Abstract
Expression of the tna operon of Escherichia coli and of Proteus vulgaris is induced by l-tryptophan. In E. coli, tryptophan action is dependent on the presence of several critical residues (underlined) in the newly synthesized TnaC leader peptide, WFNIDXXL/IXXXXP. These residues are conserved in TnaC of P. vulgaris and of other bacterial species. TnaC of P. vulgaris has one additional feature, distinguishing it from TnaC of E. coli; it contains two C-terminal lysine residues following the conserved proline residue. In the present study, we investigated l-tryptophan induction of the P. vulgaris tna operon, transferred on a plasmid into E. coli. Induction was shown to be l-tryptophan dependent; however, the range of induction was less than that observed for the E. coli tna operon. We compared the genetic organization of both operons and predicted similar folding patterns for their respective leader mRNA segments. However, additional analyses revealed that l-tryptophan action in the P. vulgaris tna operon involves inhibition of TnaC elongation, following addition of proline, rather than inhibition of leader peptide termination. Our findings also establish that the conserved residues in TnaC of P. vulgaris are essential for l-tryptophan induction, and for inhibition of peptide elongation. TnaC synthesis is thus an excellent model system for studies of regulation of both peptide termination and peptide elongation, and for studies of ribosome recognition of the features of a nascent peptide.
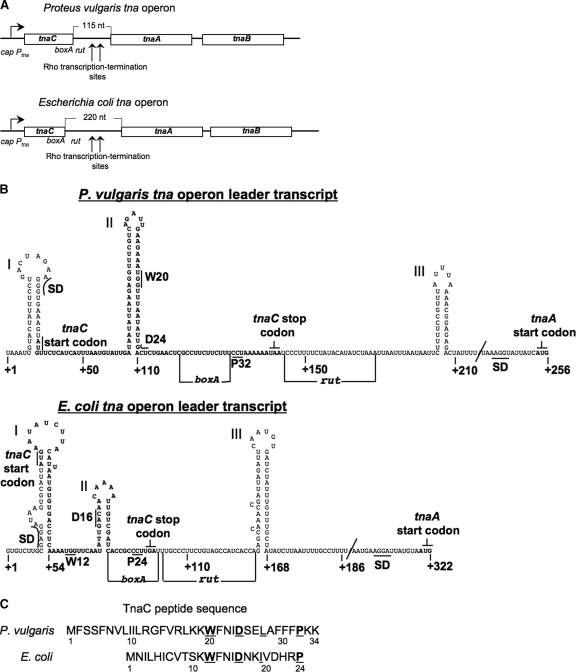
The tryptophanase (tna) operon of Escherichia coli, as well as that of Proteus vulgaris, consists of a promoter regulated by catabolite repression, a free l-tryptophan (l-Trp) responsive leader peptide coding region, tnaC, followed by two structural genes, tnaA and tnaB (Fig. 1A) (6, 18). tnaA encodes the enzyme tryptophanase, which degrades l-Trp to indole, pyruvate and ammonia. Pyruvate and ammonia can then be used as carbon and nitrogen sources, and indole can serve as a volatile signal molecule in quorum sensing or in stimulating biofilm formation (17). In E. coli, these events may play a role in bacterial colonization and possibly in bacterial pathogenesis (7, 16). tnaB encodes an l-Trp-specific permease (6, 18). The tnaC open reading frames of E. coli and P. vulgaris encode a 24-residue peptide and a 34-residue peptide, respectively (Fig. 1B). In both tna operons, the tnaC sequence is followed by a boxA site, a Rho termination factor-binding site (rut), and a noncoding region where Rho factor-dependent transcription termination can occur (Fig. 1A and C) (18). In the absence of inducing levels of l-Trp, Rho factor interacts with boxA and rut sequences in the tna transcript, instructing the transcribing RNA polymerase to terminate transcription prematurely in the noncoding region between tnaC and tnaA (Fig. 1A) (11). In the presence of l-Trp, Rho factor does not act. This inhibition of Rho factor action allows the tnaA and tnaB coding regions to be transcribed and translated (18).
FIG. 1.
