2645. The numbers of filaments that were measured ranged from 15 for PG2645 to 36 for the S460A mutant. Filaments from all three mutants were significantly shorter than those from PG2645 (P < 0.001; marked by asterisks).
FIG. 4.
Glycosylation sites in FlaA of C. jejuni 81-176. (A) Positions of the glycosylation sites of FlaA within the inferred domains of FlaA, based on the domains of Salmonella enterica serovar Typhimurium flagellin (42). There is a single glycosylation site at S206; the remaining 18 sites lie between S342 and T481. All sites, except T481, are predicted to occur within the D2-D3 domain. (B) Primary amino acid sequence of the FlaA flagellin from 81-176. The positions of the 19 sites of O-linked glycosylation are in bold and are also indicated by a black square below each serine or threonine (38).
Phase variation of FlgR and/or FlgS, the two-component system that regulates the middle, σ54-regulated flagellar genes, has been shown to affect motility in C. jejuni 81-176 (12, 15, 39). To confirm that the observed motility defects were not due to phase variation of this two-component regulatory system, whole cells of the S425A, S454A, and S460A mutants were immunoblotted with anti-FlgR and anti-FlgS antibodies. The results, shown in Fig. 5, confirmed that FlgR and FlgS were expressed in all three mutants. However, it remained possible that other unidentified genes could have undergone phase variation that affected formation of the flagellar apparatus. To confirm that a functional flagellar apparatus was synthesized in the S425A, S454A, and S460A mutants, supernatants from cultures of each mutant were immunoblotted with antibodies to two proteins that are secreted through the filament. These were flagellin and FspA, a σ28-regulated protein that is not part of the flagellar structure but which is secreted through the flagellar filament (29). Figure 5 shows that flagellin was present in the supernatant of the wild type and the S425A, S454A, and S460A mutants, but not in the supernatant of PG2112. Moreover, FspA could be detected in the supernatant of all strains examined. These data confirm that a functional flagellar secretory apparatus was being expressed in these three site-directed mutants and that the loss of glycosylation at these three sites did not prevent secretion of flagellin.
FIG. 5.
Immunoblots. Whole cells or concentrated supernatants of C. jejuni strains were immunodetected with rabbit polyclonal antibody made to recombinant FlgR, FlgS, and FspA (29) or to native flagellin purified from C. jejuni 81-176. Anti-FlgR and anti-FlgS antisera were used at a final dilution of 1:10,000, antiflagellin was used at 1:200,000, and the antibodies were detected colorimetrically. Anti-FspA antibody was used at a final dilution of 1:5,000 and detected by chemiluminescence (29). 425, 454 and 460 refer to the S425A, S454A, and S460A site-directed mutants, respectively. The predicted masses of the proteins are as follows: FlgR, 49.2 kDa; FlgS, 38.4 kDa; FlaA, 65.5 kDa; and FspA, 18 kDa.
AAG of site-directed mutants.
Since both loss of flagella and changes in sugars decorating flagellin have been shown to affect AAG of C. jejuni (5, 9, 26), all 19 mutants were tested for AAG. The results are shown in Fig. 6. The nonmotile mutant, PG2112, failed to autoagglutinate, as expected (27) (P = 0.0001). The S425A and S460A mutants, which produced truncated filaments, also showed significant reductions in AAG (P = 0.0001). However, the S454A mutant showed no AAG defect, despite the fact that it produced a truncated filament. There were five other fully motile mutants that displayed significant differences in AAG compared to PG2645. The S417A, S457A, and T481A mutants showed defects similar to that of PG2112 (P = 0.0001 compared to PG2645); the S436A, S440A, and S451A mutants showed intermediate, but still significant defects (P ≤ 0.0011).
FIG. 6.
AAG of C. jejuni. AAG was determined as described in Materials and Methods. The data represent means and standard deviations from 2 to 10 experiments. The strains are labeled as follows: wt, wild-type 81-176; M, PG2112; C, PG2645; other strains are designated by the mutated flagellin amino acid. Asterisks denote mutants that showed a statistically significant difference from PG2645.
