Abstract
Porphyromonas gingivalis is present in dental plaque as early as 4 h after tooth cleaning, but it is also associated with periodontal disease, a late-developing event in the microbial successions that characterize daily plaque development. We report here that P. gingivalis ATCC 33277 is remarkable in its ability to interact with a variety of initial, early, middle, and late colonizers growing solely on saliva. Integration of P. gingivalis into multispecies communities was investigated by using two in vitro biofilm models. In flow cells, bacterial growth was quantified using fluorescently conjugated antibodies against each species, and static biofilm growth on saliva-submerged polystyrene pegs was analyzed by quantitative real-time PCR using species-specific primers. P. gingivalis could not grow as a single species or together with initial colonizer Streptococcus oralis but showed mutualistic growth when paired with two other initial colonizers, Streptococcus gordonii and Actinomyces oris, as well as with Veillonella sp. (early colonizer), Fusobacterium nucleatum (middle colonizer), and Aggregatibacter actinomycetemcomitans (late colonizer). In three-species flow cells, P. gingivalis grew with Veillonella sp. and A. actinomycetemcomitans but not with S. oralis and A. actinomycetemcomitans. Also, it grew with Veillonella sp. and F. nucleatum but not with S. oralis and F. nucleatum, indicating that P. gingivalis and S. oralis are not compatible. However, P. gingivalis grew in combination with S. gordonii and S. oralis, demonstrating its ability to overcome the incompatibility when cultured with a second initially colonizing species. Collectively, these data help explain the observed presence of P. gingivalis at all stages of dental plaque development.
Removal of dental plaque by routine oral hygiene procedures is followed by a repetition of a species succession that starts with initially colonizing streptococci and actinomyces (5, 16). Other species follow as early, middle, and late colonizers, which establishes the following developmental process: successive attachment of saliva-suspended species to already attached bacteria and formation of multispecies communities.
Attachment is a critical event essential to preventing the bacteria from being swallowed by salivary flow. Initial colonizers bind to host-derived receptors in the salivary pellicle coating of the tooth enamel. The remainder of typical plaque development occurs by accretion of saliva-suspended species and growth of attached bacteria, thereby increasing the microbial diversity. Adherence of suspended single cells to attached cells is called coadhesion (1). Some suspended cells are already coaggregated and adhere to attached cells as coaggregates; coaggregation is defined as the specific cell-to-cell recognition and adherence of genetically distinct cell types (8). All human oral bacterial species exhibit coaggregation. For example, Streptococcus oralis coaggregates with Streptococcus gordonii (intrageneric coaggregation). Both species pair with Actinomyces oris (intergeneric coaggregation), and all of them coaggregate with Fusobacterium nucleatum (multigeneric coaggregation). Multispecies communities composed of coaggregating species characterize dental plaque biofilms in vivo (3, 17, 18).
To increase our understanding of interactions among species, we have employed two in vitro model systems and are testing numerous combinations of seven species for their ability to grow on saliva as their sole nutritional source (20, 21). First, we reported that F. nucleatum (middle colonizer) failed to grow when paired with S. oralis but grew well when A. oris was included in the three-species biofilm (20), indicating specificity by F. nucleatum for the presence of a particular initial colonizer. Recently, we showed that Aggregatibacter actinomycetemcomitans (late colonizer and periodontopathogen) exhibited mutualistic relationships with F. nucleatum and Veillonella sp. (early colonizer and commensal organism), illustrating the ability of commensals and pathogens to grow together (21).
Porphyromonas gingivalis, another periodontopathogen, forms three-species communities with F. nucleatum and S. gordonii (11). Proteomics of P. gingivalis in this three-species community revealed a broad increase in proteins involved in protein synthesis, suggesting that a multispecies relationship is advantageous for the porphyromonad (11). This research group had previously reported the presence of differentially regulated porphyromonad genes when P. gingivalis and S. gordonii were together in biofilms (22). Thus, P. gingivalis responds to the presence of other oral species.
P. gingivalis is detected in dental plaque samples within 6 h after professional tooth cleaning (5, 13), and its numbers increase in periodontally diseased sites (15). It forms biofilms with S. gordonii but not with Streptococcus mutans (12) or Streptococcus cristatus (23). P. gingivalis required a preformed streptococcal substratum for its incorporation into a biofilm (12). Partner specificity was also noted among four fresh isolates of P. gingivalis, which showed no coaggregation with a variety of oral actinomyces, aggregatibacteria, capnocytophagae, and streptococci (9) but coaggregated with F. nucleatum (7, 10). We show here that P. gingivalis exhibits widespread mutualism with initial, early, middle, and late colonizers but also shows specificity with initially colonizing streptococci, which could help explain its early appearance in the development of dental plaque biofilms. The relationship of porphyromonads with initial, early, middle, and later colonizers during biofilm growth on saliva as a sole nutritional source has not been explored previously. We hypothesize that the ability of P. gingivalis to coaggregate with S. gordonii and A. oris (initial colonizers), Veillonella sp. (early colonizer), F. nucleatum (middle colonizer), and A. actinomycetemcomitans (late colonizer) allows these bacteria to form multispecies biofilm communities.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. oralis 34, S. gordonii DL1, and A. oris ATCC 43146 were routinely cultured in Todd-Hewitt broth (THB; Difco Laboratories, Detroit, MI) or on THB agar. Veillonella sp. PK1910 was grown in THB supplemented with 0.6% lactic acid. A. actinomycetemcomitans JP2 (kind gift from D. Demuth, University of Louisville, Louisville, KY) was grown in brain heart infusion medium (BHI; Difco) supplemented with 0.04% sodium bicarbonate and 2% yeast extract. F. nucleatum ATCC 10953 was grown in BHI supplemented with 0.25% l-glutamic acid. P. gingivalis ATCC 33277 (kind gift from R. J. Lamont, University of Florida, Gainesville, FL) was grown in BHI supplemented with 0.1% yeast extract, 0.5% hemin, and 0.001% menadione. All species were grown in a Bactron anaerobic (N2:CO2:H2, 90:5:5) environmental chamber (Sheldon Manufacturing Inc., Cornelius, OR) at 37°C.