Comparison of the organizational features of the tna operon of P. vulgaris and of E. coli with special emphasis on their leader regulatory regions. (A) Schematic representation and comparison of the tna operon of P. vulgaris with that of E. coli. Boxes represent the three open reading frames within each operon. The arrow at the left indicates the direction of transcription initiated at the promoter/cap site of the tna operon. Transcription of these operons is modulated by catabolic repression involving interaction of the catabolite repressor protein with the cap site (15, 18). Vertical arrows indicate the operon regions where transcription is terminated prematurely under the influence of the Rho protein and the boxA and rut sequences. The lengths of the spacer regions separating the tnaC and tnaA genes are indicated. Numbers identify nucleotide positions in the mRNAs. (B) Schematic representation of the predicted secondary structure of the mRNA leader RNAs of both tna operons, from the nucleotide at position +1 to the G nucleotide of the tnaA start codon. Letters in boldface type indicate the nucleotides corresponding to the tnaC open reading frame. The positions of the Shine-Dalgarno (SD) sequence and start codon for the tnaC and tnaA genes are indicated. Some codons analyzed in this study as well as the stop codon of tnaC are also shown. The RNA segments boxA and rut involved in transcription termination induced by Rho factor are identified by horizontal lines. The structures were obtained using the Mfold web server (20). (C) Amino acid sequence of the TnaC peptides encoded by the tna operons. The letters in boldface type identify residues shown to be essential for induction. Numbers indicate residue positions in the peptide.
In vivo and in vitro molecular studies using the E. coli tna operon as a model system have shown that when l-Trp is bound to the translating ribosome, it inhibits release factor 2 (RF2) cleavage of the nascent peptidyl-tRNA, TnaC-tRNAPro, at the tnaC stop codon (11). This results in ribosome stalling, allowing the translating ribosome to block access of the Rho termination factor to its rut binding site, thereby preventing transcription termination in the leader region of the operon (11, 18). This ribosome stalling has been shown to depend on specific residues of the TnaC leader peptide (3-5, 12). These include a tryptophan residue at position 12 (W12); an aspartic residue at position 16 (D16); a leucine residue at position 19 (L19); the last amino acid of the TnaC peptide, proline 24 (P24); and the spacing between W12 and P24 (Fig. 1C) (5, 12). Interestingly, all of these residues, and their positions, are highly conserved in the tna operons of diverse eubacterial species—among them, P. vulgaris (5).
Many bacterial species contain a tna operon in which leader peptide translation is probably used in regulating tna operon expression (5). In this study, we have analyzed the features of tnaC of P. vulgaris that are required for Trp induction. Mutagenesis analyses, and expression studies with a reporter gene, establish that the conserved essential residues in TnaC detected previously in E. coli are also essential for induction of the P. vulgaris tna operon. In vitro translation assays with the P. vulgaris leader RNA region demonstrated that l-Trp induces the production of a stalled ribosome complex containing TnaC-tRNAPro. Since tnaC of P. vulgaris contains two lysine (Lys) codons following the proline codon that corresponds to the observed ribosome stalling position for E. coli, we conclude that l-Trp induction inhibits translation elongation—not translation termination—during synthesis of the TnaC peptide of P. vulgaris.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli K-12 strains used in this study were derived from strain W3110 (our stock). pAVK9 is a plasmid containing the P. vulgaris tna operon promoter and leader region, followed by a fusion of the first nine codons of the P. vulgaris tnaA gene to the E. coli lacZ gene (18). Expression of this TnaA-LacZ fusion protein is controlled by the promoter and leader regions of the P. vulgaris tna operon (15). pPDG100 is a plasmid containing the E. coli tna operon promoter and leader region, with a fusion of the first nine codons of E. coli tnaA to the E. coli lacZ gene. Expression of this gene/protein fusion is regulated by the promoter and leader regions of the E. coli tna operon (9). To obtain the tnaC gene variants used in this study (see Table 2; Fig. 2), nucleotides in tnaC of plasmids pAVK9 and pPDG100 were changed using a QuikChange mutagenesis PCR kit (Stratagene) and a specific complementary pair of deoxyoligonucleotide sequences (Table 1), as previously described (5).
TABLE 2.
