Abstract
Although a variety of bacterial species have been reported to use the interspecies communication signal autoinducer-2 (AI-2) to regulate multiple behaviors, the molecular mechanisms of AI-2 recognition and signal transduction remain poorly understood. To date, two types of AI-2 receptors have been identified: LuxP, present in Vibrio spp., and LsrB, first identified in Salmonella enterica serovar Typhimurium. In S. Typhimurium, LsrB is the ligand binding protein of a transport system that enables the internalization of AI-2. Here, using both sequence analysis and structure prediction, we establish a set of criteria for identifying functional AI-2 receptors. We test our predictions experimentally, assaying key species for their abilities to import AI-2 in vivo, and test their LsrB orthologs for AI-2 binding in vitro. Using these experimental approaches, we were able to identify AI-2 receptors in organisms belonging to phylogenetically distinct families such as the Enterobacteriaceae, Rhizobiaceae, and Bacillaceae. Phylogenetic analysis of LsrB orthologs indicates that this pattern could result from one single origin of the functional LsrB gene in a gammaproteobacterium, suggesting possible posterior independent events of lateral gene transfer to the Alphaproteobacteria and Firmicutes. Finally, we used mutagenesis to show that two AI-2-interacting residues are essential for the AI-2 binding ability. These two residues are conserved in the binding sites of all the functional AI-2 binding proteins but not in the non-AI-2-binding orthologs. Together, these results strongly support our ability to identify functional LsrB-type AI-2 receptors, an important step in investigations of this interspecies signal.
Autoinducer-2 (AI-2) is a small molecule produced and secreted by a large number of bacterial species belonging to very widespread branches within the kingdom Bacteria (15, 46, 64). AI-2 or its synthase, LuxS, has been implicated in the regulation of many bacterial behaviors including biofilm formation, virulence, competence, and the production of secondary metabolites like antibiotics (17, 60, 64). While in some cases, AI-2 is clearly acting through a canonical quorum-sensing mechanism (61), in others a role in central metabolism has been proposed (62). One of the obstacles to an understanding of the function of AI-2 in any given species is a lack of knowledge of the molecular mechanisms of AI-2 recognition, signal transduction, and/or processing.
Undoubtedly, one of the major difficulties in identifying AI-2 receptors is the complexity of the chemistry of this signal molecule. The product of the reaction catalyzed by LuxS is 4,5-dihydroxy-2,3-pentadione (DPD), which, in solution, spontaneously rearranges into a variety of chemically distinct forms collectively called AI-2 (31, 46). We have shown that these forms are in equilibrium and can thus interconvert and that the availability of the different forms of AI-2 is highly dependent on the chemistry of the environment (31). Additionally, different organisms recognize distinct forms of this molecule (12).
So far, two types of AI-2 receptors have been identified and are classified by their ability to bind chemically distinct DPD derivatives: the LuxP and LsrB types of receptors characterized first for Vibrio harveyi and Salmonella enterica serovar Typhimurium, respectively (12, 31). The crystal structure of the V. harveyi LuxP-AI-2 complex revealed that the ligand recognized by this receptor is a furanosyl borate diester (12), a cyclic form of DPD bound to borate, while crystal structures of the LsrB-AI-2 complexes from S. Typhimurium and Sinorhizobium meliloti show that these species recognize a DPD adduct that does not contain boron and has a different stereochemistry [(2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF)] (31, 37). The structures of the LsrB-type receptors bound to AI-2 further showed that six residues were responsible for hydrogen bonding with AI-2 and that these residues were completely conserved between the two species (31, 37). These residues are distinct from those in the LuxP AI-2 binding site, contributing to the specificity of each receptor for the form of AI-2 recognized by a given species.
LuxP is a periplasmic binding protein that, upon binding to AI-2, modulates the activity of a membrane sensor histidine kinase, LuxQ. Together, LuxPQ regulate a signal transduction cascade that controls the AI-2 quorum-sensing regulon in organisms belonging to the Vibrionales such as V. harveyi, Vibrio cholerae, and Vibrio anguillarum (5, 13, 32, 33); to date, however, LuxP-type receptors have not been found outside of the Vibrionales.
The LsrB-type receptors also belong to the large family of periplasmic binding proteins but have a low homology to LuxP (the sequence identity between the V. harveyi LuxP and the S. Typhimurium LsrB AI-2 receptors is only approximately 11%). The function of the LsrB protein has been characterized for the two closely related enteric bacteria, S. Typhimurium (56, 57) and Escherichia coli (65), the plant symbiont S. meliloti (37), and the oral pathogen Aggregatibacter (Actinobacillus) actinomycetemcomitans (48). In all these organisms it is thought that LsrB acts as the substrate binding protein of an ATP binding cassette (ABC) transport system responsible for AI-2 internalization. Due to the homology with other ABC transport systems, it is predicted that the Lsr transporter is composed of LsrB, two transmembrane proteins (LsrC and LsrD) that form a channel, and a cytoplasmic protein (LsrA) that contains an ABC binding motif and is thought to be responsible for ATP hydrolysis during transport. Once inside the cell, AI-2 is phosphorylated by the kinase LsrK and further processed by the enzymes LsrG and LsrF (56, 66). The genes encoding these proteins (with the exception of LsrK) are all in the same operon, which is regulated by the repressor LsrR. In the absence of phospho-AI-2 (P-AI-2), LsrR represses the transcription of the lsr operon; however, when AI-2 is internalized and phosphorylated by LsrK, P-AI-2 binds LsrR, causing the derepression of the operon. Thus, the increased expression of the Lsr system leads to increased AI-2 import, resulting in a rapid depletion of AI-2 from the extracellular medium.
It does not appear that the AI-2 taken up by this system is used as a carbon source, since cultures of S. Typhimurium and S. meliloti were unable to grow when AI-2 was used as the sole carbon source (37, 57). Rather, AI-2 removal via the Lsr system enables these organisms to terminate their own AI-2 signaling system and to regulate the AI-2-dependent gene expression of other organisms in the vicinity. Thus, in cultures composed of different species, bacteria with a functional Lsr system are capable of interfering with the AI-2-mediated group behaviors of the other species (63).
Recently, two studies have undertaken database sequence analysis to identify LsrB orthologs (41, 50). Those studies showed that orthologs of the Lsr system are not broadly conserved across the kingdom Bacteria while identifying hypothetical LsrB receptors in some organisms belonging to very distinct families such as the Enterobacteriaceae, Pasteurellaceae, Rhizobiaceae, Rhodobacteraceae, and Bacillaceae.
Here, we expand upon previous bioinformatic studies (41, 50) with additional analysis based not only on sequence but also on structure prediction that allow us to establish a set of criteria for predicting which orthologs of LsrB are functional AI-2 receptors. We then present experimental evidence that confirms a set of these predictions and demonstrates the presence of functional AI-2 receptors in the families Enterobacteriaceae, Rhizobiaceae, and Bacillaceae.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used are listed in Table 1. Bacteria from the family Enterobacteriaceae (E. coli MG1655 and uropathogenic E. coli [UPEC] UTI89) and the family Bacillaceae (Bacillus cereus ATCC 10987 and vaccine strain Bacillus anthracis Sterne 34F2) were grown in Luria-Bertani (LB) medium with shaking at 37°C. The bacteria from the family Rhizobiaceae (S. meliloti Rm1021, Agrobacterium tumefaciens C58, Rhizobium etli CFN42, and Rhizobium leguminosarum bv. viciae 3841) were cultured with shaking at 30°C in their optimal cultured medium, LBMC (LB medium supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2), LB medium, YEM (10 g liter−1 mannitol, 0.5 g liter−1 yeast extract, 0.2 g liter−1 MgSO4·7H2O, and 1 g liter−1 NaCl), and TYC (5 g liter−1 tryptone, 3 g liter−1 yeast extract, 0.5 g liter−1 CaCl2), respectively.
TABLE 1.
