Abstract
Bacillus subtilis CodY protein is a DNA-binding global transcriptional regulator that responds to branched-chain amino acids (isoleucine, leucine, and valine) and GTP. Crystal structure studies have shown that the N-terminal region of the protein includes a GAF domain that contains a hydrophobic pocket within which isoleucine and valine bind. This region is well conserved in CodY homologs. Site-directed mutagenesis was employed to understand the roles of some of the residues in the GAF domain and hydrophobic pocket in interaction with isoleucine and GTP. The F40A, F71E, and F98A forms of CodY were inactive in vivo. They were activatable by GTP but to a much lesser extent by branched-chain amino acids in vitro. The CodY mutant R61A retained partial repression of target promoters in vivo and was able to respond to GTP in vitro but also responded poorly to branched-chain amino acids in vitro unless GTP was simultaneously present. Thus, the GAF domain includes residues essential for full activation of CodY by branched-chain amino acids, but these residues are not critical for activation by GTP. Binding studies with branched-chain amino acids and their analogs revealed that an amino group at position 2 and a methyl group at position 3 of valine are critical components of the recognition of the amino acids by CodY.
Bacteria have evolved numerous mechanisms to adapt to reduced nutrient availability. The gram-positive bacterium Bacillus subtilis has an array of such adaptive responses, including chemotaxis and motility to migrate to an environment richer in nutrients, secretion of macromolecule-degrading enzymes, induction of transport systems for the uptake of amino acids, peptides, and other available nutrients, activation of intracellular catabolic systems, production and secretion of antibiotics that could aid in reducing the competition for limited resources, and development of competence that enables the intake of exogenous DNA (for nutrition or for developing a genetic advantage). If these adaptive responses do not improve the nutritional status of the cells, B. subtilis cells can induce sporulation, resulting in the production of dormant spores that can withstand long-term, severe environmental conditions.
The B. subtilis global regulatory protein CodY controls the expression of several hundred genes, including many of the genes involved in the adaptive responses listed above. CodY, a highly conserved protein in the low-G+C gram-positive bacteria (29, 39), was originally discovered as a repressor of the B. subtilis dipeptide permease (dpp) operon (34, 41, 46-48). The full breadth of the B. subtilis CodY regulon was revealed by studies of many individual genes regulated by CodY (6, 9, 13, 14, 24, 28, 40, 44, 46, 48, 52) and by global analyses of transcription and CodY binding in vivo (36). While CodY acts as a repressor of many genes, it can also activate transcription of some genes, including the acetate kinase gene (ackA) (43). In addition to regulating adaptive response genes, CodY controls as yet undetermined aspects of sporulation. While wild-type cells do not sporulate efficiently in a nutrient-rich medium, codY mutant cells sporulate readily in the same medium, indicating that CodY senses nutrient excess and prevents sporulation in wild-type cells (16, 39). Further, there is mounting evidence that indicates a role for CodY in the regulation of virulence gene expression in many pathogenic bacteria, including Streptococcus pyogenes (32), Listeria monocytogenes (5), Clostridium difficile (11), Staphylococcus aureus (31), Streptococcus pneumoniae (21), Bacillus cereus (22) and Bacillus anthracis (51a).
B. subtilis CodY is a dimer of 29-kDa, 259-amino-acid polypeptides (7, 29, 46). During growth of cells in rich medium, CodY is highly active, repressing genes whose products enable cells to adapt to nutrient limitation and activating expression of some central metabolism genes (43). When the cells become limited for nutrients, CodY activity diminishes. The activity of CodY, at least in vitro, depends on two effector molecules, GTP and branched-chain amino acids (BCAAs) (leucine, isoleucine, and valine) (39, 44, 45), whose intracellular concentrations drop at the transition from rapid growth to stationary phase (49). While the in vitro binding ability of B. subtilis CodY is enhanced by both types of effectors (GTP and BCAAs), CodY proteins of Lactococcus lactis and S. pneumoniae were found to respond to BCAAs alone (10, 18, 21). In fact, the first evidence that CodY senses BCAAs came from studies of L. lactis (18). GTP and the BCAAs act synergistically to increase the affinity of B. subtilis CodY for its DNA targets (20, 25, 36, 39, 44).
Crystallography has revealed that CodY is a two-domain protein. The C-terminal domain includes a winged helix-turn-helix motif (29) that contains specific residues that are required for high-affinity DNA binding (25, 41). Binding of CodY to BCAAs results in a conformational change that correlates with increased affinity of CodY for its target DNA (29, 30).
The N-terminal region of CodY comprises a GAF domain (named GAF for cyclic GMP [cGMP]-stimulated phosphodiesterase, adenylate cyclase and bacterial transcription regulator FhlA) within which isoleucine and valine bind (29). Such domains are commonly found in signaling proteins (23). In the presence of BCAAs, the residues M62, M65, F71, P72, Y75, and P99 form a hydrophobic cavity that encloses the isobutyl group of isoleucine and the isopropyl group of valine. The α-amino and α-carboxylate groups of isoleucine and valine form polar interactions with the protein and solvent. While the α-amino group forms charge-dipole interactions with the main chain carbonyl groups of T96 and F98, the α-carboxylate group forms a two-pronged ion-pairing interaction with the guanidinium group of R61 (29). Comparison of the structures of the liganded and unliganded forms of the GAF domain revealed a substantial reorganization coincident with binding of isoleucine or valine (30).
Although a crystal structure of GTP-bound CodY has not yet been obtained, a prediction from the available structure is that GTP could potentially bind within the GAF domain, which, in eukaryotic signaling systems, is associated with nucleotide binding (23, 29). Thus, in phosphodiesterase GAF domain complexes with 3′,5′-cGMP, the nucleotide is more deeply buried in the ligand-binding cavity than the BCAA ligand in CodY is (33). Simple modeling suggests that a guanine nucleotide could be accommodated without steric hindrance in the GAF domain of CodY at a position corresponding to that of cGMP in the phosphodiesterases (29). If this is true, F40 and F98 would be well placed for ring-stacking interactions with the guanine base of GTP. F98 is invariant and F40 is well conserved in all of the CodY proteins mentioned above, except for L. lactis and S. pneumoniae CodY proteins, which do not respond to GTP (10, 21, 38). R61 and the residues of CodY that appear to be involved in forming the hydrophobic pocket are highly conserved in Bacillus, Listeria, Enterococcus, Staphylococcus, Streptococcus, and Lactococcus species, suggesting that many CodY homologs recognize and bind BCAAs using these residues (29). As R61, F71, and F98 appear to be involved in ionic or hydrophobic interactions with isoleucine and both F40 and F98 were predicted to be involved in interacting with GTP (29), these residues were subjected to site-directed mutagenesis to understand their roles in interactions with the corepressors. We report the effects of such mutations on the regulation of two CodY target promoters in vivo and on effector interactions and the DNA-binding ability of CodY in vitro.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The B. subtilis and Escherichia coli strains used in this study are described in Tables 1 and 2, respectively.