Saliva preparation.
Saliva from 6 to 10 healthy individuals was collected on ice, pooled on ice, and treated with 2.5 mM dithiothreitol for 10 min, with stirring to reduce salivary protein aggregation. The saliva was then centrifuged and processed as previously described (19). Briefly, the supernatant was diluted with distilled water to produce 25% saliva and then was filtered through a 0.22-μm-pore-size SFCA low-protein-binding filter (Nalge Nunc International, Rochester, NY) and stored at −20°C. Prior to use, saliva was thawed and centrifuged to remove any precipitate that resulted from freezing and thawing.
Flow cell preparation.
Two tracks (each track was 40 mm long, 3 mm wide, and 2 mm deep) were milled into a high-density polyethylene block, resulting in two chambers, each with a 240-μl volume. A glass coverslip, which serves as the attachment substratum for the growing biofilm, was secured to the reusable flow cells with a silicone adhesive. The flow cells were cleaned overnight with 0.1 M HCl, rinsed with 5 ml of distilled water, injected with 70% ethanol, and incubated 20 min. The flow cells were then treated with 25% sterile human saliva for 15 min at 37°C in an anaerobic chamber to condition the glass surface with salivary components.
Biofilm growth conditions in flow cells.
Overnight bacterial cultures were harvested by centrifugation and washed twice with 25% sterile human saliva, and the optical density at 600 nm was adjusted to 0.1, which was equivalent to about 1 × 107 to 3 × 107 cells/ml. Flow cells were inoculated with one, two, or three species. Two-species and three-species inoculations were first coaggregated (by mixing 0.1-ml portions of appropriate combinations of P. gingivalis, S. oralis, S. gordonii, A. oris, Veillonella sp., F. nucleatum, and A. actinomycetemcomitans [equivalent to about 1 × 106 to 3 × 106 cells of each species]), and the flow cells were incubated in the anaerobic chamber to provide an environment favorable to P. gingivalis, Veillonella sp., and F. nucleatum, which are strict anaerobes. The sole nutritional source was sterile 25% saliva supplied at a flow rate of 0.2 ml/min, which approximated unstimulated salivary flow in the mouth (4).
Biofilm staining.
Staining of cells varied in the different experiments. When strains were grown as monocultures, they were visualized by primary immunofluorescence with Alexa Fluor 546- (Invitrogen), Alexa Fluor 488- (Invitrogen), Alexa Fluor 546- (Invitrogen), Alexa Fluor 488- (Invitrogen), Alexa Fluor 633- (Invitrogen), Alexa Fluor 488- (Invitrogen), and Alexa Fluor 546- (Invitrogen) conjugated immunoglobulin G of a polyclonal antiserum to P. gingivalis, S. gordonii, Veillonella sp., A. actinomycetemcomitans, S. oralis, A. oris, and F. nucleatum, respectively. When multiple species were inoculated, visualization of species was by primary immunofluorescence with Alexa Fluor 546-, Alexa Fluor 633-, Alexa Fluor 488-, Alexa Fluor 488-, Alexa Fluor 633-, Alexa Fluor 488-, and Alexa Fluor 488-conjugated immunoglobulin G of a polyclonal antiserum to P. gingivalis, S. oralis, S. gordonii, A. oris, Veillonella sp., F. nucleatum, and A. actinomycetemcomitans, respectively. Immunofluorescence was performed by injecting the antibody (5 μg/ml in filter-sterilized phosphate-buffered saline containing 1% [wt/vol] bovine serum albumin) into the appropriate flow cell track and incubating for 20 min. A final wash with phosphate-buffered saline preceded confocal laser scanning microscopy. No cross-reactivity with nonhomologous strains was observed, based on staining by antibodies.
Image and statistical analysis.
A TCS-SP2 confocal microscope (Leica, Exton, PA) with a 40×, 1.25-numerical aperture oil immersion lens was used to record confocal image stacks in five random locations near the center of the flow cells, after which biofilm biovolumes were determined by volumetric analyses (IMARIS version 5.71; Bitplane AG, Zurich, Switzerland). Fluorescence intensity thresholds were set manually for red, green, and blue pixels with cubic voxels used for the biovolume determination. Five confocal datasets were analyzed for each time point, and the mean and standard deviations were calculated. A one-way analysis of variance at the 95% confidence level with a nonparametric Tukey's pairwise comparison test was used to determine if means were statistically different between the 4-h and 18-h time points for each condition (one, two, and three species) in the flow cell studies and between the 12-h, 24-h, and 36-h time points for each condition (one, two, and three species) in the peg studies with static saliva. All images presented in the figures are maximum projections of the entire confocal image stack as produced by the Leica TCS software (Leica).
