Abstract
GTP cyclohydrolase I (GCYH-I) is an essential Zn2+-dependent enzyme that catalyzes the first step of the de novo folate biosynthetic pathway in bacteria and plants, the 7-deazapurine biosynthetic pathway in Bacteria and Archaea, and the biopterin pathway in mammals. We recently reported the discovery of a new prokaryotic-specific GCYH-I (GCYH-IB) that displays no sequence identity to the canonical enzyme and is present in ∼25% of bacteria, the majority of which lack the canonical GCYH-I (renamed GCYH-IA). Genomic and genetic analyses indicate that in those organisms possessing both enzymes, e.g., Bacillus subtilis, GCYH-IA and -IB are functionally redundant, but differentially expressed. Whereas GCYH-IA is constitutively expressed, GCYH-IB is expressed only under Zn2+-limiting conditions. These observations are consistent with the hypothesis that GCYH-IB functions to allow folate biosynthesis during Zn2+ starvation. Here, we present biochemical and structural data showing that bacterial GCYH-IB, like GCYH-IA, belongs to the tunneling-fold (T-fold) superfamily. However, the GCYH-IA and -IB enzymes exhibit significant differences in global structure and active-site architecture. While GCYH-IA is a unimodular, homodecameric, Zn2+-dependent enzyme, GCYH-IB is a bimodular, homotetrameric enzyme activated by a variety of divalent cations. The structure of GCYH-IB and the broad metal dependence exhibited by this enzyme further underscore the mechanistic plasticity that is emerging for the T-fold superfamily. Notably, while humans possess the canonical GCYH-IA enzyme, many clinically important human pathogens possess only the GCYH-IB enzyme, suggesting that this enzyme is a potential new molecular target for antibacterial development.
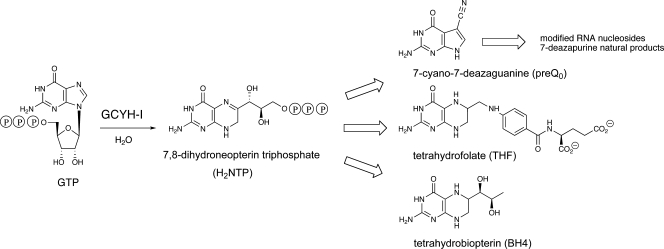
The Zn2+-dependent enzyme GTP cyclohydrolase I (GCYH-I; EC 3.5.4.16) is the first enzyme of the de novo tetrahydrofolate (THF) biosynthesis pathway (Fig. 1) (38). THF is an essential cofactor in one-carbon transfer reactions in the synthesis of purines, thymidylate, pantothenate, glycine, serine, and methionine in all kingdoms of life (38), and formylmethionyl-tRNA in bacteria (7). Recently, it has also been shown that GCYH-I is required for the biosynthesis of the 7-deazaguanosine-modified tRNA nucleosides queuosine and archaeosine produced in Bacteria and Archaea (44), respectively, as well as the 7-deazaadenosine metabolites produced in some Streptomyces species (33). GCYH-I is encoded in Escherichia coli by the folE gene (28) and catalyzes the conversion of GTP to 7,8-dihydroneopterin triphosphate (55), a complex reaction that begins with hydrolytic opening of the purine ring at C-8 of GTP to generate an N-formyl intermediate, followed by deformylation and subsequent rearrangement and cyclization of the ribosyl moiety to generate the pterin ring in THF (Fig. 1). Notably, the enzyme is dependent on an essential active-site Zn2+ that serves to activate a water molecule for nucleophilic attack at C-8 in the first step of the reaction (2).
FIG. 1.
Reaction catalyzed by GCYH-I, and metabolic fate of 7,8-dihydroneopterin triphosphate.
A homologous GCYH-I is found in mammals and other higher eukaryotes, where it catalyzes the first step of the biopterin (BH4) pathway (Fig. 1), an essential cofactor in the biosynthesis of tyrosine and neurotransmitters, such as serotonin and l-3,4-dihydroxyphenylalanine (3, 52). Recently, a distinct class of GCYH-I enzymes, GCYH-IB (encoded by the folE2 gene), was discovered in microbes (26% of sequenced Bacteria and most Archaea) (12), including several clinically important human pathogens, e.g., Neisseria and Staphylococcus species. Notably, GCYH-IB is absent in eukaryotes.
The distribution of folE (gene product renamed GCYH-IA) and folE2 (GCYH-IB) in bacteria is diverse (12). The majority of organisms possess either a folE (65%; e.g., Escherichia coli) or a folE2 (14%; e.g., Neisseria gonorrhoeae) gene. A significant number (12%; e.g., B. subtilis) possess both genes (a subset of 50 bacterial species is shown in Table 1), and 9% lack both genes, although members of the latter group are mainly intracellular or symbiotic bacteria that rely on external sources of folate. The majority of Archaea possess only a folE2 gene, and the encoded GCYH-IB appears to be necessary only for the biosynthesis of the modified tRNA nucleoside archaeosine (44) except in the few halophilic Archaea that are known to synthesize folates, such as Haloferax volcanii, where GCYH-IB is involved in both archaeosine and folate formation (13, 44).
TABLE 1.
Distribution and candidate Zur-dependent regulation of alternative GCYH-I genes in bacteriaa
| Organismc | Presence of: |
|
|---|---|---|
| folE | folE2 | |
| Enterobacteria | ||
| Escherichia coli | + | − |
| Salmonella typhimurium | + | − |
| Yersinia pestis | + | − |
| Klebsiella pneumoniaeb | + | +a |
| Serratia marcescens | + | +a |
| Erwinia carotovora | + | − |
| Photorhabdus luminescens | + | − |
| Proteus mirabilis | + | − |
| Gammaproteobacteria | ||
| Vibrio cholerae | + | − |
| Acinetobacter sp. strain ADP1 | + | +a |
| Pseudomonas aeruginosa | + | +a |
| Pseudomonas entomophila L48 | + | +a |
| Pseudomonas fluorescens Pf-5 | + | +a |
| Pseudomonas syringae | + | +a |
| Pseudomonas putida | + | +a |
| Hahella chejuensis KCTC 2396 | + | +a |
| Chromohalobacter salexigens DSM 3043 | + | +a |
| Methylococcus capsulatus | + | +a |
| Xanthomonas axonopodis | + | +a |
| Xanthomonas campestris | + | +a |
| Xylella fastidiosa | + | +a |
| Idiomarina loihiensis | − | + |
| Colwellia psychrerythraea | + | + |
| Pseudoalteromonas atlantica T6c | + | +a |
| Pseudoalteromonas haloplanktis TAC125 | + | + |
| Alteromonas macleodi | + | − |
| Nitrosococcus oceani | + | + |
| Legionella pneumophila | + | − |
| Francisella tularensis | + | − |
| Betaproteobacteria | ||
| Chromobacterium violaceum | + | − |
| Neisseria gonorrhoeae | − | + |
| Burkholderia cepacia R18194 | + | + |
| Burkholderia cenocepacia AU 1054 | + | + |
| Burkholderia xenovorans | + | − |
| Burkholderia mallei | + | − |
| Bordetella pertussis | − | + |
| Ralstonia eutropha JMP134 | + | − |
| Ralstonia metallidurans | + | + |
| Ralstonia solanacearum | + | − |
| Methylobacillus flagellatus | − | + |
| Nitrosomonas europaea | − | + |
| Azoarcus sp. | + | + |
| Bacilli/Clostridia | ||
| Bacillus subtilisd | + | + |
| Bacillus licheniformis | + | + |
| Bacillus cereus | + | − |
| Bacillus halodurans | + | + |
| Bacillus clausii | + | − |
| Geobacillus kaustophilus | + | − |
| Oceanobacillus iheyensis | − | + |
| Staphylococcus aureus | − | + |
Genes that are preceded by candidate Zur binding sites.
Zur-regulated cluster is on the virulence plasmid pLVPK.
Examples of organisms with no folE genes are in boldface type.
Zn-dependent regulation of B. subtilis folE2 by Zur was experimentally verified (17).
Expression of the Bacillus subtilis folE2 gene, yciA, is controlled by the Zn2+-dependent Zur repressor and is upregulated under Zn2+-limiting conditions (17). This led us to propose that the GCYH-IB family utilizes a metal other than Zn2+ to allow growth in Zn2+-limiting environments, a hypothesis strengthened by the observation that an archaeal ortholog from Methanocaldococcus jannaschii has recently been shown to be Fe2+ dependent (22). To test this hypothesis, we investigated the physiological role of GCYH-IB in B. subtilis, an organism that contains both isozymes, as well as the metal dependence of B. subtilis GCYH-IB in vitro. To gain a structural understanding of the metal dependence of GCYH-IB, we determined high-resolution crystal structures of Zn2+- and Mn2+-bound forms of the N. gonorrhoeae ortholog. Notably, although the GCYH-IA and -IB enzymes belong to the tunneling-fold (T-fold) superfamily, there are significant differences in their global and active-site architecture. These studies shed light on the physiological significance of the alternative folate biosynthesis isozymes in bacteria exposed to various metal environments, and offer a structural understanding of the differential metal dependence of GCYH-IA and -IB.
MATERIALS AND METHODS
Enzyme activity assays.