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype | Source, reference, or strain constructiona |
|---|---|---|
| JH642 | trpC2 pheA1 | J. A. Hoch |
| FJS107 | trpC2 | 46 |
| PJB14 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) | 25 |
| PJB15 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY::(erm::spc) | P. Joseph |
| PS251 | trpC2 codY::(erm::spc) | P. Serror |
| BB1888 | lacA::tet | 26 |
| BB2505 | lacA:tet ΔamyE::(ΦyhdG283-lacZ erm) | B. R. Belitsky, unpublished data |
| BB2511 | lacA::tet ΔamyE::spc | 3 |
| BB2611 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(R61A) | PJB14 Χ pBB1451 |
| BB2613 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(R61E) | PJB14 Χ pBB1453 |
| BB2614 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(F71A) | PJB14 Χ pBB1457 |
| BB2615 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(F71R) | PJB14 Χ pBB1455 |
| BB2616 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(F71E) | PJB14 Χ pBB1456 |
| BB2619 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(R61K) | PJB14 Χ pBB1459 |
| LH4 | trpC2 codY(F98A) | FJS107 Χ pLH9 |
| LH7 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(F98A) | LH4 Χ PJB15 DNA |
| LH11 | trpC2 codY(F40A) | FJS107 Χ pLH15 |
| LH12 | trpC2 ΔamyE::(ΦilvBΔT′-lacZ erm) codY(F40A) | LH11 Χ PJB15 DNA |
| LH15 | trpC2 ΔamyE::(ΦyhdG283-lacZ erm) | FJS107 Χ BB2505 DNA |
| LH16 | trpC2 ΔamyE::(ΦyhdG283-lacZ erm) codY::(erm::spc) | PS251 Χ BB2505 DNA |
| LH17 | trpC2 ΔamyE::(Φyhd283-lacZ erm) codY(F40A) | LH11 Χ BB2505 DNA |
| LH20 | trpC2 ΔamyE::(ΦyhdG283-lacZ erm) codY(F98A) | LH4 Χ BB2505 DNA |
| LH22 | trpC2 ΔamyE::spc codY(R61A) | BB2611 Χ BB2511 DNA |
| LH23 | trpC2 ΔamyE::(ΦyhdG283-lacZ erm) codY(R61A) | LH22 Χ BB2505 DNA |
| LH24 | trpC2 ΔamyE::spc codY(F71E) | BB2616 Χ BB2511 DNA |
| LH25 | trpC2 ΔamyE::(ΦyhdG283-lacZ erm) codY(F71E) | LH24 Χ BB2505 DNA |
To construct strain BB2611, plasmid pBB1451 was integrated by a single-crossover homologous recombination event at the codY locus of B. subtilis strain PJB14 by transformation. All strain constructions listed were by transformation of the indicated recipient strain with the indicated plasmid or chromosomal DNA.
TABLE 2.
E. coli strains used in this study
| Strain | Genotype or description | Reference |
|---|---|---|
| JM107 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14− (mcrA) Δ(lac-proAB) thi gyrA96 (Nalr) endA1 hsdR17 (rK− mK+) relA1 supE44 | 53 |
| JM107(pBAD30) | Arabinose-inducible expression vector | 19 |
| JM107(pJPM1) | Integrative vector for B. subtilis | 37 |
| JM107(pMAD) | bgaB+ integrative vector for B. subtilis | 1 |
| JM107(pBB544) | neo gene cloned in pBluescript SK(−) | 4 |
| JM107(pLH2) | bgaB gene inserted in pBB544 | This study |
| JM107(pKT1) | codY+ cloned in pBAD30 | 28 |
| JM107(pEAV20) | codY(R61A) cloned in pBAD30 | This study |
| JM107(pEAV22) | codY(F71E) cloned in pBAD30 | This study |
| JM107(pLH12) | codY(F98A) cloned in pBAD30 | This study |
| JM107(pLH16) | codY(F40A) cloned in pBAD30 | This study |
| JM107(pBB1451) | codY(R61A) cloned in pJPM1 | This study |
| JM107(pBB1453) | codY(R61E) cloned in pJPM1 | This study |
| JM107(pBB1455) | codY(F71R) cloned in pJPM1 | This study |
| JM107(pBB1456) | codY(F71E) cloned in pJPM1 | This study |
| JM107(pBB1457) | codY(F71A) cloned in pJPM1 | This study |
| JM107(pBB1459) | codY(R61K) cloned in pJPM1 | This study |
| JM107(pLH9) | codY(F98A) cloned in pLH2 | This study |
| JM107(pLH15) | codY(F40A) cloned in pLH2 | This study |
| JM107(pRPS5) | ilvB-lacZ fusion plasmid | 44 |
Culture media and DNA manipulations.
E. coli strains, all derivatives of JM107, were grown in Luria-Bertani (LB) medium (35) in the presence of ampicillin (50 μg ml−1) or chloramphenicol (25 μg ml−1) when appropriate. B. subtilis strains were grown in DS medium (15) or in TSS minimal medium (2) as indicated. Plasmid DNA was isolated from E. coli by using a QIAprep spin miniprep kit (Qiagen). Preparation and transformation of electrocompetent E. coli cells were as described previously (12). B. subtilis chromosomal DNA isolation, competent-cell preparation, and transformation were performed as described previously (15).
Site-directed mutagenesis.
Mutations in codY were introduced by a two-step overlapping PCR method. For mutations targeting R61 and F71, 0.35-kb fragments containing the 3′ end of codX and the 5′ part of codY were synthesized using Platinum HiFi Taq polymerase (Invitrogen), chromosomal DNA of strain JH642 as a template, oligonucleotide oBB272 as a forward primer and the mutagenic oligonucleotide oBB273, oBB274, oBB278, oBB280, oBB282, or oBB284 as a reverse primer (all oligonucleotide primers used in this study are listed in Table S1 in the supplemental material). In a similar manner, 0.35-kb fragments containing the internal part of codY were synthesized using the mutagenic oligonucleotide oBB275, oBB276, oBB279, oBB281, oBB283, or oBB285 as a forward primer and oligonucleotide oBB277 as a reverse primer (see Table S1 in the supplemental material). The mutation-containing PCR products were used in a second splicing step of PCR mutagenesis as overlapping templates to generate modified 0.7-kb fragments containing the 3′ end of codX and the 5′ part of codY using oligonucleotides oBB272 and oBB277 as forward and reverse PCR primers, respectively.
The spliced PCR products were digested with EcoRI and HindIII and cloned in pJPM1, a plasmid that replicates in E. coli but not in B. subtilis, where it functions as an integrative vector conferring chloramphenicol resistance (37). The following plasmids were generated, each carrying a desired mutation in codY: pBB1451 (the R61A mutation), pBB1459 (the R61K mutation), pBB1453 (the R61E mutation), pBB1457 (the F71A mutation), pBB1455 (the F71R mutation), and pBB1456 (the F71E mutation). Using transformation, these plasmids were each integrated by single-crossover homologous recombination events at the codY locus of B. subtilis strain PJB14 [amyE::(ilvBΔT-lacZ)] (25). (This fusion contains six mutations in its ilvB-derived sequences, but those mutations do not alter CodY-dependent regulation [3].) Chloramphenicol-resistant transformants were initially selected and purified and then grown for multiple generations in the absence of chloramphenicol. Chloramphenicol-sensitive colonies that would have arisen by plasmid excision and loss were isolated. The presence, as a result of gene replacement, of codY mutations in the chromosome of plasmidless strains was established using PCR with primers diagnostic for each individual mutation and by sequencing PCR products corresponding to the entire chromosomal copy of the codY gene. The resulting strains BB2611, BB2619, and BB2613 to BB2616 contained mutations leading to production of CodY(R61A), CodY(R61K), CodY(R61E), CodY(F71A), CodY(F71R), and CodY(F71E), respectively (Table 1).
For mutations targeting residues F40 and F98 of CodY, the two-step overlapping PCR method described above was followed using B. subtilis FJS107 genomic DNA as the template. For F40A, two PCR fragments were created using (i) oligonucleotide oLH5 as the forward primer and mutagenic oligonucleotide oLH7 as the reverse primer, and (ii) mutagenic oligonucleotide oLH6 as the forward primer and oligonucleotide oLH8 as the reverse primer. In the splicing step of mutagenesis, the final PCR product was created using primers oLH5 and oLH8. For F98A, the first PCR fragment was created using forward primer oLH17 and mutagenic oligonucleotide oLH20 as the reverse primer, while the second PCR fragment was created using mutagenic oligonucleotide oLH19 as the forward primer and oLH18 as the reverse primer. In the splicing step, the final PCR product was synthesized using primers oLH17 and oLH18. The final PCR products containing the mutations corresponding to F40A and F98A were cloned in pCR2.1-TOPO (Invitrogen) and were subsequently introduced into either the EcoRI site (for F40A) or between the EcoRI and BamHI sites (for F98A) of pLH2 to create pLH15 and pLH9, respectively (Table 2). pLH2 was derived by insertion of the bgaB gene of pMAD (1) into the integrative, neomycin resistance-conferring plasmid pBB544 (4) (Table 2). Plasmids pLH15 (for the F40A mutation) and pLH9 (for the F98A mutation) were introduced into B. subtilis strain FJS107 by transformation, and neomycin-resistant transformants were initially selected, purified, and then grown for multiple generations in the absence of neomycin. Neomycin-sensitive colonies that would have arisen by plasmid excision and loss were isolated. The presence, as a result of gene replacement, of the desired codY mutations in the plasmidless strains LH11 (F40A) and LH4 (F98A) was established using PCR with primers diagnostic for each individual mutation and by sequencing PCR products corresponding to the entire chromosomal copy of the codY gene. The F40A mutant also had a silent mutation (TTA to CTA) at the codon corresponding to residue L110.
