Abstract
The chlorosome envelope of Chlorobaculum tepidum contains 10 proteins that belong to four structural motif families. A previous mutational study (N.-U. Frigaard, H. Li, K. J. Milks, and D. A. Bryant, J. Bacteriol. 186:646-653, 2004) suggested that some of these proteins might have redundant functions. Six multilocus mutants were constructed to test the effects of eliminating the proteins of the CsmC/CsmD and CsmB/CsmF motif families, and the resulting strains were characterized physiologically and biochemically. Mutants lacking all proteins of either motif family still assembled functional chlorosomes, and as measured by growth rates of the mutant strains, light harvesting was affected only at the lowest light intensities tested (9 and 32 μmol photons m−2 s−1). The size, composition, and biogenesis of the mutant chlorosomes differed from those of wild-type chlorosomes. Mutants lacking proteins of the CsmC/CsmD motif family produced smaller chlorosomes than did the wild type, and the Qy absorbance maximum for the bacteriochlorophyll c aggregates in these chlorosomes was strongly blueshifted. Conversely, the chlorosomes of mutants lacking proteins of the CsmB/CsmF motif family were larger than wild-type chlorosomes, and the Qy absorption for their bacteriochlorophyll c aggregates was redshifted. When CsmH was eliminated in addition to other proteins of either motif family, chlorosomes had smaller diameters. These data show that the chlorosome envelope proteins of the CsmB/CsmF and CsmC/CsmD families play important roles in determining chlorosome size as well as the assembly and supramolecular organization of the bacteriochlorophyll c aggregates within the chlorosome.
Green sulfur bacteria (GSB; phylum Chlorobi) are obligate photolithoautotrophs that utilize chlorosomes for light harvesting (2, 13). Chlorosomes additionally occur in some green-nonsulfur bacteria, also known as filamentous anoxygenic phototrophs (phylum Chloroflexi), and in a recently discovered chlorophototrophic member of the phylum Acidobacteria, “Candidatus Chloracidobacterium thermophilum” (2, 3). Chlorosomes are the largest known light-harvesting organelles and can contain up to 250,000 bacteriochlorophyll (BChl) molecules (13, 29, 30, 39). They do not have a fixed stoichiometric ratio of the major pigment, which may be BChl c, d, or e, to any protein component, and as a result they are highly variable in size, shape, and composition. In spite of this structural heterogeneity (34), the detailed molecular and supramolecular structures of the BChls in chlorosomes of Chlorobaculum tepidum were recently solved by combining systems biology, solid-state nuclear magnetic resonance (NMR), cryo-electron microscopy, and molecular modeling (22). The fundamental structural units were found to be syn-anti monomer stacks that form coaxial nanotubes, which have a 2.1-nm spacing between the adjacent BChl layers. In addition to the major BChl species, chlorosomes contain carotenoids, isoprenoid quinones, wax esters, and a small quantity of BChl a. BChl a is known to be associated with CsmA, the most highly conserved protein in chlorosomes (13).
Although the structural organization of the BChl molecules in all chlorosomes may be similar (4, 22, 25, 37, 38), with the exception of CsmA, the composition and sequences of the envelope proteins of chlorosomes of the phyla Chlorobi, Chloroflexi, and Acidobacteria are not well conserved. Blankenship (1) suggested that lateral gene transfer might have been responsible for the presence of the genes for chlorosome biogenesis among some of these three groups of bacteria. However, because chlorosomes are found in each of three, early-diverging bacterial lineages that contain chlorophototrophs, two of which additionally contain homodimeric type 1 reaction centers (2, 3), it is possible that chlorosomes represent one of the earliest types of photosynthetic antennae and were present in a common ancestor of these phyla.
A protein-stabilized, glycolipid envelope surrounds the chlorosome BChls, and this membrane can be considered to be an asymmetric bilayer membrane in which glycolipids form the outer leaflet and the hydrophobic tails of BChls form the inner leaflet (13, 24, 50, 53). In C. tepidum, a genetically tractable model GSB, this envelope contains 10 proteins, which are designated CsmA, CsmB, CsmC, CsmD, CsmE, CsmF, CsmH, CsmI, CsmJ, and CsmX (6-8, 14, 47, 50). The structural organization of these proteins has been studied by cross-linking and immunoblotting, which led to a model for the organization of these proteins in the chlorosome envelope (28, 50, 53). CsmA, the only protein for which any detailed structural information is available, probably binds both BChl a and carotenoids (13, 23, 31, 35, 40) and forms a large, paracrystalline array known as the “baseplate” (8, 23, 28, 35, 36, 42). The structure for apo-CsmA in an organic solvent was recently determined by NMR spectroscopy, and a model for the structural organization of CsmA in the chlorosome baseplate of C. tepidum was proposed (35, 36).
Sequence comparisons suggest that the chlorosome envelope proteins can be assigned to four motif families: 1, CsmA/CsmE; 2, CsmB/CsmF (CsmH); 3, CsmC/CsmD (CsmH); and 4, CsmI/CsmJ/CsmX (48, 50). CsmA and CsmE are 49% identical and are both synthesized as precursors, which are proteolytically processed by the removal of ∼20 amino acids at their carboxy termini to generate the mature polypeptides (7, 8). CsmB and CsmF are 29% identical and 63% similar in sequence (6, 50). Moreover, the amino-terminal domain of CsmH is related in sequence to these two proteins (50). The CsmC and CsmD proteins are 26% identical and 45% similar in sequence, and these two proteins additionally share sequence similarity to the carboxyl-terminal region of CsmH. The other three chlorosome proteins (CsmI, CsmJ, and CsmX) share some sequence similarities to the precursor forms of CsmA and CsmE in their carboxyl-terminal regions, while their amino-terminal domains are obviously related to adrenodoxin-type [2Fe-2S] ferredoxins (47-50). These sequence relationships strongly imply that gene duplication and divergence have occurred among a small number of ancestral gene types, and these observations additionally suggest that some of these proteins might be functionally redundant (47, 50). This view was supported by mutational studies that showed that only CsmA was essential for the viability of C. tepidum. Mutants lacking any other single chlorosome protein still assembled functional chlorosomes that were similar in pigment composition and functionality to those of the wild type (14).
Because of the possible functional redundancy of chlorosome proteins of the different motif classes, double, triple, and quadruple mutants were constructed to study the roles of the CsmC/CsmD/CsmH and CsmB/CsmF/CsmH protein motif families in chlorosome biogenesis and structure. Mutants lacking CsmI, CsmJ, and CsmX, which form the Fe/S motif family of envelope proteins, were also constructed, and these mutants will be described in detail elsewhere (27; H. Li, N.-U. Frigaard, and D. A. Bryant, unpublished data). The results presented here show that functional chlorosomes assemble in the complete absence of proteins of the CsmC/CsmD or CsmB/CsmF motif families, but the size, shape, and composition of the resulting chlorosomes are altered. The results suggest that the chlorosome envelope proteins may also influence the structural organization of the BChls in chlorosomes and thus help to define chlorosome assembly and shape.
MATERIALS AND METHODS
Plasmid construction in Escherichia coli.
Escherichia coli DH10B cells [genotype F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG] were used for recombinant DNA manipulations. Plasmids pCT841 and pCT720 (see Fig. S1 in the supplemental material), which contain DNA fragments carrying csmC and csmB interrupted by the aadA resistance marker fragments, respectively, were previously described (5, 14). Plasmid pUCB::aacCI was constructed from pCT841 by deletion of the aadA resistance marker and insertion of the aacCI gentamicin resistance marker (Gmr) at the same PstI restriction sites. Plasmid pUCC::aacCI was constructed from pCT720 by insertion of the aacCI resistance marker (Gmr) at the NsiI restriction site within the coding sequence of the csmC gene. For cloning of the csmH gene, the sequence was modified by PCR with the primers 5′ CAA ATC AAA CCC ATA TGG CTA CCG AA (NdeI) and 5′ CGA ATG CCT TGG ATC CGT TCT ATT CA (BamHI), which introduced NdeI and BamHI sites at the 5′ and 3′ ends of the gene, respectively (mutated bases are indicated in bold, and the introduced restriction sites are underlined). Plasmid pUCH13 (see Fig. S1 in the supplemental material) was constructed by cloning the modified csmH gene into the pUC19 vector at the NdeI and BamHI restriction sites, and the plasmid pUCH::cat-ermC was constructed by inserting the cat-ermC erythromycin resistance marker (Emr) into the pUCH13 NheI restriction site within the coding sequence of the csmH gene.
Mutant construction and verification in C. tepidum.
All mutations were constructed in C. tepidum strain WT2321 (51), which is a plating strain derived from the type strain ATCC 49652 (52). C. tepidum cells were grown in CL medium or on CP plates as described previously (12, 17). Manipulations and small-scale batch growth studies were performed at 42°C in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) with an N2-CO2-H2 atmosphere (85:10:5, vol/vol/vol). Large-volume cultures were grown in tightly sealed 2-liter bottles in a water bath at 42 to 45°C. For growth rate studies, the cells were grown in CL medium in 20-ml screw-cap glass tubes on a radial rotator at 45°C. The optical density at 600 nm (OD600) of cell cultures was measured as a function of time for growth rate calculations.