β-Galactosidase levels of E. coli strains with plasmids containing tnaC of P. vulgaris or tnaC of E. coli, followed by a corresponding tnaA-lacZ gene fusiona
| tnaC source | tnaC change | β-Galactosidase activity (Miller units)b |
Induction ratio (+1MTrp/ −1MTrp) | |
|---|---|---|---|---|
| −1MTrp | +1MTrp | |||
| Proteus vulgaris | wt | 300 ± 70 | 2,300 ± 76 | 8 |
| K33R | 320 ± 66 | 2,500 ± 66 | 8 | |
| K33I | 340 ± 73 | 3,000 ± 54 | 9 | |
| K33R-K34R | 310 ± 56 | 2,450 ± 86 | 8 | |
| K33I-K34I | 340 ± 60 | 3,400 ± 65 | 10 | |
| ΔK33-34 | 550 ± 80 | 7,000 ± 75 | 13 | |
| TAA35TGA | 320 ± 76 | 2,500 ± 66 | 8 | |
| W20R | 90 ± 26 | 110 ± 76 | 1 | |
| D24A | 100 ± 26 | 200 ± 20 | 2 | |
| L27A | 110 ± 26 | 670 ± 23 | 6 | |
| P32A | 200 ± 26 | 250 ± 26 | 1 | |
| Escherichia coli | wt | 300 ± 760 | 12,300 ± 106 | 41 |
| W12R | 120 ± 36 | 110 ± 26 | 1 | |
| D16A | 150 ± 30 | 200 ± 20 | 1 | |
| P24A | 110 ± 32 | 170 ± 23 | 1 | |
Cultures of the E. coli W3110 strain containing the pAVK9 (tnaC of Proteus vulgaris) or pPDG100 (tnaC of E. coli) plasmid or its variants were grown in minimal medium plus 0.2% glycerol and 0.05% acid hydrolyzed casein, with (+1MTrp) or without (−1MTrp) 100 μg/ml dl-1-methyl-tryptophan, at 37°C. wt, wild type.
β-Galactosidase assays (see Materials and Methods) were performed on samples obtained from three independent experiments.
FIG. 2.
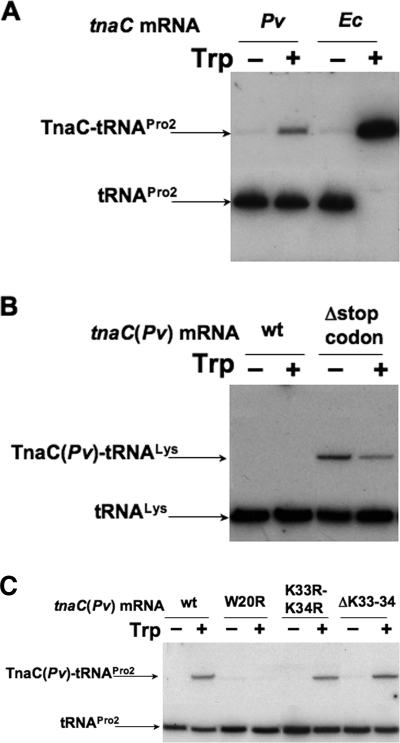
Analysis of the peptidyl-tRNA, TnaC-tRNA, accumulated in vitro in translation assays. Translation reactions were carried out in vitro using an E. coli S-30 preparation and various mRNAs containing the genetic changes indicated. The mRNA containing tnaC of P. vulgaris with the change Δstop codon corresponds to a truncated mRNA with a 3′ end at the third nucleotide of the Lys34 codon. Northern blot assays were performed using a 32P-labeled oligonucleotide complementary to E. coli tRNAPro (A and C) or tRNALys (B) sequences. Accumulation of the peptidyl-tRNA was determined from reaction mixtures containing the mRNAs indicated, in the presence or absence of 2 mM l-Trp. The positions of the peptidyl-tRNAs, TnaC-tRNAPro, and TnaC-tRNALys, as well as their corresponding free tRNAs, are indicated with arrows. Pv, P. vulgaris; Ec, E. coli.
TABLE 1.