Biofilm growth conditions on polystyrene pegs (peg biofilms).
Biofilms were grown in 25% human saliva on pegs (Nunc-Immuno transferable solid phase; Nunc 445497) (2, 14) mounted in U96 microwell plates (Nunc 163320) (2, 14, 20). Microtiter plate wells were filled with 200 μl of 25% human saliva, and pegs were then inserted for 30 min at room temperature to build a conditioning film. Overnight cultures (∼20 μl) of P. gingivalis ATCC 33277, S. oralis 34, Veillonella sp. PK1910, A. actinomycetemcomitans JP2, and F. nucleatum ATCC 10953 were added to the 200 μl of saliva in the wells; this equals about 1 × 107 to 3 × 107 cells of each species in the microtiter well; plates were placed in a humidity chamber and incubated anaerobically at 37°C for 12, 24, or 36 h. Once the wells were inoculated and the pegs placed in the saliva, the pegs were not transferred and thus remained submerged in the same saliva for 12, 24, or 36 h.
DNA extraction and quantification.
DNA was extracted from biofilms by a modified alkaline lysis protocol (3, 6, 20). Biofilm-covered pegs were immersed in 40 μl of sterile ultrapure water plus 160 μl of 0.05 M sodium hydroxide incubated at 60°C for 60 min, after which 18.4 μl of 1 M Tris-HCl (pH 7.0) was added to neutralize the pH. This extract was used as the template DNA for the quantitative real-time PCR (q-PCR) analyses (3, 6, 20). Bacterial genomic DNA for standard curves was extracted from overnight cultures of P. gingivalis ATCC 33277, A. actinomycetemcomitans JP2, Veillonella sp. PK1910, and F. nucleatum ATCC 10953 with a DNA extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Genomic DNA was stored at −20°C.
q-PCR quantification of species in biofilms.
Species-specific primers were designed with the Primer 3 program (http://www.justbio.com). No cross-hybridization of primers with nonhomologous species was observed. The primers specific for P. gingivalis were F, TGGGTTTAAAGGGTGCGTAG, and R, CAATCGGAGTTCCTCGTGAT; the annealing temperature was 60°C. The primers specific for A. actinomycetemcomitans JP2 were F, GGACGGGTGAGTAATGCTTG, and R, CCCTTACCCCACCAACTACC; the annealing temperature was 60°C. The primers specific for Veillonella sp. PK1910 were F, CCGTGATGGGATGGAAACTGC, and R, CCTTCGCCACTGGTGTTCTTC; the annealing temperature was 60°C. The primers specific for F. nucleatum ATCC 10953 were F, CTTAGGAATGAGACAGAGATG, and R, TGATGGTAACATACGAAAGG; the annealing temperature was 56°C. Quantification of P. gingivalis, A. actinomycetemcomitans, and F. nucleatum cells in the biofilms was performed with q-PCR using the SYBR green dye to detect the 16S rRNA gene amplicons. Each reaction (final reaction volume, 20 μl) contained 3 μl of template, 1.0 μl of diethyl pyrocarbonate-treated ultra pure water, 10 μl of fast SYBR green master mix (Applied Biosystems, Foster City, CA), and 3 μl each of forward and reverse primers at 375 nM. q-PCR was performed in an MX3005P thermocycler (Stratagene, La Jolla, CA) using the following thermocycle recommended for fast SYBR green master mix: 95°C for 20 s, 40 cycles of 3 s at 95°C, and 30 s at 60°C. Dissociation curves were generated by incubating reaction products at 95°C for 1 min and 56°C for 30 s and incrementally increasing the temperature to 95°C. Fluorescence data were collected at the end of the 60°C primer annealing step for 40 amplification cycles and throughout the dissociation curve analysis. The analysis of the melting curves with both primer sets showed a single sharp peak. DNA concentrations (ng/ml) were calculated based on standard curves obtained by using 10-fold serial dilutions of bacterial DNA isolated with a DNA extraction kit (Qiagen) and quantified with a PicoGreen fluorescence assay (Invitrogen). To convert ng DNA to cell numbers, the following weights and genome sizes were used: 2.34 fg/genome and 2.3 Mb for porphyromonads, 2.05 fg/genome and 2.1 Mb for aggregatibacteria, 3.05 fg/genome and 3.0 Mb for veillonellae, and 2.41 fg/genome and 2.4 Mb for fusobacteria (http://www.homd.org). The data presented are from three independent biofilms.
RESULTS
Flow cells inoculated with one species.