Bacterial strains used in this study
| Strain | Source and/or reference |
|---|---|
| Salmonella Typhimurium | ATCC 14028 |
| Escherichia coli K-12 MG1655 | 7 |
| Escherichia coli UTI89 (UPEC) | Jeffrey I. Gordon (40) |
| Bacillus anthracis Sterne 34F2 (vaccine strain) | Martin J. Blaser (21) |
| Bacillus cereus (ATCC 10987) | Adriano O. Henriques |
| Sinorhizobium meliloti Rm1021 | 29 |
| Agrobacterium tumefaciens C58 | James P. Shapleigh (3) |
| Rhizobium leguminosarum bv. viciae 3841 | Gladys Alexandre (30) |
| Rhizobium etli CFN42 | ATCC 51251 |
Databases analysis.
The KEGG SSDB (Kyoto Encyclopedia of Genes and Genomes Sequence Similarity Database [http://www.genome.jp/kegg/ssdb/]) was used to search for protein orthologs of LuxS and the proteins encoded by the lsr operon from S. Typhimurium LT2 in January 2009. This database provides amino acid sequence similarities between all protein-encoding genes in the complete genomes in the GENES database, and all possible pairwise genome comparisons were performed by use of the SSEARCH program (36) (available at http://www.genome.jp/kegg/ssdb/). In this study, we have selected gene pairs that were the best bidirectional hits and had a Smith-Waterman similarity score of at least 100. To be considered a best bidirectional hit, the relationship of gene x in genome A with gene y in genome B must be such that when x is compared against all genes in genome B, y is found as the top scoring and the reverse is also true. Pairs that met these criteria were scored as orthologous proteins.
Structure prediction.
All LsrB protein orthologs were submitted to the fold recognition server PHYRE (22) for structure prediction. In the majority of cases, S. Typhimurium LsrB was identified as one of the top 10 fold templates, and thus, the server returned a structure-based sequence alignment between LsrB and the query sequence. Alignments were examined to determine if residues previously shown to form hydrogen bonds with R-THMF in S. Typhimurium LsrB (K35, D116, D166, Q167, P220, and A222) (31) were conserved in the predicted structure. For the one-third of group II orthologs where PHYRE did not return an alignment with LsrB, simple sequence alignments were calculated using NCBI-blastp (1, 18) and checked for the conservation of the residues listed above. Such cases are noted in Table S1 in the supplemental material.
AI-2 activity in bacterial cultures.
To monitor AI-2 activity in E. coli and Bacillus cell cultures during growth, cultures were grown overnight to saturation and diluted (1:100) into 25 ml of LB medium in 250-ml Erlenmeyer flasks. In Rhizobiaceae species, cultures in the exponential phase were diluted to an optical density at 600 nm (OD600) of 1 into the appropriate medium with 80 μM chemically synthesized AI-2 (47, 66). In both cases, aliquots were collected at the indicated times, and cell-free culture fluids were prepared by the filtration of liquid cultures (51, 52), which were analyzed in duplicate for AI-2 activity using the V. harveyi BB170 bioluminescence reporter assay, as described previously (4, 5). AI-2 activity is reported as the induction of light production compared with the background light obtained with the appropriate growth medium (as previously explained in reference 37).
Protein expression and purification.
The genes encoding LsrB orthologs in R. etli, R. leguminosarum, A. tumefaciens, E. coli MG1655, and E. coli UTI89 were cloned from genomic DNA into plasmid pProEX HTb for expression as polyhistidine-tagged proteins. The B. anthracis LsrB ortholog was cloned into plasmid pET151/D-TOPO using the Champion pET Directional TOPO expression kit (Invitrogen) for expression as a polyhistidine-tagged fusion protein. N-terminal signal peptides for secretion, as determined by the program SignalP 3.0 (6), were excluded from the constructs. Plasmids were transformed into E. coli strains BL21 and FED101 (BL21 luxS null mutant), and expression was induced with 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) when the cultures reached an OD595 of 0.9. The bacteria were harvested after expression for 5 h at 22°C. Pellets were resuspended in a solution containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, 1 mM dithiothreitol, 0.36 mg/ml leupeptin, 0.36 mg/ml aprotinin, and 0.36 mg/ml DNase and lysed using an M-110Y microfluidizer (Microfluidics). The lysate was centrifuged, and the tagged protein was purified using Ni-nitrilotriacetic acid affinity chromatography (Qiagen). The protein was eluted from the column using a solution containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, and 250 mM imidazole, and the buffer was then swapped using Sephadex-G25 agarose into a solution containing 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, and 1 mM dithiothreitol. Purified protein was concentrated to 10 mg ml−1. The genes encoding the S. Typhimurium and B. cereus LsrB orthologs were cloned into pGEX-4T1, transformed, expressed, and purified as described previously (31, 37). The primers used for the cloning of the respective genes are listed in Table 2.
TABLE 2.
Primers used in this study
| Construct (purpose) | 5′ (sense) sequence | 3′ (antisense) sequence |
|---|---|---|
| R. leguminosarum/pPro (PCR) | CGCGGATCCGCCGACATCAAGATCGGT | CCGCTCGAGCGTCAGAAGACCTTGGAGAACTG |
| A. tumefaciens/pPro (PCR) | CGCGGATCCGCAGACGTCAAGATCGC | CCGCTCGAGCAATCTTCGAGAACTGATCGAT |
| R. etli/pPro (PCR) | CGCGGATCCAAGGACATCAAGATCGGC | CCGCTCGAGTCAGAAGACCTTGGAGAACTG |
| UPEC/pPro (PCR) | CGGGATCCGCGGAAAAAGTCG | CCGCTCGAGTTAATAAAGTGAGTCGATATTGTC |
| E. coli MG1655/pPro (PCR) | CGCGGATCCGCAGAGCGTATTGCATTT | CCGCTCGAGTCAGAAATCGTATTTGCCGAT |
| B. anthracis/pET151 (D171N) | CTCTAGTCCAACAGTAACGAATCAAAACCAATGGGTAAC | GTTACCCATTGGTTTTGATTCGTTACTGTTGGACTAGAG |
| B. anthracis/pET151 (A227T) | TATTAATGCAGTCATTTGTCCGGATACGACGGCACTTCCAG | CTGGAAGTGCCGTCGTATCCGGACAAATGACTGCATTAATA |
| S. Typhimurium/pGEX (PCR) | —a | |
| B. cereus/pGEX (PCR) | CGGGATCCAAGAAAAAAGCTGATGATGT | GGAATTCCTAATCAATATTATCCTTCGTAAATACGAC |
| B. anthracis/pET151 (PCR) | CACCGATAAGAAAAAAGCGGA | CTAAAAATTATATTTATCAATAT |
See reference 31.
AI-2 binding assay.
Proteins tested for AI-2 binding were denatured (70°C for 10 min) to release any bound ligand and pelleted (12). V. harveyi strain BB170 was used to test for the presence or absence of AI-2 in the resulting supernatants as previously described (4, 5).
B. anthracis mutagenesis.
The mutations D171N and A227T were introduced into two separate B. anthracis/pET151 constructs using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). Primers used for creating the mutations are given in Table 2. The same kit was then used to create the double mutant D171N A227T. The mutant proteins were expressed and purified as described above for the B. anthracis wild-type LsrB ortholog.
Phylogenetic analyses.