All chemicals were of analytical, metal-free grade from Sigma or Fisher. All solutions were prepared with Milli-Q water (18.2 MΩ/cm) and treated with Chelex-100 (Bio-Rad) to remove contaminating metals. Glass and plastic ware were soaked in nitric acid (10%), then in EDTA (5 mM), and rinsed liberally with Chelex-treated Milli-Q water before use. The genes encoding B. subtilis and N. gonorrhoeae GCYH-IB were overexpressed in E. coli and the recombinant proteins purified as previously described (12). Protein concentrations were determined spectrophotometrically using an extinction coefficient of 25,590 M−1 calculated according to Gill and von Hippel (19). Fluorescence-based activity assays were conducted as described previously (12), using either variable concentrations of metal or EDTA (5 mM), and 0.1 mM GTP. Fluorescence was measured with a SpectraMax Gemini XPS spectrofluorometer (Molecular Devices).
Steady-state kinetic analysis of B. subtilis GCYH-IB was carried out in the presence of 100 mM HEPES (pH 8.0), 100 mM KCl, 0.5 mM MnCl2, 1 mM dithiothreitol (DTT), 0.5 μM protein, and variable concentrations of GTP (3 to 50 μM) under initial velocity conditions.
Apoenzyme preparation.
Metal ions were removed from B. subtilis and N. gonorrhoeae GCYH-IB by incubating a solution (0.5 ml) of the purified, recombinant protein in 50 mM Tris-HCl (pH 8.0), 50 mM KCl, and 5 mM EDTA for 1 h at room temperature, followed by performing dialysis thrice against a 4-liter solution of 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 5 mM EDTA, and a few grams of Chelex-100. The resulting apoenzyme was dialyzed thrice against 3 liters of 50 mM Tris-HCl (pH 8.0), 50 mM KCl, and Chelex-100 to remove EDTA, then passed through Chelex-100 resin (5 ml protein solution per 3 ml resin) prior to activity assays and metal analysis. Metal content was determined by inductively coupled plasma mass spectrometry (HP-4500 ICP-MS) at the Trace Element Laboratory in the Department of Geology (Portland State University).
Metal activation studies.
The B. subtilis apoenzyme (2 μM) was incubated with various concentrations (0.1 μM to 4 mM) of metal chlorides (MnCl2, ZnCl2, MgCl2, NiCl2, CaCl2, CdCl2, CoCl2, CuCl2, CoCl3, FeCl3) or Fe(SO4) in standard buffer (100 mM HEPES [pH 8.0], 100 mM KCl) for 10 min at 37°C, then assayed for activity as described previously (12). All assays involving Fe2+ were conducted in an anaerobic chamber with degassed and N2-purged buffers.
Reconstitution of B. subtilis GCYH-IB with Mn2+ or Zn2+.
The apoenzyme was incubated in the presence of 7 molar equivalents of MnCl2 or ZnCl2 in standard buffer containing 2 mM DTT for 60 min at 25°C. Loosely bound metal was removed either by dialysis twice against 50 mM Tris-HCl (pH 8.0), 50 mM KCl, and 1 mM DTT for 4 h at 4°C, gel filtration on a Sephadex G-25 column, or by extensive washing with buffer in an Amicon ultra-centrifugal device. The enzyme was assayed for activity as described above, and the metal content was determined by ICP-MS.
Gel filtration chromatography of N. gonorrhoeae GCYH-IB.
The native molecular mass of recombinant, wild-type N. gonorrhoeae GCYH-IB (affinity tag removed) was determined by high-performance liquid chromatography (HPLC) on a BioSep-SEC-S4000 gel filtration column (300 mm by 7.8 mm; Phenomenex Inc.). GCYH-IB and the standard proteins (300 μg) were eluted in a buffer of 20 mM sodium phosphate (pH 7.2) and 100 mM NaCl at a flow rate of 1 ml/min, using a Hitachi HPLC system comprised of an L-7100 pump, L-4500A detector, L-7300 column oven, and LaChrome software. β-Amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa) were used as molecular weight (MW) standards, and the void volume was determined from the elution volume of blue dextran. Elution of the proteins from the column was monitored by absorbance at 280 nm.
Crystallization and X-ray data collection.
Selenomethionine (SeMet)-labeled enzyme was overproduced in the E. coli methionine auxotroph B834(DE3) (Novagen) following standard methods (24). The enzyme was purified and the His6 tag removed as described previously (12). Selenium substitution was verified with mass spectrometry.
SeMet-labeled GCYH-IB was crystallized at 20°C by vapor diffusion in 2 μl sitting drops containing enzyme [9 mg/ml in 50 mM Tris-acetate, 100 mM KCl, β-mercaptoethanol (BME; pH 8.0)], polyethylene glycol 6000 (10 to 16%), LiCl (1 to 1.4 M), Tris (50 mM, pH 9.0), and Tris-HCl (50 mM, pH 7.0). The Mn-reconstituted enzyme (10 mg/ml in 50 mM Tris-HCl, 50 mM KCl, 1 mM DTT [pH 8.0]) was crystallized under similar conditions but using metal-free reagents (Chelex-100-pretreated Milli-Q water and ultra-pure reagents from Hampton, Inc.) with 1 to 10 mM MnCl2 and 12 mM sodium azide (as a preservative) added to the protein solution prior to crystallization.
Crystals were cryo-protected in mother liquor plus ethylene glycol (25%) and flash-cooled in liquid nitrogen. For the SeMet-labeled enzyme, a three-wavelength selenium data set was collected at the Advanced Light Source (ALS, Berkeley, CA) beamline 8.2.2. For the GCYH-IB·Mn2+ complex, crystals were back-soaked in metal-free reservoir solution for 1 h prior to cryo protection and a single-wavelength data set was collected from a single crystal at Stanford Synchrotron Radiation Laboratory (SSRL, Stanford, CA) beamline 7 to 1. X-ray data were processed in HKL2000 (39).
Structure determination.
The crystal structure of GCYH-IB was determined by the multiwavelength anomalous dispersion method using Se as the anomalous scatterer. An initial heavy-atom substructure of 15 Se sites was identified using the Phenix-HySS program (1). Heavy-atom positions, occupancies, and atomic displacement parameters were refined in SHARP (11), and initial phases were calculated using all Se sites. Phases were improved by density modification with DM (9) or CCP4 (45) using data in the resolution range 50 to 2.2 Å and 47% solvent content (mean figure of merit increased from 0.34 to 0.85). A 2.2 Å FoFOM map was calculated and auto-traced in ARP/warp software (43), where 418 of the 514 residues in the asymmetric unit were built. The model was manually completed and partially rebuilt in Coot (14), utilizing the twofold noncrystallographic symmetry (NCS) relation. Subsequent crystallographic refinement was carried out with Phenix-refine software (1), using NCS restraints and twin lattice symmetry parameterization (3 domains per monomer). (Isotropic B-group constraints were applied to disordered parts of the model.) Finally, f′ and f″ were refined in Phenix to final values of −6.4 and 5.6, −5.4 and 4.9, and −5.6 and 3.5, for the peak, inflection and remote wavelength, respectively. Solvent molecules and ions were added, and the structure was further refined in Crystallography and NMR Systems (CNS) (4) after removal of NCS restraints and was validated using Procheck (54). The electron density for residues 146 to 160 and 188 to 210 of monomer A was weak with refined temperature factors of >60 Å2, indicating that this region is disordered.
The crystal structure of the GCYH-IB·Mn2+ complex was determined by direct difference Fourier calculation in which a protein model based on the crystal structure of the Zn2+-metallated enzyme was used to calculate phases. The structure was rigid-body-refined in CNS, then refined in refmac (36) from CCP4 while applying NCS restraints and solvent flattening. NCS restraints were removed in a second refmac run. Ligands and solvent molecules were modeled in Coot (version 0.3.3) (14), and final refinement was completed in refmac.
Bioinformatics.
Analysis of the folE/folE2 gene distribution was performed using the SEED annotation environment. Results are made available in the “Folate biosynthesis” subsystem at http://theseed.uchicago.edu/FIG/index.cgi. Candidate Zur binding sites were identified using the Genome Explorer software (41) by scanning bacterial genomes with two position-specific weight matrices constructed using the training sets of two different sets of known Zur binding sites in proteobacteria and firmicutes (42).
B. subtilis strain construction and growth conditions.
All strains were derived from the B. subtilis 168 trpc2 attSPβ wild-type strain, CU1065, and were grown in a defined minimal medium as previously described (17) or in LB. Growth curves were done using a Bioscreen CMBR system for 24 h with measurements of optical density at 600 nm every 10 min. Liquid overnight cultures with antibiotics were used to start precultures that were diluted at mid-log to a normalized optical density at 600 nm of 0.01 in a volume of 200 μl in a 100-well honeycomb microtiter plate, at which point the growth curve was started. Cultures were incubated at 37°C with shaking at 200 rpm. For selection, antibiotics were added at the following concentrations: 1 μg/ml erythromycin and 25 μg/ml lincomycin (for selecting for macrolide-lincosamide-streptogramin B resistance), 100 μg/ml spectinomycin, and 15 μg/ml kanamycin.
folE, folE2, and zur mutants were constructed using long-flanking-homology PCR as previously described (6). Long-flanking-homology PCR and chromosomal DNA transformation were used to generate strains expressing various combinations of GCYH-I enzymes. Specifically, strain HB6788 (CU1065 folE::mls) lacks GCYH-IA while its GCYH-IB is repressed, but its gene is still present (designated the ΔfolE mutant). This strain was used for construction of other strains. Strain HB6783 (CU1065 folE::mls zur::kan) expresses only GCYH-IB in minimal medium (designated the ΔfolE Δzur mutant). Strain HB6852 (CU1065 folE::mls zur::kan folE2::spc) lacks both GCYH-I isozymes (designated the ΔfolE Δzur ΔfolE2 mutant).
Protein structure accession numbers.
