Abstract
We redemonstrate that SwrA is essential for swarming motility in Bacillus subtilis, and we reassert that laboratory strains of B. subtilis do not swarm. Additionally, we find that a number of other genes, previously reported to be required for swarming in laboratory strains, are dispensable for robust swarming motility in an undomesticated strain. We attribute discrepancies in the literature to a lack of reproducible standard experimental conditions, selection for spontaneous swarming suppressors, inadvertent genetic linkage to swarming mutations, and auxotrophy.
Many species of bacteria are capable of flagellum-mediated swimming motility in liquid broth. Of those species, a subset is also capable of a related, but genetically separable, form of flagellum-mediated surface movement called swarming motility (17). Examples of swarming-proficient species include Proteus mirabilis, Vibrio parahaemolyticus, Serratia marcescens, Escherichia coli, Salmonella enterica, and Bacillus subtilis (1, 15, 16, 20, 28). In general, swarming requires a surfactant or wetting agent to reduce surface tension, an increase in flagellar number per cell, and other genetic features that are distinct from swimming (7, 14).
There is confusion in the literature concerning the genetic requirements of the swarming phenotype of B. subtilis. It is generally accepted that the ancestral undomesticated strain B. subtilis 3610 exhibits robust swarming motility (18, 20, 33). Swarming motility of strain 3610 requires the production of a secreted surfactant, called surfactin (6, 20), to reduce surface tension and permit surface spreading, and it also requires the protein SwrA to activate flagellar biosynthesis gene expression and increase the number of flagella on the cell surface (5, 20). Some reports claim that domesticated derivatives of 3610, such as the commonly used laboratory strain 168, are also swarming proficient (10, 18, 19, 24). Strain 168, however, is defective in both surfactin production (9, 25) and SwrA (5, 21, 31), and thus, swarming 168 strains challenge the genetic definition of swarming motility. Our lab has never observed swarming in laboratory strains, and here we investigated swarming motility in a reportedly swarming-proficient 168 strain.
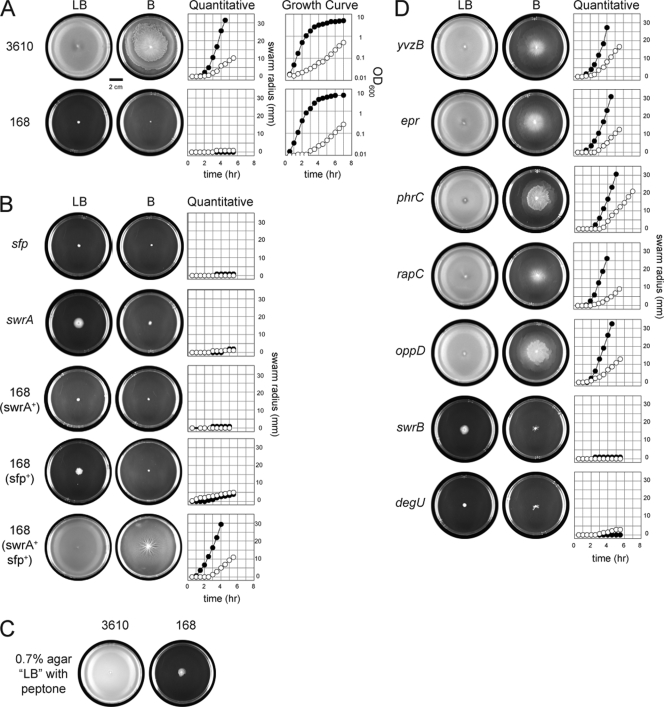
We obtained a reportedly swarming-proficient 168 strain (13) (generous gift of Simone Séror, Orsay University, Paris-Sud, France) (Table 1) and compared its swarming phenotype to that of 3610 under our standard conditions (20). Swarm plates were prepared one day prior to use with 25 ml of LB medium (10 g Bacto tryptone, 5 g Bacto yeast extract, 5 g NaCl per liter) fortified with 0.7% Bacto agar. To minimize water on the agar surface and thus minimize the potentially confounding influence of swimming motility, plates were dried 20 min prior to inoculation and 10 min postinoculation open-faced in a laminar flow hood. For qualitative swarm assays, plates were centrally inoculated with cells from a freshly grown overnight colony using a sterile stick. For quantitative swarm expansion assays, 1 ml of cells grown to mid-exponential phase (optical density at 600 nm [OD600], 0.5) was resuspended in PBS buffer (8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4 per liter, pH 7.0) containing 0.5% India ink (Higgins) to an OD600 of 10 and centrally spotted (10 μl). Swarm expansion was measured at 0.5-h intervals along a transect on the plate. Plates were incubated at 37°C in 20 to 30% humidity. Whereas strain 3610 was swarming proficient, strain 168 (Orsay) was swarming deficient (Fig. 1A). Thus, strain 168 (Orsay) appeared to behave similarly to all other laboratory strains we have tested previously (20, 21).