DISCUSSION
Although glycosylation is essential for filament biogenesis in C. jejuni, there have been no studies examining the interactions of the glycosylation system and the flagellar genes. It has been postulated that flagellin glycosylation occurs either in the cytoplasm, in close association with the flagellar machinery, or within the basal body (22). We have demonstrated here that some, but not all, of the enzymatic glycosylation machinery is localized to the poles of the cells, consistent with a possible association with the flagellar basal body/export apparatus. FlhF is a signal recognition particle-like protein that affects flagellar assembly in several polarly flagellated bacteria (19, 27), and flhF mutants of C. jejuni are also nonmotile (14). Here we have confirmed that FlhF is also localized to the poles of C. jejuni, as are two enzymatic components of the glycosylation apparatus, PseE and PseC. However, polar localization of the latter two proteins occurs independently of FlhF by an undetermined mechanism. Spatial localization of some bacterial proteins is critical to many cellular functions and virulence (17, 18, 20, 36). The process of polar localization is not well understood (31), although recent data suggest that some polar localization is due to polar enrichment of cardiolipin content (32). Clearly, additional studies will be required to understand the nature and localization of all of the components of the glycosylation machinery within C. jejuni. Despite the polar localization of some of the enzymatic machinery, glycosylation of flagellin appears to occur in the absence of a functional export apparatus. Thus, flagellins of a mass that is consistent with full glycosylation could be detected in mutants blocked at early and middle levels of the flagellar hierarchy. These data suggest that glycosylation occurs prior to export of flagellin and independently of the flagellar regulon.
When each of the 19 serine or threonine residues that can be glycosylated in 81-176 flagellin was changed to an alanine residue, most mutants (11/19) had no obvious phenotype. The remaining eight mutants fell into two classes that revealed distinct functions of the glycans. Five mutants were fully motile but defective in AAG: the S417A, S436A, S440A, S451A, and T481A mutants. Salmonella flagellin consists of four linearly connected domains, D0, D1, D2, and D3 (42). Assuming that the basic structure of C. jejuni flagellin is similar, all of the glycans, with the exception of T481, map in the region of the molecule that corresponds to D2-D3, which forms the projection on the filament surface (42) (Fig. 4). This is consistent with early immunogold electron microscopy studies that indicated that at least some of the modified amino acids were surface exposed in the Campylobacter filament (30) and with the observation that the flagellar glycans are involved in AAG (9). S417 is modified with PseAcOAc, and T481 is modified with PseAc, but the glycans on the other sites have not been determined due to the high degree of clustering of the glycans on the tryptic peptides examined by mass spectrometry (38). The involvement of these specific residues in AAG would suggest that these residues are surface exposed in the assembled filament and are available to interact with adjacent filaments from other C. jejuni cells. These residues may also be involved in interaction with specific ligands on eukaryotic cells, a hypothesis that is currently under investigation.
Three other mutants (mutations S425A, S454A, and S460A) displayed a distinct phenotype. All three showed significant reductions in motility and produced truncated flagellar filaments. All three were FlgR+ and FlgS+ by immunoblotting, indicating that the reduction in motility was not due to phase variation of either of these two regulatory proteins (12, 13). All three mutants were capable of secreting both FlaA and the nonflagellar protein, FspA, through the flagellar opening to the supernatant, indicating that the defect was not due to phase variation of another structural component of flagella or to defects in secretion of flagellin. Although some filament was assembled in all three mutants, the filaments appeared more fragile than that composed of a fully glycosylated flagellin. Earlier data had shown that C. coli VC167 flagella decorated exclusively with PseAc (lacking all LegAm) were more readily solubilized in detergent than filaments composed of flagellins modified with both LegAm and PseAc, which also suggested a role for the glycans in subunit interactions (7). Since flagellin is secreted from the S425A, S454A, and S460A mutants, and in all cases some filament is assembled, the defect appears to be in filament assembly, suggesting that these glycans on these residues play a role in subunit interactions. The S425A and S460A mutants were defective in AAG, which is consistent with production of a truncated flagellin, but the S454A mutant showed no AAG defect. This likely reflects the observations that the S454A mutant was more motile than the S425A or S460A mutant and synthesized somewhat longer filaments.