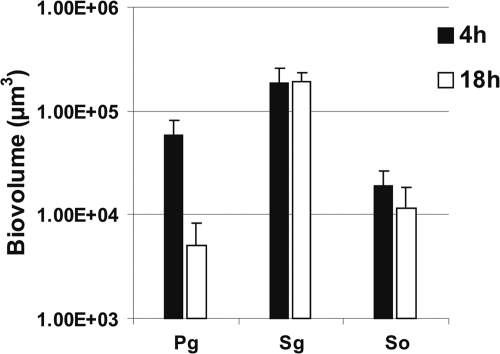
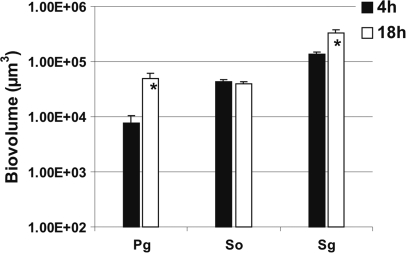
P. gingivalis, S. gordonii, and S. oralis were inoculated into separate flow cells. Images of the biofilms formed were obtained after 4 h and 18 h of incubation and used to quantify biofilm cells as biovolume (Fig. 1). Each species showed attachment to the saliva-conditioned surface at 4 h. After 18 h of incubation, the number of P. gingivalis cells decreased 10-fold, whereas S. gordonii and S. oralis cells remained nearly constant, indicating their retention but no obvious growth. We reported earlier that Veillonella sp. and A. actinomycetemcomitans also did not grow as single species on saliva (21) and that F. nucleatum monospecies cell numbers decreased (20). Only A. oris grew consistently as a single species on saliva (20).
FIG. 1.
Time-resolved change in biovolume (μm3 per field of view) of P. gingivalis (Pg), S. gordonii (Sg), and S. oralis (So) following 4 h and 18 h of incubation as single species in 25% saliva-fed flow cells.
Flow cells inoculated with two species.
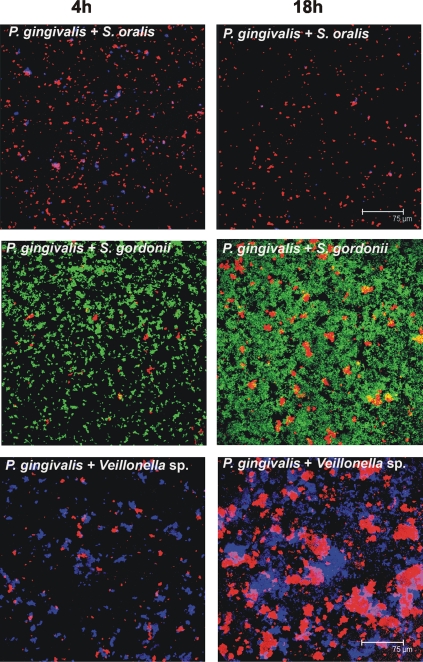
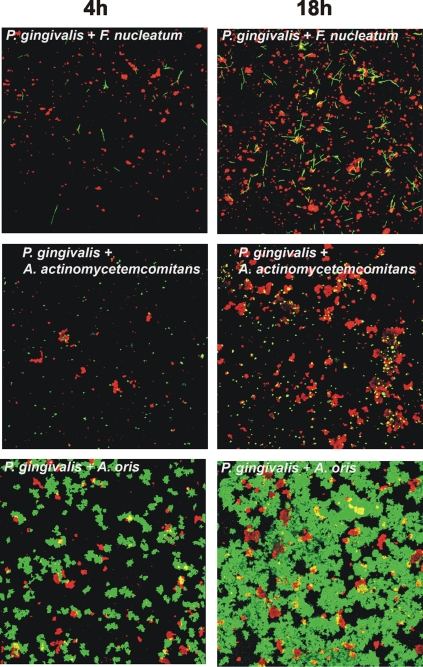
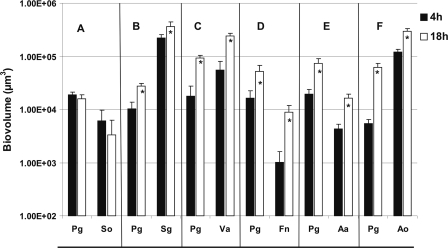
To investigate the possibility of mutualism between P. gingivalis and the other six species, each of the six pairwise mixtures of P. gingivalis and a coaggregation partner were inoculated into a saliva-fed flow cell. The biofilms formed were viewed by confocal laser scanning microscopy (Fig. 2 and 3), and the biovolumes of each species were quantified (Fig. 4). Neither species of the S. oralis-P. gingivalis pair showed growth (Fig. 4A and 2, top panels). In sharp contrast, mutualistic growth was seen with P. gingivalis and partners S. gordonii (Fig. 4B and 2, middle panels), Veillonella sp. (Fig. 4C and 2, bottom panels), F. nucleatum (Fig. 4D and 3, top panels), and A. actinomycetemcomitans (Fig. 4E and 3, middle panels), and the P. gingivalis-A. oris pair grew commensally (Fig. 4F and 3, bottom panels). These results document the favorable interaction of P. gingivalis with only certain initial colonizers (S. gordonii and A. oris) and show that P. gingivalis exhibits a broad range of favorable interactions with early, middle, and late colonizers as well.
FIG. 2.
Representative confocal micrographs of 4-h (left column) and 18-h (right column) biofilms showing no growth (top panels, P. gingivalis + S. oralis) and mutualistic growth (middle panels, P. gingivalis + S. gordonii; bottom panels, P. gingivalis + Veillonella sp.) in flow cells inoculated with two species. Bacterial cells were stained with species-specific fluorophore-conjugated immunoglobulin G (red, P. gingivalis; blue, S. oralis/Veillonella sp., green, S. gordonii), and cell-cell contact between species is evident.