TABLE 1.
Strains
| Strain | Genotypea |
|---|---|
| 168 | trpC2 swrA sfp (13) |
| 3610 | Wild type |
| DS72 | yvzB::tet (21) |
| DS2268 | epr::kan |
| DS2415 | ΔswrA |
| DS2509 | ΔswrB |
| DS3337 | sfp::mls |
| DS3649 | ΔdegU |
| DS3903 | phrC::spec |
| DS4978 | rapC::spec |
| DS4979 | oppD::kan |
| DS5106 | 168 trpC2 swrA sfp amyE::PswrA-swrA cat |
| DS5758 | 168 trpC2 swrA sfp amyE::sfp+ cat |
| DS5759 | 168 trpC2 swrA sfp amyE::PswrA-swrA cat thrC::sfp+ mls |
All strains are in the 3610 genetic background unless otherwise indicated.
FIG. 1.
Swarming motility on LB and B media. In qualitative plate images, colonized agar appears white and uncolonized agar appears black on LB and B media, as indicated. Swarming cells colonize a larger surface area than nonswarming cells. All strains are derivatives of strain 3610 unless otherwise indicated. Bar, 2 cm. (A) Quantitative swarm expansion assays on solid medium and growth in liquid medium of the indicated strains on LB medium (closed symbols) and on B medium (open symbols). To indicate variability in a particular experiment, we have reproduced the quantitative swarm expansion assay of strain 3610 on LB and B media with error bars in Fig. S5 in the supplemental material. (B) Quantitative swarm expansion assays on LB (closed symbols) and B (open symbols) media. The following strains were used: DS3337 (sfp), DS2415 (swrA), DS5106 (168 swrA+), DS5758 (168 sfp+), and DS5759 (168 swrA+ sfp+). In all assays, B medium was made according to reference 2 except for strain DS5759, for which B medium was supplemented with 780 μM threonine to compensate for thrC auxotrophy. (C) Swarm plates of the indicated strains on LB medium made with equal parts peptone instead of tryptone. (D) Quantitative swarm expansion assays of the indicated 3610-derived mutant strains on LB medium (closed symbols) and on B medium (open symbols). The following strains were used: DS72 (yvzB), DS2268 (epr), DS3903 (phrC), DS4978 (rapC), DS4979 (oppD), DS2509 (swrB), and DS3649 (degU). All points are averages for three replicates.