The evolutionary history of the lsrB gene was studied by analyzing the phylogenetic relationship of the functional orthologs identified in this study and contrasting it with the phylogeny of rpoB (RNA polymerase β-subunit). rpoB is generally accepted to provide a good representation of the phylogenetic relationships among the Bacteria (11), as it provides a phylogenetic resolution comparable to that of 16S rRNA with the advantage of being a single-copy gene. To construct the organismal tree, the rpoB gene sequences from all the organisms in Table S1 in the supplemental material and representative species of all major phyla of the Bacteria were downloaded from the KEGG database and aligned with ClustalW (59) using the translated protein sequences. Alignments were carried out with default parameters and visually inspected by use of Molecular Evolutionary Genetics Analysis (MEGA), version 4 (58). Hypervariable regions with ambiguous alignment were excluded from the analysis. The lsrB gene tree was made with all the sequences identified as being functional lsrB orthologs (group I, Table 3), and the tree was inferred using maximum likelihood in PAUP* 4.0b10 (53) using heuristic searches, 10 random taxon additions, and TBR (tree bisection and reconnection) branch swapping. MrBayes 3.1.2 (44) was used to infer branch support by running two simultaneous sets of four Markov chains for 1 million generations sampled every 100 generations. The distribution of the log likelihoods was used to evaluate the stationarity of this parameter and to determine burn-in values. Modeltest 3.7 (38) and MrModeltest 2.2 (34) were used to select the best-fitting evolutionary models for phylogenetic analyses. The rpoB phylogeny was estimated with a total data set of 83 species. This data set was translated to amino acids and analyzed using neighbor joining (45) with Poisson correction distances (68) and a gamma distribution rate variation among sites. Nodal support was estimated with nonparametric bootstrapping (1,000 replicates). The rpoB trees were rooted with Thermotoga maritima (Thermotogales). These analyses were carried out using MEGA.
TABLE 3.
Orthologs of the LuxS and Lsr proteins from S. Typhimurium present in the complete genomes of the KEGG database (January 2009)
| Species | Presence of orthologa |
% LsrB identityb | No. of binding-site residuesc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LuxS | LsrB | LsrA | LsrC | LsrD | LsrK | LsrR | LsrG | LsrF | |||
| Group I | |||||||||||
| Salmonella Typhimurium LT2 | + | + | + | + | + | + | + | + | + | 100 | 6 |
| Salmonella enterica (13 strains) | + | + | + | + | + | + | + | + | + | 100 | 6 |
| Escherichia coli (11 strains) | + | + | +d | + | + | + | + | + | + | 85 | 6 |
| Escherichia fergusonii | + | + | + | + | + | + | + | + | + | 85 | 6 |
| Yersinia pestis (7 strains) | + | + | + | + | + | + | + | + | + | 84 | 6 |
| Yersinia pseudotuberculosis (4 strains) | + | + | + | + | + | + | + | + | + | 84 | 6 |
| Yersinia enterocolitica | + | + | + | + | + | + | + | + | + | 83 | 6 |
| Klebsiella pneumoniae (2 strains) | + | + | + | + | + | + | + | + | + | 82 | 6 |
| Photorhabdus luminescens | + | + | + | + | + | + | + | + | + | 82 | 6 |
| Enterobacter sp. strain 638 | + | + | + | + | + | + | + | + | + | 82 | 6 |
| Pasteurella multocida | + | + | + | + | + | + | + | + | + | 80 | 6 |
| Haemophilus influenzae PittEE | + | + | + | + | + | + | + | + | + | 80 | 6 |
| Haemophilus somnus (2 strains) | + | + | + | + | + | + | + | + | + | 76 | 6 |
| Sinorhizobium meliloti | + | + | + | + | + | + | + | + | 72 | 6 | |
| Rhodobacter sphaeroides (2 strains) | + | + | + | + | + | + | + | 72 | 6 | ||
| Bacillus anthracis (4 strains) | + | + | + | + | + | + | + | + | + | 63 | 6 |
| Bacillus cereus (7 strains) | + | + | + | + | + | + | + | + | + | 63 | 6 |
| Bacillus thuringiensis (2 strains) | + | + | + | + | + | + | + | + | + | 63 | 6 |
| Group II | |||||||||||
| Rubrobacter xylanophilus | + | + | + | + | 36 | 3 | |||||
| Ochrobactrum anthropi | + | + | + | + | + | 35 | 4 | ||||
| Sinorhizobium medicae | + | + | + | + | + | + | 35 | 4 | |||
| Roseobacter denitrificans | + | + | + | + | 34 | 4 | |||||
| Mesorhizobium loti | + | + | + | + | 34 | 4 | |||||
| Agrobacterium tumefaciens C58 (2 strains) | + | + | + | + | + | 33 | 4 | ||||
| Leptothrix cholodnii | + | + | + | + | 33 | 4 | |||||
| Dinoroseobacter shibae | + | + | + | + | + | + | 33 | 4 | |||
| Verminephrobacter eiseniae | + | + | + | 33 | 4 | ||||||
| Burkholderia phytofirmans | + | + | + | + | 33 | 4 | |||||
| Gluconacetobacter diazotrophicus PAl 5 (JGI) | + | + | 33 | 2 | |||||||
| Rhizobium leguminosarum | + | + | + | + | + | + | 33 | 4 | |||
| Rhizobium leguminosarum bv. trifolii WSM2304 | + | + | + | + | + | 33 | 4 | ||||
| Gluconacetobacter diazotrophicus PAl 5 (Brazil) | + | + | 33 | 1 | |||||||
| Rhodococcus sp. strain RHA1 | + | + | + | + | + | 33 | 4 | ||||
| Streptomyces coelicolor | + | + | + | + | + | + | 33 | 4 | |||
| Burkholderia xenovorans | + | + | + | + | + | 32 | 3 | ||||
| Rhizobium etli | + | + | + | + | + | 32 | 4 | ||||
| Dictyoglomus thermophilum | + | + | 32 | 4 | |||||||
| Rhizobium etli CIAT 652 | + | + | + | + | 32 | 4 | |||||
| Jannaschia sp. strain CCS1 | + | + | + | 32 | 4 | ||||||
| Dictyoglomus turgidum | + | + | 32 | 4 | |||||||
| Acidiphilium cryptum JF-5 | + | + | 31 | 2 | |||||||
| Streptomyces avermitilis | + | + | + | + | 31 | 4 | |||||
| Burkholderia phymatum | + | + | + | + | + | 31 | 4 | ||||
| Deinococcus geothermalis | + | + | + | + | + | 31 | 4 | ||||
| Burkholderia ambifaria MC40-6 | + | + | + | + | + | + | 31 | 4 | |||
| Syntrophomonas wolfei | + | + | + | + | 30 | 1 | |||||
| Chloroflexus aggregans | + | + | + | 27 | 4 | ||||||
| Escherichia coli O1 (avian pathogenic) | + | + | + | + | + | 27 | 0 | ||||
| Escherichia coli UTI89 (UPEC) | + | + | + | + | + | 27 | 1 | ||||
Orthologs of both group I and group II are defined as a complete match in the bidirectional best hits and are denoted with a +.
Percentage of identity using S. Typhimurium LsrB as a reference.
Number of conserved residues in the binding site based on structure prediction using S. Typhimurium LsrB as a reference.
LsrA from Escherichia coli E24377A is truncated.
RESULTS
LsrB orthologs in completely sequenced bacterial genomes.
To search for orthologs of LsrB, we carried out a reciprocal best-hit analysis against all 809 completely sequenced bacterial genomes present in the KEGG database as of January 2009 using the protein sequence of LsrB from S. Typhimurium (STM4077). The reciprocal best-hit strategy of sequence similarity comparisons was employed previously for this type of study because it allows the distinction between orthologs and paralogs (10). The organisms with proteins identified as being orthologs are shown in Table 3 (KEGG protein identities and E values are provided in Table S1 in the supplemental material). Sorting these organisms in order of percentage of identity of the LsrB orthologs with the S. Typhimurium AI-2 receptor clearly revealed two distinct groups of LsrB orthologs, a first group with a high percentage of identity (>60%; E value below 1E−103) and a second group with a percentage of identity below 36% (E value higher than 1E-44), which we termed group I and group II, respectively.
We then performed the reciprocal best-hit analysis against all genomes using each LsrB protein sequence from group I as a reference (i.e., instead of LsrB from S. Typhimurium). In all cases, the only hits with greater than 57% identity were the other protein sequences included in group I from the first analysis. Thus, the group I orthologs are consistent regardless of the LsrB sequence used as a reference.