Overexpression of mutant CodY proteins in E. coli.
The mutant codY genes were amplified from B. subtilis chromosomal DNA by PCR using oRPS32 and oRPS33 primers (see Table S1 in the supplemental material) and cloned in E. coli to produce pure CodY for in vitro analysis. Oligonucleotide oRPS32 anneals upstream of the codY gene and contains the codY Shine-Dalgarno sequence and a SacI restriction site. oRPS33 anneals to the 3′ end of the codY gene and appends five additional histidine codons to the C-terminal histidine codon of the codY gene, followed by a stop codon and an SphI restriction site. The PCR products were digested using restriction enzymes SacI and SphI and ligated to SacI- and SphI-digested vector pBAD30 (19), placing the codY gene under the control of the araBAD promoter. The ligated product was introduced into E. coli strain JM107 by electroporation. The integrity of each of the mutant codY genes was verified by sequencing.
Purification of CodY.
E. coli strains carrying plasmid pBAD30 in which the wild-type or mutant versions of the codY gene were cloned with C-terminal six-histidine tags (Table 2) were grown in LB medium containing ampicillin until the optical density at 600 nm (OD600) reached ∼0.6. Arabinose (0.2% final concentration) was added, and growth was continued for 4 to 5 h. Cells were then harvested by centrifugation and broken by sonication. CodY proteins were purified by Talon metal affinity resin (Clontech, Mountain View, CA) chromatography using 75 mM imidazole for elution by a previously described method (44). The elution fractions were free of detectable contaminating proteins by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis. The various CodY proteins were quantified by the Bio-Rad protein assay (Bio-Rad, Hercules, CA).
DNase I footprinting.
A 453-bp fragment of the ilvB promoter region was amplified from plasmid pRPS5 (44) by PCR using Platinum Taq High Fidelity DNA polymerase and primers oRPS7 and oRPS6 (44). The latter had been labeled with [γ-32P]ATP by treatment with T4 polynucleotide kinase (Invitrogen) (27). DNase I footprinting assays were performed as described before (44). DNase I digestion products were separated on an 8 M urea-6% polyacrylamide gel by electrophoresis at 1200 V for 5 h. Gels were dried under vacuum and exposed to a phosphorimager screen. Screens were scanned with a Molecular Dynamics Storm 860 imager (GE Healthcare Bio-Sciences, Piscataway, NJ), and densitometric quantitation was performed using ImageQuant TL software (GE Healthcare Bio-Sciences). The ilvB sequence in pRPS5 contains four mutations, but these mutations have no significant effect on binding of CodY in vitro (20) or on expression of an ilvB-lacZ fusion in vivo (3).
β-Galactosidase assays.
B. subtilis strains carrying fusions to the E. coli lacZ gene were grown overnight at 37°C in TSS minimal medium supplemented with 0.5% glucose, 0.2% glutamine, 0.02% MgSO4, FeCl3-Na citrate (40 μg ml−1), and a 16-amino-acid mixture that contains the BCAAs or a 13-amino-acid mixture that lacks the BCAAs (2). A sample (0.15 ml) of the overnight culture was used to inoculate 28 ml of the same medium and grown at 37°C until the OD600 reached 0.1 to 0.2. Three or more samples of each culture were removed over the course of the next 60 min of exponential-phase growth and assayed for β-galactosidase activity (44).
Expression of mutant CodY proteins in B. subtilis.
B. subtilis strains carrying the codY point mutations described above or a codY null mutation or the wild-type codY gene were grown in 60 ml of TSS medium supplemented with 16 amino acids (as described above) to an OD600 of approximately 0.8. The cell pellets from 10-ml culture samples were resuspended in 0.5 ml solution A (41) containing 1 mM phenylmethylsulfonyl fluoride and 2 μM pepstatin A. The cells were sonicated on ice for two cycles of a 30-s pulse with a 30-s break between pulses. The lysates were then centrifuged at 13,000 rpm at 4°C. The quantity of total protein present in the supernatant fluid of the samples was determined using the Bio-Rad protein assay reagent. Two micrograms of total soluble protein was loaded on a 12% SDS-polyacrylamide gel after boiling for 5 min in Laemmli loading buffer (62.5 mM Tris [pH 6.8], 2% SDS, 10% glycerol, 0.015% bromophenol blue) containing 2.5% β-mercaptoethanol. The samples were subjected to electrophoresis at a constant voltage of 110 V for 1 h. The proteins were then transferred to Immobilon-P membranes (Millipore) at 4°C under constant voltage of 100 V in electrotransfer buffer (192 mM glycine, 25 mM Tris, 15% methanol). Immunoblotting was performed as described previously (39) using rabbit polyclonal antibody to CodY prepared by Biodesign International (Kennebunkport, ME).
RESULTS
Site-directed mutagenesis of R61, F71, F40, and F98 residues.
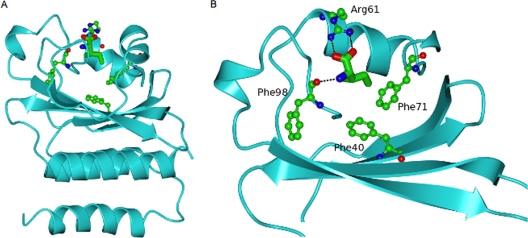
In the crystal structure, the R61 and F98 residues of B. subtilis CodY are involved in forming polar/ionic interactions with isoleucine, and F71 forms hydrophobic interactions with isoleucine (29) (Fig. 1). All these residues are highly conserved in CodY homologs from several bacterial species. Furthermore, residues F40 and F98 may be involved in interaction with GTP (29). To test the functional importance of these residues in interacting with BCAAs and GTP, they were subjected to site-directed mutagenesis. The positively charged R61 residue was changed to a nonpolar alanine (R61A), a negatively charged glutamate (R61E), or a positively charged lysine (R61K) residue. The nonpolar aromatic ring-containing F71 was changed to a nonpolar alanine (F71A), a positively charged arginine (F71R), or a negatively charged glutamate (F71E) residue. The nonpolar aromatic ring-containing F40 and F98 residues were changed to alanine (F40A and F98A, respectively). Note that the F98A alteration replaces the phenyl group side chain with a methyl group but would not be expected to affect the potential polar interactions of the main-chain carbonyl group with isoleucine (29).
FIG. 1.
Ribbon diagram of the GAF domain of B. subtilis CodY with the bound isoleucine ligand (cylinder representation) surrounded by residues whose side chains have been mutated in this study. (A) The whole GAF domain is shown. (B) Close-up of the ligand-binding pocket in which the lower helical region has been omitted and residues 70 to 96 have been left out for clarity of view. Polar interactions with the isoleucine ligand are indicated by dashed lines.
Introduction of mutations into the chromosomal codY gene.
To study the effects of alteration of R61, F71, F40, and F98, the mutations were introduced into the chromosomal codY gene by a two-step allelic replacement method (see Materials and Methods). The resultant strains had a single (mutant) copy of the codY gene. Only strains that had acquired the expected mutation and no other mutation were used in subsequent studies (with the exception of strain LH11, the F40A codY mutant, which had a silent mutation at codon 110). To rule out the possibility that potential effects on CodY target gene expression were due to instability of the mutant CodY proteins, each of the mutant B. subtilis strains was tested by immunoblotting using antibodies raised against CodY protein. The results indicate that all CodY mutants were produced as stable proteins in B. subtilis at levels comparable to those for wild-type CodY (see Fig. S1 in the supplemental material) (data not shown for strains LH4 and LH11).
Effects of CodY point mutations on ilvB expression.
The ilvB operon, which encodes enzymes involved in the biosynthesis of BCAAs, is a well-characterized B. subtilis target of CodY (36, 44, 45, 51). To test the effects of the mutations on CodY activity in vivo without interference by an independent mode of regulation via transcription termination in the leader region of the mRNA, we used a strain that carries an ilvBΔT-lacZ fusion at the nonessential amyE locus. This fusion lacks the T-box region that is involved in leucine-responsive transcription termination (17, 44).