The csmF::aadA, csmD::aadA, and csmED::aadA mutants of C. tepidum were previously described (14). For the construction of the csmB csmF, csmC csmD, and csmC csmED mutants, cells were grown to late exponential phase and harvested by centrifugation for natural transformation (12, 17) with plasmid pUCB::aacC or pUCC::aacC that had been digested with AhdI. Pelleted cells (from 1 ml of culture) and linearized DNA (10 μg) were mixed in CL medium (100 μl), spotted on a nonselective CP plate, and incubated for 10 to 18 h. The cell patch was scraped off and streaked on CP plates containing the appropriate antibiotics, and individual dark-green, transformant colonies appeared after incubation for 5 to 6 days. After two additional rounds of streaking on selective medium, single colonies were inoculated into CL medium and grown into dense cultures.
The segregation status of wild-type and mutant alleles for each gene was determined by PCR with specific primers (27) for the csm or antibiotic resistance genes and template DNAs derived from the transformants (data not shown). After the csmB csmF, csmC csmD, and csmC csmED mutant strains were verified, cells of these strains were subsequently transformed with pUCH::cat-ermC to produce csmB csmF csmH, csmC csmD csmH, and csmC csmED csmH strains. The six mutant strains were stored as frozen stocks at −80°C in 15% (vol/vol) glycerol.
Chlorosome isolation.
Chlorosomes were isolated on sucrose gradients by ultracentrifugation as described previously (28, 50). Cells (2-liter batch cultures) were grown to late exponential phase, and cells were disrupted by three passages through a French pressure cell (124 MPa, 4°C) in isolation buffer (2 M NaSCN, 10 mM Tris-HCl, 5 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, pH 7.5). Chlorosome-enriched fractions were pelleted by ultracentrifugation at 220,000 × g for 2 h at 4°C, and chlorosomes were then purified further on 7 to 47% (wt/vol) continuous sucrose gradients by ultracentrifugation at 220,000 × g for 18 h at 4°C. The chlorosome fraction was collected, diluted fourfold with phosphate-buffered saline (PBS; 10 mM KH2PO4, 50 mM NaCl, pH 7.0), and centrifuged at 240,000 × g for 1.5 h. The resulting pellet was resuspended in PBS and centrifuged again, and the firmly pelleted chlorosomes were resuspended in a small volume of PBS containing 1 mM phenylmethanesulfonyl fluoride and 2 mM dithiothreitol. The purified chlorosomes were aliquoted into microcentrifuge tubes and stored at −80°C until used.
Analysis of proteins.
Protein composition was analyzed on 16% T and 3.3% C polyacrylamide gels in the presence of sodium dodecyl sulfate (SDS-PAGE) with the Tris-Tricine electrophoresis system previously described (28, 41). The proteins from chlorosome samples containing 10 μg of BChl c were loaded for silver staining, and the proteins from samples containing 100 μg BChl c were loaded for immunodetection. Silver staining and immunoblotting were performed as described previously (28).
The Lowry protein assay was used for determination of total protein amounts in C. tepidum cells and subcellular fractions. The assay was performed using the procedure described by Sigma (procedure P5656; Sigma, St. Louis, MO) with slight modifications. Proteins were precipitated from whole cells (0.5 to 1.0 ml of dense culture) or chlorosomes (10 to 20 μl) with methanol or acetone and pelleted by centrifugation at 13,000 × g for 15 min. Bovine serum albumin (0 to 400 μg; Pierce, Rockford, IL) was used as the standard.
Spectroscopy and pigment content determination.
Pigments were extracted by diluting chlorosomes 1:100 in acetone-methanol (7:2, vol/vol) or by resuspending pelleted cells in an equal volume of acetone-methanol (7:2, vol/vol). Denatured proteins were removed by centrifugation at 13,000 × g at 4°C for 10 min. The extracted pigments were diluted 1:100 (chlorosome pigments) or 1:10 (cell pigments) in organic solvents, and the concentration of BChl c was determined by absorption spectroscopy using the following specific absorption coefficients: 86 liters g−1 cm−1 at 669 nm for methanol (43), 92.6 liters g−1 cm−1 at 662.5 nm for acetone (43), or 91 liters g−1 cm−1 at 666 nm for acetone-methanol (7:2, vol/vol) (33).
For high-performance liquid chromatography (HPLC) analysis, the extracted pigment mixture was centrifuged, filtered, and mixed with 0.1 volume of 1 M ammonium acetate before immediate injection onto the HPLC as previously described (18). The HPLC system used was an Agilent 1100 series system (Agilent Technologies, Palo Alto, CA) with a 4.5-mm × 25-cm Discovery C18 column (Supelco, Bellefonte, PA) under the control of Agilent ChemStation software. Pigment contents were determined by absorption spectroscopy by using the following absorption coefficients: BChl c, 20 liters g−1 cm−1 at 635 nm (43); BChl a, 60 liters g−1 cm−1 at 770 nm (46); carotenoids, 265 liters g−1 cm−1 at 491 nm (24); chlorobiumquinone, 17 liters g−1 cm−1 at 270 nm (21); menaquinone 7, 26 liters g−1 cm−1 at 270 nm (10).
Absorption spectra of chlorosomes and whole cells were measured from 350 to 900 nm with Visionlite Scan software (Thermo Electronic, Madison, WI) on a Genesys 10 spectrophotometer (ThermoSpectronic, Rochester, NY). Chlorosomes were diluted at the ratio of 1:10,000, and cells were diluted at the ratio of 1:10 in PBS before the absorption spectra were taken. Fluorescence emission spectra of chlorosomes or whole cells were measured under reducing conditions with an SLM-Aminco 8100 series 2 spectrofluorometer (SLM Instruments, Urbana, IL) with 460-nm excitation. Chlorosomes or cells were diluted in KH2PO4 buffer (10 mM KH2PO4, pH 7.0) to a maximum OD of 0.2 (at ∼750 nm) and incubated with 100 mM sodium dithionite in closed cuvettes for at least 1 h, and the fluorescence emission spectra were recorded with 460-nm excitation provided by an ELXE-500 xenon lamp at 22°C from 700 to 850 nm.
Electron microscopy.
Chlorosomes were diluted with distilled water to a maximum OD of 2 to 3 (at ∼750 nm) and adsorbed onto Formvar-coated Cu grids (400 mesh) by incubation at room temperature for 3 min. The chlorosomes were stained with uranyl acetate (2%, wt/vol) for 30 s in the dark, and images were recorded with a JEOL 1200 EXII transmission electron microscope (Peabody, MA).
RESULTS
Growth studies of chlorosome mutants.
Table 1 shows the growth rates of the wild type and six C. tepidum mutants lacking combinations of proteins of the CsmC/CsmD (±CsmE) motif family or of the CsmB/CsmF motif family at four different light intensities (9, 32, 80, and 210 μmol photons m−2 s−1). At saturating (∼80 μmol photons m−2 s−1) or higher (210 μmol photons m−2 s−1) light intensities, the growth rates of the mutants were nearly identical to that of the wild-type strain. At the lowest light intensity tested (9 μmol photons m−2 s−1), the csmC csmD and csmC csmED mutants had similar growth rates, and the csmC csmD csmH and csmC csmED csmH mutants also had similar growth rates. These results agreed with previous results showing that CsmE is not an essential chlorosome component (14). The csmC csmD csmH and csmC csmED csmH mutants had lower growth rates than did the wild type at low light intensity but had growth rates similar to that of the wild type at high light intensities (Table 1). Similarly, the growth of the csmB csmF and csmB csmF csmH mutants differed from the wild type at the lowest light intensity tested. Under these conditions, the csmB csmF mutant had a growth rate that was ∼75% of that of the wild type, but the csmB csmF csmH mutant grew only about half as fast as did the wild type. When the light intensity was increased to 32 μmol photons m−2 s−1, the growth rates of these two mutants were nearly the same as that of the wild type (95% and 92% of the wild-type growth rate, respectively). These data showed that mutants lacking three or even four chlorosome envelope proteins still assembled functional chlorosomes that are capable of efficient light harvesting.
TABLE 1.
Growth rates of the wild type and mutants lacking proteins of the CsmC/CsmD or CsmB/CsmF motifs under different light intensities
| Strain | Growth rate (h−1)a at growth light intensity (μmol photons m−2 s−1): |
|||
|---|---|---|---|---|
| 9 | 32 | 80 | 210 | |
| Wild type | 0.036 ± 0.004 | 0.214 ± 0.004 | 0.244 ± 0.005 | 0.264 ± 0.007 |
| csmC csmD mutant | 0.032 ± 0.003 | 0.200 ± 0.004 | 0.246 ± 0.007 | 0.270 ± 0.005 |
| csmC csmED mutant | 0.029 ± 0.004 | 0.206 ± 0.005 | 0.245 ± 0.011 | 0.271 ± 0.008 |
| csmC csmD csmH mutant | 0.028 ± 0.002 | 0.184 ± 0.005 | 0.232 ± 0.008 | 0.241 ± 0.005 |
| csmC csmED csmH mutant | 0.027 ± 0.005 | 0.189 ± 0.002 | 0.238 ± 0.002 | 0.254 ± 0.004 |
| csmB csmF mutant | 0.028 ± 0.003 | 0.202 ± 0.007 | 0.252 ± 0.005 | 0.280 ± 0.004 |
| csmB csmF csmH mutant | 0.020 ± 0.004 | 0.198 ± 0.004 | 0.236 ± 0.002 | 0.254 ± 0.008 |
The growth rates (mean values and standard deviations) are the results from at least two independent measurements.