The genomes of the organisms with LsrB orthologs were further analyzed to identify orthologs of the other proteins encoded by the lsr operon. As shown in Table 3, all species of group I have orthologs of all the proteins encoded by the lsr operon (with the exception of LsrF, a putative AI-2-processing protein, in Rhodobacter sphaeroides), whereas none of the group II organisms have orthologs of the complete operon, lacking at least two proteins encoded by genes from this operon in all cases. LsrE was not included in this analysis because the protein seems to be exclusive to the Salmonella genus, and an LsrE knockout mutant in S. Typhimurium showed no phenotype related to the regulation of the lsr operon or AI-2 production (56, 57).
Reasoning that a conservation of the residues that formed hydrogen bonds with AI-2 (31) would be crucial to LsrB function, we next used a fold recognition-based server to predict structures for the LsrB orthologs. The sequences of the LsrB orthologs were submitted to the PHYRE Web server (22), which returned structure predictions and structure-based alignments based on each of the 10 best-scoring template Protein Data Bank structures available in the PHYRE library. For all of the orthologs in group I and two-thirds of the orthologs in group II, the structure of S. Typhimurium LsrB was returned as one of these top 10 templates. The alignments with S. Typhimurium LsrB were then examined to determine if residues previously shown to form hydrogen bonds with R-THMF in S. Typhimurium LsrB (K35, D116, D166, Q167, P220, and A222) (31) were predicted to be structurally conserved. Strikingly, as shown in the last column of Table 3, these six residues were completely conserved in all of the orthologs in group I and differed in at least two positions in all cases for group II. Residue D166 (numbering based on S. Typhimurium LsrB) was not conserved in any of the group II orthologs, most typically being replaced with an asparagine. The other most common substitution was A222T (a full listing of the nonconserved amino acids is given in Table S1 in the supplemental material).
Based on these results, we hypothesize that the species in group I, which have >60% identity, orthologs to the proteins encoded by the lsr operon, and all six AI-2 binding-site residues conserved, have functional LsrB-like AI-2 receptors, whereas group II proteins are likely to have a different function.
Profiles of AI-2 removal from extracellular medium.
Previous studies of S. Typhimurium (57), E. coli (65), S. meliloti (37), and A. actinomycetemcomitans (48) revealed that the lsr operons in these organisms encode proteins involved in an ABC transport system that imports extracellular AI-2. Thus, in the presence of these organisms, AI-2 does not persist in the extracellular medium but is internalized by the cells and further modified. Our analysis, described above, indicated that all the organisms predicted to have functional LsrB receptors (group I, Table 3) also had orthologs to all the proteins encoded by the lsr operon. Thus, we predicted that the organisms in group I have a functional Lsr system for AI-2 internalization and that these organisms would rapidly remove AI-2 from culture fluids. In contrast, for organisms from group II, which lack orthologs to some of the proteins encoded by the lsr operon and presumably do not have a functional AI-2 transport system, we predicted that AI-2 would persist in the extracellular media. To test these predictions, we compared the profiles of AI-2 removal for a set of organisms from groups I and II.
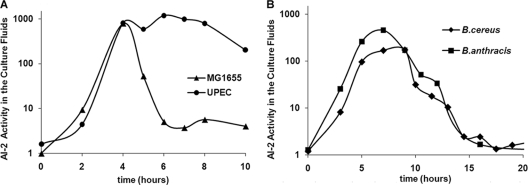
Our analysis revealed that almost all E. coli strains (11 out of 13) analyzed belong to group I. However, two E. coli strains (avian-pathogenic E. coli and UPEC UT189) have LsrB orthologs with very low sequence identity and lack orthologs to several of the proteins from the lsr operon, and they are therefore classified as members of group II (Table 3). We tested an E. coli strain (MG1655) from group I for AI-2 uptake and found, as was previously shown (65), that this strain removed AI-2 from culture fluids (Fig. 1A). We then compared the AI-2 removal profile of UPEC strain UT189 (from group II) with the profile from E. coli strain MG1655 and observed that while E. coli MG1655 efficiently cleared AI-2 from culture fluids by 6 h, UPEC strain UT189 cleared little, if any, AI-2 by 10 h (Fig. 1A). This supports our prediction that UPEC strain UT189, although belonging to the same species as MG1655, is a member of group II and accordingly does not have a functional Lsr transport system for AI-2 uptake.
FIG. 1.
AI-2 removal profile for bacteria producing AI-2. Shown is extracellular AI-2 activity in cell-free culture fluids from LuxS+ strains E. coli MG1655 (triangles) and UPEC UTI89 (circles) (A) and B. cereus (diamonds) and B. anthracis (squares) (B) cultures. Aliquots were taken at the specified times. AI-2 activity is reported as the change from the induction of light produced by V. harveyi BB170.
Like E. coli MG1655, two Bacillus strains, B. cereus (ATCC 1087) and B. anthracis (vaccine strain Sterne 34F2), have orthologs classified as belonging to group I. Putative AI-2 receptors were identified in these species previously (41, 50) but not confirmed experimentally. We tested these strains for AI-2 removal, and as expected, they were able to completely remove AI-2 from culture fluids (Fig. 1B), supporting the premise that organisms in group I have functional AI-2 transporters.
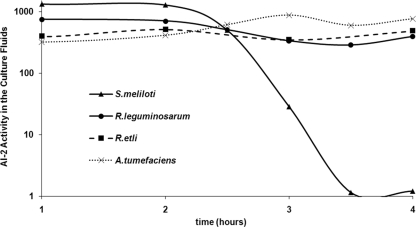
To further test the premise that group II organisms are unable to incorporate AI-2, we compared AI-2 removal profiles for organisms from the family Rhizobiaceae from group II (R. etli, R. leguminosarum, and A. tumefaciens) with AI-2 in the only member of the Rhizobiaceae from group I (S. meliloti). None of the members of the Rhizobiaceae shown in Table 3 has LuxS orthologs, and thus, we expected that none of these species would produce AI-2. This was confirmed by the fact that cell-free culture fluids collected from these bacteria produced only low levels of bioluminescence induction in a V. harveyi BB170 bioassay (data not shown). However, as we have previously shown in the case of S. meliloti, non-AI-2-producing species can still be capable of taking up AI-2 produced synthetically or by other species (37). Thus, in order to compare AI-2 removal profiles for these species, we cultured these bacteria to the same cell density (OD600 of 1), supplied chemically synthesized AI-2, and measured AI-2 activity in the culture fluids over time (Fig. 2). Over the time of the measurements, S. meliloti effectively removed the exogenously provided AI-2, while the other three species did not, supporting the prediction that the bacteria tested from group II (R. etli, R. leguminosarum, and A. tumefaciens), and likely all group II species, do not have Lsr systems capable of taking up AI-2.
FIG. 2.
Removal of exogenously supplied AI-2. S. meliloti (triangles), R. leguminosarum (circles), R. etli (squares), and A. tumefaciens (crosses) were cultured to an OD600 of 1 in their optimal culture media (LBMC, TYC, YEM, and LB, respectively). Chemically synthesized AI-2 was then added to all the cultures, and aliquots were taken at the specified times. AI-2 activity in cell-free culture fluids is reported as the change from the induction of light produced by V. harveyi BB170.
In vitro AI-2 binding to LsrB orthologs.
