Abstract
Methanosarcina species possess three operons (mtaCB1, mtaCB2, and mtaCB3) encoding methanol-specific methyltransferase 1 (MT1) isozymes and two genes (mtaA1 and mtaA2) with the potential to encode a methanol-specific methyltransferase 2 (MT2). Previous genetic studies showed that these genes are differentially regulated and encode enzymes with distinct levels of methyltransferase activity. Here, the effects of promoter strength on growth and on the rate of methane production were examined by constructing strains in which the mtaCB promoters were exchanged. When expressed from the strong PmtaC1 or PmtaC2 promoter, each of the MtaC and MtaB proteins supported growth and methane production at wild-type levels. In contrast, all mtaCB operons exhibited poorer growth and lower rates of methane production when PmtaC3 controlled their expression. Thus, previously observed phenotypic differences can be attributed largely to differences in promoter activity. Strains carrying various combinations of mtaC, mtaB, and mtaA expressed from the strong, tetracycline-regulated PmcrB(tetO1) promoter exhibited similar growth characteristics on methanol, showing that all combinations of MtaC, MtaB, and MtaA can form functional MT1/MT2 complexes. However, an in vitro assay of coupled MT1/MT2 activity showed significant variation between the strains. Surprisingly, these variations in activity correlated with differences in protein abundance, despite the fact that all the encoding genes were expressed from the same promoter. Quantitative reverse transcriptase PCR and reporter gene fusion data suggest that the mtaCBA transcripts show different stabilities, which are strongly influenced by the growth substrate.
Methanosarcina species, such as M. acetivorans and M. barkeri, are strictly anaerobic archaea that derive their energy for growth via methanogenesis, the production of methane (CH4) (35). These organisms utilize various substrates, including one-carbon (C1) compounds (CO, CO2, formate, methanol, and methylamines), acetate, and CO2, as terminal electron acceptors in an energy-conserving electron transport chain that ultimately results in the production of methane (3, 7, 22). Four distinct methanogenic pathways can be found in various methanogens, which alternatively allow the reduction of CO2, the reduction of methyl moieties from C1 compounds, the splitting of acetate, and the disproportionation of C1 compounds, known as methylotrophic methanogenesis.
In the methylotrophic pathway for methanogenesis, methanol, methylamines, or methylsulfides are disproportionated into CO2 and CH4, with the reducing equivalents from oxidation of one molecule of the substrate being used to reduce three additional molecules to CH4 (reviewed in reference 14). Biochemical studies have shown that these methylated compounds are initially activated via methylation of 2-mercaptoethanesulfonic acid (coenzyme M [CoM]). The conversion of methanol to methyl-CoM is mediated by the concerted effort of at least three proteins that interact cooperatively (28, 31-33). To form methyl-CoM, the methyl moiety from methanol is first transferred to a methyl-accepting corrinoid protein, MtaC, in a reaction catalyzed by the MtaB protein (methanol:5-hydroxy-benzimidazolyl-cobamide methyltransferase). MtaC and MtaB form a tight complex denoted MT1. The MtaA protein (comethyl-benzimidazolyl-cobamide:2-mercaptoethane sulfonic acid methyltransferase), referred to as MT2, then transfers the methyl group to CoM-HS to produce methyl-CoM. MtaC and MtaB are encoded by an operon, mtaCB (27), while MtaA is encoded by the monocistronic mtaA gene (12).
The genomes of M. acetivorans (10), M. barkeri (18), M. mazei (8), and M. thermophila (9) each carry three copies of the mtaCB operon, designated mtaCB1, mtaCB2, and mtaCB3, and two copies of mtaA, designated mtaA1 and mtaA2. The conservation of genes encoding the multiple isozymes argues that each plays a specific role in Methanosarcina metabolism. Nevertheless, studies conducted to date have not provided a clear rationale to support this idea. The ability of M. acetivorans double mutants with one of the three mtaCB copies to grow on methanol, coupled with the inability of a triple mutant to utilize this substrate, clearly shows that each operon encodes a functional methanol-specific MT1 (24). In contrast, mtaA1 mutants are incapable of growth on methanol, whereas mtaA2 mutants have no measurable growth phenotype (5). Thus, only the mtaA1 gene is capable of supporting growth on methanol, and no obvious role for mtaA2 is apparent.
Gene regulation studies also suggest that each isozyme plays a specific role in the cell because the genes are differentially regulated. Both mtaCB1 and mtaCB2 are highly expressed on methanol, while mtaCB3 is poorly expressed on this substrate. The mtaCB operons are also regulated by growth phase: the mtaCB1 promoter is expressed only during the exponential phase on methanol, while the mtaCB2 and mtaCB3 promoters are expressed before the exponential phase (5, 6). At least five distinct transcriptional regulators are involved in the differential expression of the three mtaCB operons, which further strengthens the argument that each plays a different role during growth (4, 6). Gene fusion studies showed that mtaA1 is highly induced, while mtaA2 is poorly expressed on methanol (5), supporting the former's essentiality and the latter's dispensability on this substrate.