Cellular absorption and fluorescence profiles and pigment contents.
Figure S2 in the supplemental material shows the absorption spectra of cell suspensions of the wild-type and mutant strains lacking proteins of the CsmC/CsmD or the CsmB/CsmF motif families. These spectra were recorded at equal cell densities based on OD600, and the cultures had been grown under the same light intensity and to a similar OD. The absorption spectrum of the wild type (shown as solid lines in all panels of Fig. S2 in the supplemental material) exhibited two major absorption maxima associated with BChl c: the Qy and Soret absorption peaks at about 752 nm and 462 nm, respectively. Carotenoid absorbance was observed as a shoulder at ∼520 nm, and BChl a absorption was observed as a weak shoulder at about 810 nm. As judged from the amplitudes of the Qy absorption bands for BChl c, the csmC csmD, csmC csmED, csmC csmD csmH, and csmC csmED csmH mutants (shown as dashed lines in Fig. S2A to D in the supplemental material) apparently contained less BChl c per cell or had chlorosomes with BChl c aggregated differently, thereby causing a lower molar absorptivity than that for the wild type. The Qy absorption maxima of the BChl c aggregates in the csmC csmD and csmC csmED mutants occurred at 748 nm, a 4-nm blueshift relative to that of the wild type at 752 nm. The absorbance properties of the csmB csmF and csmB csmF csmH mutant cells were very similar to those of the wild type, except that the Qy absorption maximum of BChl c was slightly redshifted to 756 nm for the csmB csmF mutant and to 753 nm for the csmB csmF csmH mutant. Fluorescence emission spectra of all strains were recorded under reducing conditions, and all spectra were very similar (Table 2). The fluorescence emission maximum of BChl c was detected at about 771 nm (at 768 nm for the csmC csmD and csmC csmED mutants), and the emission maximum for BChl a occurred at ∼805 nm in all cases.
TABLE 2.
Properties of cells of the wild-type strain and mutants lacking proteins of the CsmC/CsmD or CsmB/CsmF motif familiesa
| Strain | Maximum (nm) |
Level of pigment (mg/g of cellular protein) |
||||||
|---|---|---|---|---|---|---|---|---|
| Absorption | Fluorescenceb | BChl c | BChl a | Carotenoids | Menaquinone 7 | Chlorobiumquinone | Total quinones | |
| Wild type | 752 ± 0 | 770 ± 1 | 232 ± 18 | 2.5 ± 0.1 | 13.6 ± 0.7 | 9 ± 1 | 3.0 ± 0.4 | 12 ± 2 |
| csmC csmD mutant | 748 ± 1 | 768 ± 2 | 191 ± 18 | 2.4 ± 0.1 | 12.8 ± 0.7 | 11 ± 0 | 2.1 ± 0.3 | 13 ± 1 |
| csmC csmED mutant | 748 ± 1 | 768 ± 1 | 203 ± 16 | 2.2 ± 0.1 | 13.0 ± 0.5 | 11 ± 1 | 2.4 ± 0 | 13 ± 2 |
| csmC csmD csmH mutant | 751 ± 0 | 770 ± 2 | 144 ± 6 | 2.2 ± 0.1 | 14.4 ± 0.5 | 8 ± 1 | 2.5 ± 0.1 | 11 ± 1 |
| csmC csmED csmH mutant | 751 ± 1 | 770 ± 1 | 123 ± 8 | 2.1 ± 0.1 | 12.5 ± 0.1 | 8 ± 1 | 3.4 ± 2.1 | 11 ± 3 |
| csmB csmF mutant | 756 ± 2 | 771 ± 1 | 191 ± 37 | 2.2 ± 0.1 | 11.8 ± 0.3 | 10 ± 0 | 1.9 ± 0.3 | 12 ± 1 |
| csmB csmF csmH mutant | 753 ± 1 | 771 ± 1 | 192 ± 31 | 2.5 ± 0.2 | 12.0 ± 0.2 | 10 ± 1 | 3.2 ± 0.8 | 13 ± 2 |
The cells were grown at a light intensity of about 30 μmol photons m−2 s−1 to the mid-exponential phase (OD600 of ∼1.0). Reported values are the means and standard deviations of at least two measurements from independent cell cultures.
Fluorescence emission spectra were taken under reducing conditions in the presence of 10 mM sodium dithionite.
HPLC analyses were performed to compare the total cellular pigment contents in the wild type and the mutant strains. These measurements were performed at least twice with cells from independent, mid-exponential-phase cultures (OD600, ∼1.0), and the average values are presented in Table 2. Cellular protein contents were used as the reference point for comparisons of the pigment contents in the wild type and the mutant strains. Per g of cellular protein, wild-type cells grown at ∼30 μmol photons m−2 s−1 contained 232 mg of BChl c, 2.5 mg of BChl a, 13.6 mg of carotenoids, and 12 mg of quinones (Table 2). The BChl a and quinone contents varied slightly in the mutant strains. This may have been due to minor differences in the growth stage or growth conditions or perhaps to some variability in the extraction procedure or its efficiency. The carotenoid contents decreased very slightly, if at all, from 12.0 to 11.8 mg per g of cellular protein in the mutants lacking proteins of the CsmB/CsmF motif (the csmB csmF and csmB csmF csmH mutants). The BChl c contents in the csmC csmD, csmC csmED, csmB csmF, and csmB csmF csmH mutants were slightly reduced (∼10 to 15%) compared to that of the wild type. However, the BChl c contents were much lower, by 40 to 45% (to 144 and 123 mg of BChl c per g of cellular protein), in the csmC csmD csmH and csmC csmED csmH mutants, respectively. Therefore, both the whole-cell absorption spectra (see Fig. S2C and D in the supplemental material) and HPLC pigment analyses (Table 2) consistently showed that the csmC csmD csmH and csmC csmED csmH mutant cells contained less BChl c than did the wild type.
Chlorosome isolation and protein analyses.
Chlorosomes were isolated from batch cultures of each mutant strain. After cell disruption and low-speed centrifugation to remove the cell debris, the supernatant containing the chlorosomes was subjected to ultracentrifugation, which yielded a chlorosome-enriched pellet and a supernatant enriched in soluble proteins. After ultracentrifugation, an orange fraction was observed in the uppermost portion of the supernatants for all of the mutants lacking multiple proteins of the CsmC/CsmD or CsmB/CsmF motif families. Except for the csmC mutant, no similar fraction was observed for the wild type or any of the previously characterized single mutants (14). Extraction of the pigments from this orange fraction into 100% methanol showed that this fraction mostly contained carotenoids (λmax = 460 nm and 490 nm) as well as smaller amounts of BChl c (λmax = 669 nm) and BChl a (λmax = 770 nm). The ratio of carotenoids to BChl c in this fraction was about 20-fold higher than that for wild-type chlorosomes. SDS-PAGE showed that, together with a large number of other cellular proteins, CsmA and CsmB were present in this orange fraction. The properties of this fraction were similar to those of carotenosomes, which accumulate when BChl c biosynthesis is blocked (15). Thus, this orange fraction may contain intermediates of chlorosome biogenesis near the beginning of BChl c incorporation. As estimated from the carotenoid and BChl a contents, this fraction represented only ∼1% of the total chlorosomes.
The chlorosome-containing pellet from the first ultracentrifugation step was resuspended and loaded onto the top of sucrose gradients. For the wild-type strain, the chlorosome fraction appeared as a highly concentrated, dark green zone in the upper quarter of the sucrose gradient (see Fig. S3 in the supplemental material). A faint, light-green band, which contained mainly membrane fragments, was observed at about 35% (wt/vol) sucrose, and other cell debris was observed at the bottom of the tube as a brownish pellet. The gradients for the csmC csmD and csmC csmED mutants were similar in appearance to those for the wild-type strain, but the gradients for the csmC csmD csmH, csmC csmED csmH, csmB csmF, and csmB csmF csmH mutants were obviously different. For these mutants the chlorosome band was observed at the normal position, but an additional green fraction was present, and it occurred just below the major chlorosome-containing fraction on the sucrose gradients. This additional fraction, here designated fraction 2, occurred at sucrose concentrations of ∼27 to 37% (wt/vol) and overlapped with the fraction containing the membrane fragments. For the csmC csmD csmH and csmC csmED csmH mutants, fraction 2 was also a well-defined band with relatively distinct boundaries (see Fig. S3 in the supplemental material). Fraction 2 was processed in the same manner as were chlorosomes: fractions were collected from the sucrose gradients, pelleted by two more rounds of ultracentrifugation, and resuspended in PBS.