Coordinates and structure factors for GCYH-IB and GCYH-IB·Mn crystal structures have been deposited into the Protein Data Bank (PDB) under accession numbers 3D1T and 3D2O, respectively.
RESULTS
GCYH-IB functionally replaces GCYH-IA in B. subtilis under zinc-limiting conditions.
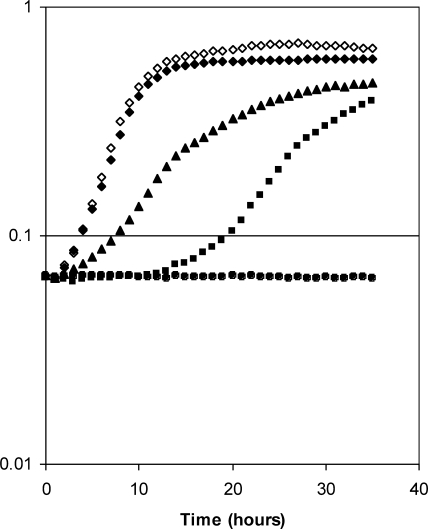
B. subtilis has both folE and folE2 genes (also known as mtrA and yciA, respectively). To determine whether GCYH-IB can functionally replace GCYH-IA, we constructed a B. subtilis ΔfolE mutant and investigated its complementation with GCYH-IB. In a defined, low-Zn2+ minimal medium (no added Zn2+), cells expressing GCYH-IA grew well (Fig. 2). However, the ΔfolE mutant had a significant lag time and a reduced logarithmic growth rate (Fig. 2). This lag is consistent with the repression of GCYH-IB expression by Zur in the beginning of the growth when, even with no added Zn2+, there is sufficient contaminating Zn2+ to activate Zur. We speculate that the transition to more-rapid growth results from derepression of GCYH-IB when Zn2+ is depleted and Zur repression is released. Indeed, a ΔfolE Δzur double mutant, in which GCYH-IB is constitutively expressed, displayed a greatly reduced lag phase (Fig. 2). However, the constitutive expression of GCYH-IB did not support wild-type levels of growth in this minimal medium, although this strain did grow at the same rate as the wild-type strain in rich (LB) medium (data not shown). As expected, the ΔfolE Δzur ΔfolE2 triple mutant strain, expressing neither GCYH-IA nor GCYH-IB, was unable to grow in either minimal (Fig. 2) or rich (data not shown) medium. Taken together, these results indicate that GCYH-IB can functionally replace GCYH-IA in B. subtilis under Zn2+-limiting conditions.
FIG. 2.
GCYH-IB-dependent complementation of the B. subtilis ΔfolE mutant strain. Growth curves in defined minimal medium for wild-type (WT) and mutant B. subtilis strains expressing either or both types of GCYH-I. Results for the wild-type strain expressing GCYH-IA (⧫), the ΔfolE2 strain expressing GCYH-IA (◊), the ΔfolE strain with delayed expression of GCYH-IB (▪), the ΔfolE Δzur strain expressing GCYH-IB (▴), and the ΔfolE2 ΔfolE Δzur strain with no GCYH-IA or IB expression (•) are shown.
Zur-dependent regulation of folE2 is conserved across bacterial species.
The expression of GCYH-IB in B. subtilis is controlled by the Zn-dependent Zur repressor and is thus upregulated under Zn-limiting conditions (17), consistent with a metal ion other than Zn as the physiological metal for this family of cyclohydrolases. To investigate whether orthologs of folE2 in other bacteria are similarly subject to Zur-mediated control, we analyzed bacterial regulons by the use of a comparative genomics approach (18, 48). Annotation of the folE and folE2 genes in available genomes had been previously performed (3). Two position-specific weight matrices were constructed for known Zur binding sites from gram-positive and gram-negative bacteria (42) and were used to scan for candidate Zur binding sites in 5′ untranslated regions of folE2 and neighbor genes in bacterial genomes. To account for possible operon structures, we also analyzed upstream regions of genes that are likely cotranscribed with the folE2 gene (i.e., chains of adjacent genes in the same orientation with intergenic distances not exceeding 100 nucleotides).
Strong Zur operator sites were identified upstream of folE2 genes in 21 bacterial genomes (Table 1). This unique regulatory feature is conserved across phyla, including examples in 15 gammaproteobacteria (Pseudomonadales, Xanthomonadales, some enterobacteria, and other lineages), three betaproteobacteria (some Burkholderiales), and three Bacillus species. Notably, all organisms that have the Zur-regulated folE2 gene also contain a copy of folE (e.g., B. subtilis). Similarly, no Zur regulatory sites were found upstream of folE2 genes in those organisms that lack the Zn2+-dependent GCYH-IA isozyme (e.g., N. gonorrhoeae). These results show that Zur regulation of GCYH-IB is widespread and suggest that in bacteria possessing both folE and folE2, like B. subtilis, the GCYH-IB isozyme is expressed under Zn2+-limiting conditions to replace the Zn-dependent GCYH-IA.
GCYH-IB and GCYH-IA have different metal requirements in vitro.
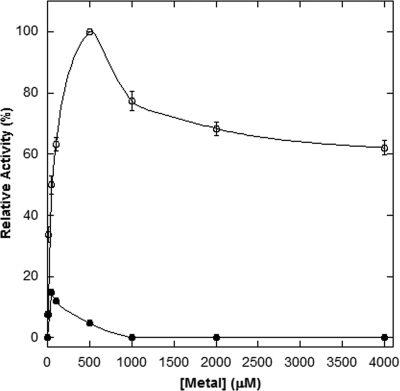
It is well established that GCYH-IA uses a bound Zn2+ ion for activity (2). To investigate the metal dependence of GCYH-IB, the purified recombinant B. subtilis enzyme was assayed for activity in the presence of a variety of metal ions or EDTA. While no activity was observed in the presence of EDTA, the presence of several metal ions supported catalysis (data not shown). To obtain unambiguous, quantitative data on the effect of various metal ions on catalytic activity, the enzyme was first demetallated by dialyzing against an EDTA/Chelex-containing buffer to generate the apoenzyme, which was then assayed in the presence of specific metal ions over a concentration range of 100 nM to 4 mM. Catalysis is supported to various extents by Mn2+, Fe2+, Mg2+, Co2+, Zn2+, and Ni2+ (Table 2 and Fig. 3), with optimal catalytic activity observed with a metal concentration ranging from 50 μM (Zn2+) to 1.0 mM (Fe2+), while Ca2+, Cd2+, Cu2+, Co3+, and Fe3+ fail to support activity at any concentration. Notably, although the enzyme exhibits some activity in the presence of Zn2+, catalysis is significantly higher in the presence of Mn2+ and, to a lesser extent, Fe2+ and Mg2+.
TABLE 2.
Metal dependence of B. subtilis GCYH-IB enzymatic activity
| Metal | Metal concn (μM)a | Relative activity (%) |
|---|---|---|
| No metal | 0 | 0 |
| Mn(II) | 500 | 100 |
| Fe(II) | 1000 | 75 ± 8 |
| Mg(II) | 100 | 43 ± 4 |
| Co(II) | 100 | 24 ± 3 |
| Zn(II) | 50 | 14 ± 1 |
| Ni(II) | 100 | 9.8 ± 1.3 |
| Ca(II) | NA | 0 |
| Cd(II) | NA | 0 |
| Cu(II) | NA | 0 |
| Co(III) | NA | 0 |
| Fe(III) | NA | 0 |
Metal concentration for optimal activity. NA, no activity detected regardless of metal concentration.
FIG. 3.
B. subtilis GCYH-IB enzymatic activity versus Mn2+ (○) or Zn2+ (•). Each data point represents the average results for four sets of triplicate assays (error bars correspond to the standard deviation in the data).
Steady-state kinetic analysis of the enzyme in the presence of 0.5 mM MnCl2 provided a kcat of 0.0011 s−1 and a Km of 9.87 μM with GTP as substrate. Although the Km is comparable to the values determined for the IA enzyme isolated from other bacteria, the kcat is at the low end of values reported for the IA enzyme (e.g., 0.0035 s−1 and 0.05 s−1 for the Thermus thermophilus [51] and E. coli [2] IA enzymes, respectively).
While Mn2+ is clearly more effective in supporting catalysis, it appears to bind with lower affinity than does Zn2+ (Table 2 and Fig. 3). However, as suggested by the absence of enzymatic activity in the presence of EDTA, neither metal appears to bind tightly to the enzyme. Indeed, when the enzyme is analyzed for metal content after purification, only substoichiometric amounts of metal are detected by ICP-MS (metal/protein mole ratios of 0.048 and 0.002 for Zn2+ and Mn2+, respectively). Furthermore, even when the apoenzyme is remetallated and then subjected to conditions designed to remove loosely bound metals (e.g., gel filtration), again only substoichiometric quantities of metal are detected (metal/protein mole ratios of 0.043 and 0.002 for Zn2+ and Mn2+, respectively). These results are in marked contrast to those observed for GCYH-IA, in which Zn2+ was observed to bind very tightly to the human enzyme (50) and incubation of the Zn2+-metallated enzyme with a variety of metal chelators failed to demetallate the enzyme and had no effect on catalytic activity. The results of those studies also showed that the enzyme was catalytically active with metal-free GTP, not metallated GTP. Our data do not allow us to conclude whether the enzyme binds free GTP or metallated GTP, but given the low affinity of metals for GCYH-IB, the enzyme may be active with either, depending on the ambient metal and GTP concentrations.
Native MW of N. gonorrhoeae GCYH-IB.