Oligodeoxynucleotides used to change specific codons in the tnaC genes examined in this study
| tnaC source | Change | Oligodeoxynucleotide sequence |
|---|---|---|
| Proteus vulgaris | K33R | 5′-CGCCTTCTTCTTTCCTCGTAAATAATCCCTTTTCTATAC-3′ |
| 5′-GTATAGAAAAGGGATTATTTACGAGGAAAGAAGAAGGCG-3′ | ||
| K33I | 5′-CGCCTTCTTCTTTCCTATAAAATAATCCCTTTTC-3 | |
| 5′-GAAAAGGGATTATTTTATAGGAAAGAAGAAGGCG-3′ | ||
| K33R-K34R | 5′-CGCCTTCTTCTTTCCTCGTCGTTAATCCCTTTTCTATAC-3′ | |
| 5′-GTATAGAAAAGGGATTAACGACGAGGAAAGAAGAAGGCG-3′ | ||
| K33I-K34I | 5′-CGCCTTCTTCTTTCCTATAATATAATCCCTTTTCTATAC-3′ | |
| 5′-GTATAGAAAAGGGATTATATTATAGGAAAGAAGAAGGCG-3′ | ||
| ΔK33-34 | 5′-CGCCTTCTTCTTTCCTTAATCCCTTTTCTATAC-3′ | |
| 5′-GTATAGAAAAGGGATTAAGGAAAGAAGAAGGCG-3′ | ||
| TAA35TGA | 5′-CTTCTTCTTTCCTAAAAAATGATCCCTTTTCTATACATATC-3′ | |
| 5′-GATATGTATAGAAAAGGGATCATTTTTTAGGAAAGAAGAAG-3′ | ||
| W20R | 5′-CGTCAGATTGAAGAAACGGTTTAATATTGA-3′ | |
| 5′-GAGTCAATATTAAACCGTTTCTTCAATCTGACG-3′ | ||
| D24A | 5′-GAAATGGTTTAATATTGCCTCTGAACTCGCCTTC-3′ | |
| 5′-GAAGGCGAGTTCAGAGGCAATATTAAACCATTTC-3′ | ||
| L27A | 5′-ATATTGACTCTGAAGCCGCCTTCTTCTTTCC-3′ | |
| 5′-GGAAAGAAGAAGGCGGCTTCAGAGTCAATAT-3′ | ||
| P32A | 5′-GAACTCGCCTTCTTCTTTCCTAAAAAATAATCCCTTTTC-3′ | |
| 5′-GAAAAGGGATTATTTTTTAGGAAAGAAGAAGGCGAGTTC-3′ | ||
| Escherichia coli | W12R | 5′-GTGTGACCTCAAAATGGTTCAATATTGACAAC-3′ |
| 5′-GTTGTCAATATTGAACCATTTTGAGGTCACAC-3′ | ||
| D16A | 5′-CTCAAAATGGTTCAATATTGCCAACAAAATTGTCGATCACC-3′ | |
| 5′-GGTGATCGACAATTTTGTTGGCAATATTGAACCATTTTGAG-3′ | ||
| P24A | 5′-GTCGATCACCGCGCTTGATTTGCCTTC-3′ | |
| 5′-GAAGGCAAATCAAGCGCGGTGATCGAC-3′ |
Growth conditions and β-galactosidase assays.
In the experiments described in Table 2, strains or plasmid-containing strains were grown with shaking at 37°C in minimal medium plus 0.2% glycerol, 0.05% acid casein hydrolysate, and 100 μg/ml ampicillin, with or without 100 μg/ml dl-1-methyl-Trp. When each culture reached a density of 100 Klett units (660 filter), the cells were harvested and disrupted by sonication, and β-galactosidase activity (in Miller units) was determined using standard procedures (4, 5).
Detection of TnaC-tRNAPro and TnaC-tRNALys in vitro.