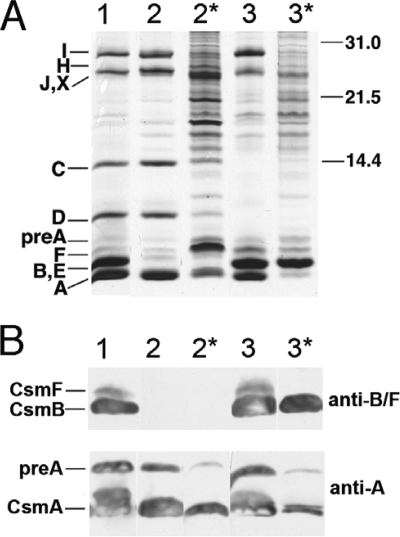
The protein profiles of chlorosomes and fraction 2 samples from the wild type and the csmB csmF and the csmC csmD csmH mutants were analyzed by SDS-PAGE, and proteins were detected by silver staining and immunoblotting (Fig. 1). Silver staining revealed many proteins between 14 kDa and 31 kDa in the fraction 2 samples from the csmB csmF and the csmC csmD csmH mutants (Fig. 1A). Most of these proteins were probably derived from the contaminating membrane fragments in the csmB csmF sample. CsmA, the most abundant protein in wild-type chlorosomes, was present in fraction 2 from the csmB csmF mutant (Fig. 1B, lane 2*), but CsmA was only a minor component of fraction 2 of the csmC csmD csmH mutant (Fig. 1B, lane 3*). The most abundant protein in fraction 2 of the csmC csmD csmH mutant is a protein migrating at the same position as CsmB and CsmE. Figure 1B shows results of immunoblotting analyses of chlorosomes and fraction 2 with anti-CsmA and anti-CsmF antibodies (the latter cross-react with both CsmF and CsmB) (28, 50). Immunoblotting with anti-CsmF antibodies confirmed that the abundant protein in fraction 2 of the csmC csmD csmH mutant was CsmB (Fig. 1B, lane 3*). The amount of CsmB in this sample greatly exceeded the amount of CsmA. Interestingly, the most abundant protein in fraction 2 of the csmB csmF mutant was a protein with an apparent mass slightly smaller than that of pre-CsmA (Fig. 1, lane 2*). This abundant polypeptide did not cross-react with antibodies to CsmA, CsmB (Fig. 1B), or any other chlorosome protein (data not shown). This polypeptide was subsequently identified by mass spectrometry as the product of open reading frame CT0426, whose product is similar in sequence to the Flp/Fap pilin components of various bacteria (data not shown).
FIG. 1.
SDS-PAGE analysis and immunoblotting of chlorosome fractions from wild-type and mutant strains. (A) Silver-stained SDS-polyacrylamide gels showing the protein composition of chlorosomes and fraction 2 samples. Csm polypeptides are identified at the left, and the migration positions of molecular mass markers (sizes in kDa) are indicated at the right. Lane 1, chlorosomes from the wild type; lane 2, chlorosomes from the csmB csmF mutant; lane 2*, fraction 2 from the csmB csmF mutant; lane 3, chlorosomes from the csmC csmD csmH mutant; lane 3*, fraction 2 from the csmC csmD csmH mutant. Samples containing 2 μg BChl c were loaded for lanes 2* and 3*, and samples containing 10 μg BChl c were loaded for lanes 1, 2, and 3. (B) Immunoblots probed with anti-CsmF and anti-CsmA polyclonal antibodies. Polyclonal antibodies for CsmF cross-react with both CsmF and CsmB. Lane 1, chlorosomes from the wild type; lane 2, chlorosomes from the csmB csmF mutant; lane 2*, fraction 2 from the csmB csmF mutant; lane 3, chlorosomes from the csmC csmD csmH mutant; lane 3*, fraction 2 from the csmC csmD csmH mutant. Samples containing 20 μg of BChl c were loaded for lanes 2* and 3*, and samples containing 100 μg BChl c were loaded for lanes 1, 2, and 3.
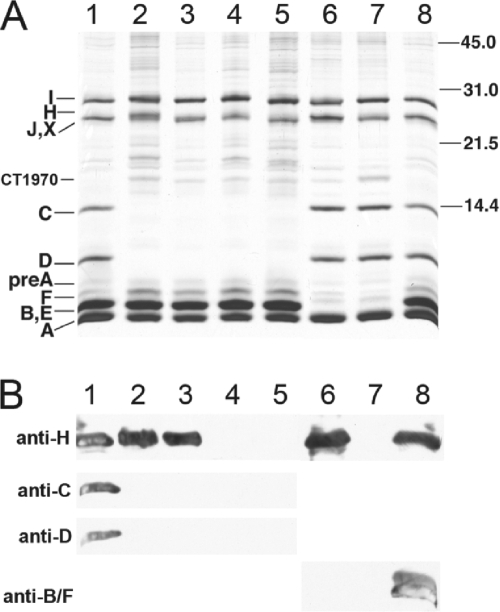
In addition to the fraction 2 samples of the various mutants described above, the chlorosomes of each mutant strain were also analyzed by SDS-PAGE, and proteins were detected and identified by silver staining and immunoblotting (Fig. 2). Immunoblotting with antibodies against chlorosome proteins verified that the CsmB, CsmC, CsmD, CsmF, and CsmH proteins were completely missing as expected in the appropriate double and triple mutant strains (Fig. 2B). Silver staining of the chlorosome proteins showed that the relative amounts of the remaining proteins (i.e., CsmA, CsmC, CsmD, CsmI, and CsmJ) in the csmB csmF and csmB csmF csmH chlorosomes were present at about the same levels as in wild-type chlorosomes (Fig. 2A, lanes 6 and 7). However, the relative amounts of CsmF and CsmB seemed to increase slightly in chlorosomes from the csmC csmD, csmC csmED, csmC csmD csmH, and csmC csmED csmH mutants, and there also appeared to be a slight decrease in the relative CsmA content (compare lanes 2, 3, 4, and 5 in Fig. 2A).
FIG. 2.
Protein composition analysis of chlorosomes isolated from the wild-type strain and the mutants lacking proteins of the CsmC/CsmD or CsmB/CsmF motif families. (A) SDS-PAGE with proteins detected by silver staining. Csm polypeptides are identified at the left, and the migration positions of molecular mass markers (sizes in kDa) are indicated at the right. The proteins from chlorosome samples containing 10 μg of BChl c were loaded for each lane. (B) Immunoblots probed with anti-CsmH, anti-CsmC, anti-CsmD, and anti-CsmF polyclonal antibodies (the anti-CsmF polyclonal antibodies cross-react with both CsmF and CsmB). Chlorosomes containing 100 μg of BChl c were loaded for each lane. Lanes 1 and 8, wild-type chlorosomes; lanes 2 to 5, chlorosomes from the csmC csmD, csmC csmED, csmC csmD csmH, and csmC csmED csmH mutants; lanes 6 and 7, chlorosomes from the csmB csmF and csmB csmF csmH mutants, respectively.
Absorption profile and pigment contents of the chlorosomes.
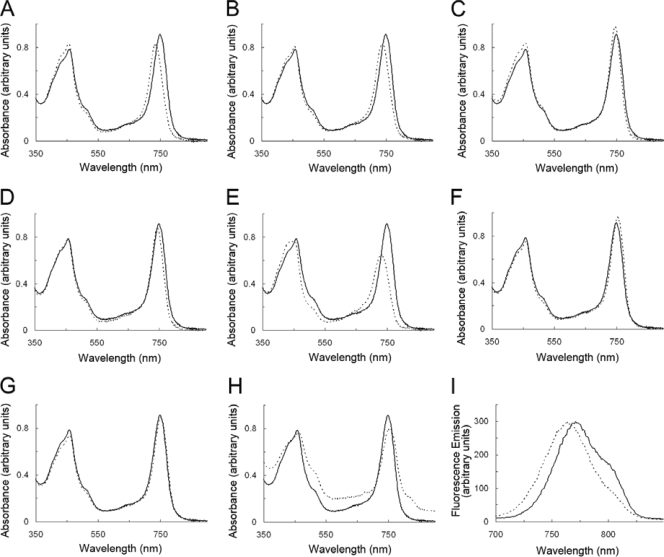
The absorption spectra of wild-type chlorosomes (solid lines) and chlorosomes (or fraction 2 samples) from the various mutants (dashed lines) are shown in Fig. 3A to H. The absorption spectra were taken in aqueous buffer and have been normalized on the basis of their contents of monomeric BChl c (OD669 in 100% methanol = 1). The Qy absorption maximum for BChl c in the wild-type chlorosomes was about 750 nm. Isolated chlorosomes from the csmC csmD and csmC csmED mutants had Qy absorption maxima that were blueshifted by about 12 nm (Fig. 3A and B). The Qy absorption maximum for BChl c was also blueshifted about 5 nm for chlorosomes from the csmC csmD csmH and csmC csmED csmH mutants (Fig. 3C and D). In contrast to the csmC csmD csmH chlorosomes, the fraction 2 sample from the csmC csmD csmH mutant had a Qy absorption maximum at 732 nm, which was blueshifted by 18 nm relative to that of the wild-type chlorosomes (Fig. 3E). In addition, the amplitude of the Qy absorption peak of this second fraction sample was much smaller than that for any other chlorosome sample, although all spectra were normalized to the same concentration of BChl c. These large deviations in properties relative to wild-type chlorosomes clearly indicate that the BChl c organization was modified in the fraction 2 sample from the csmC csmD csmH mutant. The Qy absorption maxima for BChl c in the csmB csmF and csmB csmF csmH chlorosomes were redshifted by 6 and 3 nm compared to the wild type (Fig. 3F and G). Similarly, the Qy absorption maximum of BChl c in the fraction 2 sample from the csmB csmF mutant also was redshifted about 4 nm (Fig. 3H). A shoulder at 810 nm was observed for this sample, which was probably caused by CsmA-bound BChl a and some Fenna-Matthews-Olson BChl a-binding protein and reaction centers from the contaminating membranes.