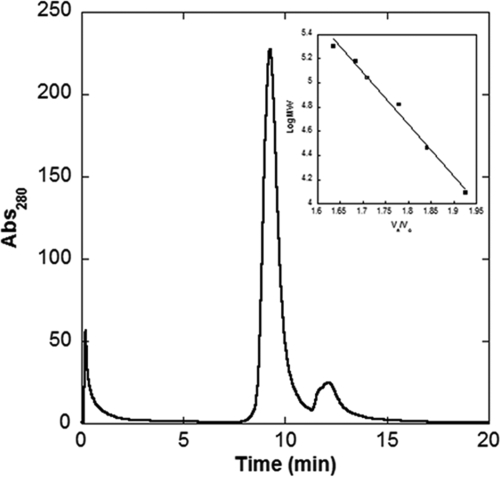
Determination of the native MW of GCYH-IB was done by gel filtration (Fig. 4) after calibration of the column with five standard proteins (Fig. 4, inset). Gel filtration analysis of recombinant, wild-type N. gonorrhoeae GYCH-IB provided an elution time of 9.2 min (Fig. 4), corresponding to an apparent MW of 108,559 ± 2,975. Given that the calculated MW of GCYH-IB is 28,568, the data are consistent with a tetramer quaternary structure (3.8 subunits calculated from the data) for the enzyme in the solution. A tetramer structure is consistent with the structures for the other known bimodular members of the T-fold superfamily (8).
FIG. 4.
HPLC gel filtration analysis of N. gonorrhoeae GCYH-IB. GCYH-IB elutes at 9.22 min. The broad trailing peak at ∼12 min is due to a small molecule contaminant (retention time is after cytochrome c). The inset graph shows the standard curve presented as log MW versus Ve/V0 where Ve is the measured elution volume and V0 is the volume of a completed excluded solute. The protein standards β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa) are plotted as closed squares (▪). The N. gonorrhoeae GCYH-IB data are plotted as open squares (□). Each data point represents the average of two trials. The protein standard data fit the linear regression as follows: y = 12.335 − 4.26661x (R = 0.99494). Abs280, absorbance at 280 nm.
Crystal structure of GCYH-IB reveals a T-fold.
To understand the structural basis of the metal requirement of GCYH-IB, we determined its crystal structure in two forms—one for the recombinant enzyme purified in solutions lacking added metal (GCYH-IB) and one for the apoenzyme remetallated with manganese (GCYH-IB·Mn). Enzymes from both B. subtilis and N. gonorrhoeae were pursued for crystallization, but only the N. gonorrhoeae ortholog produced diffracting crystals. The two orthologs are 64% similar and 35% identical in sequence and possess similar biochemical properties in vitro (data not shown).
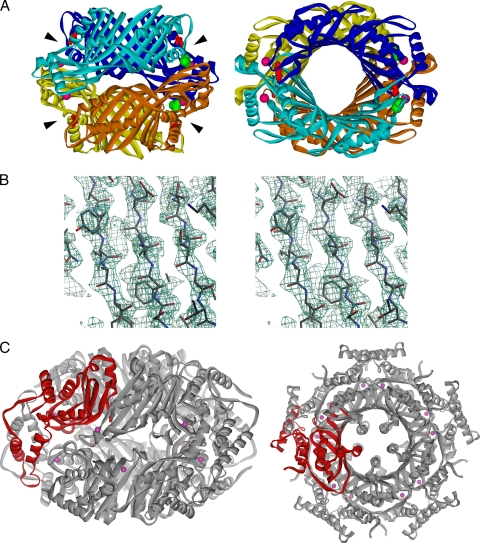
The crystal structure of GCYH-IB was determined by seleno-MAD methods (Fig. 5 and Table 3). Two monomers, A and B, were identified in the asymmetric unit. The final structure is missing residues 1 to 13 of monomer A and residues 1 to 14 of monomer B of the total 257 amino acids in each monomer, and contains two Zn2+ ions and one acetate molecule. A homotetrameric complex representing the probable biological state of the enzyme could be generated from the asymmetric-unit dimer by a crystallographic twofold rotation operation (Fig. 5A). The monomer is a globular subunit composed of nine β-strands and six α-helices (Fig. 6A), of which eight sequential antiparallel β-strands (β1 to β6, β8, and β9) and four antiparallel α-helices (α1, α2, α4, and α6) form a core with the classic T-fold architecture characteristic of bimodular pterin and purine binding enzymes (8). In this core, a highly twisted eight-stranded β-sheet is layered on its concave side with four antiparallel α-helices. The dimer of the asymmetric unit is constructed by a cyclic arrangement of the two eight-stranded β-sheets from the two monomers to form a 16-stranded antiparallel β-barrel. In the biological homotetramer, two β-barrels join together head-to-head to form a central tunnel that is 60 Å long and 17 Å in diameter (Fig. 5A).
FIG. 5.
Overall structure of GCYH-IB. (A) Ribbon diagram showing a side view (left) and a top view (right) of the GCYH-IB biological tetramer. The two dimers are colored in shades of orange and blue, respectively. The four active centers harboring the metal binding sites and GTP binding pockets are located at the intersubunit interfaces and are indicated with arrowheads. Zinc ions, acetate ligands, and the conserved T-fold substrate anchor Glu216 in the active sites are shown as magenta balls, green Corey-Pauling-Koltun model, and red sticks, respectively. (B) Stereoview of a figure of merit (FOM)-weighted experimental electron density map (resolution, 2.2 Å; contour level, 1.5 σ), calculated after solvent flattening in the β-sheet region and superimposed on the refined model. The figure was prepared with Bobscript (15). (C) Ribbon diagram showing a side view (left) and a top view (right) of the homodecamer of GCYH-IA. The monomer is shown in red.
TABLE 3.
X-ray data collection, phasing, and structure refinement statistics
| Parameter | Value(s) for: |
|||
|---|---|---|---|---|
| GCYH-IB | GCYH-IB·Mn2+ | |||
| Structure refinement | ||||
| Resolution range (Å) | 45.9-2.2h | 30.33-2.04 | ||
| No. of reflections (working/free) | 42,833i/4,631 | 31,937/1,710 | ||
| No. of atoms | ||||
| Protein/water | 3,829/133 | 3,829/261 | ||
| Active-site metal ions | 2 | 2 | ||
| Other ions | 1 | 2 | ||
| Other ligands | 4 | 3 | ||
| Rcryst/Rfreej | 0.20/0.25 | 0.20/0.26 | ||
| rmsd bond length (Å) | 0.007 | 0.006 | ||
| rmsd bond angle (°) | 0.825 | 0.993 | ||
| Ramachandran plot—residues in: | ||||
| Favored regions (%) | 93.2 | 96.9 | ||
| Allowed regions (%) | 6.8 | 3.1 | ||
| Wilson B factor (Å2) | 40.3 | 37.4 | ||
| Data collection | ||||
| Space group | C2221 | C2221 | ||
| Data set | Sepeak | Seinflection | Seremote | |
| Unit cell (Å) | 91.7, 100.3, 114.1 | 91.7, 100.2, 114.0 | 91.8, 100.4, 114.1 | 92.2, 100.4, 113.9 |
| Wavelength (Å) | 0.9793 | 0.9795 | 0.9747 | 0.9761 |
| Resolution (Å) | 50.0-2.20 | 50.0-2.20 | 50.0-2.20 | 30.33-2.04 |
| Unique reflections | 26,718 (2,616)a | 26,761 (2,656) | 26,490 (2,136) | 33,675 (3,314) |
| Completeness (%) | 99.8 (99.0) | 99.8 (100.0) | 99.0 (92.0) | 99.8 (99.9) |
| Redundancy | 5.7 (5.0) | 5.8 (5.7) | 5.4 (3.5) | 4.5 (4.5) |
| Rmerge (%)b | 8.7 (61.5) | 7.5 (36.9) | 8.4 (89.1) | 7.1 (42.8) |
| I/σ(I) | 16.3 (2.4) | 17.6 (4.2) | 15.0 (3.5) | 11.0 (2.8) |
| Phasing statistics (29-2.30 Å) | ||||
| Rderc | 0.074d | 0.137 | ||
| Ranome | 0.069 | 0.059 | 0.058 | |
| RCullisf (isod/ano) | 0.49/0.73 | 0.83/0.72 | 0.84/0.86 | |
| Phasing powerg | ||||
| Centric | 0.06 | 0.11 | 0.16 | |
| Acentric | ||||
| isod | 0.06 | 0.09 | 0.15 | |
| ano | 1.20 | 1.46 | 1.10 | |
| FOM, overall | 0.34 | |||
| After DM | 0.83 | |||
| After Solomon | 0.85 | |||
| No. of Se sites | 15 | |||
Highest-resolution shell (2.2-2.28 Å for GCYH-IB, and 2.04-2.11 Å for GCYH-IB·Mn2+) information in parentheses.
Rmerge = 100 × (ΣhΣi|I(h) − I(h)i|)/ΣhΣiI(h)i, where I(h)i is the ith observation of reflection h and I(h) is the mean intensity of all observations of reflection h.
Rder = Σh|FPH − FP|/Σh|FP|, where |FP| and |FPH| are the observed structure factor amplitudes of the native and the derivative, respectively.
The dispersive differences were treated as isomorphous replacement information where the data collected at a wavelength of 0.9747 Å are treated as native data.
Ranom = Σh|FPH+ − FPH−|/Σh|FPH|, where |FPH+| and |FPH−| are the Friedel pair observed structure factor amplitudes of the derivative at a given wavelength, and FPH is their average.
RCullis = Σh[|FH| − (|FPH| − |FP|)]/Σh(|FPH| − |FP|), where |FH| is the calculated heavy-atom structure factor for reflection h.