One possible explanation for the differential regulation of the mtaCB operons is that they encode isozymes with enzymatic properties tailored for different growth conditions. Initial biochemical studies showed significant differences in the methyltransferase activities of mutants carrying only one of the three MT1 isozymes: strains carrying only mtaCB1 exhibited methyltransferase activity similar to that of the wild type, while the methyltransferase activities of strains carrying only mtaCB2 or mtaCB3 were two and four times lower than that of the wild type, respectively (24). While such differences are probably at least partially due to the regulated expression of the genes on methanol and thus to unequal protein abundances, it remains possible that the isozymes possess different enzymatic properties. Moreover, it is clear that the different isozymes are coexpressed under some conditions and growth phases. Thus, it is likely that the different subunits have the opportunity to form additional isozymes with mixed compositions, further increasing the complexity of the methanol methyltransferase system.
In this study, we constructed strains in which the native promoters of the mtaCB operons were exchanged for those corresponding to their isozymes to determine the contribution of promoter strength to characteristics of growth on methanol. We also constructed synthetic operons of mtaCBA, in all possible combinations, to determine if the different isozymes could interact with each other to support growth of M. acetivorans on methanol. In these operons, the expressions of the mta genes were under the control of the same promoter: the strong, tetracycline-regulated PmcrB(tetO1) (11). These data show that all Mta subunits are functional and can interact in any combination. However, interpretation of the data with respect to biochemical activity is not possible with these strains, due to the unanticipated presence of posttranscriptional gene control elements within some or all of the methyltransferase genes. Strikingly, it appears that the stability of the mta transcripts is strongly affected by the growth substrate in a physiologically relevant manner.
MATERIALS AND METHODS
Media and growth conditions.
Methanosarcina acetivorans strains were grown with single-cell morphology (30) at 37°C in HS broth or 1.5% agar medium containing either 125 mM methanol or 50 mM trimethylamine (TMA) (2). Solid-medium plates were incubated in an intrachamber anaerobic incubator (20). Puromycin (CalBiochem, San Diego, CA) was added from a sterile, anaerobic stock at a final concentration of 2 μg ml−1 to select M. acetivorans strains carrying the puromycin transacetylase gene (pac) (25). 8-ADP (8-aza-2,6- diaminopurine; Sigma, St. Louis, MO) was added from a sterile, anaerobic stock at a final concentration of 20 μg ml−1 to select against hypoxanthine phosphoribosyltransferase (encoded by hpt) (25). When appropriate, tetracycline (100 μg ml−1; Sigma, St. Louis, MO) was added from a sterile, anaerobic stock to induce expression from the PmcrB(tetO1) promoter. Standard conditions were employed for growth of Escherichia coli (34).
DNA methods and transformation.
Standard methods were used to isolate and manipulate plasmid DNA from E. coli strains (1). E. coli WM3118 (11) was used as a host for all pJK027A derivatives. E. coli DH5α/λpir (21) was used as host for all pir-dependent replicons. E. coli DH10B (Stratagene, La Jolla, CA) was used for all other plasmid replicons. The DNA sequences of all cloning constructs were confirmed by DNA sequencing at the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois at Urbana-Champaign, Urbana, IL. E. coli strains were transformed by electroporation using an E. coli Gene Pulser as recommended (Bio-Rad, Hercules, CA). Liposome-mediated transformation was employed for M. acetivorans as previously described (19).
Integration of mtaCBA operons and promoter swaps in M. acetivorans.
All M. acetivorans strains used in this study are presented in Table 1. The PmcrB-Tetr and PmcrB-φC31int alleles were introduced into the chromosome of WWM147 via pJK024 as previously described (25) in order to construct WWM148. pAB5 was introduced in WWM13 to construct WWM390. The mtaCBA operons constructed in pJK027A and derivatives of pJK200 were integrated in the genomes of WWM148 and WWM390, respectively, via φC31-mediated site-specific recombination (11). All strains were shown to carry the indicated mtaCBA operons by using PCR primers specific to the individual mta genes. Strains were isolated and maintained on TMA to prevent accumulation of mutations that might confer improved ability to grow on methanol. All genetic manipulations of Methanosarcina strains were performed under strictly anaerobic conditions in an anaerobic glove box.
TABLE 1.