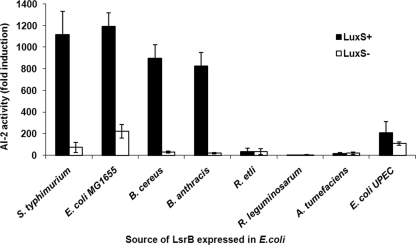
While the above-described results support our ability to identify species with functional AI-2 transporters, they do not directly show that the identified LsrB ortholog is responsible for AI-2 binding. In order to directly test for AI-2 binding ability, we cloned the LsrB orthologs from the same organisms tested as described above (three belonging to group I and four belonging to group II) and compared their abilities to bind AI-2 with that of LsrB from S. Typhimurium. The candidate proteins were overexpressed in both an E. coli strain that produces AI-2 and, as a negative control, a luxS mutant E. coli strain that does not make AI-2. These proteins were then purified and tested for the ability to bind AI-2 using a previously developed assay (12) in which the protein is heat denatured to release any bound ligand. The denatured protein is then pelleted, and the resulting supernatants are added to a reporter strain of V. harveyi that bioluminesces in response to AI-2. As shown in Fig. 3, all three orthologs from group I (i.e., that have >60% identity, a complete set of orthologs to the lsr genes, and the six amino acids from the binding pocket conserved), E. coli MG1655, B. cereus, and B. anthracis, showed a LuxS-dependent AI-2 binding ability similar to that observed for the previously characterized S. Typhimurium LsrB protein (Fig. 3). Conversely, no AI-2 binding activity was detected in the candidates from group II (R. etli, R. leguminosarum, A. tumefaciens, and UPEC UT189) (Fig. 3). Thus, as predicted from sequence analysis and structure prediction (above), LsrB orthologs from group I demonstrate AI-2 binding ability, while group II orthologs lack this ability.
FIG. 3.
Binding of AI-2 to potential LsrB-like orthologs. Proteins were expressed in either LuxS+ (black bars) or LuxS− (white bars) E. coli strains (BL21 and FED101, respectively), purified, and denatured to release the ligand. The released ligand was added to a V. harveyi AI-2 reporter strain (BB170) to determine AI-2 activity. AI-2 activity is reported as the change in the induction of light production by V. harveyi BB170 supplemented with protein supernatant compared to that of the appropriate buffer. Error bars represent the standard deviations for three independent cultures.
The amino acids aspartate 166 and alanine 222 are required for AI-2 binding.
Based on predicted structure-based sequence alignments (see above), the amino acids that form hydrogen bonds with AI-2 are completely conserved in all of the LsrB orthologs that demonstrated the ability to bind AI-2. In contrast, all the proteins that were unable to bind AI-2 in our in vitro assays lacked at least two of these residues. Specifically, in R. etli, R. leguminosarum, and A. tumefaciens, there are predicted to be two substitutions: D166N and A222T (numbering follows that of LsrB from S. Typhimurium). Indeed, the majority of the proteins in group II have these substitutions, although other substitutions are observed (see Table S1 in the supplemental material for detailed information). The complete conservation of AI-2 hydrogen binding residues in orthologs of group I but not group II is apparent in a multiple-sequence alignment of all of the LsrB orthologs for which we have experimental data (purple in Fig. S1 in the supplemental material). It is worth noting that 29 non-binding-site residues are completely conserved across all groups in this alignment (yellow in Fig. S1 in the supplemental material). However, structural analysis shows that these residues are not clustered. Moreover, these residues are disproportionately Gly and Pro (10 and 4 conserved occurrences, respectively), suggesting that unlike the six residues in the binding site, these residues are conserved for structural rather than functional reasons.
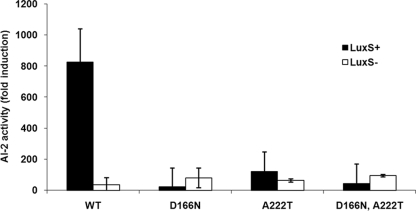
We interpreted this to indicate that residues D166 and A222 are essential for the AI-2 binding ability, and to test this idea, we introduced the mutations described above (D166N and A222T) into the B. anthracis LsrB ortholog, both individually and together, and assayed for AI-2 binding ability. As shown in Fig. 4, while the wild-type protein is capable of binding AI-2, no AI-2 activity was present in the binding pockets of any of the mutants as measured by the V. harveyi bioassay. As a complementary experiment, we tested the ability to create AI-2 binding capacity in the distantly related LsrB ortholog of R. etli by mutating the putative binding-site residues to mimic the binding site of the proteins from group I. These mutants failed to show AI-2 binding in the V. harveyi bioassay (data not shown), indicating that these proteins have already diverged to such a degree that other aspects of the protein structure important for AI-2 binding are missing.
FIG. 4.
Binding of AI-2 by wild-type B. anthracis and mutant D166N and A222T LsrB-like proteins. B. anthracis wild-type (WT) and mutant D166N and A222T proteins were expressed in either LuxS+ (black bars) or LuxS− (white bars) cultures as explained in the legend of Fig. 3. AI-2 activity is reported as the change in the induction of light production by V. harveyi BB170 supplemented with protein supernatant compared to that supplemented with the appropriate buffer. Error bars represent the standard deviations for three independent cultures.
These results show that D166 and A222, conserved in all the LsrB orthologs we have shown to bind AI-2, are necessary (though not sufficient) for the ability of these proteins to bind AI-2 and thus provide a useful criterion for the identification of other LsrB-like AI-2 receptors. It is possible that more conservative mutations would still allow AI-2 binding, but such mutations were not observed in our list of orthologs. Furthermore, these results support the hypothesis that the proteins in group II are incapable of AI-2 binding and are therefore very unlikely to function as AI-2 receptors in vivo.
Evolution of functional LsrB-like AI-2 receptors.
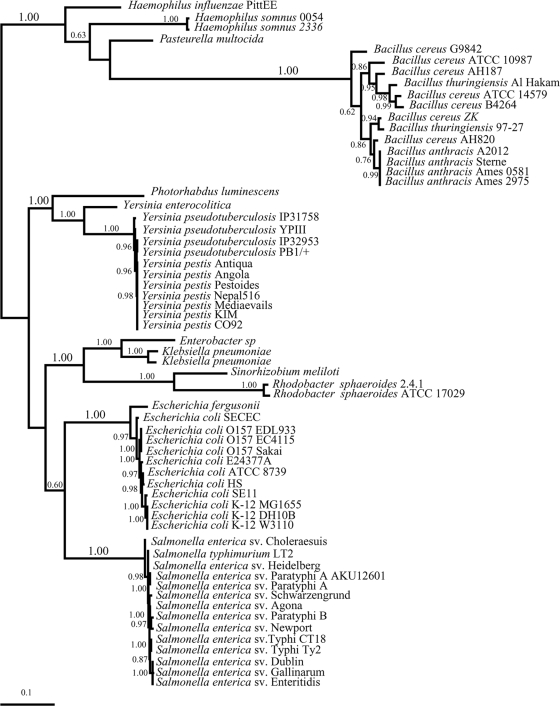
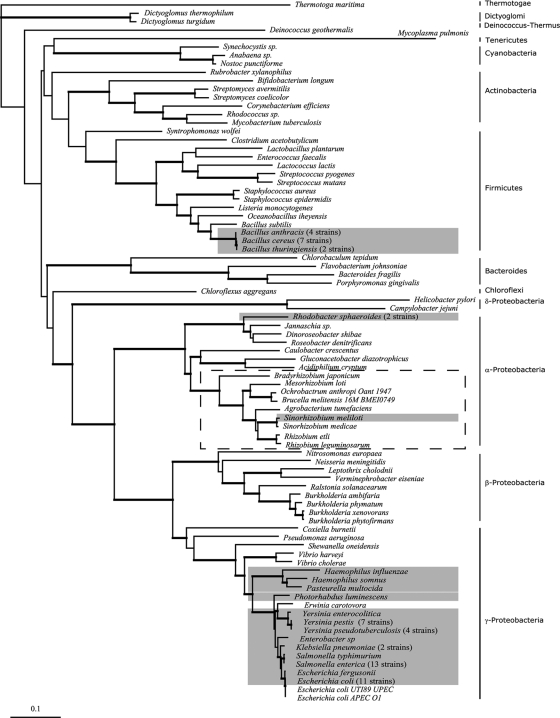
Our sequence/structural and functional studies lead us to predict that all the organisms from group I have LsrB orthologs that function as LsrB-AI-2 receptors. This group contains members from the evolutionarily distant orders of the Enterobacteriales, Pasteurellales, Rhizobiales, Rhodobacterales, and Bacillales. To infer the evolutionary history of the lsrB gene, we determined the phylogenetic tree of all the lsrB gene orthologs from group I (Fig. 5) and compared it to the rpoB housekeeping gene organismal tree constructed with representatives of all major phyla of the Bacteria (Fig. 6). Importantly, the organismal tree recovers all major phyla and classes with high bootstrap support. The relationship among phyla has a lower bootstrap support, but this does not influence our analysis because the phylogenetic relationship between all species with functional lsrB genes (Fig. 6) is also well supported in this tree.