Strains carrying mutant or wild-type versions of the codY gene were grown in a defined medium (TSS) supplemented with 0.5% glucose, 0.2% glutamine, and a mixture of 16 amino acids that includes the three BCAAs (2). CodY is known to be active in cells grown in this medium (14, 44). Indeed, the wild-type strain showed 16-fold repression of ilvBΔT-lacZ expression compared to the codY null strain, PJB15 (Table 3). These results are consistent with previous reports (25, 44). The various codY point mutant strains showed a significant loss of repressing activity in the BCAA-containing medium, in most cases approaching the level of derepression seen for the null mutant (Table 3).
TABLE 3.
ilvBΔT-lacZ fusion activity in codY point mutantsa
| Growth medium | Strain (genotype or substitution in CodY) | Μean β-galactosidase sp act ± SD (fold derepression)b | % of codY null activity |
|---|---|---|---|
| With BCAAs | PJB14 (Wild-type) | 46 ± 15 (1) | 6 |
| PJB15 (codY) | 722 ± 77 (15.7) | 100 | |
| BB2611 (R61A) | 490 ± 67 (10.7) | 68 | |
| BB2613 (R61E) | 649 ± 52 (14.1) | 90 | |
| BB2619 (R61K) | 436 ± 77 (9.5) | 60 | |
| BB2614 (F71A) | 199 ± 39 (4.3) | 28 | |
| BB2615 (F71R) | 649 ± 101 (14.1) | 90 | |
| BB2616 (F71E) | 685 ± 67 (14.9) | 95 | |
| LH12 (F40A) | 671 ± 35 (14.6) | 93 | |
| LH7 (F98A) | 609 ± 33 (13.2) | 84 | |
| Without BCAAs | PJB14 (Wild-type) | 482 ± 43 (1) | 66 |
| PJB15 (codY) | 728 ± 55 (1.5) | 100 | |
| BB2611 (R61A) | 628 ± 64 (1.3) | 86 | |
| BB2613 (R61E) | 683 ± 49 (1.4) | 94 | |
| BB2619 (R61K) | 658 ± 45 (1.3) | 90 | |
| BB2614 (F71A) | 494 ± 34 (1) | 68 | |
| BB2615 (F71R) | 663 ± 99 (1.3) | 91 | |
| BB2616 (F71E) | 702 ± 43 (1.4) | 96 | |
| LH12 (F40A) | 661 ± 34 (1.4) | 91 | |
| LH7 (F98A) | 629 ± 33 (1.3) | 86 |
Cells were grown to mid-exponential phase in TSS minimal medium supplemented with glucose, glutamine, and a 13-amino-acid mixture with or without added BCAAs.
β-Galactosidase activities (in Miller units) represent the averages of three samples taken during subsequent exponential growth at OD600 values ranging from 0.2 to 0.9 in three independent cultures. The change in derepression (fold derepression) represents the β-galactosidase activity in a mutant strain divided by the activity in the wild-type strain.
When the positively charged R61 residue was changed to alanine (R61A) or to lysine (R61K), the resultant mutant strains retained some repressing activity, but changing R61 to glutamate (R61E) resulted in nearly complete loss of repression (Table 3). Since changing arginine to lysine (both of which are polar, positively charged residues) resulted in a protein with R61K that is only partially active, the guanidinium group of arginine appears to provide a specific interaction with BCAAs, GTP, or DNA.
When F71 was changed to alanine (F71A), the protein retained 72% of its repressing activity (Table 3), even though the side chain of alanine is significantly different from the side chain of phenylalanine. However, when F71 was changed to arginine (F71R) or glutamate (F71E), loss of repressing activity was essentially complete. We also saw strong derepression in the BCAA-containing medium when residues F40 and F98 were changed to alanine (F40A and F98A, respectively), indicating that these mutations strongly affect the repressing activity of CodY (Table 3).
Since some of the mutant strains showed residual repression, we tested whether such repression was in response to the BCAAs. That is, the same panel of strains was grown in the medium used above but without the three BCAAs. In this medium, wild-type CodY is only partially active, and the level of ilvBΔT-lacZ expression is about 70% of that of a codY null mutant (Table 3). (This extent of repression in BCAA-free medium is presumably due to endogenous synthesis of BCAAs, GTP, or both.) All of the point mutants (as well as the wild-type strain) showed virtually complete loss of repression (Table 3). The simplest explanation is that the R61A, R61K, and F71A mutants are partially responsive to BCAAs. Alternatively, amino acid limitation caused by the lack of BCAAs might induce the stringent response, which would lower the GTP pool in the cell (8), leading to reduced CodY activity and greater derepression of ilvB-lacZ expression.
Effects of CodY point mutations on yhdG expression.
To ensure that the observations made with the ilvBΔT-lacZ fusion strains were not specific to ilvB regulation, we also tested the effects of these mutations on regulation of an additional CodY target, yhdG, a gene shown to be strongly negatively regulated by CodY in microarray analyses and yhdG-lacZ fusion assays (36; B. R. Belitsky and A. L. Sonenshein, unpublished data). In addition, gel mobility shift and DNase I footprinting assays have shown that CodY binds to the yhdG promoter region in vitro (36; B. R. Belitsky and A. L. Sonenshein, unpublished data). In medium containing the BCAAs (Table 4), the wild-type strain showed strong repression of yhdG-lacZ expression compared to the codY null strain, but the various point mutant strains showed a significant loss of repressing activity. The F71E, F40A, and F98A mutants were strongly derepressed, while the R61A mutant showed partial derepression (21% of codY null behavior) (Table 4). To test whether the mutants retained any ability to respond to BCAAs, they were grown in TSS minimal medium plus 13 amino acids. With the exception of the R61A mutant, which showed an additional 3.4-fold derepression, the remaining mutants showed the same level of derepression as that seen in the 16-amino-acid medium (Table 4).
TABLE 4.
yhdG-lacZ fusion activity in codY point mutantsa
| Growth medium | Strain (genotype or substitution in CodY) | Mean β-galactosidase sp act ± SD (fold derepression) | % of codY null activity |
|---|---|---|---|
| With BCAAs | LH15 (Wild-type) | 0.3 ± 0.1(1) | 0.001 |
| LH16 (codY) | 185 ± 6 (617) | 100 | |
| LH23 (R61A) | 38 ± 10 (127) | 21 | |
| LH25 (F71E) | 166 ± 16 (553) | 90 | |
| LH17 (F40A) | 170 ± 19 (567) | 92 | |
| LH20 (F98A) | 151 ± 27 (503) | 82 | |
| Without BCAAs | LH15 (Wild-type) | 17 ± 4 (1) | 11 |
| LH16 (codY) | 158 ± 8 (9.3) | 100 | |
| LH23 (R61A) | 99 ± 19 (5.8) | 63 | |
| LH25 (F71E) | 152 ± 12 (8.9) | 96 | |
| LH17 (F40A) | 157 ± 8 (9.2) | 100 | |
| LH20 (F98A) | 151 ± 13 (8.9) | 96 |
Cells were grown and assayed for β-galactosidase activity as described in Table 3, footnotes a and b.
Effects of codY mutations on interaction with effectors.
To resolve the question of how well mutant CodY proteins interact with DNA and respond to GTP and BCAAs, wild-type and mutant His-tagged CodY proteins were expressed in E. coli, purified by cobalt affinity chromatography, and tested in DNase I footprinting assays. To make detailed analysis manageable, we focused on three mutant proteins (with the F40A, F71E, and F98A changes) that retained little or no repressing activity and one mutant (R61A) that retained some repressing activity in vivo.