Strains used in this study
| Straina | Genotype |
|---|---|
| WWM1 | Δhpt |
| WWM13 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt |
| WWM82 | Δhpt::PmcrB-φC31int-attP |
| WWM92 | Δhpt::PmcrB(tetO1)::uidA |
| WWM147 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt |
| WWM148 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP |
| WWM168 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC1B2A1 |
| WWM169 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC1B3A2 |
| WWM170 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC2B2A1 |
| WWM171 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC2B2A2 |
| WWM172 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC2B3A1 |
| WWM173 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC2B3A2 |
| WWM174 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC3B2A1 |
| WWM175 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC3B2A2 |
| WWM176 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC1B1A1 |
| WWM177 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC1B2A2 |
| WWM178 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC2B1A2 |
| WWM179 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC3B1A1 |
| WWM180 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC3B1A2 |
| WWM181 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC1B1A2 |
| WWM182 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC2B1A1 |
| WWM183 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC3B3A2 |
| WWM184 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC3B3A1 |
| WWM185 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 ΔmtaA1 ΔmtaA2 Δhpt::PmcrB-Tetr-φC31int-attP::PmcrB(tetO1)-mtaC1B3A1 |
| WWM390 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP |
| WWM392 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP::PmtaC1-mtaCB1 |
| WWM395 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attR::PmtaC1-mtaCB2 |
| WWM398 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attR::PmtaC1-mtaCB3 |
| WWM393 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP::PmtaC2-mtaCB1 |
| WWM396 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP::PmtaC2-mtaCB2 |
| WWM399 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP::PmtaC2-mtaCB3 |
| WWM394 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP::PmtaC3-mtaCB1 |
| WWM397 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP::PmtaC3-mtaCB2 |
| WWM400 | ΔmtaCB1 ΔmtaCB2 ΔmtaCB3 Δhpt::φC31int-attP::PmtaC3-mtaCB3 |
Measurement of growth characteristics.
To determine the growth characteristics of strains carrying different mtaCBA operons, 0.2 ml of a late-exponential-phase (optical density at 600 nm [OD600] = ca. 0.5) culture grown on HS-TMA plus tetracycline was inoculated into either 10 ml of HS-TMA plus tetracycline, or HS-methanol plus tetracycline. To determine the growth parameters of the promoter swap strains, 0.3 ml of a TMA-grown culture (OD600 = 0.4 to 0.5) was inoculated into 10 ml of HS-TMA or HS-methanol. All cultures were incubated at 37°C and growth was monitored by measuring light scattering at 600 nm using a Bausch & Lomb Spectronic 21 spectrophotometer. At least three replicates were performed for all experiments.
Rate of methane production.
The rates at which various strains produced methane from methanol were measured using cell suspensions as previously described (5).
Methyltransferase activity assay.
Actively growing (OD600 = 0.35 to 0.45) HS-TMA or HS-methanol cultures, both with tetracycline, were harvested anaerobically by centrifugation at 5,000 rpm for 10 min. Cell pellets were washed with equal volumes of HS-MOPS (400 mM NaCl, 13 mM KCl, 54 mM MgCl2·6H2O, 2 mM CaCl2·2H2O, 50 mM MOPS [morpholinepropanesulfonic acid], pH 7.0), repelleted by centrifugation, and resuspended in 50 mM MOPS (pH 7.0) containing 1× protease inhibitor cocktail (Complete, Mini, EDTA-free; Roche Diagnostic, Indianapolis, IN). The cells were lysed by two 10-s sonication treatments with a Sonifier S125 (Branson Sonic Power Co., Danbury, CT). The protein concentration was measured using the Coomassie Plus Bradford assay kit (Pierce, Rockford, IL) with bovine serum albumin as the standard. Assay components were assembled in an anaerobic chamber under N2-H2 (95%/5%) and included 4 mg protein extract, 100 mM methanol or TMA, 1.5 mM Ti(III) citrate (36), 10 mM ATP, 20 mM MgCl2, 3.2 mM BES (bromoethanesulfonic acid), 2 mM HS-CoM (2-mercaptoethanesulfonic acid), and 50 mM MOPS, pH 7.0. The total volume of the assays was 1 ml. Assays were incubated under N2-H2 (95%/5%) at 37°C for up to 3 h. The methylation of CoM was determined by measuring the consumption of the free thiol of CoM with Ellman's reagent as previously described (24).
β-Glucuronidase activity assay.
M. acetivorans WWM92 was grown as described above for cultures used in methyltransferase assays. Cell extract preparation and β-glucuronidase activities were measured as previously described (25).
Overexpression and purification of His6-tagged mtaA1.
N-terminally His6-tagged mtaA1 was PCR amplified (N-terminally His6-tagged mtaA1 primers 5′-GGCGCGCCCATATGGGCAGCAGCCATCATCATCATCATCACAGCAGCGGCCTGGTGCCGCGCGGCAGCCACATGACCGATATGAGCGAATT-3′ [forward] and GGCGCGCCAAGCTTTTAGGCGTAGAATTCGTTTCTTGC [reverse]) and cloned into the NdeI and HindIII sites (under lined) of pJK027A. The N-terminally His6-tagged mtaA1 gene was then subcloned into pET11a in order to construct pRO011 and then transformed in E. coli BL21(DE3). To overexpress the tagged MtaA1 protein, E. coli BL21(DE3)/pRO011 was grown aerobically at 37°C on Luria-Bertani medium containing ampicillin (100 μg ml−1) to an OD600 of ca. 0.7 and subsequently induced by adding 0.2 mM (1 mM for mtaB1) isopropyl β-d-1-thiogalactopyranoside.