FIG. 3.
Representative confocal micrographs of 4-h (left column) and 18-h (right column) biofilms showing mutualistic growth (top panels, P. gingivalis + F. nucleatum; middle panels, P. gingivalis + A. actinomycetemcomitans) and commensal growth (P. gingivalis + A. oris) in flow cells inoculated with two species. Bacterial cells were stained with species-specific fluorophore-conjugated immunoglobulin G (red, P. gingivalis; green, F. nucleatum/A. actinomycetemcomitans/A. oris), and cell-cell contact between species is evident.
FIG. 4.
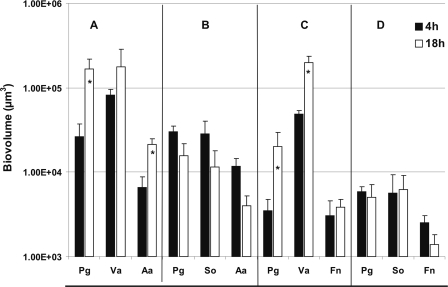
Time-resolved changes in biovolumes (μm3 per field of view) of two-species flow cells inoculated with coaggregates of P. gingivalis (Pg) and S. oralis (So) (A), S. gordonii (Sg) (B), Veillonella sp. (Va) (C), F. nucleatum (Fn) (D), A. actinomycetemcomitans (Aa) (E), or A. oris (Ao) (F). An asterisk indicates statistically significant increases (P < 0.05) in bacterial growth.
Flow cells inoculated with three species.
The inability of P. gingivalis to grow with S. oralis was explored further by including a third species and examining the formation of multispecies communities in saliva-fed flow cells. Also, substituting Veillonella sp. for S. oralis in the triple-species biofilms was investigated. The biovolume of P. gingivalis increased sixfold between 4 h and 18 h of incubation, when P. gingivalis was inoculated as a coaggregate with Veillonella sp. and A. actinomycetemcomitans; Veillonella sp. biovolume increased twofold and A. actinomycetemcomitans biovolume increased threefold, indicating a productive interaction in this triple-species community (Fig. 5 and 6A). In contrast, substituting S. oralis for Veillonella sp. inhibited growth of all species, although initial binding to the saliva-coated substratum was excellent (Fig. 6B, 4-h time point); reduced numbers of the three species were seen at 18 h (Fig. 6B). The same trend was observed in triple-species inocula containing P. gingivalis, Veillonella sp., and F. nucleatum (Fig. 6C) versus P. gingivalis, S. oralis, and F. nucleatum (Fig. 6D). The biovolumes of porphyromonads, veillonellae, and fusobacteria increased (Fig. 6C), but the biovolumes of porphyromonads, streptococci, and fusobacteria did not (Fig. 6D), indicating that Veillonella sp. is a favorable partner for later colonizers, whereas S. oralis is not. Moreover, S. oralis appears to hinder growth of all four gram-negative species.
FIG. 5.
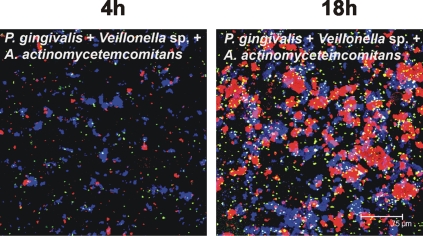
Representative confocal micrographs of biofilm from three-species-inoculated flow cell. Communities following 4 h and at 18 h of incubation show intimate interaction of Veillonella sp. (blue) with A. actinomycetemcomitans (green) and P. gingivalis (red) to form mutualistic multispecies communities.
FIG. 6.
Time-resolved changes in biovolumes (μm3 per field of view) of three-species-inoculated flow cell following 4-h (black bar) and 18-h (open bar) incubation showing growth enhanced by Veillonella sp. and growth inhibited by S. oralis. (A) P. gingivalis (Pg) + Veillonella sp. (Va) + A. actinomycetemcomitans (Aa); (B) P. gingivalis (Pg) + S. oralis (So) + A. actinomycetemcomitans (Aa); (C) P. gingivalis (Pg) + Veillonella sp. (Va) + F. nucleatum (Fn); and (D) P. gingivalis (Pg) + S. oralis (So) + F. nucleatum (Fn). An asterisk indicates statistically significant increases (P < 0.05) in bacterial growth.
The specificity of P. gingivalis for growth with S. gordonii but not S. oralis was investigated by inoculating the three species into a flow cell. All species attached well to the saliva-coated substratum; P. gingivalis biovolume increased sixfold, and S. gordonii biovolume increased twofold, but S. oralis failed to grow between 4 h and 18 h (Fig. 7). Thus, P. gingivalis is capable of significant growth with a suitable initial colonizer (S. gordonii), even in the presence of an unfavorable initial colonizer (S. oralis).
FIG. 7.
Time-resolved changes in biovolume (μm3 per field of view) of three-species-inoculated flow cell showing specificity by P. gingivalis (Pg) for S. gordonii (Sg) in the presence of S. oralis (So) following 4-h (black bar) and 18-h (open bar) incubation. An asterisk indicates statistically significant increases (P < 0.05) in bacterial growth.
q-PCR quantification of species in static biofilms on pegs immersed in saliva.