FIG. 3.
Absorption spectra (A to H) and fluorescence emission spectra (I) of chlorosomes from the wild type (solid lines) and the mutants lacking proteins of the CsmC/CsmD or the CsmB/CsmF motif families (dashed lines). Spectra were normalized such that the absorption shown is that for cells containing an equivalent amount of monomeric BChl c (OD669 in methanol = 1). (A) Chlorosomes from the csmC csmD mutant; (B) chlorosomes from the csmC csmED mutant; (C) chlorosomes from the csmC csmD csmH mutant; (D) chlorosomes from the csmC csmED csmH mutant; (E) fraction 2 from the csmC csmD csmH mutant; (F) chlorosomes from the csmB csmF mutant; (G) chlorosomes from the csmB csmF csmH mutant; (H) fraction 2 from the csmB csmF mutant; (I) fluorescence emission spectra of wild-type chlorosomes (solid line) and fraction 2 from the csmC csmD csmH mutant (dashed line).
Fluorescence emission spectra of the above-described samples were recorded under reducing conditions with 10 mM sodium dithionite (data not shown). The BChl c emission maximum was blueshifted by 2 to 6 nm in the samples from the csmC csmD, csmC csmED, csmC csmD csmH, and csmC csmED csmH mutants and redshifted by ∼2 nm in the samples from the csmB csmF and csmB csmF csmH mutants. The emission maximum of BChl a appeared as a shoulder at about 805 nm in all the samples; however, consistent with the low content of CsmA in this fraction, this 805-nm emission was barely detected in the fraction 2 sample from the csmC csmD csmH mutant (Fig. 3I). The absorption and fluorescence emission maxima of the chlorosome samples from the wild-type and mutant strains are summarized in Table S1 in the supplemental material.
HPLC analyses were used to determine the pigment contents for the chlorosomes and the fraction 2 samples from the wild-type and mutant strains (see Table S1 in the supplemental material). Using BChl c content as the basis for comparison, the BChl a content was 10 to 12 mg per g of BChl c in the wild-type and most of the mutant chlorosomes, but the BChl a content was notably higher (1.7-fold and 5-fold) in the fraction 2 samples from the csmC csmD csmH and csmB csmF mutants. The increased BChl a contents in these samples were probably due to contamination from cytoplasmic membranes, which are enriched in Fenna-Matthews-Olson BChl a-binding protein and reaction centers. The BChl a contents of the csmC csmD csmH and the csmC csmED csmH chlorosomes, as well as fraction 2 from the csmC csmED csmH mutant, were ∼50% higher than those of wild-type chlorosomes. Considering the decreased cellular BChl c contents in the csmC csmD csmH and csmC csmED csmH mutants, the relatively high levels of BChl a in these samples were possibly due to the decreased cellular (and thus chlorosome) BChl c contents. Wild-type chlorosomes contained 57 mg of carotenoids per g of BChl c, and this value was about 30% greater in the chlorosomes of the csmC csmD csmH and csmC csmED csmH mutants. In contrast, chlorosomes from the csmB csmF and csmB csmF csmH mutants contained lower levels of carotenoids (∼25% less than that of the wild type). The quinone contents (menaquinone 7 and chlorobiumquinone) were similar in the wild-type and mutant samples except for the twofold increase in menaquinone contents in the chlorosomes of the csmC csmD csmH and csmC csmED csmH mutants and in fraction 2 from the csmC csmED csmH mutant (see Table S1 in the supplemental material).
Electron microscopy of chlorosomes.
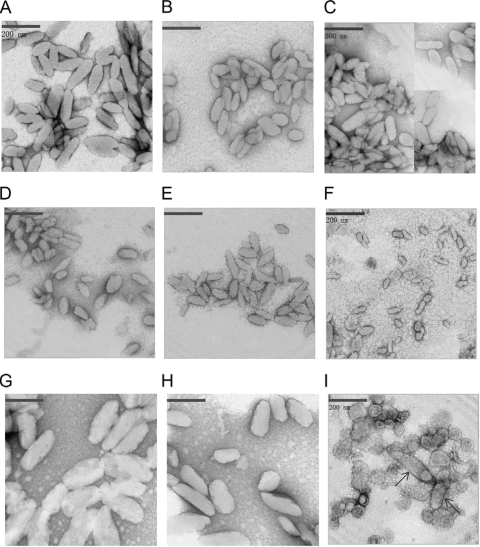
Electron microscopy revealed obvious size and shape differences among the chlorosomes isolated from the wild-type and the mutant strains lacking proteins of the CsmC/CsmD or CsmB/CsmF motif families (see Fig. 4 and Table 3). The average dimensions of wild-type chlorosomes were 165 ± 60 nm (length) and 60 ± 25 nm (width), with a length-to-width ratio of 2.7 ± 0.6 (Fig. 4A; Table 3). Chlorosomes from the csmC csmD, csmC csmED, csmC csmD csmH, and csmC csmED csmH mutants were smaller than wild-type chlorosomes; these chlorosomes were about 100 ± 30 nm in length and 45 ± 15 nm in width (Fig. 4B to E). The length-to-width ratio of chlorosomes from the csmC csmD and csmC csmED mutants, 2.0 ± 0.6 and 2.0 ± 0.5, respectively, differed significantly from that of wild-type chlorosomes. However, this difference was less obvious in chlorosomes from the csmC csmD csmH and csmC csmED csmH mutants (length-to-width ratios, 2.4 ± 0.7 and 2.6 ± 0.8, respectively). Thus, the additional removal of CsmH partially reversed the change in chlorosome shape produced by the absence of CsmC and CsmD. Fraction 2 of the csmC csmD csmH mutant mostly contained very small chlorosomes (Fig. 4F), which were only 55 ± 10 nm in length and 27 ± 7 nm in width and which were thus ∼85% smaller than wild-type chlorosomes.
FIG. 4.
Transmission electron micrographs of negatively stained chlorosomes isolated from the wild type and mutants lacking proteins of the CsmC/CsmD or CsmB/CsmF motif families. (A) Chlorosomes from the wild type; (B) chlorosomes from the csmC csmD mutant; (C) chlorosomes from the csmC csmED mutant; (D) chlorosomes from the csmC csmD csmH mutant; (E) chlorosomes from the csmC csmED csmH mutant; (F) fraction 2 from the csmC csmD csmH mutant; (G) chlorosomes from the csmB csmF mutant; (H) chlorosomes from the csmB csmF csmH mutant; (I) fraction 2 from the csmB csmF mutant. The samples were stained with 2% (wt/vol) aqueous uranyl acetate. Bars, 200 nm.
TABLE 3.
Dimensions of negatively stained chlorosomes from C. tepidum mutants lacking proteins of the CsmC/CsmD motif or the CsmB/CsmF motif classesa
| Strain | Length (nm) | Width (nm) | Length-to-width ratio |
|---|---|---|---|
| Wild type | 165 ± 60 | 60 ± 25 | 2.7 ± 0.6 |
| csmC csmD mutant | 104 ± 35 | 52 ± 19 | 2.0 ± 0.6 |
| csmC csmED mutant | 112 ± 30 | 56 ± 25 | 2.0 ± 0.5 |
| csmC csmD csmH mutant | 99 ± 30 | 42 ± 15 | 2.4 ± 0.7 |
| csmC csmD csmH fraction 2 mutant | 55 ± 10 | 27 ± 7 | 2.0 ± 0.4 |
| csmC csmED csmH mutant | 90 ± 30 | 35 ± 15 | 2.6 ± 0.8 |
| csmB csmF mutant | 229 ± 79 | 82 ± 26 | 3.0 ± 0.6 |
| csmB csmF fraction 2 mutant | 200 ± 45 | 65 ± 20 | 3.1 ± 0.6 |
| csmB csmF csmH mutant | 181 ± 65 | 75 ± 27 | 2.4 ± 0.7 |
The values shown are mean values and standard deviations for measurements on 25 to 60 negatively stained chlorosomes from one chlorosome preparation for each strain.
Conversely, chlorosomes from the csmB csmF mutant were both longer and wider than those from the wild-type strain (length, 229 ± 79 nm; width, 82 ± 26 nm), with a length-to-width ratio of 3.0 ± 0.6 (Fig. 4G). The dimensions of the chlorosomes from the csmB csmF csmH mutant were more similar to those of wild-type chlorosomes (Fig. 4H). The fraction 2 sample from the csmB csmF mutant contained a few chlorosomes with sizes similar to those of the chlorosomes from the csmB csmF mutant strain (shown by the arrows), but this fraction was mostly enriched in irregularly shaped objects with diameters ranging from 50 to 150 nm (Fig. 4I). Considering the BChl a content (see Table S1 in the supplemental material) and the polypeptide complexity (Fig. 1B) of this fraction, these irregularly shaped objects probably were membrane fragments, vesicles, and other cellular debris.