To purify the recombinant MtaA1, the cell pellet from 6 liters of E. coli BL21(DE3)/pRO011 culture was resuspended in a buffer containing 50 mM sodium phosphate (pH 7.0) and 300 mM NaCl and broken three times through a French pressure cell at 20,000 lb/in2. After centrifugation at 15,000 rpm for 30 min at 4°C to remove cell debris, the supernatant was loaded onto a 1-ml HisTrap HP column (Amersham Biosciences, GE Healthcare UK Limited, Buckinghamshire, United Kingdom). For the removal of unbound proteins, the column was washed with 10 column volumes of a buffer containing 50 mM sodium phosphate (pH 6.0), 300 mM NaCl, 10% (vol/vol) glycerol, and 10 mM imidazole. MtaA1 was eluted with 17.5 ml of buffer containing 50 mM sodium phosphate (pH 6.0), 300 mM NaCl, 10% (vol/vol) glycerol, and 500 mM imidazole. Imidazole and sodium chloride were removed by dialysis at 4°C by using a Slide-A-Lyzer dialysis cassette (Thermo Scientific, Rockford, IL) in a buffer containing 50 mM sodium phosphate (pH 6.0) and 10% (vol/vol) glycerol.
Western blot analysis.
Polyclonal antibodies were generated at the Immunological Resource Center, University of Illinois at Urbana-Champaign, Urbana, IL. Rabbits were immunized with a synthetic conserved peptide of MtaC isozymes (C-CGGGAVNQDFVSQ-NH2; amino acids 209 to 221) to generate anti-MtaC antibody. Chickens were immunized with purified His6-tagged mtaA1 to produce anti-MtaA. The indicated amounts of total cell protein from various strains were run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and then electrophoretically transferred to Hybond ECL membranes (Amersham Biosciences, GE Healthcare UK Limited, Buckinghamshire, United Kingdom). Anti-MtaC was used at a 1:3,000 dilution, while anti-MtaA was used at a 1:5,000 dilution. Peroxidase-labeled anti-rabbit antibody (Amersham Biosciences, GE Healthcare UK Limited, Buckinghamshire, United Kingdom) and horseradish peroxidase-conjugated anti-chicken/turkey immunoglobulin G (Zymed, San Francisco, CA) were used at 1:20,000 and 1:2,000 dilutions, respectively. The membranes were blocked with 5% nonfat dried milk in phosphate-buffered saline (pH 7.5) with 0.1% (vol/vol) Tween 20 for at least 1 h at ambient temperature. After incubation with each of the primary and secondary antibodies, the membranes were washed as recommended by the manufacturer (Amersham Biosciences, GE Healthcare UK Limited, Buckinghamshire, United Kingdom) but with twofold-longer incubations and twice the number of washes. The antigen-primary antibody complex was detected with the peroxidase-conjugated secondary antibody by using the ECL Western blotting detection system as described by the manufacturer (Amersham Biosciences, GE Healthcare UK Limited, Buckinghamshire, United Kingdom) and exposed on Biomax XAR film (Kodak, Rochester, NY) at ambient temperature.
Total RNA extraction.
Total RNA was isolated from cultures used to assay for methyltransferase activity and β-glucuronidase activity as described above. Cells were aseptically withdrawn at 37°C by using an anaerobic syringe and immediately transferred to a sterile tube containing 3 volumes of TRIzol LS reagent (Invitrogen, Carlsbad, CA). Following phase separation with chloroform, the total RNA was precipitated with 70% ethanol and purified by use of the RNeasy kit as prescribed by the manufacturer (Qiagen Corp., CA). RNA was eluted with RNase-free water and subsequently treated with Turbo DNA-free DNase (Ambion, Austin, TX) according to the manufacturer's instructions. The concentration and purity of the RNA were quantified using a spectrophotometer (NanoDrop, Wilmington, DE).
Quantitative reverse transcriptase PCR (RT-PCR).
Gene-specific primers were designed from the genome of M. acetivorans C2A by using the Primer Express v2.0 software (Applied Biosystems, Foster City, CA) at the Functional Genomics Unit of the W. M. Keck Center, University of Illinois, Urbana, IL.
A one-step RT-PCR was performed using the ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Reaction mixes (20 μl total) contained 1× SYBR green reaction mix with ROX (Invitrogen, Carlsbad, CA), 0.5 μl SuperScript III (Invitrogen, Carlsbad, CA), 2.5 ng RNA, and 10 pmol (each) of primers. Synthesis of cDNA from RNA and subsequent amplification were performed as follows: 50°C for 5 min and 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. To confirm PCR product specificity, a melting curve analysis was performed at 95°C, 60°C, and finally 95°C, all for 15 s. Two negative controls were run simultaneously with the RNA samples to check for nucleic acid contamination: (i) reaction solution that contained RNA template but without RT and (ii) reaction solution that contained RT without RNA template.
The relative standard curve method was used to quantify the expression of the uidA and mta genes (ABI PRISM 7900HT sequence detection system user bulletin 2). Standard curves relating CT values to the log amount of RNA were constructed for each gene, including the reference gene, rpoA1, by using RNA isolated from WWM82 grown on TMA (OD600 = 0.45 to 0.50). The average input for each gene from triplicate measurements of at least two biological test samples was calculated using the linear regression of the standard curve and normalized to the rpoA1 value to obtain the expression level of each gene.