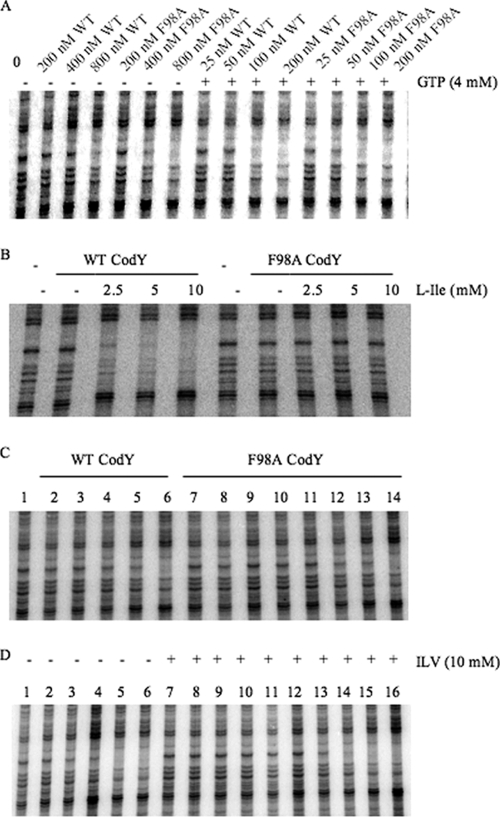
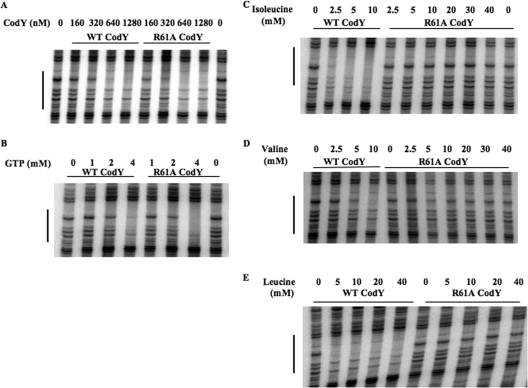
An alanine substitution for phenylalanine at position 98 (F98A) of CodY had no deleterious effect on corepressor-independent binding or on the ability of GTP (4 mM) to stimulate binding to DNA (Fig. 2A). Both the wild-type and F98A forms of CodY in the presence of GTP had an apparent dissociation constant (Kd), defined as the concentration of CodY that leads to protection of 50% of the probe molecules, of 50 to 100 nM (Fig. 2A and Table 5). On the other hand, F98A CodY (at 125 nM) was not stimulated by isoleucine (Fig. 2B and Table 6) or at any CodY concentration tested, by a mixture of isoleucine, valine, and leucine (ILV) (Fig. 2C and D). The F98A alteration increased the apparent Kd for DNA binding in the presence of 10 mM ILV from 25 to 50 nM seen for wild-type CodY to 200 to 400 nM (Fig. 2C and D), a value close to the Kd for effector-independent binding (Fig. 2A and D).
FIG. 2.
DNase I footprinting analysis of wild-type and F98A CodY binding to the ilvB promoter in the presence and absence of effectors. CodY proteins were incubated with an end-labeled DNA fragment that includes the previously described high-affinity binding site II within the ilvB promoter region (44) and then challenged with DNase I before electrophoresis under denaturing conditions (see Materials and Methods for details). (A) Increasing concentrations of wild-type (WT) and F98A CodY proteins were incubated with the ilvB probe in the absence (−) or presence (+) of 4 mM GTP as indicated above the gel. (B) Mutant and wild-type (WT) CodY proteins (125 nM) were incubated with the ilvB probe in the presence of increasing concentrations of l-isoleucine as indicated above the gel (−, no l-isoleucine). (C) Increasing concentrations of mutant and wild-type CodY proteins were incubated with the ilvB probe in the presence of 10 mM each of isoleucine, leucine, and valine. The concentrations (in nanomolar) of wild-type CodY protein (lanes 1 to 6) and F98A CodY protein (lanes 7 to 14) in the various lanes were as follows: 0 (lane 1), 6 (lane 2), 12 (lane 3), 25 (lane 4), 50 (lane 5), 100 (lane), 6 (lane 7), 12 (lane 8), 25 (lane 9), 50 (lane 10), 100 (lane 11) 200 (lane 12), 400 (lane 13), and 800 (lane 14). (D) Increasing concentrations of F98A mutant CodY protein was incubated with the ilvB probe in the presence or absence of 10 mM each of isoleucine, valine, and leucine (ILV) as indicated above the gel. The concentration (in nanomolar) of CodY in the various lanes were as follows: 0 (lane 1), 100 (lane 2), 200 (lane 3), 400 (lane 4), 800 (lane 5), 1,600 (lane 6), 0 (lane 7), 6 (lane 8), 12 (lane 9), 25 (lane 10), 50 (lane 11), 100 (lane 12), 200 (lane 13), 400 (lane 14), 800 (lane 15), and 1,600 (lane 16).
TABLE 5.
Binding of wild-type and mutant CodY proteins to the ilvB promoter regiona
| CodY protein | Approximate Kd (nM) for binding to ilvB site II |
||
|---|---|---|---|
| No effectors | GTP (4 mM) | BCAAs (10 mM) | |
| Wild-type | 400-800 | 50-100 | 25-50 |
| F98A | ∼400 | 50-100 | ∼200 |
| F71E | 400-800 | 85-100 | >160b |
| F40A | ∼400 | 50-100 | >100b |
| R61A | ∼320 | ∼50 | ∼160 |
The results presented reflect analyses of DNase I footprinting experiments displayed in Fig. 2 to 6 and other experiments (not shown) in which the concentration of CodY was varied but the concentrations of the effectors (when present) were saturating. The CodY concentration that gave 50% protection was taken to be an approximation of the Kd for binding.
At the concentrations shown, F71E CodY and F40A CodY were not able to bind to the ilvB probe at BCAA concentrations as high as 40 mM. Higher concentrations of mutant CodY were not tested.
TABLE 6.
Activation of wild-type and mutant CodY proteins by individual BCAAsa
| CodY concn (nM) | CodY type | Concn (mM) of effector that gave 50% protection of ilvB site II |
||
|---|---|---|---|---|
| Ile | Val | Leu | ||
| 85 | Wild-type | <2.5 | ||
| F71E | >40 | |||
| R61A | >40 | |||
| 160 | Wild-type | 2.5-5.0 | <5.0 | |
| F71E | >40 | >40 | ||
| R61A | >40 | >40 | ||
| 100 | Wild-type | <2.5 | ||
| F40A | >10 | |||
| 125 | Wild-type | <2.5 | ||
| F98A | >10 | |||
DNase I footprinting experiments shown in Fig. 2 to 6 and other experiments (not shown) were analyzed to determine the concentrations of BCAAs that gave 50% protection of the high-affinity CodY binding site II in the ilvB promoter region at the fixed CodY concentrations tested. CodY concentrations were chosen on the basis of titration experiments with wild-type CodY (which varied slightly from day to day and batch to batch) so as to exclude effector-independent binding while permitting detection of BCAA-induced binding. The mutant forms of CodY were tested at the same concentrations as wild-type CodY.
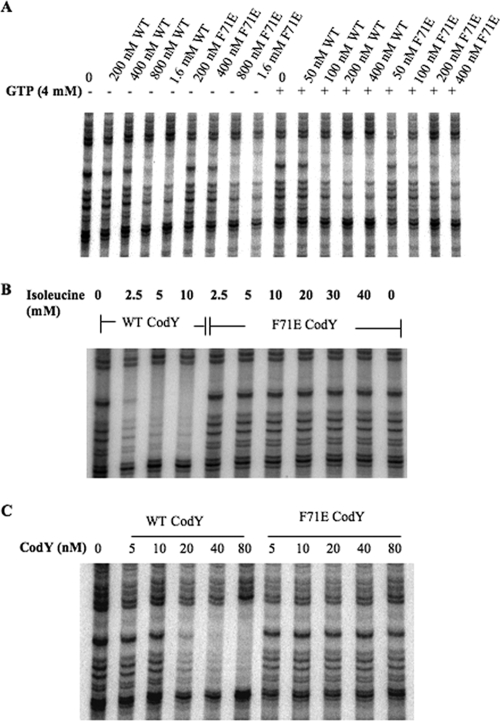
The F71E mutant protein had no defect in intrinsic (effector-independent) binding to DNA and was stimulated by 4 mM GTP to almost the same extent as the wild-type CodY (Fig. 3A and Table 5). F71E CodY (at 85 nM), however, was not activated for DNA binding by the presence of isoleucine, even at a concentration as high as 40 mM (Fig. 3B and Table 6). Moreover, the affinity of F71E CodY for DNA in the presence of 10 mM ILV was at least fourfold lower than that of wild-type CodY (Fig. 3C), even when 2 mM GTP was also present. (Had we used concentrations of CodY higher than 80 nM in Fig. 3C, we would presumably have seen binding by F71E CodY at 100 to 200 nM due to the presence of GTP [Fig. 3A].)
FIG. 3.