DISCUSSION
Other than the involvement of CsmA (and probably the closely related CsmE) in binding BChl a (23, 31, 35, 40), and the essential role of CsmA in forming the structurally important chlorosome baseplate (9, 14), the roles of chlorosome envelope proteins have remained enigmatic. Previous studies have shown that chlorosomes retain their size and shape when they are treated with low concentrations of SDS or other detergents, under which conditions all proteins except CsmA are completely extracted from the chlorosome envelope (23). Thus, once the BChl c aggregates and the baseplate have assembled, no chlorosome protein other than possibly CsmA is required to maintain the shape and integrity of chlorosomes (23). A similar conclusion was reached from mutational studies of C. tepidum in which nine single mutants, each lacking one chlorosome envelope protein, were constructed and characterized (14). No dramatic phenotypic changes could be discerned after exhaustive analyses of most of these nine mutants. The mutant lacking CsmB, the second most abundant protein in the chlorosome envelope (6, 14), contained ∼25% lower levels of carotenoids, and chlorosomes from a csmH mutant were ∼25% shorter on average than wild-type chlorosomes. Chlorosomes from a csmC mutant were also slightly smaller than those of the wild type, and the Qy absorption maximum of the BChl c aggregates in this mutant was blueshifted to 743 nm (9, 14). This csmC mutant grew slightly more slowly (∼10%) than did the wild type, but only at a limiting light intensity. However, the csmB, csmD, csmF, and csmH single mutant strains exhibited no growth defects at any tested light intensity. These mutational studies established that C. tepidum chlorosomes are remarkably robust structures, which can tolerate considerable changes in their pigment content and envelope protein composition. To explain these surprising observations, it was hypothesized that proteins belonging to the same chlorosome motif family might be functionally redundant and that the absence of observable phenotypic effects for most of the single mutants was due to the relatively large number (n = 10) of functionally redundant chlorosome envelope proteins in C. tepidum (14, 50).
To test this hypothesis in the studies reported here, six additional, multilocus C. tepidum mutants lacking proteins of the CsmC/CsmD or CsmB/CsmF motif families were constructed by targeted, insertional gene inactivation, and the resulting mutants were physiologically and biochemically characterized. Supporting the hypothesis of functional redundancy, growth defects were detected in mutants lacking multiple chlorosome proteins of the CsmC/CsmD and CsmB/CsmF motif families. These multilocus mutants were impaired in light harvesting and grew more slowly at very low light intensities. However, because no growth defects were observed at higher light intensities that were limiting for growth of the wild type (see reference 20), these mutants still assemble chlorosomes that function relatively normally in light harvesting.
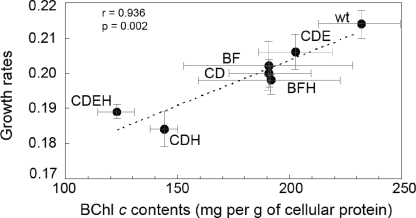
Mutants lacking the proteins of the CsmB/CsmF motif (i.e., the csmB csmF and csmB csmF csmH mutants) had cellular and chlorosome carotenoid contents that were slightly lower than those of the wild type, and the cellular and chlorosome contents of BChl c were lower than those in the wild-type strain in the csmC csmD csmH and csmC csmED csmH mutants and slightly reduced (∼10%) in four other mutants (csmC csmD, csmC csmED, csmB csmF, and csmB csmF csmH strains). The lower pigment contents of these mutant cells might arise from defects in chlorosome biogenesis. Such defects could cause an increase in the pool of unassembled BChls and carotenoids, and this in turn might decrease the overall flux through these biosynthetic pathways by end product inhibition of key regulatory enzyme activities. Interestingly, a strong correlation was observed between growth rates at low light intensity and cellular BChl c content (per g of cellular protein; see Fig. 5). This correlation strongly suggested that the lower levels of BChl c, which is the major light-harvesting pigment in C. tepidum, were likely to be responsible for the lower growth rates that were observed for the mutants. Carotenoids also participate in light harvesting, energy transfer, and triplet quenching in C. tepidum (16, 25), and the slower growth of the csmB csmF and csmB csmF csmH mutants at the lowest light intensities might be partly due to their decreased carotenoid contents (Table 1). The csmB csmF csmH mutant had a much larger growth rate defect at low light intensity than did either the csmB csmF or the csmC csmD mutant (Table 1), although these three strains had nearly identical BChl c contents per cell (Table 2). Cryo-electron microscopic studies of these chlorosomes are planned in hopes of determining whether differences in the organization of the BChls in these mutants could explain the growth rate differences.
FIG. 5.
Correlation between growth rate at a limiting light intensity and the cellular BChl c content of C. tepidum strains. Correlation (r = 0.936, P = 0.002) of growth rate (Table 1) and cellular BChl c contents (Table 2) of cells grown at a light intensity of 32 μmol photons m−2 s−1. The plotted points represent the wild type (wt) or the mutants lacking proteins of the CsmC/CsmD or the CsmB/CsmF motif families. The label for each data point indicates the missing chlorosome proteins, and error bars indicate the standard deviations of values for growth rate and BChl c content.
When the studies reported here were initiated, only very limited information was available concerning the distribution of chlorosome proteins in green bacteria, but this has changed rapidly and dramatically because of genome sequencing studies for Chlorobi and Chloroflexi. Of 10 chlorosome proteins identified in C. tepidum, only three, CsmA, CsmC, and CsmX, are universally encoded in the genomes of 12 sequenced Chlorobi strains. The csmJ gene is missing only from Chlorobium chlorochromatii CaD3, and the csmB, csmE, and csmH genes are missing only in Chloroherpeton thalassium. However, it should also be noted that CsmH is highly variable in both length and sequence. Because the csmD gene is found only in three strains (C. tepidum, Chlorobaculum parvum, and Prosthecochloris aestuarii DSM 217), it may have arisen and diverged relatively recently following a gene duplication event. The csmF gene is found in all of the Chlorobi genomes except C. thalassium, C. chlorochromatii CaD3, Chlorobium luteolum DSM 273, and Chlorobium phaeovibrioides DSM 265. This gene probably arose by duplication of csmB after the ancestor of strains related to C. thalassium diverged from the lineage leading to other Chlorobi strains, and csmF may have subsequently been lost from the genomes of the other three strains, which have relatively small genomes. Finally, csmI clearly arose by duplication from csmJ or csmX, and this gene is missing from the genomes of C. thalassium, C. phaeovibrioides, and Chlorobium ferrooxidans. Collectively, these observations strongly suggest that several gene duplications have occurred, which have significantly increased the complexity of the complement of chlorosome envelope proteins in some but not all members of the Chlorobi. These distribution data are in excellent agreement with the working hypothesis that the proteins of the major chlorosome motif families in C. tepidum play redundant roles in chlorosome biogenesis and function.
Early studies suggested that chlorosomes from Chloroflexus aurantiacus contained only three proteins: CsmA, CsmM, and CsmN (11, 32). CsmA, an abundant 5.7-kDa polypeptide, is structurally and functionally orthologous to CsmA in GSB, and this protein is similarly synthesized as a precursor with a carboxyl-terminal extension of 27 amino acids (8, 45). CsmA of C. aurantiacus has been implicated in BChl a binding and is likely to form the paracrystalline baseplate that is also observed in chlorosomes of this organism (23, 31, 40). CsmM and CsmN, with apparent molecular masses of 11 kDa and 18 kDa, respectively, are related to one another and are possibly more distantly related in sequence to the CsmC and CsmD proteins of C. tepidum (32, 48, 50). Tang et al. (44) have recently reported that AcsF, the oxygen-dependent ring cyclase of BChl biosynthesis, may be a component of the C. aurantiacus chlorosome envelope. However, no evidence was presented to show that this protein uniquely colocalizes with other chlorosome proteins, and no other enzymes of BChl biosynthesis have been detected in other similar studies with chlorosomes (15, 19, 48; A. M. Garcia Costas, Y. Tsukatani, and D. A. Bryant, unpublished data).
At least three other proteins are likely to occur in chlorosomes of C. aurantiacus (19). CsmO (Caur_1311) is a member of the CsmB/CsmF protein family and occurs in three sequenced Chloroflexi strains, and the predicted sequence matches the N-terminal sequence of a protein in chlorosomes (19, 26). Frigaard et al. (19) additionally identified CsmP (Caur_0142; 17.2 kDa) and CsmY (Caur_0356; 22.0 kDa) in chlorosomes prepared from C. aurantiacus j-10-fl. The csmP gene lies just downstream of the genes encoding CsmM and CsmN, is probably cotranscribed with them, and is found in genomes of all chlorosome-containing Chloroflexi but does not appear to occur in the genomes of other chlorosome-producing organisms. CsmY is predicted to be an Fe/S protein; has sequence similarity to CsmI, CsmJ, and CsmX; and is found in all chlorosome-producing Chloroflexi, but CsmY is not found in Roseiflexus spp. These observations indicate that the chlorosome envelope of C. aurantiacus contains proteins of the same four motif families as those in C. tepidum and that these two types of chlorosomes are more similar in polypeptide composition than has generally been recognized. Interestingly, six proteins have recently been identified in chlorosome preparations of the phototrophic acidobacterium “Candidatus Chloroacidobacterium thermophilum,” but only CsmA and an Fe/S protein have obvious homologs in other chlorosome-containing organisms (3; Garcia Costas et al., unpublished).