RESULTS
Construction and characterization of the promoter swap strains.
To examine the effect of promoter activity on methanol methyltransferase function, we constructed plasmids in which the native PmtaC promoters were exchanged for one another. These plasmids were then integrated into the chromosome of a strain from which all three native mtaCB operon copies were deleted. The resulting strains thus carried only a single mtaCB copy expressed from one of the three promoters. Each strain was then examined for the ability to utilize methanol as a growth substrate and for methanol-dependent methane formation (Table 2).
TABLE 2.
Growth characteristicsa and rates of methane production on methanol of the promoter swap strains
| mtaCB operons(s) | Mean ± SD |
|||
|---|---|---|---|---|
| Generation time (h) | Lag time (h) | Cell yield (OD600) | Sp actb | |
| All present | 11.4 ± 0.5 | 77 ± 3 | 0.77 ± 0.01 | 150 ± 3 |
| All deleted | NGc | NG | NG | NG |
| PmtaC1-mtaCB1 | 10.6 ± 0.5 | 29 ± 2 | 0.77 ± 0.01 | 165 ± 25 |
| PmtaC1-mtaCB2 | 14.3 ± 2.2 | 180 ± 7 | 0.82 ± 0.01 | 155 ± 5 |
| PmtaC1-mtaCB3 | 18.2 ± 1.1 | 152 ± 3 | 0.83 ± 0.01 | 143 ± 18 |
| PmtaC2-mtaCB1 | 14.0 ± 0.3 | 38 ± 3 | 0.80 ± 0.00 | 116 ± 15 |
| PmtaC2-mtaCB2 | 15.5 ± 0.6 | 110 ± 2 | 0.85 ± 0.00 | 80 ± 8 |
| PmtaC2-mtaCB3 | 12.4 ± 0.3 | 33 ± 3 | 0.80 ± 0.00 | 179 ± 6 |
| PmtaC3-mtaCB1 | NG | NG | NG | NG |
| PmtaC3-mtaCB2 | 54.4 ± 6.9 | 495 ± 43 | 0.72 ± 0.01 | 87 ± 3 |
| PmtaC3-mtaCB3 | 49.2 ± 4.1 | 288 ± 31 | 0.34 ± 0.03 | 89 ± 11 |
Growth was measured as described in Materials and Methods by using strains WWM1, WWM390, and WWM392 to WWM400. The lag time represents the time needed to reach one-half the maximum OD600. Values represent the means and standard deviations of triplicate measurements.
Activity is reported in mU (see the text). Values represent the means and standard errors of four independent measurements.
NG, no growth.
The results indicate that all three methanol-dependent MT1 enzymes are fully functional when expressed from the relatively strong PmtaC1 and PmtaC2 promoters. Moreover, they suggest that the previously observed poor growth of a strain carrying only mtaCB3 can be attributed largely to the PmtaC3 promoter. Accordingly, when PmtaC1 and PmtaC2 were used, the growth rates, lag times, and growth yields from methanol were similar to the wild-type values, regardless of which mtaCB coding region was used. In contrast, when the PmtaC3 promoter was used, growth was severely affected. Under these conditions, mtaC1 could not support growth on methanol, whereas mtaCB2 supported very slow growth and long lag times, and mtaCB3 supported reduced growth yields with the latter. The strains showed no measurable differences when grown on media with TMA as the substrate, indicating that the observed effects are methanol specific (data not shown). The rate at which methane was produced from methanol by resting cell suspensions was also comparable to the wild-type rate when PmtaC1 and PmtaC2 controlled the expression of the three mtaCB operons, consistent with the growth characteristics of these strains on methanol (Table 2). When driven by PmtaC2 and PmtaC3, somewhat lower rates of methane production supported by the mtaCB2 operon were observed: both promoters reduced the rate to about half that observed when this operon was under PmtaC1. The activity of mtaC3 when driven by its native promoter was only half that observed when PmtaC1 and PmtaC2 controlled the expression of this operon.
Construction of synthetic tetracycline-regulated mtaCBA operons.
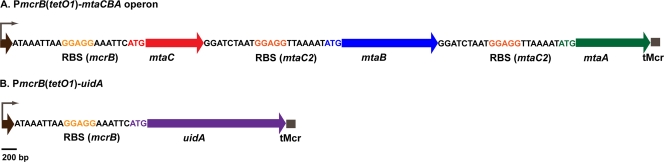
To remove the effects of differential transcription from our results and to test whether the different MtaA, MtaB, and MtaC proteins could functionally interact, we constructed a series of synthetic mtaCBA operons that utilize the same tetracycline-regulated promoter (Fig. 1). Each operon is transcribed from the strong, tetracycline-regulated promoter PmcrB(tetO1) (11). The tetracycline-induced expression of PmcrB(tetO1) is about twofold higher than methanol-induced expression from the native PmtaC1 promoter and fourfold higher than expression from PmtaA1, based on reporter gene studies (5, 6, 11). Thus, ample expression of the synthetic operons should be readily achieved by tetracycline induction. In the absence of tetracycline, reporter gene activity expressed from PmcrB(tetO1) is below the limit of detection (11). In all constructs, the putative ribosome binding site (RBS) of mcrB from M. barkeri Fusaro was used for the translation of mtaC, while the mtaC2 RBS sequence was used for the translation of mtaB and mtaA (Fig. 1). Thus, as far as possible, the synthetic operons maintained the same translational controls as well. Each operon was integrated in a single copy into the chromosome of a mutant lacking all three mtaCB operons and both mtaA genes (5, 24). This strain is incapable of growth on methanol; thus, any utilization of this substrate can be attributed to the integrated synthetic mtaCBA operon.