We next explored the genetic basis for the swarming defect we observed in strain 168 (Orsay). As with other laboratory strains, colonies of strain 168 (Orsay) failed to produce the transparent ring normally indicative of surfactin production, due to a mutation of the gene sfp (25). Complementation with the wild-type sfp gene in 168 was sufficient to restore surfactin production but was insufficient to restore swarming motility (Fig. 1B) (20). Laboratory strains also fail to swarm because of a loss-of-function frameshift mutation in the gene encoding SwrA (5, 21). Sequencing of the swrA gene confirmed that strain 168 (Orsay) contained the frameshift mutation, but introduction of a swrA complementation construct at an ectopic site in the chromosome (amyE::PswrA-swrA) was also insufficient to restore swarming motility (Fig. 1B). Swarming motility was fully rescued, however, when sfp and swrA were simultaneously complemented in the 168 strain (Fig. 1B) or when the swrA frameshift mutation was repaired in spontaneous suppressors isolated from 168 complemented with sfp alone (see Fig. S1 in the supplemental material). Furthermore, mutation of either sfp or swrA in the 3610 genetic background abolished swarming (Fig. 1B). We conclude that Sfp and SwrA are necessary for swarming. We further conclude that, with respect to swarming motility, strain 168 (Orsay) is genetically no different from any other laboratory strain we have tested, as it fails to swarm due to simultaneous defects in Sfp and SwrA (21). We infer that the apparent swarming observed in some laboratory strains is not due to genetic differences but rather due to differences in experimental conditions.
In our swarming assays, we take steps to minimize surface water. In some cases of the reported swarming of strain 168, plates were poured 1 h before use, dried for 5 min, and incubated at 60 to 70% humidity (13). When 0.7% agar LB plates were freshly poured and not dried, we noticed that toothpick inoculation of the cells disturbed the agar surface and caused a pool of water to well forth from the agar (see Fig. S2 in the supplemental material). Pools of water emerged even when the plates were dried for 5 or 10 min prior to inoculation, but water did not emerge when the plates were dried for 15 min or longer (see Fig. S2 in the supplemental material). The colony size of strain 168 was proportional to the amount of water extracted from the agar, but the cells did not exhibit swarming motility (see Fig. S2 in the supplemental material). We conclude that excess water was not sufficient to promote swarming of the laboratory strain. Nonetheless, we recommend drying plates for 20 min prior to inoculation to minimize any contribution of swimming motility to apparent surface migration.
Another difference in experimental conditions may concern the nutritional content of the medium. Some labs have tested swarming motility on LB medium in which tryptone was replaced by an equal amount of peptone (13). We reproduced the “LB” medium containing peptone and found that whereas strain 3610 was swarming proficient, strain 168 was swarming deficient (Fig. 1C). Thus, the peptone substitution did not promote swarming in lab strains.
Some labs have also reported swarming of laboratory strains on a defined medium called B medium [15 mM (NH4)2SO4, 8 mM MgSO4·7H2O, 27 mM KCl, 7 mM sodium citrate·H2O, 50 mM Tris·HCl (pH 7.5), 2 mM CaCl2·2H2O, 1 μM FeSO4·7H2O, 10 μM MnSO4·4H2O, 0.6 mM KH2PO4, 4.5 mM glutamic acid, 860 μM lysine, 780 μM tryptophan, and 0.5% glucose) (2, 13, 18, 19). In our hands, 3610 was swarming proficient on B medium, but strain 168 was swarming deficient (Fig. 1A). We conclude that altering medium composition was insufficient to promote swarming of laboratory strains. Furthermore, mutation of either sfp or swrA rendered strain 3610 nonswarming on B medium, and complementation of sfp and swrA restored B medium swarming to strain 168 (Fig. 1B). We conclude that the genetic requirements for swarming are the same for both LB and B medium.
On undefined rich LB medium, strain 3610 swarmed rapidly as a featureless monolayer, whereas on defined B medium, it swarmed in a branched dendritic pattern (18, 20) (Fig. 1A). In addition, the growth rate of 3610 in liquid B medium and swarm rate on solid B medium were both reduced fivefold relative to comparable assays with LB (Table 2), suggesting that the rate of swarming and the rate of growth were related. To further explore the connection between growth rate and swarming rate, we performed swarm expansion assays at lower temperatures. At 30°C, the growth rate in LB broth was reduced 2.5-fold relative to 37°C, and the swarming rate on LB agar was reduced 2.5-fold as well (Table 2; also, see Fig. S3 in the supplemental material). We conclude that swarming rate is correlated with growth rate. We infer that differences in growth may account for differences in swarm patterns (11). We note that regardless of the medium composition or the growth rate, the duration of the lag prior to swarming initiation was relatively constant.