In vitro translation assays were performed as previously described (4) using mRNA obtained from each one of the pAVK9 or pPDG100 constructs. DNA fragments containing a T7 promoter before a tnaC open reading frame were generated by PCR, using as templates the pAVK9 or pPDG100 plasmid variants carrying changes in their tnaC codons. The following oligonucleotides were used with the pAVK9 plasmid constructs: forward, 5′-GTGAATTATTAATACGACTCACTATAGGGTAAATTGTACTATTTC-3′, containing the T7 promoter (residues in boldface type); reverse, 5′-GATATGTATAGAAAAGGGATTA-3′; or 5′-TTATTTTTTAGGAAAGAAGAA-GG-3′ for the deletion stop codon (Δstop codon) construct (Fig. 2). The following oligodeoxynucleotides were used with the pPDG100 plasmid constructs: forward, 5′-GTGAATTATTAATACGACTCACTATAGGGTGTCTTGCGAGGATAAGTG-3′, containing the T7 promoter (residues in boldface type); reverse, 5′-GGTGATGGCTACTGAAGGGCAAATCA-3′. The PCR DNA fragments obtained were used as templates to produce mRNAs in vitro by using T7 RNA polymerase (4). In vitro translation reactions were performed with a cell extract obtained from E. coli strain A19 RNase I− (trpR ΔlacZ ΔtrpEA2 tnaA bgl::Tn10) (13). In the experiments described in the legend for Fig. 2, the in vitro translation reactions were performed using 2 μg of tnaC mRNA per 50 μl of reaction mixture. The resulting extract reactions were resolved by electrophoresis on 10% tricine-sodium dodecyl sulfate gels, and the bands were transferred to nylon membranes. The TnaC-tRNAPro and TnaC-tRNALys bands were detected by hybridizing the membranes using a 5′-32P-labeled deoxynucleotide primer, 5′-CCTCCGACCCCCGACACCCCAT-3′, that was complementary to the tRNAPro sequence, or a 5′-CCTGCGACCAATTGATTAAA-3′ primer, that was complementary to the tRNALys sequence (2, 8).
RESULTS AND DISCUSSION
Comparison of the tna operon features of E. coli and P. vulgaris.
In many eubacterial species, l-Trp induction of expression of the tna operon appears to have similar features. Interestingly, l-Trp induction of the tna operon of P. vulgaris is not as efficient as is induction of the tna operon of E. coli (15). We compared the structural features of the tna operons of P. vulgaris and E. coli with the objective of providing greater insight into the regions of the operon that are essential for l-Trp induction and how they function. As shown in Fig. 1A, the genetic organizations of the tna operons of P. vulgaris and E. coli are quite similar. In addition, both operons appear to have the same regulatory elements (Fig. 1A). Since RNA structures and sequences in the tna leader region play a significant role in l-Trp-induced expression of this operon (11, 14), the predicted secondary structures of both leader RNA segments were deduced (Fig. 1B). It can be seen in this figure, that despite existing nucleotide sequence differences, our predictions reveal three similar stem-loop structures in both leader RNA sequences. (i) The initial structure, structure I, whose function is unknown, is close to the 5′ end of the tna operon mRNA and contains the Shine-Dalgarno and start codon sequences for the leader peptide, TnaC. (ii) A second potential structure contains the tnaC leader sequence (Fig. 1B, structure II). In E. coli, this structure has been shown to be important in synchronizing translation of the leader peptide coding region with transcription of the structural genes of this operon (14). (iii) The third stem-loop structure (Fig. 1B, structure III), formed by sequences in between tnaC and tnaA, is believed to be involved in the transcription termination event performed by Rho factor (1, 11). These observations suggest that the mRNA leader segments of these operons could fold similarly, forming structures that play the same critical roles in tna operon expression in both organisms. Most interestingly, comparison of the two tnaC coding sequences revealed several differences, as well as similarities (Fig. 1C). Thus, tnaC of P. vulgaris encodes a peptide that would be 10 residues longer than TnaC of E. coli (Fig. 1C). Despite this size difference, TnaC of P. vulgaris contains a core of residues—W20, D24, L/I27, and P32 (shown in bold and italics)—that are also present in TnaC of E. coli, KWFNIDXX(L/I)XXXXP, and are essential for induction, both in P. vulgaris (Table 2) and in E. coli (5). Interestingly, the number of residues separating the Trp and aspartic residues, and their distance from the proline residue, are the same in both organisms (Fig. 1C). This implies that the mechanisms of peptide-mediated induction could be similar in these two organisms. However, TnaC of P. vulgaris contains two Lys residues following its conserved Pro32 residue. If, as has been shown for E. coli, the action of l-Trp on the ribosome translating tnaC of P. vulgaris is to inhibit release factor catalysis of the translation termination reaction, then the stalled TnaC-peptidyl-tRNA would have two Lys residues following its crucial Pro residue. Alternatively, if l-Trp induction inhibits TnaC elongation at the TnaC Pro32 residue, or at the first Lys residue, then peptide elongation rather than peptide termination would be the event inhibited by the presence of inducing levels of l-Trp. In either case, the resulting stalled ribosome would protect the boxA and rut sequences, thereby inhibiting Rho binding and action, as shown for E. coli (11). Since each of these two possibilities is feasible, they required experimental investigation (see below).