Although glycosylation of polar flagella appears to be the norm rather than the exception (7, 22, 33), Campylobacter and Helicobacter, both members of the epsilon division of the Proteobacteria, have been shown to require glycosylation to assemble a flagellar filament (22). The other trait common to the flagellins of the epsilon division of Proteobacteria is the absence of Toll-like receptor 5 (TLR5) recognition sites (1). The TLR5 activation residues in Salmonella enterica serovar Typhimurium flagellin map primarily to the D1 domain, although additional contribution from the D2-D3 domain and the C-terminal D1 domain is also required (1, 35). The D1 residues that are critical for TLR5 activation map to the convex surface of the flagellin monomer in the filament and form the contact point between the convex surface of one subunit and the concave surface of an adjacent monomer (35). It has been suggested that loss of these sites in the Epsilonproteobacteria required a complex series of compensatory mutations that allowed for subunit interactions in the absence of TLR5 binding sites (1). Moreover, it has recently been shown that the quaternary structure of C. jejuni flagellar filaments is composed of 7 protofilaments per turn, in contrast to the 11 protofilaments found in the Salmonella filament (4). The data presented here suggest that specific glycans may contribute to the compensatory changes that allow these bacteria to assemble a flagellar filament but avoid the innate immune response.
Acknowledgments
We thank Scarlett Goon for construction of the flhF and flhB mutants, Rob Williams for electron microscopy, and Gary Majam and Dawn Pattarini for technical assistance.
This study was supported by RO1 AI043559 from NIAID and Work Unit 6000.RAD1.DA3.A0308 from the Military Infectious Disease Research Program.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government. P.G. is a civilian employee of the U.S. Government. This work was prepared as part of her official duties.
Footnotes
Published ahead of print on 11 September 2009.
REFERENCES
- 1.Andersen-Nissen, E., K. D. Smith, K. L. Strobe, S. L. R. Barrett, B. T. Cookson, S. M. Logan, and A. Aderem. 2005. Evasion of toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA 102:9247-9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 3.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karylshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galkin, V. E., X. Yu, J. Bielnick, J. Heuser, C. P. Ewing, P. Guerry, and E. H. Egelman. 2008. Divergence of quaternary structures among bacterial flagellar filaments. Science 320:382-385. [DOI] [PubMed] [Google Scholar]
- 5.Golden, N. J., and D. W. K. Acheson. 2002. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70:1761-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goon, S., C. P. Ewing, M. Lorenzo, D. Pattarini, G. Majam, and P. Guerry. 2006. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect. Immun. 74:769-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50:659-671. [DOI] [PubMed] [Google Scholar]
- 8.Guerry, P., C. M. Szymanski, M. M. Prendergast, T. E. Hickey, C. P. Ewing, D. L. Pattarini, and A. P. Moran. 2001. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerry, P., C. P. Ewing, M. Schirm, M. Lorenzo, J. Kelly, D. Pattarini, G. Majam, P. Thibault, and S. M. Logan. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60:299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerry, P., R. Yao, R. A. Alm, D. H. Burr, and T. J. Trust. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 235:474-481. [DOI] [PubMed] [Google Scholar]
- 12.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646-1659. [DOI] [PubMed] [Google Scholar]
- 13.Hendrixson, D. R. 2008. Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol. Microbiol. 70:519-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 15.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54 dependent but not σ28 dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagella secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 16.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaumouille, V., O. Francetic, P. J. Sansonetti, and G. T. Van Nhieu. 2008. Cytoplasmic targeting of IpaC to the bacterial pole directs polar type III secretion in Shigella. EMBO J. 27:447-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kocks, C., R. Hellio, P. Gounon, H. Ohayon, and P. Cossart. 1993. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J. Cell Sci. 105:699-710. [DOI] [PubMed] [Google Scholar]