TABLE 2.
Growth rates and swarm ratesa
| Medium | Temp (°C) | Swarm rate (mm/h) | Growth rate (generations/h) | Reduction inb: |
|
|---|---|---|---|---|---|
| Swarm rate | Growth rate | ||||
| LB | 37 | 15 | 3.5 | 1 | 1 |
| LB | 30 | 6 | 1.4 | 2.5 | 2.5 |
| B | 37 | 3 | 0.8 | 5 | 5 |
Strain 3610 was used to generate all data.
Relative to cells cultured in LB at 37°C (standard conditions).
Ultimately we were unable to reproduce swarming in laboratory strains, and we reassert that laboratory strains are defective for swarming-motility. It is difficult to explain reports of swarming-proficient laboratory strains, because these cells are defective for both surfactin and swrA. Thus, the apparent swarming of strain 168 must be due to poorly reproducible environmental factors and/or selection for genetic revertants.
Testing genes reported to be required for swarming.
Laboratory strains have been used to identify new genetic determinants for swarming (5, 10, 13). We hypothesize that studying swarming motility in laboratory strains that do not swarm gives rise to false identification of swarming genes. Here we reinvestigated the swarming behavior of various mutants of the robust swarming strain 3610 under our standard conditions. The mutations in 3610 were confirmed by PCR product length polymorphism (see Fig. S4 in the supplemental material).
YvzB is a putative protein that would be 76% identical to the C-terminal region of the Hag flagellin protein and was reported to be necessary for swarming in laboratory strains (13). However, we found that a yvzB mutant swarmed like wild-type strain 3610 (Fig. 1D). Furthermore, it is difficult to identify a ribosome binding site and translational start site for the yvzB gene, there is no published evidence that yvzB is translated, and yvzB may, in fact, be a pseudogene. Regardless, we conclude that yvzB is not required for swarming motility.
Epr is a minor extracellular protease reportedly required for swarming motility in laboratory strains (10, 29). It has been proposed that Epr has a C-terminal domain which, when proteolytically cleaved, acts as an extracellular signal that promotes swarming behavior (12, 24). Mutation of epr in 3610, however, resulted in no impairment of swarming on LB or B medium, consistent with other reports (6) (Fig. 1D). We conclude that Epr is not required for swarming motility.
Competence-stimulating factor (CSF) is a secreted pentapeptide, encoded by the phrC gene, which stimulates competence for DNA uptake and was reportedly required for swarming in laboratory strains (13, 30). CSF is imported by the oligopeptide permease that is encoded by oppD, and it antagonizes RapC, a phosphatase encoded by rapC that inhibits competence (8, 30). Strains of 3610 mutated for phrC, oppD, or rapC displayed swarming motility indistinguishable from that of the wild type (Fig. 1D). We conclude that CSF, RapC, and the oligopeptide permease are not required for swarming motility.
YvjD is encoded by the yvjD gene, located immediately downstream of swrA. Research in laboratory strains suggested that yvjD was required for swarming motility despite the fact that the parent was unable to spread across the surface of the plate (5, 27). Furthermore, because a reverse transcriptase PCR product spanning the swrA and yvjD genes was obtained, it was concluded that swrA and yvjD constituted an operon (6). To reflect the similar function and coregulation, swrA was renamed swrAA and yvjD was renamed swrAB (5). Recently, however, two groups have shown that yvjD is expressed separately from swrA and that YvjD is a topological determinant of cell division that restricts the MinCD cell division site selection machinery (4, 26). Consequently, the YvjD protein was found to be functionally unrelated to SwrA, and YvjD was renamed MinJ. We conclude that both reasons (similar function and obligate coexpression) for renaming SwrA SwrAA are invalid, and we suggest that the protein simply be referred to as SwrA, according to the original nomenclature (21). Furthermore, we note that use of the SwrAA nomenclature can create confusion with SrfAA (surfactin synthase) and has also indirectly caused another protein required for swarming motility, SwrB, to be confused with MinJ (SwrAB).