PPdisp= (1/Nrefl)Σh[||FPH| − |FP||/∫02π(|FPH − FPHcalc|) P(ϕ) d(ϕ)], where P(ϕ) is the probability of a phase value of ϕ for reflection h. PPano = (1/Nrefl)Σh[|ΔobsANO|/∫02π(|ΔobsANO − ΔcalcANO|) P(ϕ) d(ϕ)], where ΔobsANO and ΔcalcANO are the Friedel pair differences in the observed and calculated structure factor amplitudes, respectively, for reflection h.
During density modification, structure factors were calculated for remote-wavelength data in the resolution range 2.3 to 2.2 Å.
Anomalous pairs treated as separate reflections.
Crystallographic R factor = 100 × (Σh||Fobs(h)| − |Fcalc(h)||)/Σh|Fobs(h)|, where Fobs(h) and Fcalc(h) are the observed structure factor amplitude and the structure factor amplitude calculated from the model, respectively. The free R factor was monitored with 10% and 5% of the data excluded from refinement for GCYH-IB and GCYH-IB·Mn2+, respectively.
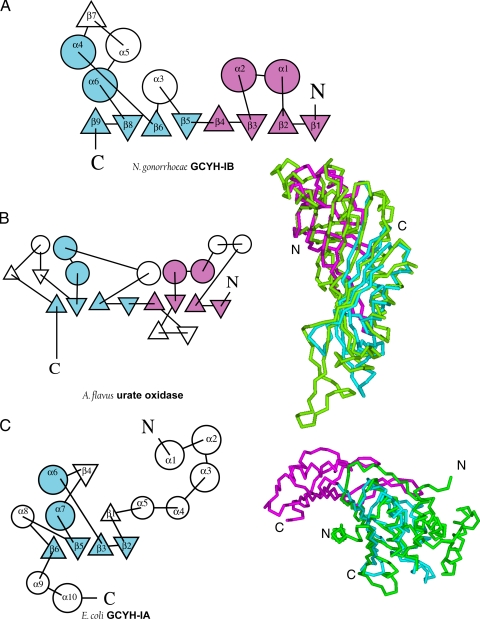
FIG. 6.
Topology of the GCYH-IB monomer and structural comparison with other T-fold enzymes. Topology diagrams of N. gonorrhoeae GCYH-IB (A), A. flavus urate oxidase (B), and E. coli GCYH-IA (C) calculated with the program TOPS (34). The N- and C-terminal modules of the core bimodular T-fold are shown in magenta and cyan, respectively. Cα trace superpositions of the N-terminal (magenta) and C-terminal (cyan) modules of GCYH-IB with corresponding enzymes (green) are shown in panels B and C. Secondary structure nomenclature is shown for the two isozymes.
A search for similar structures was done using the DALI search engine and the FSSP database (fold classification based on structure-structure alignment of proteins) (26). Several bimodular T-fold enzymes were identified, as well as GCYH-IA (Fig. 6C), a unimodular T-fold enzyme. The best fit was to an Aspergillus flavus urate oxidase (PDB identification [ID] 1UOX; root mean square deviation [rmsd], 3.4 Å over 197 Cα atoms) (Fig. 6B) (47) that follows a similar β8α4 topology. Furthermore, this search identified Nitrosomonas europaea DUF198, an ortholog of N. gonorrhoeae GCYH-IB that is annotated in the PDB as a protein of unknown function. The two proteins exhibit 56% sequence identity and identical tertiary and quaternary structures (PDB ID 2R5R; rmsd, 1.6 Å over 238 Cα atoms).
Comparison of GCYH-IA and GCYH-IB structures.
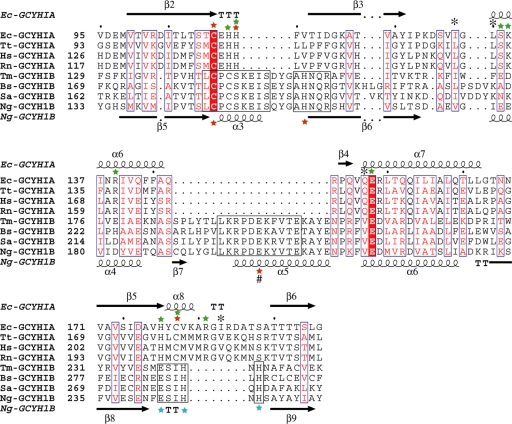
GCYH-IB is a homotetramer built around a bimodular β8α4 T-fold core (Fig. 5A and 6A) while GCYH-IA is a homodecamer β4α2 unimodular T-fold enzyme (Fig. 5C and 6C). Pairwise structural comparison of the monomeric subunits of GCYH-IB and E. coli GCYH-IA (PDB ID 1FBX) (46) using the DaliLite server (25) yielded an alignment strictly in the four β-strands and two α-helices (β2, β3, β5, β6, α6, and α7 of GCYH-IA, and β5, β6, β8, β9, α4, and α6 of GCYH-IB) of the T-fold (rmsd, 3.2 Å over 94 Cα atoms; Fig. 6C). Based on this alignment, a multisequence alignment of the two enzyme subfamilies in the shared core region was generated (15 sequences of each; Fig. 7). The new alignment differs significantly from our previously reported alignment that was based on sequence information and predicted tertiary structure (12). In the current alignment, the two subfamilies exhibit 28% sequence similarity and only two invariant residues—the metal and substrate liganding residues Cys147 and Glu216 (N. gonorrhoeae numbers), respectively. This unusually low primary structural identity, compared with 7 to 17% sequence identity and 39 to 53% sequence similarity between T-fold enzymes in general, explains the improper annotation of GCYH-IB genes in genomic databases and limited success in previous attempts to generate a model of the tertiary structure by sequence-based homology modeling (12).
FIG. 7.
Structure-guided multisequence alignment of GCYH-IA and GCYH-IB in the shared T-fold region. For clarity, only four sequences from each subfamily are shown. Residue labels and secondary structure elements (nomenclature same as in Fig. 6) are shown above and below the sequence alignment for subfamilies A and B, respectively. Residues conserved between the two subfamilies are in blue boxes, with the invariant residues (the substrate-binding Glu and metal-coordinating Cys) highlighted in red. Conserved regions within the GCYH-IB subfamily are in black boxes. The metal ion liganding side chains in each subfamily are labeled with red stars. The # symbol indicates a metal liganding side chain from the neighboring subunit. Additional GCYH-IB-specific, strictly conserved active-site residues are labeled with blue stars. Residues known to interact via their side chains or backbones with the GTP substrate in crystal structures of GCYH-IA are labeled with green stars and black asterisks, respectively. Ec, Escherichia coli; Tt, Thermus thermophilus; Hs, Homo sapien; Rn, Rattus norvegicus; Tm, Thermotoga maritima; Bs, Bacillus subtilis; Sa, Staphylococcus aureus; Ng, Neisseria gonorrhoeae.
In addition to the absence in GCYH-IB of the N- and C-terminal domains found in GCYH-IA (Fig. 6C), the following functionally important differences are seen in the T-fold domain, specifically in the active site. (i) There is an insertion in GCYH-IB of a two-turn α-helix (α3) between the first two strands (β5 and β6) of the common T-fold β-sheet (Fig. 6A and C and 7). This change results in the loss of the Zn2+ binding loop C110EHH113 found in GCYH-IA (E. coli numbering), although the Cys residue is retained (Fig. 7). (ii) There is an insertion in GCYH-IB of a β-strand (β7) and α-helix (α5) between the two α-helices (α4 and α6) of the T-fold (Fig. 6A and C and 7). β7 and α5 are part of the intersubunit interface that harbors the active site. Significantly, α5 provides the strictly conserved Glu201 as a carboxylate ligand to the active-site metal ion in GCYH-IB (see below). (iii) There is a deletion in GCYH-IB of helix α8 found in the T-fold domain of GCYH-IA (Fig. 6A and C and 7). Importantly, α8 contains the second Cys ligand to Zn2+ in GCYH-IA. This Cys is missing in GCYH-IB, and the helix is replaced with a β-turn containing the strictly conserved residues Glu243, His246, and His248.
Rearranged metal binding site and accommodation of Mn2+.
As in GCYH-IA, the active site of GCYH-IB is located at the interface between three subunits—two from one β-barrel and one from the head-to-head-facing β-barrel of the biological multimer. Two of the four active sites in the GCYH-IB homotetramer are partially disordered in the crystal. Each of the four active sites encompasses the metal binding site and the putative GTP binding pocket with the conserved Glu216 that serves to anchor the substrate guanine moiety, a characteristic feature of all T-fold proteins (8).
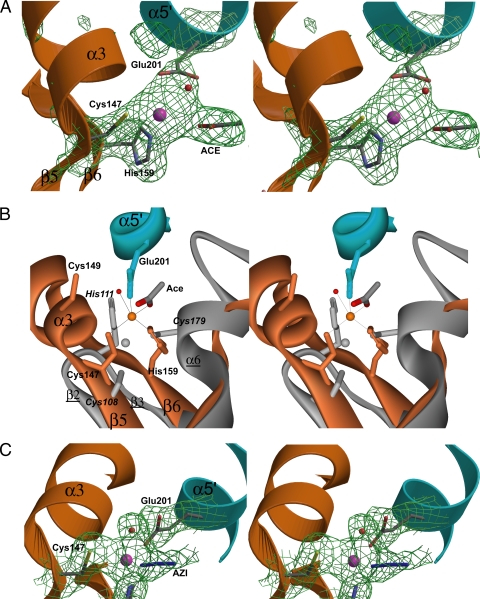
The presence of bound Zn2+ in the GCYH-IB crystal was confirmed by an X-ray fluorescence scan near the Zn absorption edge (see the figure in the supplemental material) and by an omit Fo-Fc map (Fig. 8A). Zn2+ is bound in a trigonal bipyramidal geometry and is coordinated by the thiol group of Cys147, a side chain oxygen atom of Glu201 from the neighboring subunit, and an oxygen atom from a bound acetate molecule (present in the protein sample at 50 mM) as equatorial ligands, and Nɛ of His159 and a water molecule as the bottom and top axial ligands, respectively (Fig. 8A and B). The average Zn2+-to-ligand distance is 2.4 Å. Although the active site as a whole putatively involves three subunits, the metal binding site involves residues from only two subunits, one from each β-barrel.