DNase I footprinting analysis of wild-type CodY and F71E CodY binding to the ilvB promoter region in the presence and absence of effectors. (A) Increasing concentrations of wild-type (WT) or F71E CodY protein were incubated with the ilvB probe in the presence (+) or absence (−) of 4 mM GTP as indicated above the gel. (B) The reaction mixtures contained 85 nM wild-type CodY or F71E CodY with increasing concentrations of isoleucine as indicated above the gel. (C) All samples contained 2 mM GTP and 10 mM ILV. The sample in lane 1 did not contain any protein. The concentrations of wild-type CodY and F71E CodY in each reaction mixture are indicated above the gel.
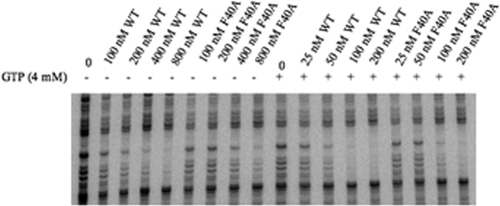
An alanine substitution for phenylalanine at position 40 (F40A) of the CodY protein caused a slight (twofold) reduction in effector-independent binding (i.e., intrinsic binding efficiency) to the ilvB probe but caused no significant difference in the stimulation of binding by GTP (Fig. 4). In another experiment, F40A CodY, at a concentration of 100 nM, did not respond to isoleucine (10 mM) (Table 6).
FIG. 4.
DNase I footprinting analysis of wild-type and F40A CodY binding to the ilvB promoter. Increasing concentrations of the wild-type (WT) or F40A CodY proteins were incubated with the ilvB probe in the absence (−) or presence (+) of 4 mM GTP as indicated above the gel.
The greatly reduced responses of the F98A, F71E, and F40A forms of CodY to BCAAs correlate well with results from in vivo studies in which strains producing these mutant CodY proteins showed nearly complete derepression of the ilvBΔT-lacZ and yhdG-lacZ fusions both in the presence and absence of BCAAs in the growth medium (Tables 3 and 4). A possible interpretation of these results is that these alterations prevent any physiologically significant interaction of CodY with BCAAs. To explain the failure of GTP to activate the mutant CodY proteins in vivo, we postulate that interaction with BCAAs is a prerequisite for stimulation by GTP at its physiological concentration or, less likely, that our in vivo growth conditions do not provide a test for stimulation of CodY by GTP.
R61A CodY.
Unlike strains carrying F98A, F71E, or F40A CodY, an R61A mutant strain retained partial repressing activity in medium containing BCAAs (Tables 3 and 4). We sought to understand this behavior through in vitro studies of CodY-effector interaction. The R61A CodY protein showed no defect in intrinsic binding to DNA (Fig. 5A) and responded to GTP to the same extent as the wild-type protein (Fig. 5B and Table 5). Thus, the altered phenotype of the mutant in vivo presumably reflects a change in interaction with other effectors.
FIG. 5.
DNase I footprinting analysis of wild-type CodY and R61A CodY binding to the ilvB promoter region. (A) The binding reaction mixtures contained the indicated concentrations of purified wild-type (WT) or mutant CodY proteins. No effectors were present during these incubations. (B) The reaction mixtures contained 85 nM wild-type CodY or R61A CodY with increasing concentrations of GTP as indicated above the gel. (C) The reaction mixtures contained 85 nM wild-type CodY and R61A CodY with increasing concentrations of isoleucine as indicated above the gel. (D) The reaction mixtures contained 160 nM wild-type or R61A CodY and various concentrations of valine as indicated above the gel. (E) The reaction mixtures contained 160 nM wild-type or R61A CodY and various concentrations of leucine as indicated above the gel.
To ascertain whether the R61A mutant protein could be stimulated for DNA binding by the BCAAs, footprinting assays were performed in the presence of each BCAA individually. We used a concentration of wild-type CodY (85 nM) that was below the concentration needed for effector-independent protection and found that this concentration was sufficient to give complete protection of the ilvB site in the presence of 2.5 mM isoleucine or 4 mM GTP (Fig. 5B and C). On the other hand, R61A CodY (85 nM) did not respond to isoleucine at concentrations as high as 40 mM (Fig. 5C and Table 6). In similar experiments, 160 nM wild-type CodY was stimulated for DNA binding in the presence of 2.5 to 5 mM valine or <5 mM leucine, while 160 nM R61A CodY was not stimulated by either of these compounds even at 40 mM (Fig. 5D and E and Table 6). These results indicate that R61A CodY is generally defective in responding to BCAAs in vitro.
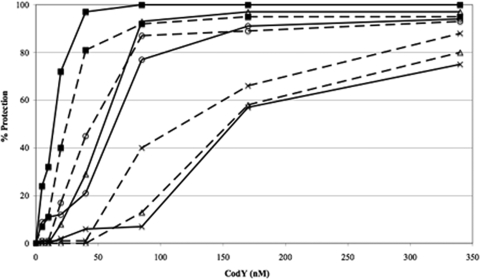
GTP and BCAAs are known to have a synergistic effect in enhancing the binding of wild-type CodY to the ilvB promoter region in gel shift and footprinting assays (42, 44). Moreover, the presence of one of the effectors lowers the concentration of the second effector required to stimulate CodY for DNA binding (20). To investigate whether R61A CodY responds to isoleucine in the presence of GTP, we performed a series of in vitro footprinting assays using increasing concentrations of wild-type CodY and R61A CodY with no effectors or in the presence of 4 mM GTP, 10 mM isoleucine, or both effectors. In the absence of effectors, the affinities of wild-type and R61A CodY for DNA were similar (Fig. 6). In the presence of 4 mM GTP, the affinities of the two proteins for DNA were equally stimulated (two- to threefold) by GTP. In the presence of 10 mM isoleucine, the affinity of wild-type CodY for DNA was increased by threefold, while that of R61A CodY was slightly reduced (Fig. 6). Most importantly, in the presence of both 4 mM GTP and 10 mM isoleucine, the affinity of wild-type CodY for DNA was increased 10-fold, while that for R61A CodY was increased by 4- to 5-fold (Fig. 6), suggesting that R61A CodY retains the ability to respond to isoleucine, albeit less efficiently than wild-type CodY, as long as GTP is present. This property of R61A CodY may explain its retention of partial repressing activity in vivo.
FIG. 6.
DNase I footprinting analysis of the additive effects of GTP and isoleucine on wild-type and R61A CodY binding to the ilvB promoter. The concentrations of wild-type and R61A CodY are indicated on the x axis (in nanomolar), and the relative protection is indicated on the y axis (as a percentage) as determined by densitometry analysis. The extent of protection provided by the wild-type protein in the presence of both effectors (4 mM GTP and 10 mM isoleucine) was set at 100% protection, and the relative percentage protection values were estimated by comparison. Wild-type CodY (solid line) and R61A CodY (broken line) are shown. The various conditions are represented by the symbols indicated as follows: no effectors (×), 4 mM GTP (○), 10 mM isoleucine (▵), and 4 mM GTP plus 10 mM isoleucine (▪).
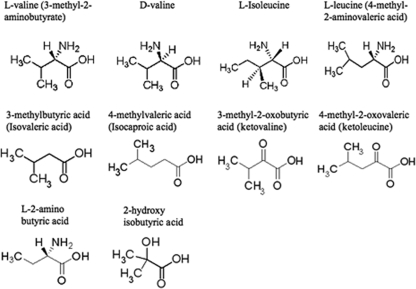
Interaction of CodY with BCAAs and BCAA-related compounds.