We conclude that the use of laboratory strains is not a reliable approach for identifying genes required for swarming motility. In comparison, genes identified in the 3610 strain as being required for swarming motility have reproducible and robust swarming effects. For example, cells mutated for swrB, encoding the membrane-associated protein of unknown function SwrB, were swarming defective under all conditions (21) (Fig. 1D). Likewise, cells mutated for degU, encoding the pleiotropic response regulator DegU, were swarming defective under all conditions (23, 33) (Fig. 1D). We recommend the use of either the ancestral strain 3610 or laboratory strains that have been genetically repaired for both surfactin biosynthesis (sfp) and swrA for the genetic analysis of swarming motility.
Pitfalls of the swarm assay.
Swarming motility is an important biological function that appears to be relatively easy to assay. Nonetheless, it is important to recognize the sensitivity of the assay and the selective pressure for swarming, and care must be taken in the preparation and execution of swarm assays to obtain reliable and reproducible results (see Fig. S5 in the supplemental material). Here we try to explain some of the conflicting results and discrepancies reported in the literature.
(i) Plate preparation and assay conditions.
Although increased surface wetness of the medium was insufficient to reproduce swarming in B. subtilis lab strains, water content plays a critical role in swarming. Cells inoculated under very soft or very wet conditions could be swimming through the water on the surface of the plate rather than exhibiting bona fide swarming, which requires cell-to-cell contact, surfactin production, and hyperflagellation (17). To complicate interpretations further, some publications internally compare and draw conclusions from the results of assays with multifactorial variation in parental strains, medium composition, incubation durations, incubation temperatures, inoculation concentrations, and growth stages of the inoculum. We suggest that modifying only one variable at a time is a good scientific practice that makes results easier to interpret and reproduce. We advise maintaining consistency in standard conditions and recommend treatments that minimize water on the agar surface.
(ii) Suppressor mutations.
Swarm assays on nonswarming strains that proceed for extended periods of time are likely to select for suppressor mutations. Rare gain-of-function point mutations that restore swarming motility may arise in as little as 48 h (4, 23). Loss-of-function missense mutations that restore swarming motility may arise in as little as 18 to 24 h (3, 28). Reversion of the swrA frameshift allele can occur in less than 12 h (21) (see Fig. S1 in the supplemental material). We note that in some studies, swarming assays were allowed to proceed for 36 h to 10 days (18, 19). Considering that B. subtilis has a doubling time of ∼20 min in rich media at 37°C and that swarming motility, once initiated, will cause cells to cover an entire petri plate in as little as 2 h, prolonged incubation seems unwise unless suppressors are the goal.
Suppressors manifest as asymmetric flares that arise from a central colony, and flares can be clearly seen in some publications (see Fig. S1 in the supplemental material) (10, 12). We are concerned that the dendritic patterning on some media, such as B medium, may resemble and obscure genetic suppressors. Indeed, it was recently claimed that swrA was not required for swarming in laboratory strains that had been complemented for sfp alone (13). We noted, however, that 168 complemented for sfp alone swarmed later than 3610, and the appearance of the dendrites was sporadic, consistent with a suppressor mutation. We isolated cells from the dendrites of a 168 sfp+ strain and found that the swrA gene had indeed reverted to the functional allele (see Fig. S1 in the supplemental material). We refute claims that swrA is not required for swarming motility. We recommend the use of quantitative swarm expansion assays that track the first 8 h after inoculation to give robust kinetics of behavior prior to the possible arrival of confounding secondary mutations. We note that genetic suppressors should be of concern not only for the study of B. subtilis but for the study of other swarming bacteria as well.
(iii) Genetic linkage.