FIG. 1.
Genetic composition and structure of the PmcrB(tetO1)-driven mtaCBA operons and uidA. The brown arrows represent PmcrB(tetO1). The gene from which the RBS sequence is derived is listed in parentheses. tMcrB represents the terminator sequence from mcrB of M. barkeri Fusaro.
Physiological characterization of strains carrying the synthetic tetracycline-regulated mtaCBA operons.
On TMA medium with tetracycline, strains carrying all 18 possible combinations of mtaC, mtaB, and mtaA exhibited growth characteristics similar to those of the parental strain (data not shown), showing that overexpression of the mta genes is not toxic to the host cells. When cells pregrown on TMA medium with tetracycline were inoculated into media with methanol plus tetracycline, all combinations of mtaCBA supported growth (Table 3). Further, the maximum growth yields of all Mta combinations were similar. Thus, functional methanol-dependent MT1/MT2 activity is possible with any combination of MtaA, MtaB, and MtaC. Consistent with the data from the promoter swap strains, the growth characteristics of the strains carrying PmcrB(tetO1)-mtaC1B1, PmcrB(tetO1)-mtaC2B2, and PmcrB(tetO1)-mtaC3B3 were indistinguishable, indicating that all three MT1 isozymes can be equally functional during growth on methanol if expressed from a strong promoter. Further, constructs with mtaA2 grew as well on methanol as constructs with mtaA1 (e.g., mtaC1B1A1 versus mtaC1B1A2, mtaC2B2A1 versus mtaC2B2A2, and mtaC3B3A1 versus mtaC3B3A2), despite the observation that mtaA2 alone was not sufficient to allow growth on methanol when expressed from its native promoter (5). Therefore, both MtaA1 and MtaA2 are fully functional methanol-dependent MT2 enzymes. However, the growth rates of the strains varied over a fivefold range, and only PmcrB(tetO1)-mtaC1B3A1 supported growth at a rate similar to those of strains in which the three operons are expressed from their native promoters. No clear trends emerge from the data to suggest which protein combinations are preferred. Surprisingly, all of the synthetic-operon strains displayed a prolonged lag phase relative to the wild type when switched from TMA to methanol, despite the fact that transcription of the genes was induced prior to inoculation into methanol medium.
TABLE 3.
Growth characteristicsa of the PmcrB(tetO1)-mtaCBA strains when grown on methanol plus tetracycline
| Relevant genotype | Mean ± SD |
||
|---|---|---|---|
| Generation time (h) | Growth yield (OD600) | Lag time (h) | |
| Wild type | 9.2 ± 0.4 | 0.69 ± 0.04 | 55 ± 1 |
| mta deletion | NGb | NG | NG |
| PmcrB(tetO1)-mtaC1B1A1 | 18.7 ± 4.7 | 0.71 ± 0.08 | 169 ± 25 |
| PmcrB(tetO1)-mtaC1B2A1 | 41.2 ± 11.9 | 0.60 ± 0.05 | 219 ± 6.0 |
| PmcrB(tetO1)-mtaC1B3A1 | 11.5 ± 0.8 | 0.69 ± 0.05 | 261 ± 8 |
| PmcrB(tetO1)-mtaC1B1A2 | 20.1 ± 2.1 | 0.68 ± 0.01 | 169 ± 22 |
| PmcrB(tetO1)-mtaC1B2A2 | 22.2 ± 4.8 | 0.68 ± 0.02 | 169 ± 46 |
| PmcrB(tetO1)-mtaC1B3A2 | 12.5 ± 1.2 | 0.64 ± 0.01 | 264 ± 13 |
| PmcrB(tetO1)-mtaC2B1A1 | 22.1 ± 1.7 | 0.67 ± 0.02 | 126 ± 10 |
| PmcrB(tetO1)-mtaC2B2A1 | 18.4 ± 5.8 | 0.66 ± 0.01 | 230 ± 17 |
| PmcrB(tetO1)-mtaC2B3A1 | 33.1 ± 3.4 | 0.61 ± 0.04 | 222 ± 25 |
| PmcrB(tetO1)-mtaC2B1A2 | 20.4 ± 2.3 | 0.68 ± 0.01 | 129 ± 15 |
| PmcrB(tetO1)-mtaC2B2A2 | 17.8 ± 3.7 | 0.68 ± 0.08 | 267 ± 40 |
| PmcrB(tetO1)-mtaC2B3A2 | 14.7 ± 2.0 | 0.66 ± 0.06 | 262 ± 46 |
| PmcrB(tetO1)-mtaC3B1A1 | 39.4 ± 14.4 | 0.66 ± 0.01 | 192 ± 19 |
| PmcrB(tetO1)-mtaC3B2A1 | 22.2 ± 7.9 | 0.63 ± 0.06 | 239 ± 35 |
| PmcrB(tetO1)-mtaC3B3A1 | 16.0 ± 3.1 | 0.67 ± 0.06 | 233 ± 3 |
| PmcrB(tetO1)-mtaC3B1A2 | 49.0 ± 5.1 | 0.61 ± 0.04 | 189 ± 8 |
| PmcrB(tetO1)-mtaC3B2A2 | 30.3 ± 4.0 | 0.62 ± 0.02 | 251 ± 3 |
| PmcrB(tetO1)-mtaC3B3A2 | 17.1 ± 2.2 | 0.64 ± 0.04 | 256 ± 14 |
Growth was measured as described in Materials and Methods by using strains WWM82, WWM148, and WWM168 to WWM185. The growth yield represents the maximum OD600 of the culture. The lag time represents the time needed to reach one-half the maximum OD600. Values represent the means and standard deviations of at least triplicate measurements.