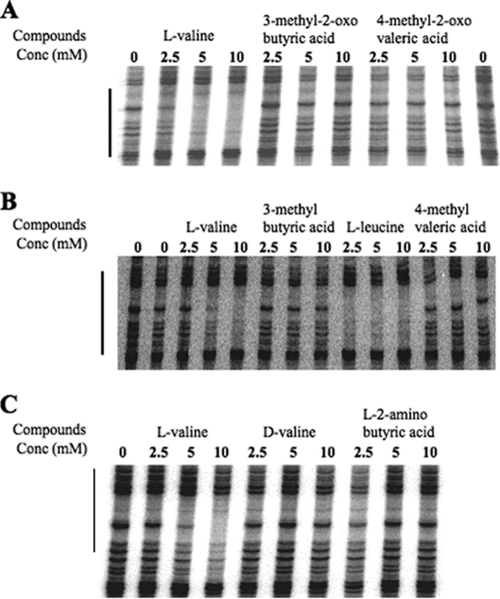
To obtain additional information about the specificity of CodY-effector interaction, we compared the abilities of the three BCAAs and analogs (Fig. 7) to stimulate binding of CodY to DNA. As expected, addition of increasing concentrations of l-valine resulted in the stimulation of DNA binding by CodY as detected by protection of the target DNA (44) (Fig. 8). However, addition of 3-methyl-2-oxobutyric acid (also known as α-ketoisovalerate or ketovaline) or 4-methyl-2-oxovaleric acid (also known as α-ketoisocaproate or ketoleucine) did not stimulate DNA binding by CodY (Fig. 8A), suggesting that the α-amino groups of valine and leucine are important for interaction with CodY. To rule out the possibility that the absence of CodY interaction with the keto acids is due to interference caused by the introduction of a 2-keto group, we also tested whether 3-methylbutyric acid and 4-methylvaleric acid can stimulate DNA binding by CodY. In fact, the addition of 3-methylbutyric acid and 4-methylvaleric acid did not stimulate binding, indicating that the α-amino group is required for interaction with CodY and cannot be replaced by a keto group (Fig. 8B). Three additional compounds closely related to l-valine were also tested in DNase I footprinting assays. In contrast to l-valine, d-valine did not stimulate DNA binding of CodY, indicating that the chirality of the α-amino group is important for interaction of l-valine with CodY (Fig. 8C). l-2-Aminobutyric acid, which differs from l-valine by the absence of a 3-methyl group, and 2-hydroxyisobutyric acid, which lacks methylene and amino groups, also failed to stimulate target DNA binding by CodY (Fig. 8C and data not shown).
FIG. 7.
BCAAs and BCAA-related compounds used in this study.
FIG. 8.
DNase I footprinting analysis of wild-type CodY binding to the ilvB promoter in the presence of BCAAs and BCAA-related compounds. All reaction mixtures contained 160 nM wild-type CodY. (A) Reaction mixtures shown in lanes 2 to 4, lanes 5 to 7, and lanes 8 to 10 contained increasing concentrations (Conc) of l-valine, 3-methyl-2-oxobutyric acid, and 4-methyl-2-oxovaleric acid, respectively, as indicated above the gel. The samples in the leftmost and rightmost lanes did not contain BCAAs or BCAA-related compounds. (B) The samples in lanes 2 and 3, lanes 4 and 5, and lanes 6 and 7 contained increasing concentrations of l-valine, 3-methyl butyric acid, and 4-methyl valeric acid, respectively, as indicated above the gel. The sample in the leftmost lane did not contain BCAAs or BCAA-related compounds. (C) The samples in lanes 2 to 4, lanes 5 to 7, and lanes 8 to 10 contained increasing concentrations of l-valine, d-valine, and l-2-amino butyric acid, respectively, as indicated above the gel. The sample in lane 1 did not contain BCAAs or BCAA-related compounds.
DISCUSSION
Interaction with GTP and BCAAs stimulates binding of CodY to DNA (39, 44). While the exact mechanisms by which these interactions stimulate the DNA-binding activity of CodY are not known, the interaction of CodY with BCAAs has been found to induce a significant conformational change that could be revealed both by partial proteolysis experiments (29) and by structural analysis of the liganded and unliganded forms of the N-terminal domain (30). Interaction with isoleucine induces a major repositioning of numerous residues, creating the BCAA-binding pocket from regions that are unstructured or differently structured in the unliganded protein. In addition, binding of isoleucine or valine in the N-terminal domain leads to a major change in the relative orientation of the two C-terminal winged helix-turn-helix domains in the CodY dimer (V. Levdikov, E. Blagova, and A. J. Wilkinson, unpublished data).
Our results indicate that residues R61, F71, and F98 are very likely to be involved in interactions with BCAAs, but not in effector-independent DNA binding. Any small differences in affinity seen in the absence of effectors could have been due to small variations in the specific activities of the various protein preparations and are unlikely to have physiological significance. In the crystal structure of the CodY-isoleucine complex, R61, F71, and F98 lie within a pocket that surrounds the isoleucine (29). The local structure appears to be stabilized by hydrophobic and ionic interactions between these residues (among others) and isoleucine. F40, however, does not participate in interactions with the isoleucine ligand but lies more deeply in the extended binding pocket within the GAF domain (29) (Fig. 1). Both F40 and F98 have been viewed as candidates for interaction with GTP (29). The fact that both the F98A and F40A forms of CodY retain the ability to be activated by GTP makes it unlikely that they contribute to GTP binding through ring stacking or that the GAF domain is the site of GTP binding. Recent biochemical experiments designed to test direct interaction of GTP with the GAF domain also seemed to exclude such interaction (30). That is, while the binding of isoleucine to the GAF domain could be demonstrated by saturation transfer difference nuclear magnetic resonance, we saw no evidence of GTP binding using this technique. Moreover, GTP did not affect the isoleucine signals in the difference spectrum in a competition experiment. Since the saturation transfer difference nuclear magnetic resonance technique is well suited to detecting weakly binding ligands (the Kd for GTP is inferred to be in the millimolar range [20]), these results led us to conclude that the GAF domain of CodY is not by itself the binding site for GTP (30).
Our in vitro work indicates that the R61A form of CodY is fully responsive to GTP and that the F71E form has at most a twofold defect in stimulation by GTP. Thus, F40, R61, F71, and F98 are not essential for CodY-GTP interaction, implying that these mutant proteins are selectively defective in the response to BCAAs. The R61A mutant CodY does respond to isoleucine in vitro, however, when GTP is present. Assuming that the in vitro binding results accurately reflect the behavior of CodY in cells, a possible explanation is that R61A CodY retains sufficient affinity for isoleucine that, when GTP is present, this affinity is increased enough to stimulate CodY. Replacing R61 with K or F71 with A also permits significant retention of repressing activity in vivo. Although we have not examined the behavior of these latter mutant proteins in vitro, they are also likely to retain some affinity for BCAAs. Replacing R61 with E results in significant loss of repressor activity. We predict that R61E retains little or no responsiveness to BCAAs.
The F40A, F71E, and F98A mutants show nearly complete loss of repressing activity in vivo. A plausible explanation for the different behavior of R61A and the three other mutants would be that the F40A, F71E, and F98A forms of CodY have less ability to interact with BCAAs even in the presence of GTP and that CodY-GTP interaction is not sufficient by itself to cause substantial repression by the mutant proteins in vivo under our growth conditions. If correct, this explanation would force us to reexamine our understanding of the roles of GTP and BCAAs in vivo. That is, while either GTP or a BCAA can activate CodY in vitro if provided at a high enough concentration, the activity of CodY in vivo may be low unless both types of effectors are bound. Perhaps the intracellular concentration of GTP is not high enough under the growth conditions we have utilized to activate CodY in the absence of adequate BCAA binding.
The behavior of the F98A CodY protein in vitro is at odds with our expectations on the basis of analysis of the crystal structure. F98 participates in BCAA binding through a charge-dipole interaction involving its main-chain carbonyl group. Although this group is retained following the replacement of phenylalanine by alanine, it is possible that the mutation is associated with a structural alteration that prevents the carbonyl group of alanine from playing the same role as that of phenylalanine at that position. Phe98 has a dramatically altered location in the unliganded GAF structure (30) and undergoes a 10-Å movement upon ligand binding, further complicating the interpretation of the effects of the F98A mutation.
While the mutational analysis described here has begun to reveal the protein residues that are important for effector binding, the study of the ability of BCAA analogs to activate CodY helps to define the critical elements of effector structure. We report here that the 2-amino group and its chirality as well as the 3-methyl group of valine are likely to be essential for CodY-amino acid interaction. These results raise the possibility that analogs of the natural BCAAs that conform to these rules but have modifications that render them nonmetabolizable in vivo might be able to bind to CodY and convert it to its active conformation. Such compounds might prove useful in reducing the virulence of bacteria in which CodY is a repressor of virulence factor genes (50).
Supplementary Material
Acknowledgments
We thank S. S. Dineen for helpful discussions. A.C.V. is very grateful to R. P. Shivers for strains and other materials and helpful advice.