As the 3610 ancestral strain is poorly competent, genetic constructs are typically first integrated into a laboratory strain and then transduced into 3610. The generalized transducing phage SPP1 packages ∼40 kb of DNA, and linkage of polymorphisms is always a concern given this strategy (32). Swarming motility is particularly susceptible to linkage effects, as the swrA polymorphism of laboratory strains is in the vicinity of a variety of other motility-related genes. For example, the yvzB gene is linked to the swrA gene by 12 kb on the B. subtilis chromosome. Thus, a swrA mutation can be cotransduced (or cotransformed) with a yvzB allele to give the false impression that yvzB is required for swarming motility. Indeed, the frameshift mutation in swrA was first mapped by linkage to an antibiotic cassette inserted within yvzB (22). One must take care to ensure that phenotypes are 100% linked to the construct of interest. We note that the problem of linkage is not specific to the 3610 strain and can complicate any genetic cross in which one parent is swrA+ and one parent is swrA.
(iv) Auxotrophy.
Auxotrophic mutants require nutritional supplements to the medium in order to grow. In undefined rich media, such as LB, auxotrophies are masked by the complex mixture of nutrients. B medium, on the other hand, is a defined medium and is susceptible to auxotrophy. A classic example of auxotrophy is the trpC2 mutation in 168 strains, the growth of which requires the addition of tryptophan to B medium. Auxotrophy may account for the lack of swarming motility reported in a number of mutants on B medium (13). A 168 laboratory strain derivative was used that had an sfp+ complementation construct integrated into the ectopic thrC gene to restore surfactin production (13). Consequently, a threonine auxotrophy was generated, and there is no report of addition of threonine to the B medium used to measure swarming motility (13). Thus, one reason the mutants tested in this genetic background may have appeared to be nonswarming on B medium was that they could not grow. We recommend the use of complex media in swarming assays to minimize the impact of auxotrophy.
Conflicting reports of genes required for swarming motility could be caused by any one, or a combination, of the pitfalls explained above. Nonetheless, we conclude that SwrA is essential for swarming motility and that laboratory strains of B. subtilis do not swarm.
Supplementary Material
Acknowledgments
We are grateful to Simone Séror for sharing her laboratory strain and genetic constructs. We thank Kris Blair, Rui Chen, Loralyn Cozy, and Yi-huang Hsueh for generating constructs.
This work was supported by NSF grant MCB-0721187 to D.B.K.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alberti, L., and R. Harshey. 1990. Differentiation of Serratia marcescens 274 into swimmer and swarmer cells. J. Bacteriol. 172:4322-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., S. Engelmann, R. Schmid, A. Sorokin, A. Lapidus, and M. Hecker. 1997. Expression of a stress- and starvation-induced dps/pexB-homologous gene is controlled by the alternative sigma factor σB in Bacillus subtilis. J. Bacteriol. 179:7251-7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair, K. M., L. Turner, J. T. Winkelman, H. C. Berg, and D. B. Kearns. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636-1638. [DOI] [PubMed] [Google Scholar]
- 4.Bramkamp, M., R. Emmins, L. Weston, C. Donovan, R. A. Daniel, and J. Errington. 2008. A novel component of the division site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70:1556-1569. [DOI] [PubMed] [Google Scholar]
- 5.Calvio, C., F. Celandroni, E. Ghelardi, G. Amati, S. Salvetti, F. Ceciliani, A. Galizzi, and S. Senesi. 2005. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J. Bacteriol. 187:5356-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connelly, M. B., G. M. Young, and A. Sloma. 2004. Extracellular proteolytic activity plays a central role in swarming motility in Bacillus subtilis. J. Bacteriol. 186:4159-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copeland, M. F., and D. B. Weibel. 2009. Bacterial swarming: a model system for studying dynamic self-assembly. Soft Matter 5:1174-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core, L., and M. Perego. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 49:1509-1522. [DOI] [PubMed] [Google Scholar]
- 9.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and D. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821-831. [DOI] [PubMed] [Google Scholar]
- 10.Dixit, M., C. S. Murudkar, and K. K. Rao. 2002. epr is transcribed from a σD promoter and is involved in swarming of Bacillus subtilis. J. Bacteriol. 184:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fall, R., D. B. Kearns, and T. Nguyen. 2006. A defined medium to investigate sliding motility in a Bacillus subtilis flagella-less mutant. BMC Microbiol. 6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta, M., and K. K. Rao. 2009. Epr plays a key role in DegU-mediated swarming motility of Bacillus subtilis. FEMS Microbiol. Lett. 295:187-194. [DOI] [PubMed] [Google Scholar]
- 13.Hamze, K., D. Julkowska, S. Autret, K. Hinc, K. Nagorska, A. Sekowska, I. B. Holland, and S. J. Séror. 2009. Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC. Microbiol. 155:398-412. [DOI] [PubMed] [Google Scholar]
- 14.Harshey, R. M. 1994. Bees aren't the only ones: swarming in gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 15.Harshey, R. M., and T. Matsuyama. 1994. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc. Natl. Acad. Sci. USA 91:8631-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauser, G. 1885. Über faül nissbacterien und derenbeziehungen zur septicämie. F.G.W. Vogel, Leipzig, Germany.