FIG. 8.
The metal site in GCYH-IB. (A) Stereoview of annealed omit Fo-Fc electron density map (1,500 K, 2.2 Å, 2.5 σ; Zn2+ and its ligands omitted from the phase calculation) superimposed on the model. (B) Three-dimensional alignment of GCYH-IB (orange and cyan; labels in roman type) and T. thermophilus GCYH-IA (gray; italic and underlined labels) in the active-site region. Strictly conserved Cys149 in the metal binding loop of GCYH-IB is shown, although it does not interact with the bound metal. (C) Mn2+-occupied metal site in the GCYH-IB·Mn2+ complex. Stereoview of annealed omit Fo-Fc electron density map (1,500 K, 2.04 Å, 2.5 σ; Mn2+ and its ligands omitted from phase calculation) superposed on the model. Enzyme subunits are colored as in Fig. 5A. Metal ions and water molecules are shown as magenta and red spheres, respectively. Secondary structure elements in GCYH-IB are labeled. The figure was prepared in Bobscript (15).
The liganding side chains, their positions in the Zn2+ coordination sphere, and the protein secondary structure elements in which they lie are different from those seen in GCYH-IA. For illustration, the three-dimensional alignment with the crystal structure of the T. thermophilus enzyme, the highest-resolution available structure of GCYH-IA, is shown in Fig. 8. In E. coli (PDB ID 1FBX [46]) and T. thermophilus GCYH-IA (PDB ID 1WM9 [51]), Zn2+ is coordinated in a distorted incomplete trigonal bipyramidal geometry by two thiol groups and a histidine side chain (Cys110, Cys181, His113 in E. coli residue numbers [Fig. 7]; Cys108, Cys179, His111 in T. thermophilus residue numbers [Fig. 8B]) as equatorial ligands all from the same subunit and by a water molecule as an axial ligand. The liganding side chains come from the loop between the first two β-strands of the T-fold (β2 and β3 in T. thermophilus GCYH-IA nomenclature in Fig. 7) and a short α-helix (α8) (Fig. 7). These Zn2+ liganding side chains and coordination geometry are preserved in all available structures of zinc-containing GCYH-IA, including the human (2) and rat (31, 32) enzymes. In contrast, in GCYH-IB, the metal binding site is formed by two GCYH-IB-specific insertions: the helix-loop insertion (α3) between the first two β-strands of the T-fold (β5 and β6) and the helical insertion α5 from the neighboring subunit. Despite a similar coordination geometry, the average Zn2+-to-ligand distance in GCYH-IA is 2.32 Å (2, 51, 31, 32), slightly shorter than that in GCYH-IB.
Crystals of GCYH-IB remetallated with Mn2+ (GCYH-IB·Mn) were grown using Zn2+-free reagents. The presence of bound Mn2+ was confirmed with an X-ray emission spectrum and fluorescence scan near the Mn2+ absorption edge following a wash and 1-h back soak of the crystal in metal-free solution (see the figure in the supplemental material). The crystal structure was determined by difference Fourier methods using model phases from the Zn2+-metallated structure (Table 3). The two structures are similar and superpose with an rmsd of 0.3 Å over 486 Cα atoms. Metal coordination and distances are also similar, except for the replacement of acetate with azide (added to the acetate-free protein sample at 12 mM concentration prior to crystallization) as an equatorial ligand to Mn2+ (Fig. 8C). Although the identity of the bound metal cannot be confirmed from the difference Fourier maps, the presence of an absorption edge for Mn2+ after a back soak of the crystal confirms the presence of bound Mn2+.
Putative GTP binding pocket.
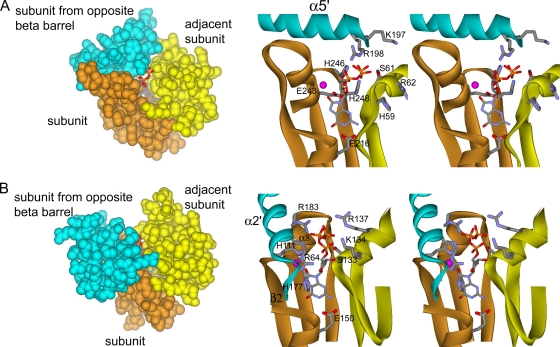
To elucidate the GTP binding site of GCYH-IB, we are conducting crystallographic analyses of complexes of N. gonorrhoeae GCYH-IB with GTP derivatives 8-oxo-GTP and 7-deaza-GTP. Meanwhile, we generated a docking model of 8-oxo-GTP onto N. gonorrhoeae GCYH-IB based on the superposition of the zinc-containing N. gonorrhoeae GCYH-IB crystal structure with that of the complex of T. thermophilus GCYH-IA with 8-oxo-GTP and zinc (PDB ID 1WUQ [51]), the only available structure of a ternary complex of GCYH-IA with zinc and a substrate derivative. Notably, the structures of GCYH-IA from a variety of sources (E. coli, T. thermophilus, human, and rat) are virtually identical (e.g., comparison of the crystal structures of human GCYH-IA without bound substrate [PDB ID 1FB1] and T. thermophilus GCYH-IA with 8-oxo-GTP yields an rmsd of 0.9 Å over 182 Cα atoms), and the active-site residues are invariant, so the comparison with T. thermophilus GCYH-IA should be relevant for all GCYH-IA members. In the T. thermophilus structure, 8-oxo-GTP, an extremely tight-binding inhibitor, presumably mimics the intermediate that forms immediately following nucleophilic attack at C-8 of GTP in the first step of catalysis (51). The superposition was applied such that the T-fold core of the N-terminal module of one N. gonorrhoeae GCYH-IB subunit and the T-fold core of the C-terminal module of the adjacent subunit in the same β-barrel align maximally with the T-fold cores of two adjacent subunits of the unimodular T. thermophilus GCYH-IA, hence aligning the intersubunit interface that harbors the active site. The superposition was done using the secondary-structure matching (SSM) protocol of Coot (29) and yielded an rmsd of 3.0 Å over 96 Cα atoms. Based on this superposition, 8-oxo-GTP was docked manually into the active site of N. gonorrhoeae GCYH-IB and subjected to energy minimization in CNS (4). The docking model indicates that, as in GCYH-IA, the substrate binds near the metal site in the cleft between three subunits, two from one β-barrel and one from the opposite β-barrel of the GCYH-IB tetramer (Fig. 9). The following similarities and differences are seen between the two enzyme subfamilies in the substrate binding pocket. (i) The substrate conformation and orientation in the active site of the two enzymes are similar. (ii) In GCYH-IA, the substrate binding cleft is deep and is covered from the front by helix α2 from the subunit in the opposite β-barrel (T. thermophilus nomenclature in Fig. 9B). In GCYH-IB, the cleft is shallow and more open and is covered from the top by helix α5 from the subunit in the opposite β-barrel (N. gonorrhoeae GCYH-IB nomenclature in Fig. 9A). (iii) One of the two side chains that interact with the guanine base in GCYH-IA is conserved in GCYH-IB (the invariant Glu150 in GCYH-IA; Glu216 in GCYH-IB). The other is His177 in helix α8 which is absent in GCYH-IB (Fig. 7 and 9). (iv) Three of the five basic residues (Arg64, His111, and Arg183) that stabilize the substrate triphosphate moiety in GCYH-IA come from secondary structure elements that are absent in GCYH-IB, namely helix α2, the β2-β3 loop, and helix α8, respectively, of GCYH-IA (Fig. 7 and 9). The other two basic side chains (Lys134 and Arg137) are not conserved and can be either acidic or neutral in GCYH-IB (Fig. 7). Instead, in GCYH-IB, the triphosphate appears to be stabilized by a patch of conserved basic residues from the adjacent subunit (His59 and Arg62) and the subunit in the opposite β-barrel (Lys197 and Arg198) (Fig. 9A). (v) The only active-site residue that interacts via its side chain with the substrate ribose moiety in GCYH-IA is Ser133, which is positionally conserved in GCYH-IB and comes from the adjacent subunit (Ser61) (Fig. 9A), although it is too distant to interact with the ribose moiety in the present model. Additionally, the side chains of the GCYH-IB-specific active-site residues Glu243, His246, and His248 are within 5 Å from the ribose moiety in the docking model, indicating that they may participate in catalysis.
FIG. 9.
Putative GTP binding pocket in GCYH-IB and comparison with GCYH-IA. (A) Docking model of the complex of N. gonorrhoeae GCYH-IB with 8-oxo-GTP shown in the active-site region. (B) Crystal structure of T. thermophilus GCYH-IA in complex with 8-oxo-GTP shown in the active-site region. Left panel shows the Corey-Pauling-Koltun model representation of the intersubunit interface showing the substrate binding cleft. Right panel shows stereoview of the substrate binding pocket showing interactions with the enzyme. Subunits are colored as in Fig. 5A. All panels depict the view of the intersubunit interface from the helical (outer) side of the barrel, i.e., looking down the arrows in Fig. 5A. Secondary structure elements mentioned in the text are labeled. 8-Oxo-GTP and the active-site side chains interacting with substrate are shown in stick model. The zinc ion is shown as a magenta ball.