FIG. 5.
Evolutionary history of genes encoding functional LsrB orthologs inferred with maximum likelihood. The lsrB gene tree was constructed with the sequences from all organisms in group I. This is an unrooted phylogram oriented to show maximum congruence with the organismal tree. Numbers on the nodes indicate posterior probability as estimated with MrBayes.
FIG. 6.
Molecular phylogeny of Bacteria estimated with the rpoB gene. The rpoB gene tree was constructed with all the organisms listed in Table 3 (groups I and II) and representative species of all major phyla of the Bacteria. This represents our best inference of the organismal tree. Gray boxes indicate species with functional lsrB genes (group I, Table 3), and the dashed box locates the species with protein sequences in Table 3 that are likely to function as a rhamnose binding protein. The numbers after species names indicate the number of strains analyzed for the respective species. Taxonomic classifications (phyla) are shown on the right. This tree was inferred with neighbor joining, and the branch lengths are scaled to the number of amino acid substitutions per site. Thickened branches indicate high bootstrap support (higher than 75%). This is a measurement of phylogenetic strength between nodes, and this value reflects a high confidence in the inferred relationships between species. Branch lengths are scaled in terms of the number of nucleotide substitutions per site.
This analysis indicates that the phylogenies of lsrB and rpoB largely overlap in their diversification patterns although with some important exceptions. The majority of the species included in group I of Table 3 clustered within the Enterobacteriales and Pasteurellales (both Gammaproteobacteria), and the diversification pattern of the lsrB gene mimics the phylogenetic relationships obtained in the rpoB organismal tree within this group (compare distributions in Fig. 5 and 6); that is, the lsrB gene tree recovers all species groups, and the relationship among members of the Enterobacteriales and Pasteurellales is largely congruent between gene trees. Additionally, the widespread occurrence of LsrB among the Enterobacteriales and Pasteurellales strongly suggests a single origin for this AI-2 receptor that occurred in an ancestor of these organisms after the diversification of the Enterobacteriales and Pasteurellales from the Vibrionales. Nonetheless, the presence of lsrB genes in the Enterobacteriales and Pasteurellales is not ubiquitous, as shown by Erwinia carotovora and two E. coli strains (UTI89 and APECO1), suggesting independent events of gene loss (Fig. 6).
The major discordance between the lsrB and rpoB phylogenies relates to the occurrence of functional LsrB in S. meliloti (Rhizobiales, Alphaproteobacteria), R. sphaeroides (Rhodobacterales, Alphaproteobacteria), and three species of Bacillus (Bacillales, Firmicutes). Specifically, lsrB genes from these species cluster with strong nodal support (Bayesian posterior probability of 1.0) (Fig. 5) with specific clades of the Enterobacteriales and Pasteurellales. Thus, these species appear “misplaced” in the lsrB gene phylogeny (Fig. 5), in contrast with the organismal phylogeny (rpoB tree) (Fig. 6). This type of incongruence is consistent with lateral gene transfer (LGT) events (9, 54).
In the case of Bacillus species, the phylogenetic pattern of the lsrB gene tree reveals that these species cluster with the Pasteurellales. Thus, the occurrence of the lsrB gene in the Bacillus lineage could be explained by a putative LGT event from bacteria of the family Pasteurellaceae. The occurrence of this gene within so many Bacillus species indicates that if such a transfer occurred, enough time has passed for the lineage to diversify into at least three different species (Fig. 5 and 6).
The two species from the Alphaproteobacteria (S. meliloti and R. sphaeroides) are nested within the Enterobacteriales, clustering with the Klebsiella and Enterobacter species. Given the phylogenetic distance that separates S. meliloti and R. sphaeroides (Fig. 6), it is surprising that lsrB gene topology clusters these two species together. The most likely explanation for this occurrence requires at least more than one LGT event. Such a pattern could be obtained if two sequential LGT events had occurred, for example, from one enterobacterium (most likely an ancestor of Klebsiella and Enterobacter) first to a Sinorhizobium species and then to a Rhodobacter species or from one Enterobacter species first to a Rhodobacter species and then to a Sinorhizobium species. However, with the data at hand, it is difficult to predict the specific order of these events. Furthermore, we predict that the proposed LGT to S. meliloti and R. sphaeroides must have been a quite recent event given that no further alphaproteobacterium species were identified with group I LsrB orthologs. Alternatively, we could postulate one LGT event to the ancestor of these Alphaproteobacteria with a massive number of gene losses, but we find this possibility to be very unlikely.
DISCUSSION
A variety of bacterial species have been shown to be capable of responding to AI-2 by the regulation of a range of niche-specific functions, but the mechanisms for AI-2 detection have been characterized in only a few cases (17, 64). This constitutes a major obstacle in work toward an understanding of the function of AI-2. While sequence analysis of bacterial genomes reveals the presence of orthologs of LsrB-like AI-2 receptors in gram-negative as well as gram-positive bacteria (41, 50; this study), establishing which orthologs are, in fact, functional as AI-2 receptors is important for determining if and how these species use AI-2 as a chemical signal. Thus, after analyzing the sequences and predicted structures of LsrB orthologs, we identified criteria for predicting which LsrB orthologs are functional AI-2 receptors and assayed the AI-2 binding ability of selected candidates to test our criteria. Our results not only support our predictions but also provide the first biochemical confirmation of the presence of functional AI-2 receptors in gram-positive bacteria, specifically in B. anthracis and B. cereus.
Our sequence and structural analyses allowed us to categorize the organisms with LsrB orthologs into two different groups. Members of group I have (i) LsrB orthologs with greater than 60% sequence identity with S. Typhimurium LsrB, (ii) orthologs to the other key transport proteins encoded by the lsr operon, and (iii) complete conservation of all six residues that hydrogen bond with AI-2 in S. Typhimurium LsrB (based on structure predictions). On the other hand, in organisms belonging to group II, the LsrB orthologs have a sequence identity below 36%, are missing orthologs to key proteins encoded by the lsr operon, and lack at least two of the six residues in the AI-2 binding pocket. These characteristics led us to hypothesize that the organisms from group I had functional AI-2 binding proteins, whereas the LsrB orthologs in group II were likely to have a different function. In all organisms where the function of either the LsrB protein or its gene has been studied, LsrB has been shown, along with other proteins that form the Lsr transport system, to participate in the uptake of AI-2 (37, 48, 57, 65); thus, we further predicted that organisms with a functional LsrB and orthologs to all the proteins from the Lsr system would take up AI-2. Accordingly, all the organisms from group I tested for the binding of AI-2 by LsrB or for in vivo AI-2 removal (S. Typhimurium, S. meliloti [37], E. coli K-12 [MG1655], B. cereus, and B. anthracis) were capable of both of these functions. None of the proteins from the organisms that we tested from group II (UPEC UT189, R. etli, R. leguminosarum, and A. tumefaciens) were capable of binding AI-2, nor were these organisms able to take up AI-2. In addition, our analysis of predicted structures of the LsrB orthologs identified key binding-site residues that are not conserved in group II organisms. Mutagenesis of the B. anthracis LsrB ortholog (classified as group I and demonstrated to bind AI-2) with the two most common group II substitutions (D166N and A222T) confirmed that these residues are critical for AI-2 binding. This result strongly supports our use of binding-site conservation as a key criterion in identifying group I orthologs.