- 17.Henrichsen, J. 1972. Bacterial surface translocation: a survey and a classification. Bacteriol. Rev. 36:478-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julkowska, D., M. Obuchowski, I. B. Holland, and S. J. Séror. 2004. Branched swarming patterns on a synthetic medium formed by wild-type Bacillus subtilis strain 3610: detection of different cellular morphologies and constellations of cells as the complex architecture develops. Microbiology 150:1839-1849. [DOI] [PubMed] [Google Scholar]
- 19.Julkowska, D., M. Obuchowski, I. B. Holland, and S. J. Séror. 2005. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: critical effects of surfactin and the composition of the medium. J. Bacteriol. 187:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 21.Kearns, D. B., F. Chu, R. Runder, and R. Losick. 2004. Genes governing swarming in Bacillus subtilis, and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 52:357-369. [DOI] [PubMed] [Google Scholar]
- 22.Kearns, D. B., and R. Losick. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66:395-409. [DOI] [PubMed] [Google Scholar]
- 24.Murudkar, C. S., P. Kodgire, and K. K. Rao. 2006. The carboxy terminal domain of Epr, a minor extracellular serine protease, is essential for the swarming motility of Bacillus subtilis 168. FEMS Microbiol. Lett. 257:24-31. [DOI] [PubMed] [Google Scholar]
- 25.Nakano, M. M., M. A. Marahiel, and P. Zuber. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patrick, J. E., and D. B. Kearns. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70:1166-1179. [DOI] [PubMed] [Google Scholar]
- 27.Senesi, S., E. Ghelardi, F. Celandroni, S. Salvetti, E. Parisio, and A. Galizzi. 2004. Surface-associated flagellum formation and swarming differentiation in Bacillus subtilis are controlled by the ifm locus. J. Bacteriol. 186:1158-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shinoda, S., and K. Okamoto. 1977. Formation and function of Vibrio parahaemolyticus lateral flagella. J. Bacteriol. 129:1266-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloma, A., A. Ally, D. Ally, and J. Pero. 1988. Gene encoding a minor extracellular protease in Bacillus subtilis. J. Bacteriol. 170:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solomon, J. M., B. A. Lazazzera, and A. D. Grossman. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 10:2014-2024. [DOI] [PubMed] [Google Scholar]
- 31.Stanley-Wall, N. R., and B. A. Lazazzera. 2005. Defining genetic differences between wild and domestic strains of Bacillus subtilis that affects poly-γ-DL-glutamic acid production and biofilm formation. Mol. Microbiol. 57:1143-1158. [DOI] [PubMed] [Google Scholar]
- 32.Tavares, P., M. A. Santos, R. Lurz, G. Morelli, H. de Lencastre, and T. A. Trautner. 1992. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 255:81-92. [DOI] [PubMed] [Google Scholar]
- 33.Verhamme, D. T., T. B. Kiley, and N. R. Stanley-Wall. 2007. DegU co-ordinates multicellular behavior exhibited by Bacillus subtilis. Mol. Microbiol. 65:554-568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.