Multispecies biofilm formation in the open, saliva-fed flowing system was compared with biofilms formed in the closed solid-phase polystyrene peg system; pegs remained submerged in the same saliva for 12, 24, or 36 h. When P. gingivalis was inoculated as a single species, its cell numbers did not increase significantly (Fig. 8A), and we reported previously that monospecies Veillonella sp. and F. nucleatum do not grow in static culture; A. actinomycetemcomitans shows significant growth only after 36 h of incubation (21). In combination with Veillonella sp., P. gingivalis increased fivefold between 12 h and 24 h and decreased slightly between 24 h and 36 h (Fig. 8B). Cell numbers of Veillonella sp. increased eightfold between 12 and 24 h. With F. nucleatum, P. gingivalis increased 5-fold between 12 h and 36 h, and F. nucleatum cell numbers increased 22-fold between 12 h and 24 h and another 2-fold by 36 h (Fig. 8C). With A. actinomycetemcomitans, P. gingivalis increased 10-fold between 12 h and 36 h, and A. actinomycetemcomitans increased 5-fold (Fig. 8D). Collectively, these results demonstrate mutualism between P. gingivalis and these three gram-negative partners.
FIG. 8.
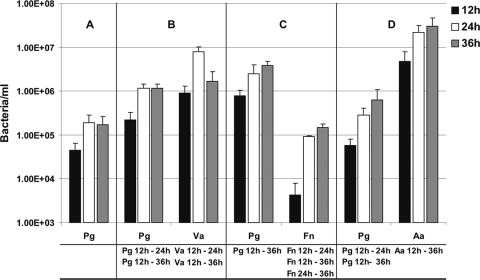
q-PCR quantification of P. gingivalis cells alone in biofilms and with coaggregation partners in two-species-inoculated biofilms grown on polystyrene pegs submerged in 25% saliva and incubated anaerobically for 12, 24, or 36 h. (A) Single species, P. gingivalis; (B) two species, P. gingivalis (Pg) + Veillonella sp. (Va); (C) two species, P. gingivalis (Pg) + F. nucleatum (Fn); and (D) two species, P. gingivalis (Pg) + A. actinomycetemcomitans (Aa). Statistically significant increases (P < 0.05) in bacterial growth on the pegs is indicated at the bottom of each panel; for example in panel B, “Pg 12 h - 24 h” indicates that significant growth of P. gingivalis occurred between 12 h and 24 h in the two-species biofilm with Veillonella sp.
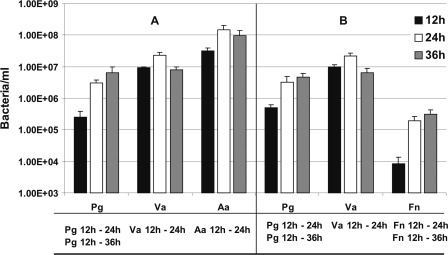
In triple-species combination with Veillonella sp. and A. actinomycetemcomitans, P. gingivalis increased 12-fold between 12 h and 24 h and an additional 2-fold between 24 h and 36 h, indicating a dramatic and flourishing interaction with these partners (Fig. 9A). Veillonella sp. increased threefold, and A. actinomycetemcomitans increased fivefold, indicating a mutualistic three-species relationship. A mutualistic relationship was also discovered by replacing A. actinomycetemcomitans with F. nucleatum: P. gingivalis numbers increased 7-fold between 12 h and 24 h (Fig. 9B); Veillonella sp. increased 2-fold, but F. nucleatum cell numbers jumped 22-fold between 12 h and 24 h and an additional 2-fold in the next 12 h. Thus, P. gingivalis is not simply a late-colonizing periodontopathogen, but rather it forms mutualistic biofilm communities with a variety of oral species spanning initial, early, middle, and late colonizers of enamel surfaces.
FIG. 9.
q-PCR quantification of three-species-inoculated biofilms grown on polystyrene pegs submerged in 25% saliva and incubated anaerobically for 12, 24, or 36 h. Pg, P. gingivalis; Va, Veillonella sp.; Aa, A. actinomycetemcomitans; and Fn, F. nucleatum. Statistically significant increases (P < 0.05) in bacterial growth on the pegs is indicated at the bottom of each panel; for example in panel A, “Pg 12 h - 24 h” indicates that significant growth of P. gingivalis occurred between 12 h and 24 h in the three-species inoculated biofilm with Veillonella sp. and A. actinomycetemcomitans.
DISCUSSION
Coupling the reported observations that P. gingivalis is present in dental plaque samples taken in the first 6 h after oral hygiene treatment (5, 13) and our results here, we clearly determined that the periodontopathogen P. gingivalis is capable of being an integral component of mutualistic multispecies oral biofilm communities at all stages of dental plaque development. We show that P. gingivalis adheres to saliva-coated glass and polystyrene but cannot grow without other species. When combined with a variety of initial (S. gordonii and A. oris), early (Veillonella sp.), middle (F. nucleatum), and late (A. actinomycetemcomitans) colonizers, P. gingivalis is an active member of these mutualistic communities.