These results offer experimental evidence that functional LsrB-AI-2 receptors are present in particular members of the Enterobacteriaceae (S. Typhimurium and E. coli), Rhizobiaceae (S. meliloti), and Bacillaceae (B. cereus and B. anthracis), and given the correlation of our experimental results with our classification scheme, we predict that all the other LsrB orthologs from group I are functional AI-2 receptors and that these organisms are competent for AI-2 uptake. Accordingly, we expect that the members of the families Pasteurellaceae and Rhodobacteraceae in group I (Table 3) also have functional AI-2 transporters. On the other hand, we believe that it is likely that all group II members have orthologs that are not involved in AI-2 transport and thus that these organisms do not take up AI-2 via an LsrB-type mechanism. The criteria described here can be used to predict the presence (or absence) of functional LsrB-like AI-2 receptors in newly sequenced species, and as new species are sequenced, we expect the number of organisms in group I to increase.
The large majority of the organisms from group I belong to the Enterobacteriales and the Pasteurellales. This, coupled with the fact that the diversification pattern of the lsrB gene largely mimics the bacterial phylogenetic relationships within this group, is consistent with a single origin for the LsrB-AI-2 receptor that likely occurred in an ancestor of these organisms after the diversification of the Enterobacteriales and the Pasteurellales from the Vibrionales. Thus, the occurrence of LsrB receptors in one species of the Rhizobiales (S. meliloti) and the Rhodobacterales (R. sphaeroides) and in three species of the Bacillales was very surprising and immediately raised the possibility of LGT. The hypothesis of LGT between organisms from the Enterobacteriales or the Pasteurellales and these three orders was supported by the comparison of the lsrB gene tree and the rpoB organismal tree. Specifically, in the lsrB gene tree, Bacillus species are clustered with the Pasteurellales, and S. meliloti and R. sphaeroides are nested within the Enterobacteriales. These are nested patterns where species appeared to be “misplaced” in the gene phylogeny and can be interpreted as an indication of events of LGT. Often, genes that have been acquired by LGT have an atypical nucleotide distribution (reflected in GC content or codon usage) compared with the rest of the genome (25). However, in this case, analysis of GC usage and codon bias provided no information to argue for or against the hypothesis of LGT (data not shown). Certainly, other occurrences such as convergent evolution by natural selection or the ancient origin of lsrB at the base of the Bacteria tree with a large number of events of gene loss could also explain the observed patterns, but since we do not have specific data to support a particular explanation over the others, we favor LGT as the most parsimonious explanation, as it requires the minimum number of assumptions. LGT events are now well accepted as a major force in the evolution of bacterial genomes (8, 23) leading to an increment in the number of genes (35) and pathways (19) and often enabling bacteria to acquire new functions, such as traits associated with pathogenicity, that allow adaptation to novel environments. In the specific cases of S. meliloti and R. sphaeroides, it is intriguing that that these organisms have acquired the AI-2 receptor but not its synthase (LuxS); thus, these organisms have potentially gained the ability to eavesdrop on their neighbor's signal, as previously suggested (37, 41). It will also be interesting to determine the adaptive value of this new function and explore its impact on the physiology of these organisms. LGT has been proposed for other autoinducer receptors and regulators from the LuxI/LuxR family of species-specific quorum-sensing proteins, where it was previously proposed that the acquisition of this family of proteins has benefited certain bacterial species by allowing them to gain an efficient mechanism for regulating virulence genes (8, 16, 26).
Interestingly, the LsrB ortholog in R. leguminosarum bv. trifolii, which we identified as belonging to group II, has been shown to be essential for rhamnose (a methyl-pentose sugar) uptake and growth in this sugar and is thus likely to be a rhamnose binding protein (42, 43). Motivated by this finding, we used the protein sequence of R. leguminosarum bv. trifolii (KEGG identification number pRL110413) to carry out a reciprocal best-hit analysis against all the genome sequences used in the previous analysis. We found that there are 12 orthologs of the R. leguminosarum binding protein (along with the proteins from the rhamnose transport operon) present in group II (shown in Table 3). Thus, these 12 binding proteins are orthologs of both LsrB of S. Typhimurium and the rhamnose binding protein of R. leguminosarum. These proteins have more than 65% sequence identity with the R. leguminosarum protein but less than 36% identity with S. Typhimurium LsrB. We interpret this as strong evidence that these 12 proteins in group II are functioning as rhamnose binding proteins, in agreement with our prediction that they are not AI-2 receptors (these proteins are highlighted in the Table S1 in the supplemental material). These 12 organisms correspond to species belonging to Alphaproteobacteria that cluster together in the organismal rpoB tree (Fig. 6). Interestingly, S. meliloti is the only organism that has an LsrB ortholog belonging to group I and also a different set of proteins that are orthologs to the R. leguminosarum proteins from the rhamnose transport operon, further corroborating our hypothesis that the acquisition of LsrB occurred by LGT in S. meliloti.
While the presence of a functional LsrB ortholog does not prove that AI-2 import is involved in the control of AI-2-mediated behavior, it is suggestive. Accordingly, the function of the Lsr system in AI-2 signaling has already been shown for a member of the Pasteurellaceae, A. actinomycetemcomitans (an organism not present in Table 3 because, to date, its genome is not present in the KEGG database). Shao and coworkers previously showed that this oral pathogen is capable of internalizing AI-2 via the Lsr system and, importantly, that LsrB is required to mediate the complete AI-2-dependent activation of biofilm formation in this organism (48, 49). In other cases such as that of Photorhabdus luminescens, an insect pathogen belonging to the Enterobacteriaceae, the transcription of the lsr operon was shown to be induced by AI-2, and AI-2 was also implicated in the regulation of biofilm formation and motility (24). However, it remains to be demonstrated whether or not the Lsr system is involved in mediating these AI-2-regulated behaviors. Likewise, it will be interesting to determine whether the Lsr system is involved in mediating AI-2 signal transduction in B. cereus and B. anthracis, where AI-2 has been implicated in regulating biofilm formation (2) and growth rate (21). Certainly, the results presented here give support to that possibility.
This study, along with the two previous studies based on sequence analysis (41, 50), also reveals that certain bacteria such as Helicobacter pylori (39), Streptococcus mutans (55), Staphylococcus epidermidis (27), Porphyromonas gingivalis (20, 67), Pseudomonas aeruginosa (14), and Bacillus subtilis (28), which have been shown to respond to AI-2, do not have either of the known types of AI-2 receptors (neither LuxP nor LsrB), and thus we expect that other receptors for AI-2 remain to be discovered. These receptors may be of entirely new classes or may be promiscuous receptors for other small molecules. Novel receptor classes are likely to be identified by approaches that rely on genetic screens to isolate mutants involved in modulating AI-2-regulated phenotypes, and as shown here, integration with approaches that use sequence analysis coupled with biochemical assays may prove very useful. Clearly, an elucidation of the proteins involved in AI-2 recognition and signal relay is essential for studying the potential functions of this class of signal molecule in intra- and interspecies cell-to-cell communication and/or intra- and intercellular signal transduction. The identification and experimental confirmation of functional LsrB receptors in this study open the door to the understanding of the molecular basis of AI-2-mediated behavioral regulation in a variety of new species.
Supplementary Material
Acknowledgments
The work performed in the laboratory of K.B.X. was supported by Fundação para a Ciência e Tecnologia, Portugal (FCT), grant PTDC/BIA-BCM/73676/2006. C.S.P. and P.H.B. were supported by FCT awards SFRH/BD/28543/2006 and SFRH/BPD/26852/2006, respectively. The laboratory of S.T.M. gratefully acknowledges grants from the NIH (grant AI074041), the Camille and Henry Dreyfus Foundation, and Swarthmore College. We also thank program FLAD/NSF project 600-10/2006 for sponsoring the stay of S.T.M. in the laboratory of K.B.X. during his sabbatical.