NG, no growth.
Biochemical characterization of strains carrying the synthetic tetracycline-regulated mtaCBA operons.
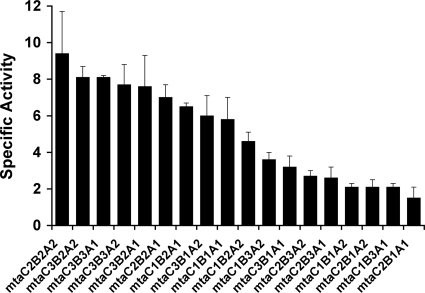
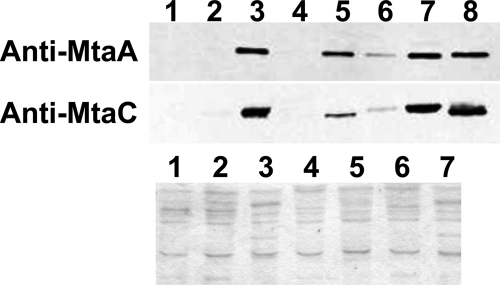
Methanol-dependent CoM methylation was assayed in extracts of strains carrying the various synthetic operons to assess the relative activities of MT1/MT2 isozymes with different compositions (Fig. 2). Significant activity was observed for all combinations of Mta proteins in this coupled-MT1/MT2 assay; however, we could not find any correlation between activity and specific isozyme combination or between activity and phenotype for growth on methanol. Because the coupled MT1/MT2 activities involve a second-order interaction of three discrete proteins, activity is strongly dependent on the concentration of each protein (29). Therefore, we assessed the levels of the MtaC and MtaA proteins in each extract by Western blotting. (MtaB was not quantified, due to our inability to generate a specific antibody [data not shown].) Surprisingly, we found dramatically different levels of the Mta proteins in each extract (Fig. 3 and data not shown). Because the same promoter was used in each construct, it is unlikely that this result is caused by differences in transcription initiation, and thus, the levels of Mta proteins appear to be regulated by a posttranscription initiation mechanism. (For ease of discussion, we refer to this as posttranscriptional regulation; however, as discussed below, this could involve regulation at the transcript elongation level.)
FIG. 2.
Overall methanol methyltransferase activities of PmcrB(tetO1)-mtaCBA operons. Assays were performed using crude cell extracts as described in Materials and Methods. Specific activities are presented in mU (see the text). The values represent the means and standard deviations of two biological replicates, each measured in triplicate.
FIG. 3.
(Upper panel) Western immunoblot analysis of MtaC and MtaA proteins expressed from synthetic PmcrB(tetO1)-mtaCBA operons. (Lower panel) Coomassie blue-stained polyacrylamide gel electrophoresis gel run with equal amounts of total protein from each extract. Lane 1, parental strain (WWM 148), from which all mta genes were deleted, grown on TMA; lane 2, PmcrB(tetO1)-mtaC2B2A1 (WWM170) strain, grown on TMA plus tetracycline; lane 3, PmcrB(tetO1)-mtaC2B2A1 (WWM170) strain, grown on methanol plus tetracycline; lane 4, PmcrB(tetO1)-mtaC1B1A1 (WWM176) strain grown on TMA plus tetracycline; lane 5, PmcrB(tetO1)-mtaC1B1A1 (WWM176) strain grown on methanol plus tetracycline; lane 6, PmcrB(tetO1)-mtaC3B3A1 (WWM184) strain, grown on TMA plus tetracycline; lane 7, PmcrB(tetO1)-mtaC3B3A1 (WWM184) strain, grown on methanol plus tetracycline; lane 8, control strain (WWM1), with all mta genes expressed from their native promoters, grown on methanol. For MtaC and MtaA detection, 4 μg and 0.5 μg total protein, respectively, were loaded in each well.