Features of the tnaC coding region of P. vulgaris essential for Trp induction of tna operon expression.
As previously mentioned, the tnaC coding region of P. vulgaris contains two Lys codons, at codon positions 33 and 34, following its Pro32 codon, and preceding its presumed stop codon (Fig. 1C). To explore how important these Lys codons, and other tnaC codons, are for Trp induction of expression of the P. vulgaris tna operon, plasmids containing coding region fusions between tnaA of P. vulgaris, or tnaA of E. coli, and an E. coli lacZ gene were used to examine the significance of their coding region differences (see Materials and Methods). These derivative plasmids were transformed into an E. coli strain lacking the tnaA and lacZ genes, and β-galactosidase activity was measured in cultures of transformed cells grown in the presence or absence of 1-methyl-Trp, as inducer. The results obtained are summarized in Table 2. Replacing the P. vulgaris Lys33 and/or Lys34 codons with Arg or Ile codons did not substantially affect induction of tnaA-lacZ fusion expression by l-Trp (Table 2, column 5; compare row 1 with rows 2 to 5). Similar results were obtained in expression studies with a construct containing a deletion removing these two Lys codons (Table 2, column 5; compare row 1 with row 6). These results establish that the Lys codons located 3′ of the Pro32 codon of tnaC are not essential for Trp induction of P. vulgaris tna operon expression. To demonstrate that this induction depends on the TnaC peptide features, we examined the effects of changing those codons corresponding to positions of residues shown to be essential for induction in E. coli (Table 2) (5). Replacing the Trp20, Asp24, or Pro32 codon of tnaC of P. vulgaris reduced induction of tnaA-lacZ expression (Table 2, column 5; compare row 1 with rows 8, 9, and 11); expression was reduced comparably to that of the corresponding E. coli tnaC constructs (Table 2, column 5; compare row 12 with rows 13 to 15). These results show that Trp induction of P. vulgaris tna operon expression depends on conserved, crucial features of the TnaC peptide (5), but not on the two Lys residues following the essential Pro32 residue of P. vulgaris TnaC.
l-Trp-induced ribosome stalling during translation of tnaC mRNA of P. vulgaris.
In E. coli, Trp induction of tna operon expression involves inhibition of translation termination during ribosome-mediated translation of tnaC mRNA (10). The stalled ribosome contains the unreleased peptidyl-tRNA, TnaC-tRNAPro, which accumulates, and can be detected in vivo or in vitro, by standard Northern blotting procedures (2, 13). Experiments were therefore performed with tnaC of P. vulgaris to determine if Trp induction also leads to the accumulation of TnaC-tRNAPro (Fig. 2A). Translation reactions were performed in vitro using mRNAs containing only the leader regions of the tna operons of P vulgaris and E. coli, in the presence or absence of l-Trp (see Materials and Methods). The final products of each reaction were resolved using gel electrophoresis, and molecules containing tRNAPro were identified using an oligodeoxynucleotide complementary to a specific tRNAPro sequence. We observed with tnaC mRNAs of P. vulgaris that the presence of l-Trp induced the accumulation of TnaC-tRNAPro (Fig. 2A, compare lane 1 with lane 2). As expected, the same result was obtained with tnaC mRNAs of E. coli (Fig. 2A, compare lane 3 with lane 4). However, the accumulation of TnaC-tRNAPro was greater using E. coli mRNAs than it was using P. vulgaris mRNAs (Fig. 2A, compare lane 2 with lane 4). Thus, ribosome stalling induced by l-Trp at the tnaC Pro32 codon of P. vulgaris is reduced relative to the stalling induced by l-Trp in tnaC of E. coli. These results agree with the induction levels observed in vivo using tnaA-lacZ fusions (Table 2). The induction level observed with P. vulgaris tnaC was eightfold higher, while the induction level observed with E. coli tnaC was approximately 41-fold higher (Table 2, column 5; compare row 1 with row 12). Similar results were obtained comparing tryptophanase induction in strains of P. vulgaris with that of E. coli (15). Therefore, we conclude that there is a correlation between the