In conclusion, the docking model suggests that the GTP binding interactions in GCYH-IB are significantly different from those in GCYH-IA, despite similar orientations of the substrate in the active site.
DISCUSSION
Zn2+ is an essential cofactor for numerous proteins. In bacteria and eukaryotes, cellular Zn2+ levels are sensed by specific transcription factors such as the repressor Zur in B. subtilis and E. coli (23, 35) and the activator Zap1p in Saccharomyces cerevisiae (30). Cellular responses to low Zn2+ conditions include increased expression of high-affinity Zn2+ transporters, such as ZnuABC, and substitution of Zn2+-dependent proteins/enzymes with alternative proteins/isozymes that do not rely on this metal ion as a cofactor. Examples of the latter strategy include Zn2+-dependent regulation of the alcohol dehydrogenase isozyme ADH4 in yeast (30) and paralogs of ribosomal protein S14 in B. subtilis (37).
Results from the present study clearly show that in B. subtilis, folE encodes the housekeeping Zn2+-dependent GCYH-IA used under normal growth conditions, when Zn2+ is readily available. In contrast, folE2, encoding the newly discovered GCYH-IB, is derepressed when Zn2+ is low, thereby allowing folate biosynthesis even under Zn2+-limiting conditions. This observation can be generalized to most organisms that contain copies of both the folE and folE2 genes, since in these organisms expression of folE2 is controlled by an upstream Zur repressor site. On the other hand, organisms that do not possess the Zn2+-dependent GCYH-IA isozyme (e.g., N. gonorrhoeae) consistently lack a Zur regulatory site upstream of the folE2 gene and rely solely on constitutively expressed GCYH-IB for folate biosynthesis (12).
The identification of the “correct” in vivo metal ion for a protein is often problematic when different metal ions support the relevant biological activity in vitro. The low affinity that both Zn2+ and Mn2+ exhibit for GCYH-IB (as well as the other metals that support catalysis) suggests that a free metal ion concentration of at least a low micromolar concentration is required in vivo in order to effectively metallate the enzyme. Thus, GCYH-IB may function physiologically with Mn2+, Fe2+, or Mg2+, all of which are present at sufficient levels in the cytosol to potentially metallate this enzyme. For example, Mn2+ supports the highest level of catalytic activity and is estimated to be present in B. subtilis at effective concentrations of 100 μM or higher (as estimated from the measured KD [equilibrium dissociation constant] value for Mn2+-dependent activation of the MntR metalloregulatory protein [20]). In contrast, free Zn2+ levels are estimated to be at most 1 pM (S. Gabriel and J. D. Helmann, unpublished observations), consistent with previous measurements in E. coli (40). Thus, despite the apparent tighter binding observed for Zn2+, it is highly unlikely that Zn2+ is involved to any significant extent in GCYH-IB activity in vivo. Indeed, absolute affinity is not necessarily the relevant measure for identification of the in vivo metal; from the Irving-Williams series (16, 27), one predicts that Zn2+ will bind to protein metal sites in preference to metals such as Mn2+ or Fe2+ even when the proteins have evolved to specifically utilize these other metals. The observation that protein metallation is determined not by absolute affinities, but by metal ion availability, is exemplified by the Mn2+-dependent proteins MntR and MncA; MntR binds Zn2+ with an affinity at least 103 greater than that for Mn2+ (20), yet functions physiologically as a manganese-specific sensor since zinc levels in the cell are far below those required for metallation of MntR. MncA, which binds Zn2+ with an affinity of ∼105 greater than that for Mn2+ (53), is observed to bind only Mn2+ in vivo, again due to the low abundance of free Zn2+.
Catalytic activity with metals other than Zn2+ has been observed with GCYH-IB orthologs from N. gonorrhoeae and M. jannaschii, which exhibit optimal activity in the presence of Mn2+ (data not shown) and Fe2+ (22), respectively. Interestingly, in the latter case the product is also different, as the archaeal GCYH-IB has been shown to produce dihydroneopterin cyclic phosphate (22), not dihydroneopterin triphosphate as observed with GCYH-IA (5) and bacterial GCYH-IB (12). Sequence conservation of key metal binding residues indicates that a metal dependence distinct from that of GCYH-IA is a property of the GCYH-IB enzyme family in general. For example, the introduction of an acidic side chain (Glu201) in the metal site of GCYH-IB allows accommodation of Mn2+, which exhibits a greater preference for oxygen ligands than for Zn2+.
The identification of the physiologically relevant cofactor for bacterial GCYH-IB enzymes will require further study and, given the ability of the B. subtilis enzyme to be activated by several metal ions, may vary between organisms or in response to changing metal ion availability. In the specific case of N. gonorrhoeae, we note that this organism survives in the highly oxidizing environment created by the host immune response and accumulates millimolar levels of Mn2+, an effective scavenger of reactive oxygen species (ROS) (49). A similar utilization of Mn2+ to fight ROS damage resulting from radiation exposure occurs in Deinococcus radiodurans (10), which also contains only GCYH-IB. In these organisms, Mn2+ is likely to be the physiologically relevant metal for GCYH-IB activation.
Structural analysis clearly demonstrates that GCYH-IB, like GCYH-IA, is a member of the T-fold family, but unlike the IA enzyme, GCYH-IB is a member of the bimodular subfamily, not the unimodular subfamily. The T-fold structural superfamily is comprised of functionally distant enzymes assembled through a common oligomerization of the T-fold, a protein fold comprised of a pair of two-stranded antiparallel β-sheets and two helices, to form a β2nαn barrel (8). In the functional enzymes, two barrels associate in a head-to-head fashion and similarly, bind planar substrates of purine/pterin structures at the interface of monomers and use a conserved Glu/Gln to anchor the substrate. Although the crystallographic asymmetric unit of GCYH-IB is a dimer forming a single barrel, the crystal structure reveals that a substantial 32% of the total monomeric surface is buried in the homotetramer (26.4% of the monomer surface area is buried in the intradimer interface and 22.3% of the dimer surface area is buried in the interdimer interface), consistent with a stable tetrameric structure. The gel filtration data demonstrate that the tetramer is the stable form of the enzyme in solution, and given that the crystal structure reveals that the active site is comprised of residues from three subunits, with the key metal binding residues originating from subunits in different barrels, it is unlikely that the dimer is the biological form of the enzyme. A similar structural organization is observed in DHNA, and in this case, the architecture leads to posttranslational regulation by metal-induced oligomerization (21). It is not clear whether metal binding induces oligomerization and formation of the GCYH-IB tetramer. In the crystal structure of the N. europaea GCYH-IB ortholog (PDB ID 2R5R [Midwest Center for Structural Genomics, unpublished data]), no bound metal is found (possibly due to use of chelating agents during protein purification) despite the conserved metal binding site. Yet, the structure reveals a homotetrameric quaternary arrangement identical to that of N. gonorrhoeae GCYH-IB, indicating that metal binding is not necessary for the formation of the biological tetramer. Taken together, these observations indicate that the probable biological form of the enzyme is the tetramer, similar to urate oxidase (47), the T-fold family member showing the highest structural similarity to GCYH-IB.
As demonstrated for GCYH-IA (51), the metal ion in GCYH-IB likely serves to activate a bound water molecule for nucleophilic attack in the first step of the reaction, and to stabilize the formyl intermediate generated after the opening of the purine ring. However, the differences in the putative GTP binding pocket and associated metal dependence between GCYH-IA and -IB suggest that differences in catalytic strategies for the two enzyme families may exist and further underscores the remarkable mechanistic plasticity of the T-fold superfamily. Indeed, this superfamily supports an extraordinarily diverse range of chemistries while exhibiting virtually identical tertiary structures for the core T-fold domain (8). Notably, the significant differences in the active-site architecture of the GCYH-IA and -IB enzymes, coupled with the occurrence of the latter enzyme in a number of clinically important human pathogens that lack GCYH-IA (Table 1) and thus depend solely on GCYH-IB for folate biosynthesis, position GCYH-IB as a potential new molecular target for the development of antibacterial agents against growing threats such as methicillin (meticillin)-resistant Staphylococcus aureus.
Supplementary Material
Acknowledgments
This work was partly supported by National Institutes of Health grants GM70641 to D.I.-R. and V.d.C.-L., GM059323 to J.D.H., and GM15539 to P.S. and by a fellowship from the National Foundation for Cancer Research. The Stanford Synchrotron Research Laboratory Structural Molecular Biology Program is supported by the Department of Energy, National Institutes of Health, and the National Institute of General Medical Sciences. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health (NIGMS).
Portions of this research were conducted at the Advanced Light Source, a national user facility operated by Lawrence Berkeley National Laboratory, on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences.
We thank A. Gonzalez and C. Smith from SSRL for help with the metal emission and fluorescence scans.