Specificity by P. gingivalis for coadherence with certain streptococci was reported by Lamont and coworkers (12) and Xie et al. (23). Biofilms were formed when S. gordonii was the partner but were not formed with either S. mutans or S. cristatus. Our results agree with and extend those observations. Although it grows mutualistically with S. gordonii, P. gingivalis cannot grow when paired with a different initial colonizer, S. oralis (Fig. 4), but P. gingivalis grows well in S. gordonii-S. oralis three-species communities (Fig. 7). Thus, P. gingivalis shows a high degree of specificity for community partnerships with streptococci, which account for 60 to 90% of initial colonizers of enamel. It has been proposed for many years that anaerobic bacteria, such as P. gingivalis, are unsuited for initial colonization and must wait for more aerotolerant species, such as streptococci, to reduce the oxygen tension to permissive levels for anaerobic growth. However, our results here and those of Lamont and coworkers (12) and Xie et al. (23) emphasize instead the importance of partner specificity among streptococci and P. gingivalis in the establishment of P. gingivalis with initial colonizers.
Inability to form productive partnerships with S. oralis is not limited to P. gingivalis. We reported that F. nucleatum, another strict anaerobe, did not grow with S. oralis (20). However, F. nucleatum paired with initial colonizer A. oris with or without S. oralis was productive. As with F. nucleatum, P. gingivalis grows commensally with A. oris. Collectively, these results indicate temporal specificity for coaggregation by P. gingivalis with certain initial colonizers of enamel. These results suggest that P. gingivalis be considered among other early-colonizing species, such as veillonellae, that also coaggregate with initial colonizers. Veillonella sp. PK1910 and P. gingivalis ATCC 33277 are coaggregation partners and grow mutualistically on saliva. Our results here (Fig. 6) indicate that Veillonella sp. PK1910 but not S. oralis 34 promotes growth of P. gingivalis in three-species communities with either A. actinomycetemcomitans or F. nucleatum. In fact, the three-species communities of S. oralis 34 and P. gingivalis ATCC 33277 with either A. actinomycetemcomitans or F. nucleatum decreased in cell numbers. Clearly, P. gingivalis and veillonellae have a productive pairwise relationship, which supports their placement as early colonizers of enamel.
Our laboratory has been interested in comparing the flow cell system and the static peg-biofilm system as models for in vitro study of multispecies communities of initial and early colonizers of enamel. Generally, the same partner specificities and mutualistic growth on saliva are seen with both systems (3, 20, 21). P. gingivalis and the other six species studied here were capable of binding to both saliva-conditioned substrata. As a single-species inoculum in the static system, P. gingivalis was retained on the polystyrene surface, although no significant growth occurred (Fig. 8A). However, initial attachment at 4 h to the saliva-coated surface in the flow cell was not maintained during the ensuing 14 h (Fig. 1). It is likely that the loss of porphyromonad biovolume is a result of the flow dynamics, whereby metabolically stressed cells are unable to maintain adherence under conditions of constant flow. Experiments with either model system were conducted under an anaerobic atmosphere, suggesting that redox potential is unlikely to be a factor.
In flow cells, P. gingivalis attached to the saliva-coated glass surface at about the same level, whether as a single species or as part of multispecies coaggregates (Fig. 1 to 7). In contrast, P. gingivalis binds at higher numbers when it is in two-species biofilms with Veillonella sp., A. actinomycetemcomitans, or F. nucleatum in static biofilm pegs (Fig. 8 and 9). With Veillonella sp., P. gingivalis cell numbers in the biofilm pegs were fivefold higher than those of P. gingivalis as a single species (Fig. 8); with A. actinomycetemcomitans, P. gingivalis biofilm cells were the same, except that a threefold higher number was noted at 36 h. However, P. gingivalis and F. nucleatum appear to be particularly fitted for each other; the numbers of P. gingivalis cells are 15- to 20-fold higher at the three time points. These results might be indicative of natural relationships that occur between species during the development of dental plaque. The static peg system is a closed system (compared to flow cells) and allows accumulation of end products of digestion of salivary components by each biofilm member species. Apparently, this accumulation is highly beneficial to community growth and results in enhanced growth of all species in these two-species biofilms.
Three-species communities with P. gingivalis, Veillonella sp., and either A. actinomycetemcomitans or F. nucleatum are even better suited for cooperation than the two-species situations. Compared with monospecies peg biofilms, P. gingivalis in combination with Veillonella sp. and A. actinomycetemcomitans ex-hibited 5-, 15-, and 35-fold greater cell numbers at 12-, 24-, and 36-h time points, respectively (Fig. 9). Likewise, P. gingivalis in combination with Veillonella sp. and F. nucleatum exhibited 12-, 15-, and 25-fold greater cell numbers at 12-, 24-, and 36-h time points, respectively. These data suggest that Veillonella sp. plays an integral role in coaggregates with P. gingivalis and the two other partners. Indeed, examination of the numbers of Veillonella sp. cells in three-species biofilms with P. gingivalis and A. actinomycetemcomitans revealed 45-, 32-, and 20-fold greater cell numbers at 12-, 24-, and 36-h time points, respectively, compared to those of veillonella single-species biofilms (data not shown). Further, the numbers of Veillonella sp. cells in three-species biofilms with P. gingivalis and F. nucleatum revealed cell numbers at 12-, 24-, and 36-h time points that were 50-, 31-, and 16-fold greater, respectively, than those for veillonella single-species biofilms (data not shown). Likewise, F. nucleatum increased 33-fold at 36 h with P. gingivalis and Veillonella sp., compared to F. nucleatum by itself (data not shown). Also, A. actinomycetemcomitans in biofilms with P. gingivalis and Veillonella sp. exhibited numbers that were 51-, 212-, and 14-fold higher than those for A. actinomycetemcomitans single-species biofilms (data not shown). Clearly, in static biofilm pegs, these species grow well together, compared to individually, and these data suggest that they are naturally compatible.