Trp-induced level of ribosome stalling at the P. vulgaris Pro32 codon position and expression of its tna operon. These findings suggest that l-Trp-induced ribosome stalling at the Pro32 position could be the mechanism used by P. vulgaris in regulating tna operon expression. However, the two Lys codons following the Pro32 codon of P. vulgaris could also serve as sites of ribosome stalling. To distinguish between these possibilities, Northern blot analyses were performed on translation reactions carried out in vitro, using an oligodeoxynucleotide complementary to a unique segment of the tRNALys sequence (Fig. 2B). Translation of tnaC mRNA of P. vulgaris did not lead to the accumulation of any peptidyl-tRNALys in the presence or absence of l-Trp (Fig. 2B, compare lane 1 with lane 2). Thus, l-Trp does not induce ribosome stalling at either the Lys33 or Lys34 codon. To verify this approach, translation of a truncated tnaC mRNA of P. vulgaris was also examined, which did not contain a stop codon (the mRNA's 3′ end was at the third nucleotide of the Lys34 codon). As expected, translation resulted in the accumulation of TnaC-tRNALys in the presence or absence of added l-Trp (Fig. 2B, lanes 3 and 4). These results demonstrate that translation of the tnaC sequence of P. vulgaris can proceed beyond its Pro32 codon. Most interestingly, the accumulation of TnaC-tRNALys observed during translation of this stop codon-deficient truncated tnaC mRNA was reduced in the presence of l-Trp relative to the level observed in the absence of added l-Trp (Fig. 2B, compare lane 3 with lane 4). Thus, l-Trp probably does not induce ribosome stalling at either of the Lys codons of tnaC mRNA but, rather, at the preceding Pro32 codon. That l-Trp can inhibit translation elongation during translation of tnaC mRNA of E. coli has been shown previously in studies of puromycin action (2, 11).
We also tested, in vitro, using tnaC of P. vulgaris, whether TnaC-tRNAPro accumulation is dependent on its essential tnaC codons (Fig. 2C). Replacing the conserved Trp20 codon of tnaC mRNA with an Arg codon reduced the accumulation of TnaC-tRNAPro in the presence of l-Trp (Fig. 2C, compare lane 2 with lane 4). Similar results were obtained using tnaC mRNAs with changes in the D24 and P32 codons (data not shown). However, no effect on the accumulation of TnaC-tRNAPro was observed during translation of mRNAs containing changes in the Lys codons located downstream of the P32 codon (Fig. 2C, compare lane 2 with lane 6 or 8). These findings suggest that Trp-induced accumulation of TnaC-tRNAPro during translation of tnaC mRNA of P. vulgaris depends on the conserved codons located between the W20 and P32 codons and that ribosome stalling on P. vulgaris tnaC mRNA is due to l-Trp inhibition of translation elongation, at the tnaC Pro32 codon. Thus, l-Trp-induced ribosome stalling at the Pro32 codon of P. vulgaris tnaC appears to be responsible for tna operon expression.
Conclusions.
Comparison of l-Trp induction expression of the tnaC operons of P. vulgaris and E. coli has established that these operons share many essential features (Fig. 1). However, significant differences exist. (i) l-Trp-induced expression of tnaC is greater in E. coli than in P. vulgaris (Table 1 and Fig. 2) (15). (ii) l-Trp binding to the translating ribosome inhibits translation termination in E. coli (10), but inhibits translation elongation in P. vulgaris (Fig. 2). We conclude that in both organisms, the same conserved residues of the core TnaC peptide (Table 1) instruct the translating ribosome to create an l-Trp binding site in the ribosome at which bound l-Trp inhibits translation termination or elongation. The reduced induction observed for the P. vulgaris tna operon relative to induction of the E. coli tna operon (Fig. 2) may indicate that l-Trp binding has less of an inhibitory effect on translation elongation than it does on translation termination. Alternatively, nonconserved features of the nascent TnaC peptide could affect its folding within the ribosome, influencing l-Trp binding, or alter interactions between the nascent peptide and the ribosome exit tunnel (19). Additional experiments will be needed to distinguish between these possibilities.