Footnotes
Published ahead of print on 18 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, P. D., K. Gopal, R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. K. Pai, R. J. Read, T. D. Romo, J. C. Sacchettini, N. K. Sauter, L. C. Storoni, and T. C. Terwilliger. 2004. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11:53-55. [DOI] [PubMed] [Google Scholar]
- 2.Auerbach, G., A. Herrmann, A. Bracher, G. Bader, M. Gutlich, M. Fischer, M. Neukamm, M. Garrido-Franco, J. Richardson, H. Nar, R. Huber, and A. Bacher. 2000. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. Proc. Natl. Acad. Sci. USA 97:13567-13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonafé, L., B. Thony, J. M. Penzien, B. Czarnecki, and N. Blau. 2001. Mutations in the sepiapterin reductase gene cause a novel tetrahydrobiopterin-dependent monoamine-neurotransmitter deficiency without hyperphenylalaninemia. Am. J. Hum. Genet. 69:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Burg, A. W., and G. M. Brown. 1966. The biosynthesis of folic acid. VI. Enzymatic conversion of carbon atom 8 of guanosine triphosphate to formic acid. Biochim. Biophys. Acta 117:275-278. [DOI] [PubMed] [Google Scholar]
- 6.Butcher, B. G., and J. D. Helmann. 2006. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by bacilli. Mol. Microbiol. 60:765-782. [DOI] [PubMed] [Google Scholar]
- 7.Clark, B. F., and K. A. Marcker. 1966. The role of N-formyl-methionyl-sRNA in protein biosynthesis. J. Mol. Biol. 17:394-406. [DOI] [PubMed] [Google Scholar]
- 8.Colloc'h, N., A. Poupon, and J. P. Mornon. 2000. Sequence and structural features of the T-fold, an original tunnelling building unit. Proteins 39:142-154. [DOI] [PubMed] [Google Scholar]
- 9.Cowtan, K. 1994. An automated procedure for phase improvement by density modification. Joint CCP4/ESF-EACBM Newsl. Protein Crystallogr. 31:34-38. [Google Scholar]
- 10.Daly, M. J., E. K. Gaidamakova, V. Y. Matrosova, A. Vasilenko, M. Zhai, A. Venkateswaran, M. Hess, M. V. Omelchenko, H. M. Kostandarithes, K. S. Makarova, L. P. Wackett, J. K. Fredrickson, and D. Ghosal. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025-1028. [DOI] [PubMed] [Google Scholar]
- 11.de La Fortelle, E., and G. Bricogne. 1997. Maximum-likelihood heavy atom parameter refinement in the MIR and MAD methods, p. 472-494. In C. W. Carter and R. W. Sweet (ed.), Methods in enzymology, vol. 276. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 12.El Yacoubi, B., S. Bonnett, J. N. Anderson, M. A. Swairjo, D. Iwata-Reuyl, and V. de Crécy-Lagard. 2006. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J. Biol. Chem. 281:37586-37593. [DOI] [PubMed] [Google Scholar]
- 13.El Yacoubi, B., G. Phillips, I. K. Blaby, C. E. Haas, Y. Cruz, J. Greenberg, and V. de Crécy-Lagard. 2009. A gateway platform for functional genomics in Haloferax volcanii: deletion of three tRNA modification genes. Archaea 2:211-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 15.Esnouf, R. M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 55:938-940. [DOI] [PubMed] [Google Scholar]
- 16.Frausto da Silva, J. J. R., and R. J. P. Williams. 2001. The biological chemistry of the elements: the inorganic chemistry of life, 2nd ed. Oxford University Press, Inc. New York, NY.
- 17.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelfand, M. S., P. S. Novichkov, E. S. Novichkova, and A. A. Mironov. 2000. Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform. 1:357-371. [DOI] [PubMed] [Google Scholar]
- 19.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 20.Golynskiy, M. V., W. A. Gunderson, M. P. Hendrich, and S. M. Cohen. 2006. Metal binding studies and EPR spectroscopy of the manganese transport regulator MntR. Biochemistry 45:15359-15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulding, C. W., M. I. Apostol, M. R. Sawaya, M. Phillips, A. Parseghian, and D. Eisenberg. 2005. Regulation by oligomerization in a mycobacterial folate biosynthetic enzyme. J. Mol. Biol. 349:61-72. [DOI] [PubMed] [Google Scholar]
- 22.Grochowski, L. L., H. Xu, K. Leung, and R. H. White. 2007. Characterization of an Fe(2+)-dependent archaeal-specific GTP cyclohydrolase, MptA, from Methanocaldococcus jannaschii. Biochemistry 46:6658-6667. [DOI] [PubMed] [Google Scholar]
- 23.Hantke, K. 2005. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8:196-202. [DOI] [PubMed] [Google Scholar]
- 24.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm, L., and J. Park. 2000. DaliLite workbench for protein structure comparison. Bioinformatics 16:566-567. [DOI] [PubMed] [Google Scholar]
- 26.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 27.Irving, H. M. N. H., and R. J. P. Williams. 1953. The stability of transition-metal complexes. J. Chem. Soc. 637:3192-3210. [Google Scholar]
- 28.Katzenmeier, G., C. Schmid, J. Kellermann, F. Lottspeich, and A. Bacher. 1991. Biosynthesis of tetrahydrofolate. Sequence of GTP cyclohydrolase I from Escherichia coli. Biol. Chem. Hoppe-Seyler. 372:991-997. [DOI] [PubMed] [Google Scholar]
- 29.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256-2268. [DOI] [PubMed] [Google Scholar]
- 30.Lyons, T. J., A. P. Gasch, L. A. Gaither, D. Botstein, P. O. Brown, and D. J. Eide. 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97:7957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maita, N., K. Hatakeyama, K. Okada, and T. Hakoshima. 2004. Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein. J. Biol. Chem. 279:51534-51540. [DOI] [PubMed] [Google Scholar]
- 32.Maita, N., K. Okada, K. Hatakeyama, and T. Hakoshima. 2002. Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP. Proc. Natl. Acad. Sci. USA 99:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarty, R. M., and V. Bandarian. 2008. Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem. Biol. 15:790-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalopoulos, I., G. M. Torrance, D. R. Gilbert, and D. R. Westhead. 2004. TOPS: an enhanced database of protein structural topology. Nucleic Acids Res. 32:D251-D254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, C. M., and J. D. Helmann. 2005. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8:188-195. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 37.Natori, Y., H. Nanamiya, G. Akanuma, S. Kosono, T. Kudo, K. Ochi, and F. Kawamura. 2007. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol. Microbiol. 63:294-307. [DOI] [PubMed] [Google Scholar]
- 38.Nichol, C. A., G. K. Smith, and D. S. Duch. 1985. Biosynthesis and metabolism of tetrahydrobiopterin and molybdopterin. Annu. Rev. Biochem. 54:729-764. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski, Z., and W. Minor (ed.). 1997. Processing of X-ray diffraction data collected in oscillation mode, vol. 276. Academic Press, San Diego, CA. [DOI] [PubMed]
- 40.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 41.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2000. Statistical analysis of complete bacterial genomes: palindromes and systems of restriction-modification. Mol. Biol. (Mosk). 34:246-252. (In Russian.) [PubMed] [Google Scholar]
- 42.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 100:9912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perrakis, A., R. Morris, and V. S. Lamzin. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6:458-463. [DOI] [PubMed] [Google Scholar]
- 44.Phillips, G., B. El Yacoubi, B. Lyons, S. Alvarez, D. Iwata-Reuyl, and V. de Crécy-Lagard. 2008. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: a new role for GTP cyclohydrolase I. J. Bacteriol. 190:7876-7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Project, C. C. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 46.Rebelo, J., G. Auerbach, G. Bader, A. Bracher, H. Nar, C. Hosl, N. Schramek, J. Kaiser, A. Bacher, R. Huber, and M. Fischer. 2003. Biosynthesis of pteridines. Reaction mechanism of GTP cyclohydrolase I. J. Mol. Biol. 326:503-516. [DOI] [PubMed] [Google Scholar]
- 47.Retailleau, P., N. Colloc'h, D. Vivares, F. Bonnete, B. Castro, M. El Hajji, J.-P. Mornon, G. Monard, and T. Prange. 2004. Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode. Acta Crystallogr. D Biol. Crystallogr. 60:453-462. [DOI] [PubMed] [Google Scholar]
- 48.Rodionov, D. A. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 107:3467-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seib, K. L., H. J. Wu, S. P. Kidd, M. A. Apicella, M. P. Jennings, and A. G. McEwan. 2006. Defenses against oxidative stress in Neisseria gonorrhoeae: a system tailored for a challenging environment. Microbiol. Mol. Biol. Rev. 70:344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki, T., H. Kurita, and H. Ichinose. 2004. GTP cyclohydrolase I utilizes metal-free GTP as its substrate. Eur. J. Biochem. 271:349-355. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka, Y., N. Nakagawa, S. Kuramitsu, S. Yokoyama, and R. Masui. 2005. Novel reaction mechanism of GTP cyclohydrolase I. High-resolution X-ray crystallography of Thermus thermophilus HB8 enzyme complexed with a transition state analogue, the 8-oxoguanine derivative. J. Biochem. (Tokyo) 138:263-275. [DOI] [PubMed] [Google Scholar]
- 52.Thöny, B., G. Auerbach, and N. Blau. 2000. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem. J. 347(Pt 1):1-16. [PMC free article] [PubMed] [Google Scholar]
- 53.Tottey, S., K. J. Waldron, S. J. Firbank, B. Reale, C. Bessant, K. Sato, T. R. Cheek, J. Gray, M. J. Banfield, C. Dennison, and N. J. Robinson. 2008. Protein-folding location can regulate manganese-binding versus copper- or zinc-binding. Nature 455:1138-1142. [DOI] [PubMed] [Google Scholar]
- 54.Vaguine, A. A., J. Richelle, and S. J. Wodak. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55(Pt 1):191-205. [DOI] [PubMed] [Google Scholar]
- 55.Yim, J. J., and G. M. Brown. 1976. Characteristics of guanosine triphosphate cyclohydrolase I purified from Escherichia coli. J. Biol. Chem. 251:5087-5094. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.