This work was supported by a research grant (GM042219 to A.L.S.) and a National Research Service Award (GM082155 to L.D.H.) from the U.S. National Institute of General Medical Sciences and by grants (BBS/B1213X to A.J.W.) and (082829/Z/07/Z to A.J.W.) from the BBSRC and Wellcome Trust, respectively.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belitsky, B. R., and A. L. Sonenshein. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belitsky, B. R., and A. L. Sonenshein. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, H. J., D. M. Pearce, S. Glenn, C. M. Taylor, M. Kuhn, A. L. Sonenshein, P. W. Andrew, and I. S. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453-1467. [DOI] [PubMed] [Google Scholar]
- 6.Bergara, F., C. Ibarra, J. Iwamasa, J. C. Patarroyo, R. Aguilera, and L. M. Márquez-Magaña. 2003. CodY is a nutritional repressor of flagellar gene expression in Bacillus subtilis. J. Bacteriol. 185:3118-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blagova, E. V., V. M. Levdikov, K. Tachikawa, A. L. Sonenshein, and A. J. Wilkinson. 2003. Crystallization of the GTP-dependent transcriptional regulator CodY from Bacillus subtilis. Acta Crystallogr. 59:155-157. [DOI] [PubMed] [Google Scholar]
- 8.Cashel, M. A., D. Gentry, V. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 9.Debarbouille, M., R. Gardan, M. Arnaud, and G. Rapoport. 1999. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J. Bacteriol. 181:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Hengst, C. D., P. Curley, R. Larsen, G. Buist, A. Nauta, D. van Sinderen, O. P. Kuipers, and J. Kok. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206-219. [DOI] [PubMed] [Google Scholar]
- 12.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferson, A. E., L. V. Wray, Jr., and S. H. Fisher. 1996. Expression of the Bacillus subtilis gabP gene is regulated independently in response to nitrogen and amino acid availability. Mol. Microbiol. 22:693-701. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, S. H., K. Rohrer, and A. E. Ferson. 1996. Role of CodY in regulation of the Bacillus subtilis hut operon. J. Bacteriol. 178:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouet, A., and A. L. Sonenshein. 1990. A target for carbon source-dependent negative regulation of the citB promoter of Bacillus subtilis. J. Bacteriol. 172:835-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita, M., and R. Losick. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandoni, J. A., S. B. Fulmer, V. Brizzio, S. A. Zahler, and J. M. Calvo. 1993. Regions of the Bacillus subtilis ilv-leu operon involved in regulation by leucine. J. Bacteriol. 175:7581-7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handke, L. D., R. P. Shivers, and A. L. Sonenshein. 2008. Interaction of Bacillus subtilis CodY with GTP. J. Bacteriol. 190:798-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendriksen, W. T., H. J. Bootsma, S. Estevao, T. Hoogenboezem, A. de Jong, R. de Groot, O. P. Kuipers, and P. W. Hermans. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190:590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsueh, Y. H., E. B. Somers, and A. C. Wong. 2008. Characterization of the codY gene and its influence on biofilm formation in Bacillus cereus. Arch. Microbiol. 189:557-568. [DOI] [PubMed] [Google Scholar]
- 23.Hurley, J. H. 2003. GAF domains: cyclic nucleotides come full circle. Sci. STKE 2003:PE1. [DOI] [PubMed] [Google Scholar]
- 24.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 25.Joseph, P., M. Ratnayake-Lecamwasam, and A. L. Sonenshein. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 187:4127-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, H.-J., C. Jourlin-Castelli, S.-I. Kim, and A. L. Sonenshein. 2002. Regulation of the Bacillus subtilis ccpC gene by CcpA and CcpC. Mol. Microbiol. 43:399-410. [DOI] [PubMed] [Google Scholar]
- 27.Kim, H. J., A. Roux, and A. L. Sonenshein. 2002. Direct and indirect roles of CcpA in regulation of Bacillus subtilis Krebs cycle genes. Mol. Microbiol. 45:179-190. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J. H., S. I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. 2003. Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levdikov, V. M., E. Blagova, P. Joseph, A. L. Sonenshein, and A. J. Wilkinson. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in Gram-positive bacteria. J. Biol. Chem. 281:11366-11373. [DOI] [PubMed] [Google Scholar]
- 30.Levdikov, V. M., E. Blagova, V. L. Colledge, A. A. Lebedev, D. C. Williamson, A. L. Sonenshein, and A. J. Wilkinson. 2009. Structural rearrangement accompanying ligand binding in the GAF domain of CodY from Bacillus subtilis. J. Mol. Biol. 390:1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majerczyk, C. D., M. R. Sadykov, T. T. Luong, C. Lee, G. A. Somerville, and A. L. Sonenshein. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malke, H., and J. J. Ferretti. 2007. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56:707-714. [DOI] [PubMed] [Google Scholar]
- 33.Martinez, S. E., A. Y. Wu, N. A. Glavas, X. B. Tang, S. Turley, W. G. Hol, and J. A. Beavo. 2002. The two GAF domains in phosphodiesterase 2A have distinct roles in dimerization and in cGMP binding. Proc. Natl. Acad. Sci. USA 99:13260-13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathiopoulos, C., J. P. Mueller, F. J. Slack, C. G. Murphy, S. Patankar, G. Bukusoglu, and A. L. Sonenshein. 1991. A Bacillus subtilis dipeptide transport system expressed early during sporulation. Mol. Microbiol. 5:1903-1913. [DOI] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 36.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller, J. P., G. Bukusoglu, and A. L. Sonenshein. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J. Bacteriol. 174:4361-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petranovic, D., E. Guedon, B. Sperandio, C. Delorme, D. Ehrlich, and P. Renault. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53:613-621. [DOI] [PubMed] [Google Scholar]
- 39.Ratnayake-Lecamwasam, M., P. Serror, K.-W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serror, P., and A. L. Sonenshein. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843-852. [DOI] [PubMed] [Google Scholar]
- 42.Shivers, R. P. 2005. The Bacillus subtilis ilvB operon: an intersection of global regulators. Ph.D. thesis. Tufts University, Boston, MA. [DOI] [PubMed]
- 43.Shivers, R. P., S. S. Dineen, and A. L. Sonenshein. 2006. Positive regulation of Bacillus subtilis ackA by CodY and CcpA: establishing a potential hierarchy in carbon flow. Mol. Microbiol. 62:811-822. [DOI] [PubMed] [Google Scholar]
- 44.Shivers, R. P., and A. L. Sonenshein. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599-611. [DOI] [PubMed] [Google Scholar]
- 45.Shivers, R. P., and A. L. Sonenshein. 2005. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol. Microbiol. 56:1549-1559. [DOI] [PubMed] [Google Scholar]
- 46.Slack, F. J., J. P. Mueller, and A. L. Sonenshein. 1993. Mutations that relieve nutritional repression of the Bacillus subtilis dipeptide permease operon. J. Bacteriol. 175:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slack, F. J., J. P. Mueller, M. A. Strauch, C. Mathiopoulos, and A. L. Sonenshein. 1991. Transcriptional regulation of a Bacillus subtilis dipeptide transport operon. Mol. Microbiol. 5:1915-1925. [DOI] [PubMed] [Google Scholar]
- 48.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 49.Soga, T., Y. Ohashi, Y. Ueno, H. Naraoka, M. Tomita, and T. Nishioka. 2003. Quantitative metabolome analysis using capillary electrophoresis mass spectrophotometry. J. Proteome Res. 2:488-494. [DOI] [PubMed] [Google Scholar]
- 50.Sonenshein, A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917-927. [DOI] [PubMed] [Google Scholar]
- 51.Tojo, S., T. Satomura, K. Morisaki, J. Deutscher, K. Hirooka, and Y. Fujita. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 56:1560-1573. [DOI] [PubMed] [Google Scholar]
- 51a.van Schaik, W., A. Château, M. A. Dillies, J. Y. Coppée, A. L. Sonenshein, and A. Fouet. 2009. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect. Immun. 77:4437-4445. [DOI] [PMC free article] [PubMed]
- 52.Wray, L. V., Jr., A. E. Ferson, and S. H. Fisher. 1997. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors including CodY, GlnR, TnrA, and Spo0H. J. Bacteriol. 179:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.