In flow cells with P. gingivalis, the biovolumes of F. nucleatum, A. actinomycetemcomitans, and Veillonella sp. were 6-, 11-, and 12-fold higher, respectively, than monospecies biovolumes at 18 h (data not shown). Various three-species combinations of F. nucleatum, A. actinomycetemcomitans, and Veillonella sp. with P. gingivalis yielded biovolume increases of the first three species of 3- to 15-fold compared to their respective single-species biofilm biovolume values (data not shown). Given that the two-model systems are significantly different in design (flow versus static), the high degree of correlation between them is remarkable. Even so, the increases in cell numbers in multispecies communities in flow cells are more modest than the increases in the static peg system. A priori, one might think that the flow cell would support unlimited growth because saliva is replenished continuously. However, our results suggest that a static system provides greater opportunity for the two- and three-species communities to interact metabolically and flourish. These results may reflect the importance of time-dependent signal accumulation that regulates community metabolism of saliva in the microtiter well. Mutualistic relationships built on this complex nutritional source may require temporal metabolic give and take in which each species contributes its part to enhance the growth of all species. A metabolic contribution will remain in a microtiter well, but in a flow cell, it might be swept away before it has an opportunity to contribute to the community metabolism.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health and in part by the Colgate-Palmolive Co. CRADA.
We thank N. I. Chalmers for her assistance with the q-PCR and R. J. Palmer for helpful comments on the manuscript.
Footnotes
Published ahead of print on 11 September 2009.
REFERENCES
- 1.Bos, R., H. C. van der Mei, and H. J. Busscher. 1996. Co-adhesion of oral microbial pairs under flow in the presence of saliva and lactose. J. Dent. Res. 75:809-815. [DOI] [PubMed] [Google Scholar]
- 2.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalmers, N. I., R. J. Palmer, Jr., J. O. Cisar, and P. E. Kolenbrander. 2008. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J. Bacteriol. 190:8145-8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawes, C., S. Watanabe, P. Biglow-Lecomte, and G. H. Dibdin. 1989. Estimation of the velocity of the salivary film at some different locations in the mouth. J. Dent. Res. 68:1479-1482. [DOI] [PubMed] [Google Scholar]
- 5.Diaz, P. I., N. I. Chalmers, A. H. Rickard, C. Kong, C. L. Milburn, R. J. Palmer, Jr., and P. E. Kolenbrander. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 72:2837-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoshino, T., T. Fujiwara, and M. Kilian. 2005. Use of phylogenetic and phenotypic analyses to identify nonhemolytic streptococci isolated from bacteremic patients. J. Clin. Microbiol. 43:6073-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolenbrander, P. E., and R. N. Andersen. 1989. Inhibition of coaggregation between Fusobacterium nucleatum and Porphyromonas (Bacteroides) gingivalis by lactose and related sugars. Infect. Immun. 57:3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolenbrander, P. E., R. N. Andersen, and L. V. Holdeman. 1985. Coaggregation of oral Bacteroides species with other bacteria: central role in coaggregation bridges and competitions. Infect. Immun. 48:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolenbrander, P. E., R. N. Andersen, and L. V. Moore. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect. Immun. 57:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuboniwa, M., E. L. Hendrickson, Q. Xia, T. Wang, H. Xie, M. Hackett, and R. J. Lamont. 2009. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 9:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 13.Li, J., E. J. Helmerhorst, C. W. Leone, R. F. Troxler, T. Yaskell, A. D. Haffajee, S. S. Socransky, and F. G. Oppenheim. 2004. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 97:1311-1318. [DOI] [PubMed] [Google Scholar]
- 14.Mampel, J., T. Spirig, S. S. Weber, J. A. Haagensen, S. Molin, and H. Hilbi. 2006. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl. Environ. Microbiol. 72:2885-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 16.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95:369-380. [DOI] [PubMed] [Google Scholar]
- 17.Palmer, R. J., Jr., P. I. Diaz, and P. E. Kolenbrander. 2006. Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 188:4117-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Periasamy, S., N. I. Chalmers, L. Du-Thumm, and P. E. Kolenbrander. 2009. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl. Environ. Microbiol. 75:3250-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Periasamy, S., and P. E. Kolenbrander. 2009. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect. Immun. 77:3542-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simionato, M. R., C. M. Tucker, M. Kuboniwa, G. Lamont, D. R. Demuth, G. D. Tribble, and R. J. Lamont. 2006. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect. Immun. 74:6419-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie, H., G. S. Cook, J. W. Costerton, G. Bruce, T. M. Rose, and R. J. Lamont. 2000. Intergeneric communication in dental plaque biofilms. J. Bacteriol. 182:7067-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]