- 19.Kusumoto, A., A. Shinohara, H. Terashima, S. Kojima, T. Yakushi, and M. Homma. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390-1399. [DOI] [PubMed] [Google Scholar]
- 20.Lai, E. M., U. Nair, N. D. Phadke, and J. R. Maddock. 2004. Proteomic screening and identification of differentially distributed membrane proteins in Escherichia coli. Mol. Microbiol. 52:1029-1044. [DOI] [PubMed] [Google Scholar]
- 21.Larsen, J. C., C. M. Szymanski, and P. Guerry. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logan, S. M. 2006. Flagellar glycosylation—a new component of the motility repertoire? Microbiology 152:1249-1262. [DOI] [PubMed] [Google Scholar]
- 23.Logan, S. M., I. C. Schoenhofen, and P. Guerry. 2008. O-Linked flagellar glycosylation in Campylobacter, p. 471-481. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 24.Logan, S. M., J. P. M. Hui, E. Vinogradov, A. J. Aubrey, J. E. Melanson, J. F. Kelly, H. Nothaft, and E. C. Soo. 2009. Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J. 276:1014-1023. [DOI] [PubMed] [Google Scholar]
- 25.McNally, D. J., J. P. M. Hui, A. J. Aubrey, K. K. K. Mui, P. Guerry, J.-R. Brisson, S. M. Logan, and E. C. Soo. 2006. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 281:18489-18498. [DOI] [PubMed] [Google Scholar]
- 26.Misawa, N., and M. J. Blaser. 2000. Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect. Immun. 68:6168-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, T. S., and B. I. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphrey, and J. M. Wells. 2004. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 29.Poly, F., C. Ewing, S. Goon, T. E. Hickey, D. Rockabrand, G. Majam, L. Lee, J. Phan, N. J. Savarino, and P. Guerry. 2007. Heterogeneity in a novel Campylobacter jejuni protein that is secreted through the flagellar filament. Infect. Immun. 75:3859-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Power, M. E., P. Guerry, W. D. McCubbin, C. M. Kay, and T. J. Trust. 1994. Structural and antigenic characteristics of Campylobacter coli FlaA flagellin. J. Bacteriol. 176:3303-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pugsley, A. P., and N. Buddelmeijer. 2004. Traffic spotting: poles apart. Mol. Microbiol. 53:1559-1562. [DOI] [PubMed] [Google Scholar]
- 32.Romantsov, T., S. Helbig, D. E. Culham, C. Gill, L. Stalker, and J. M. Wood. 2007. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 64:1455-1465. [DOI] [PubMed] [Google Scholar]
- 33.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 34.Schirm, M., I. Schoenhofen, S. M. Logan, K. Waldron, and P. Thibault. 2005. Identification of unusual bacterial glycosylation by tandem mass spectrometry analysis of intact proteins. Anal. Chem. 77:7774-7782. [DOI] [PubMed] [Google Scholar]
- 35.Smith, K. D., E. Andersen-Nissen, F. Hayashi, K. Strobe, M. A. Bergman, S. L. R. Barrett, B. T. Cookson, and A. Aderem. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for portofilament formation and bacterial motility. Nat. Immunol. 4:1247-1253. [DOI] [PubMed] [Google Scholar]
- 36.Steinhauer, J., R. Agha, T. Pham, A. W. Varga, and M. B. Goldberg. 1999. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol. Microbiol. 32:367-377. [DOI] [PubMed] [Google Scholar]
- 37.Taboada, E. N., R. R. Acedillo, C. D. Carrillo, W. A. Findlay, D. T. Medeiros, O. L. Mykytczuk, M. J. Roberts, C. A. Valencia, J. M. Farber, and J. H. E. Nash. 2004. Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J. Clin. Microbiol. 42:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thibault, P., S. M. Logan, J. F. Kelly, J.-R. Brisson, C. P. Ewing, T. J. Trust, and P. Guerry. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 39.Wosten, M. M. S. M., J. A. Wagenaar, and J. P. M. van Putten. 2004. The flgS/flgR two component signal transduction system regulates the fla operon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 40.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 41.Yao, R., and P. Guerry. 1996. Molecular cloning and site-specific mutagenesis of a gene involved in arylsulfatase production in Campylobacter jejuni. J. Bacteriol. 178:3335-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron microscopy. Nature 424:643-650. [DOI] [PubMed] [Google Scholar]