Recent structural studies of chlorosomes by cryo-electron microscopy and solid-state NMR provide some possible insights into how these chlorosome proteins might affect chlorosome structure. End-on views of wild-type chlorosomes (diameters, ∼60 nm) show that the organization of BChls in each wild-type chlorosome is structurally unique and that chlorosomes are composed of multiple, concentric BChl c nanotubes (34). However, chlorosomes of a bchQ bchR bchU mutant are more uniform in length and are smaller in diameter; they contain only one or sometimes two nanotubes and have diameters of 30 to 40 nm (22, 34). Structural studies have additionally shown that the syn-anti monomer stacks of BChl c are differently oriented in the chlorosomes of the bchQ bchR bchU mutant and the wild type (22). Thus, chlorosomes from the bchQ bchR bchU mutant have a higher length-to-width ratio than do wild-type chlorosomes because of differences in the organization of the BChls. In spite of these structural differences on the interior, the complement of chlorosome envelope proteins and their relative amounts are nearly identical for the wild type and the bchQ bchR bchU mutant (27).
The data in Table 3 can be interpreted on the basis of these structural observations. Chlorosomes lacking proteins of the CsmB/CsmF motif families are both longer and have a larger diameter than do wild-type chlorosomes. Thus, average chlorosomes from these mutants probably contain a larger number of BChl c molecules and have more BChl nanotubes per chlorosome, and these nanotubes are likely to be longer and have larger diameters on average than those of wild-type chlorosomes. Because the absorption of the BChl aggregates in the chlorosomes is redshifted by 3 to 6 nm, the orientation and packing of the BChls within the syn-anti monomer stacks might also differ somewhat from BChl suprastructure in wild-type chlorosomes. However, as reflected by a lower growth rate for cells grown at low light intensity, chlorosomes of the csmB csmF and csmB csmF csmH mutants are less efficient in light harvesting than are chlorosomes of the csmC csmD mutant, even though their BChl c contents are very similar (Tables 1 and 2; see Table S1 in the supplemental material).
Conversely, chlorosomes lacking CsmC and CsmD are much shorter than wild-type chlorosomes but have diameters that are only about 10% smaller than those of wild-type chlorosomes. The absorbance maximum for the BChls in these mutants was strongly blueshifted to 738 nm, and this clearly implies that the BChl c suprastructure inside the chlorosomes is different when CsmC and CsmD are missing. Because the absorption properties of the BChls resemble those in the bchQ bchR bchU mutant whose structure has been determined (4, 22), it is possible that the orientation of the BChl c suprastructure relative to the long axis of the chlorosome is altered when CsmC and CsmD are missing. However, when CsmH is additionally eliminated, the width/diameter of the chlorosomes decreases considerably (but the length stays about the same [Table 3]) and the BChl absorption is less blueshifted (see Table S1 in the supplemental material). The decrease in the diameter of the chlorosomes increases the length-to-width ratio to values that are more similar to those of the wild-type chlorosomes (Table 3). The results obtained here differ somewhat from those for the single chlorosome protein mutants described previously (14). When csmH alone was mutated, the average length of the chlorosomes decreased but the average diameter remained about the same as that for the wild-type chlorosomes. Interestingly, the mutants lacking CsmC, CsmD, and CsmH accumulated much less BChl c per cell (Table 2), and this is strongly correlated with a lower growth rate at light intensities below 80 μmol photons m−2 s−1 (Fig. 5 and Table 1). Further studies will be required to resolve whether the smaller chlorosomes in these mutants are the result of a defect in BChl import or a size limitation imposed by the missing envelope proteins.
In summary, the genetic elimination of multiple chlorosome proteins from the CsmC/CsmD and the CsmB/CsmF motif families had significant effects on pigment biosynthesis, chlorosome structure and assembly, light harvesting, and ultimately growth rate. Elimination of CsmC and CsmD led to chlorosomes that were shorter than those of the wild type, while removal of CsmB and CsmF led to chlorosomes that were both longer and of greater diameter than those of the wild type. Mutants additionally lacking CsmH produced chlorosomes with a smaller average diameter. We propose that these chlorosome envelope proteins influence the number of BChl c nanotubes per chlorosome and that they may also help to establish the orientation of BChl c suprastructure relative to the long axis of the chlorosomes. These changes could affect the overall size as well as the length-to-width ratio of chlorosomes, and the combination of changes affects the light-harvesting efficiency of these remarkable antennae.
Supplementary Material
Acknowledgments
This research was supported by grant DE-FG02-94ER20137 from the U.S. Department of Energy to D.A.B.
We thank N.-U. Frigaard for helpful suggestions and discussion.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Blankenship, R. E. 1992. Origin and early evolution of photosynthesis. Photosynth. Res. 33:91-111. [PubMed] [Google Scholar]
- 2.Bryant, D. A., and N.-U. Frigaard. 2006. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14:488-496. [DOI] [PubMed] [Google Scholar]
- 3.Bryant, D. A., A. M. Garcia Costas, J. A. Maresca, A. Gomez Maqueo Chew, C. G. Klatt, M. M. Bateson, L. J. Tallon, J. Hostetler, W. C. Nelson, J. F. Heidelberg, and D. M. Ward. 2007. “Candidatus Chloracidobacterium thermophilum”: an aerobic phototrophic acidobacterium. Science 317:523-526. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, D. A., E. V. Vassilieva, N.-U. Frigaard, and H. Li. 2002. Selective protein extraction from Chlorobium tepidum chlorosomes using detergents. Evidence that CsmA forms multimers and binds bacteriochlorophyll a. Biochemistry 41:14403-14411. [DOI] [PubMed] [Google Scholar]
- 5.Chung, S. 1995. Characterization of chlorosomes in green sulfur bacteria. Ph.D. dissertation. The Pennsylvania State University, University Park.
- 6.Chung, S., and D. A. Bryant. 1996. Characterization of csmB genes, encoding a 7.5-kDa protein of the chlorosome envelope, from the green sulfur bacteria Chlorobium vibrioforme 8327d and Chlorobium tepidum. Arch. Microbiol. 166:234-244. [DOI] [PubMed] [Google Scholar]
- 7.Chung, S., and D. A. Bryant. 1996. Characterization of the csmD and csmE genes from Chlorobium tepidum. The CsmA, CsmC, CsmD, and CsmE proteins are components of the chlorosome envelope. Photosynth. Res. 50:41-59. [DOI] [PubMed] [Google Scholar]
- 8.Chung, S., G. Frank, H. Zuber, and D. A. Bryant. 1994. Genes encoding two chlorosome components from the green sulfur bacteria Chlorobium vibrioforme strain 8327d and Chlorobium tepidum. Photosynth. Res. 41:261-275. [DOI] [PubMed] [Google Scholar]
- 9.Chung, S., G. Shen, J. Ormerod, and D. A. Bryant. 1998. Insertional inactivation studies of csmA and csmC genes of the green sulfur bacterium Chlorobium vibrioforme 8327d: the chlorosome protein CsmA is required for viability but CsmC is dispensible. FEMS Microbiol. Lett. 164:353-361. [DOI] [PubMed] [Google Scholar]
- 10.Dunphy, P. J., and A. F. Brodie. 1971. The structure and function of quinones in respiratory metabolism. Methods Enzymol. 18:407-461. [Google Scholar]
- 11.Feick, R. G., and R. C. Fuller. 1984. Topography of the photosynthetic apparatus of Chloroflexus aurantiacus. Biochemistry 23:3693-3700. [Google Scholar]
- 12.Frigaard, N.-U., and D. A. Bryant. 2001. Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl. Environ. Microbiol. 67:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frigaard, N.-U., and D. A. Bryant. 2006. Chlorosomes: antenna organelles in green photosynthetic bacteria, p. 79-144. In J. M. Shively (ed.), Microbiology monographs, vol. 2. Complex intracellular structures in prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 14.Frigaard, N.-U., H. Li, K. J. Milks, and D. A. Bryant. 2004. Nine mutants of Chlorobium tepidum each unable to synthesize a different chlorosome protein still assemble functional chlorosomes. J. Bacteriol. 186:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frigaard, N.-U., H. Li, P. Martinsson, S. K. Das, H. A. Frank, T. J. Aartsma, and D. A. Bryant. 2005. Isolation and characterization of carotenosomes from a bacteriochlorophyll c-less mutant of Chlorobium tepidum. Photosynth. Res. 86:101-111. [DOI] [PubMed] [Google Scholar]
- 16.Frigaard, N.-U., J. A. Maresca, C. E. Yunker, A. D. Jones, and D. A. Bryant. 2004. Genetic manipulation of carotenoid biosynthesis in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 186:5210-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frigaard, N.-U., Y. Sakuragi, and D. A. Bryant. 2004. Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274:325-340. [DOI] [PubMed] [Google Scholar]
- 18.Frigaard, N.-U., S. Takaichi, M. Hirota, K. Shimada, and K. Matsuura. 1997. Quinones in chlorosomes of green sulfur bacteria and their role in the redox-dependent fluorescence studied in chlorosome-like bacteriochlorophyll c aggregates. Arch. Microbiol. 167:343-349. [Google Scholar]
- 19.Frigaard, N.-U., E. V. Vassilieva, H. Li, K. J. Milks, J. Zhao, and D. A. Bryant. 2001. The remarkable chlorosome. In PS2001 proceedings. Proc. 12th Int. Congr. Photosynth., article S1-003. CSIRO Publishing, Canberra, Australia.