Acknowledgments
We are grateful to Jacqueline Moreno for her help in performing the experiments conducted in this study.
This study was supported by a grant provided to C.Y. from the National Science Foundation, MCB-0615390, and by start-up funds provided to L.R.C.-V. from the University of Alabama—Huntsville.
Footnotes
Published ahead of print on 18 September 2009.
REFERENCES
- 1.Ciampi, M. S. 2006. Rho-dependent terminators and transcription termination. Microbiology 152:2515-2528. [DOI] [PubMed] [Google Scholar]
- 2.Cruz-Vera, L. R., M. Gong, and C. Yanofsky. 2006. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl. Acad. Sci. USA 103:3598-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz-Vera, L. R., A. New, C. Squires, and C. Yanofsky. 2007. Ribosomal features essential for tna operon induction: tryptophan binding at the peptidyl transferase center. J. Bacteriol. 189:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cruz-Vera, L. R., S. Rajagopal, C. Squires, and C. Yanofsky. 2005. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell 19:333-343. [DOI] [PubMed] [Google Scholar]
- 5.Cruz-Vera, L. R., and C. Yanofsky. 2008. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of tna operon expression. J. Bacteriol. 190:4791-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeley, M. C., and C. Yanofsky. 1982. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J. Bacteriol. 151:942-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Martino, P., A. Merieau, R. Phillips, N. Orange, and C. Hulen. 2002. Isolation of an Escherichia coil strain mutant unable to form biofilm on polystyrene and to adhere to human pneumocyte cells: involvement of tryptophanase. Can. J. Microbiol. 48:132-137. [DOI] [PubMed] [Google Scholar]
- 8.Dong, H., L. Nilsson, and C. G. Kurland. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260:649-663. [DOI] [PubMed] [Google Scholar]
- 9.Gollnick, P., and C. Yanofsky. 1990. tRNA(Trp) translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J. Bacteriol. 172:3100-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong, F., K. Ito, Y. Nakamura, and C. Yanofsky. 2001. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro). Proc. Natl. Acad. Sci. USA 98:8997-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, F., and C. Yanofsky. 2002. Analysis of tryptophanase operon expression in vitro: accumulation of TnaC-peptidyl-tRNA in a release factor 2-depleted S-30 extract prevents Rho factor action, simulating induction. J. Biol. Chem. 277:17095-17100. [DOI] [PubMed] [Google Scholar]
- 12.Gong, F., and C. Yanofsky. 2002. Instruction of translating ribosome by nascent peptide. Science 297:1864-1867. [DOI] [PubMed] [Google Scholar]
- 13.Gong, F., and C. Yanofsky. 2001. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 276:1974-1983. [DOI] [PubMed] [Google Scholar]
- 14.Gong, F., and C. Yanofsky. 2003. A transcriptional pause synchronizes translation with transcription in the tryptophanase operon leader region. J. Bacteriol. 185:6472-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamath, A. V., and C. Yanofsky. 1992. Characterization of the tryptophanase operon of Proteus vulgaris. Cloning, nucleotide sequence, amino acid homology, and in vitro synthesis of the leader peptide and regulatory analysis. J. Biol. Chem. 267:19978-19985. [PubMed] [Google Scholar]
- 16.Martino, P. D., R. Fursy, L. Bret, B. Sundararaju, and R. S. Phillips. 2003. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can. J. Microbiol. 49:443-449. [DOI] [PubMed] [Google Scholar]
- 17.Ren, D., L. A. Bedzyk, R. W. Ye, S. M. Thomas, and T. K. Wood. 2004. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl. Environ. Microbiol. 70:2038-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart, V., and C. Yanofsky. 1985. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 164:731-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap, M. N., and H. D. Bernstein. 2009. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell 34:201-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]