We thank Chengetai Mahomva and Bella Liu for technical assistance. We are very grateful to Adriano O. Henriques, Gladys Alexandre, James P. Shapleigh, Jeffrey I. Gordon, and Marcus B. Jones for providing the strains used in this study. We thank Bonnie Bassler, Michiko Taga, Brian Hammer, and Isabel Gordo for critically reading the manuscript. We also thank Bonnie Bassler for receiving C.S.P. in her laboratory.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Auger, S., E. Krin, S. Aymerich, and M. Gohar. 2006. Autoinducer 2 affects biofilm formation by Bacillus cereus. Appl. Environ. Microbiol. 72:937-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek, S. H., A. Hartsock, and J. P. Shapleigh. 2008. Agrobacterium tumefaciens C58 uses ActR and FnrN to control nirK and nor expression. J. Bacteriol. 190:78-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 5.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 6.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. [DOI] [PubMed] [Google Scholar]
- 9.Brown, J. R. 2003. Ancient horizontal gene transfer. Nat. Rev. Genet. 4:121-132. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, S. B., and N. D. Young. 2003. OrthoParaMap: distinguishing orthologs from paralogs by integrating comparative genome data and gene phylogenies. BMC Bioinformatics 4:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Case, R. J., Y. Boucher, I. Dahllof, C. Holmstrom, W. F. Doolittle, and S. Kjelleberg. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73:278-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 13.Croxatto, A., J. Pride, A. Hardman, P. Williams, M. Camara, and D. L. Milton. 2004. A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum. Mol. Microbiol. 52:1677-1689. [DOI] [PubMed] [Google Scholar]
- 14.Duan, K., C. Dammel, J. Stein, H. Rabin, and M. G. Surette. 2003. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol. Microbiol. 50:1477-1491. [DOI] [PubMed] [Google Scholar]
- 15.Federle, M. J., and B. L. Bassler. 2003. Interspecies communication in bacteria. J. Clin. Investig. 112:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, K. M., and J. R. Garey. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379-2387. [DOI] [PubMed] [Google Scholar]
- 17.Hardie, K. R., and K. Heurlier. 2008. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat. Rev. Microbiol. 6:635-643. [DOI] [PubMed] [Google Scholar]
- 18.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwasaki, W., and T. Takagi. 2009. Rapid pathway evolution facilitated by horizontal gene transfers across prokaryotic lineages. PLoS Genet. 5:e1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James, C. E., Y. Hasegawa, Y. Park, V. Yeung, G. D. Tribble, M. Kuboniwa, D. R. Demuth, and R. J. Lamont. 2006. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect. Immun. 74:3834-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, M. B., and M. J. Blaser. 2003. Detection of a luxS-signaling molecule in Bacillus anthracis. Infect. Immun. 71:3914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 23.Koonin, E. V., K. S. Makarova, and L. Aravind. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krin, E., N. Chakroun, E. Turlin, A. Givaudan, F. Gaboriau, I. Bonne, J. C. Rousselle, L. Frangeul, C. Lacroix, M. F. Hullo, L. Marisa, A. Danchin, and S. Derzelle. 2006. Pleiotropic role of quorum-sensing autoinducer 2 in Photorhabdus luminescens. Appl. Environ. Microbiol. 72:6439-6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lerat, E., and N. A. Moran. 2004. The evolutionary history of quorum-sensing systems in bacteria. Mol. Biol. Evol. 21:903-913. [DOI] [PubMed] [Google Scholar]
- 27.Li, M., A. E. Villaruz, V. Vadyvaloo, D. E. Sturdevant, and M. Otto. 2008. AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombardia, E., A. J. Rovetto, A. L. Arabolaza, and R. R. Grau. 2006. A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J. Bacteriol. 188:4442-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, L. D., C. K. Yost, M. F. Hynes, and G. Alexandre. 2007. The major chemotaxis gene cluster of Rhizobium leguminosarum bv. viciae is essential for competitive nodulation. Mol. Microbiol. 63:348-362. [DOI] [PubMed] [Google Scholar]
- 31.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 32.Neiditch, M. B., M. J. Federle, S. T. Miller, B. L. Bassler, and F. M. Hughson. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 18:507-518. [DOI] [PubMed] [Google Scholar]
- 33.Neiditch, M. B., M. J. Federle, A. J. Pompeani, R. C. Kelly, D. L. Swem, P. D. Jeffrey, B. L. Bassler, and F. M. Hughson. 2006. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126:1095-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nylander, J. A. A. 2004. MrModeltest version 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 35.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 36.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira, C. S., J. R. McAuley, M. E. Taga, K. B. Xavier, and S. T. Miller. 2008. Sinorhizobium meliloti, a bacterium lacking the autoinducer-2 (AI-2) synthase, responds to AI-2 supplied by other bacteria. Mol. Microbiol. 70:1223-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 39.Rader, B. A., S. R. Campagna, M. F. Semmelhack, B. L. Bassler, and K. Guillemin. 2007. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J. Bacteriol. 189:6109-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reigstad, C. S., S. J. Hultgren, and J. I. Gordon. 2007. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J. Biol. Chem. 282:21259-21267. [DOI] [PubMed] [Google Scholar]
- 41.Rezzonico, F., and B. Duffy. 2008. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson, J. S., M. F. Hynes, and I. J. Oresnik. 2004. A genetic locus necessary for rhamnose uptake and catabolism in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 186:8433-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson, J. S., and I. J. Oresnik. 2007. l-Rhamnose transport is sugar kinase (RhaK) dependent in Rhizobium leguminosarum bv. trifolii. J. Bacteriol. 189:8437-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 45.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 46.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 47.Semmelhack, M. F., S. R. Campagna, M. J. Federle, and B. L. Bassler. 2005. An expeditious synthesis of DPD and boron binding studies. Org. Lett. 7:569-572. [DOI] [PubMed] [Google Scholar]
- 48.Shao, H., D. James, R. J. Lamont, and D. R. Demuth. 2007. Differential interaction of Aggregatibacter (Actinobacillus) actinomycetemcomitans LsrB and RbsB proteins with autoinducer 2. J. Bacteriol. 189:5559-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao, H., R. J. Lamont, and D. R. Demuth. 2007. Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect. Immun. 75:4211-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, J., R. Daniel, I. Wagner-Dobler, and A. P. Zeng. 2004. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 53.Swofford, D. L. 2001. PAUP*, phylogenetic analysis using parsimony. Sinauer Associates, Sunderland, MA.
- 54.Syvanen, M. 1994. Horizontal gene transfer: evidence and possible consequences. Annu. Rev. Genet. 28:237-261. [DOI] [PubMed] [Google Scholar]
- 55.Sztajer, H., A. Lemme, R. Vilchez, S. Schulz, R. Geffers, C. Y. Yip, C. M. Levesque, D. G. Cvitkovitch, and I. Wagner-Dobler. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190:401-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 57.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 58.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 59.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 61.Waters, C. M., and B. L. Bassler. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319-346. [DOI] [PubMed] [Google Scholar]
- 62.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53:291-396. [DOI] [PubMed] [Google Scholar]
- 63.Xavier, K. B., and B. L. Bassler. 2005. Interference with AI-2-mediated bacterial cell-cell communication. Nature 437:750-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 65.Xavier, K. B., and B. L. Bassler. 2005. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187:238-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xavier, K. B., S. T. Miller, W. Lu, J. H. Kim, J. Rabinowitz, I. Pelczer, M. F. Semmelhack, and B. L. Bassler. 2007. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem. Biol. 2:128-136. [DOI] [PubMed] [Google Scholar]
- 67.Yuan, L., J. D. Hillman, and A. Progulske-Fox. 2005. Microarray analysis of quorum-sensing-regulated genes in Porphyromonas gingivalis. Infect. Immun. 73:4146-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zuckerkandl, E., and L. Pauling. 1965. Evolutionary divergence and convergence in proteins, p. 97-166. In H. J. V. V. Bryson (ed.), Evolving genes and proteins. Academic Press, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.