- 20.Frigaard, N.-U., G. D. Voigt, and D. A. Bryant. 2002. Chlorobium tepidum mutant lacking bacteriochlorophyll c made by inactivation of the bchK gene, encoding bacteriochlorophyll c synthase. J. Bacteriol. 184:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frydman, B., and H. Rappaport. 1963. Non-chlorophyllous pigments of Chlorobium thiosulfatophilum—chlorobiumquinone. J. Am. Chem. Soc. 85:823-825. [Google Scholar]
- 22.Ganapathy, S., G. T. Oostergetel, P. K. Wawrzyniak, M. Reus, A. Gomez Maqueo Chew, F. Buda, E. J. Boekema, D. A. Bryant, A. R. Holzwarth, and H. J. M. de Groot. 2009. Alternating syn-anti bacteriochlorophylls form supramolecular helical nanotubes in chlorosomes. Proc. Natl. Acad. Sci. USA 106:8525-8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez Maqueo Chew, A., N.-U. Frigaard, and D. A. Bryant. 2007. Bacteriochlorophyllide c C-82 and C-121 methyltransferases are essential for adaptation to low light in Chlorobaculum tepidum. J. Bacteriol. 189:6176-6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holo, H., M. Broch-Due, and J. G. Ormerod. 1985. Glycolipids and the structure of chlorosomes in green bacteria. Arch. Microbiol. 143:94-99. [Google Scholar]
- 25.Ikonen, T. P., H. Li, J. Psencík, P. A. Laurinmäki, S. J. Butcher, N.-U. Frigaard, R. E. Serimaa, D. A. Bryant, and R. Tuma. 2007. X-ray scattering and electron cryomicroscopy study on the effect of carotenoid biosynthesis to the structure of Chlorobium tepidum chlorosomes. Biophys. J. 93:620-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann, R. P., R. A. Brunisholz, and H. Zuber. 1994. Structural differences in chlorosomes from Chloroflexus aurantiacus grown under different conditions support the BChl c-binding function of the 5.7 kDa polypeptide. FEBS Lett. 342:319-324. [DOI] [PubMed] [Google Scholar]
- 27.Li, H. 2006. Organization and function of chlorosome proteins in the green sulfur bacterium Chlorobium tepidum. Ph.D. thesis. The Pennsylvania State University, University Park.
- 28.Li, H., N.-U. Frigaard, and D. A. Bryant. 2006. Molecular contacts for chlorosome envelope proteins revealed by cross-linking studies with chlorosomes from Chlorobium tepidum. Biochemistry 45:9095-9103. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Planells, A., J. B. Arellano, C. M. Borrego, C. López-Iglesias, F. Gich, and J. Garcia-Gil. 2002. Determination of the topography and biometry of chlorosomes by atomic force microscopy. Photosynth. Res. 71:83-90. [DOI] [PubMed] [Google Scholar]
- 30.Montaño, G. A., B. P. Bowen, J. T. LaBelle, N. W. Woodbury, V. B. Pizziconi, and R. E. Blankenship. 2003. Characterization of Chlorobium tepidum chlorosomes: a calculation of bacteriochlorophyll c per chlorosome and oligomer modeling. Biophys. J. 85:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montaño, G. A., H. M. Wu, S. Lin, D. C. Brune, and R. E. Blankenship. 2003. Isolation and characterization of the B798 light-harvesting baseplate from the chlorosomes of Chloroflexus aurantiacus. Biochemistry 42:10246-10251. [DOI] [PubMed] [Google Scholar]
- 32.Niedermeier, G., J. A. Shiozawa, F. Lottspeich, and R. G. Feick. 1994. The primary structure of two chlorosome proteins from Chloroflexus aurantiacus. FEBS Lett. 342:61-65. [DOI] [PubMed] [Google Scholar]
- 33.Oelze, J. 1985. Analysis of bacteriochlorophylls. Methods Microbiol. 18:57-284. [Google Scholar]
- 34.Oostergetel, G. T., M. Reus, A. Gomez Maqueo Chew, D. A. Bryant, E. J. Boekema, and A. R. Holzwarth. 2007. Long-range organization of bacteriochlorophyll in chlorosomes of Chlorobium tepidum investigated by cryo-electron microscopy. FEBS Lett. 581:5435-5439. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen, M. Ø., L. Pham, D. B. Steensgaard, and M. Miller. 2008. A reconstituted light-harvesting complex from the green sulfur bacterium Chlorobium tepidum containing CsmA and bacteriochlorophyll a. Biochemistry 47:1435-1441. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen, M. Ø., J. Underhaug, J. Dittmer, M. Miller, and N. C. Nielsen. 2008. The three-dimensional structure of CsmA: a small antenna protein from the green sulfur bacterium Chlorobium tepidum. FEBS Lett. 582:2869-2874. [DOI] [PubMed] [Google Scholar]
- 37.Psencík, J., J. B. Arellano, T. P. Ikonen, C. M. Borrego, P. A. Laurinmäki, S. J. Butcher, R. E. Serimaa, and R. Tuma. 2006. Internal structure of chlorosomes from brown-colored Chlorobium species and the role of carotenoids in their assembly. Biophys. J. 91:1433-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Psencík, J., T. P. Ikonen, P. Laurinmäki, M. C. Merckel, S. J. Butcher, R. E. Serimaa, and R. Tuma. 2004. Lamellar organization of pigments in chlorosomes: the light harvesting complexes of green photosynthetic bacteria. Biophys. J. 87:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saga, Y., Y. Shibata, S. Itoh, and H. Tamiaki. 2007. Direct counting of submicrometer-sized photosynthetic apparatus dispersed in medium at cryogenic temperature by confocal laser fluorescence microscopy: estimation of the number of bacteriochlorophyll c in single light-harvesting antenna complexes chlorosomes of green photosynthetic bacteria. J. Phys. Chem. B 111:12605-12609. [DOI] [PubMed] [Google Scholar]
- 40.Sakuragi, Y., N.-U. Frigaard, K. Shimada, and K. Matsuura. 1999. Association of bacteriochlorophyll a with the CsmA protein in chlorosomes of the photosynthetic green filamentous bacterium Chloroflexus aurantiacus. Biochim. Biophys. Acta 1413:172-180. [DOI] [PubMed] [Google Scholar]
- 41.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 42.Staehelin, L. A., J. R. Golecki, and G. Drews. 1980. Supramolecular organization of chlorosomes (Chlorobium vesicles) and of their membrane attachment sites in Chlorobium limicola. Biochim. Biophys. Acta 589:30-45. [DOI] [PubMed] [Google Scholar]
- 43.Stanier, R. Y., and J. H. Smith. 1960. The chlorophylls of green bacteria. Biochim. Biophys. Acta 41:478-484. [DOI] [PubMed] [Google Scholar]
- 44.Tang, K. H., J. Wen, X. Li, and R. E. Blankenship. 2009. Role of the AcsF protein in Chloroflexus aurantiacus. J. Bacteriol. 191:3580-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theroux, S. J., T. E. Redlinger, R. C. Fuller, and S. J. Robinson. 1990. Gene encoding the 5.7-kilodalton chlorosome protein of Chloroflexus aurantiacus: regulated message levels and a predicted carboxy-terminal protein extension. J. Bacteriol. 172:4497-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuji, K., S. Takaichi, K. Matsuura, and K. Shimada. 1995. Specificity of carotenoids in chlorosomes of the green filamentous bacterium, Chloroflexus aurantiacus, p. 99-103. In P. Mathis (ed.), Photosynthesis: from light to biosphere, vol. 3. Kluwer, Dordrecht, The Netherlands. [Google Scholar]
- 47.Vassilieva, E. V., M. L. Antonkine, B. L. Zybailov, F. Yang, C. U. Jakobs, J. H. Golbeck, and D. A. Bryant. 2001. Electron transfer may occur in the chlorosome envelope: the CsmI and CsmJ proteins of chlorosomes are 2Fe-2S ferredoxins. Biochemistry 40:464-473. [DOI] [PubMed] [Google Scholar]
- 48.Vassilieva, E. V., N.-U. Frigaard, and D. A. Bryant. 2000. Chlorosomes: the light-harvesting complexes of the green bacteria. Spectrum 13:7-13. [Google Scholar]
- 49.Vassilieva, E. V., J. G. Ormerod, and D. A. Bryant. 2002. Biosynthesis of chlorosome proteins is not inhibited in acetylene-treated cultures of Chlorobium vibrioforme. Photosynth. Res. 71:69-81. [DOI] [PubMed] [Google Scholar]
- 50.Vassilieva, E. V., V. L. Stirewalt, C. U. Jakobs, N.-U. Frigaard, K. Inoue-Sakamoto, M. A. Baker, A. Sotak, and D. A. Bryant. 2002. Subcellular localization of chlorosome proteins in Chlorobium tepidum and characterization of three new chlorosome proteins: CsmF, CsmH, and CsmX. Biochemistry 41:4358-4370. [DOI] [PubMed] [Google Scholar]
- 51.Wahlund, T. M., and M. T. Madigan. 1995. Genetic transfer by conjugation in the thermophilic green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 177:2583-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wahlund, T. M., C. R. Woese, R. W. Castenholz, and M. T. Madigan. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch. Microbiol. 156:81-90. [Google Scholar]
- 53.Wullink, W., J. Knudsen, J. M. Olson, T. E. Redlinger, and E. F. J. van Bruggen. 1991. Localization of polypeptides in isolated chlorosomes from green phototrophic bacteria by immuno-gold labeling electron microscopy. Biochim. Biophys. Acta 1060:97-105. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.