Substrate-dependent, posttranscriptional regulation of mtaCBA operons.
To gain further insight into the mechanism of posttranscriptional regulation, we also examined MT1/MT2 activity after inducing transcription of selected mtaCBA operons in TMA media. When grown on TMA plus tetracycline, extracts of strains carrying PmcrB(tetO1)-mtaC1B1A1, PmcrB(tetO1)-mtaC2B2A1, and PmcrB(tetO1)-mtaC3B3A1 exhibited similar overall TMA-dependent methyltransferase activities, of ca. 45 nmol CoM consumed min−1 mg protein−1 (mU). However, these extracts displayed negligible methanol-dependent methyltransferase activity (mtaC1B1A1, 0.05 ± 0.01 mU; mtaC2B2A1, 0.02 ± 006 mU; mtaC3B3A1, 0.33 ± 0.19 mU). In contrast, when grown on methanol plus tetracycline, these strains exhibited a dramatic increase in methanol-dependent methyltransferase activity (Fig. 2). Western blots show that these low activities correlated with the relative amounts of soluble Mta proteins produced on TMA plus tetracycline (Fig. 3). To verify that this effect was not due to an unanticipated effect of growth substrate on the activity of the PmcrB(tetO1) promoter, we also assayed β-glucuronidase activity in extracts of a strain carrying a PmcrB(tetO1)-uidA fusion. The activities on TMA plus tetracycline and methanol plus tetracycline (expressed as mU as described above) were similar: that on TMA plus tetracycline was 537.7 ± 52.2 mU, and that on methanol plus tetracycline was 565.7 ± 22.0 mU.
The observed substrate-dependent differences in protein abundance could be due to the relative stabilities of either the proteins themselves or the mRNA that encodes them (or both). Quantitative RT-PCR analysis showed that mtaCBA transcript levels were ca. 10 times higher on methanol plus tetracycline than on TMA plus tetracycline, indicating that the latter explanation accounts for most, if not all, of the observed effect (Table 4). Transcript levels of the uidA gene were unaffected by growth substrate, indicating that the effect is mediated by either the mta coding sequences or the mtaC2 RBS sequence, which is unique to the synthetic operon constructs.
TABLE 4.
Relative mRNA abundances of the uidA and mta genes when strains were grown on methanol or TMA
| Operon structure | mRNA ratioa |
|||
|---|---|---|---|---|
| uidA | mtaA | mtaC | mtaB | |
| PmcrB(tetO1)-uidA | 1.0 ± 0.2 | NAb | NA | NA |
| PmcrB(tetO1)-mtaC3B3A1 | NA | 9.5 ± 2.1 | 9.5 ± 2.7 | 9.9 ± 3.3 |
| PmcrB(tetO1)-mtaC2B2A1 | NA | 10.3 ± 1.7 | 15.2 ± 6.8 | 13.6 ± 5.8 |
| PmcrB(tetO1)-mtaC1B1A1 | NA | 8.8 ± 2.6 | 7.9 ± 2.1 | 8.9 ± 2.3 |
| PmcrB(tetO1)-mtaC1B1A2 | NA | 9.2 ± 2.2 | NDc | ND |
The ratios of mRNA levels from strains grown on methanol to mRNA levels from strains grown on TMA are shown. mRNA abundance was determined by qRT-PCR as described, by using total RNA isolated from strains WWM82 [PmcrB(tetO1)-uidA], WWM184 [PmcrB(tetO1)-mtaC3B3A1], WWM170 [PmcrB(tetO1)-mtaC2B2A1], WWM176 [PmcrB(tetO1)-mtaC1B1A1], and WWM181 [PmcrB(tetO1)-mtaC1B1A2] grown on either HS-methanol or HS-TMA. Tetracycline was added to all cultures to induce expression from the PmcrB(tetO1) promoter. All primers used in this experiment are listed in the supplemental material.
NA, not applicable.
ND, not determined.
DISCUSSION
The data presented above show that each of the mtaA, mtaB, and mtaC genes encodes a fully functional methanol-dependent MT1/MT2 isozyme subunit. In general, the growth characteristics of all mta combinations are similar. Thus, previous results showing that mtaA2 was nonfunctional (5) and that strains dependent on mtaCB3 grew very poorly (24) are almost certainly due to poor expression of the genes rather than intrinsically poor activity of the proteins they encode. This idea finds support in a number of studies showing that mtaA2 and mtaCB3 are expressed at low levels relative to their homologs. Analysis of reporter gene fusions showed that mtaA2 was expressed at very low levels on all tested methanogenic substrates (5). DNA microarray analysis of M. mazei (13) and proteomic analyses of M. acetivorans (16) and M. thermophila (9) also suggested that mtaA2 was not expressed during growth on methanol. Similarly, reporter gene studies showed that PmtaC3 is expressed at levels 10- and 64-fold lower than those of PmtaC1 and PmtaC2 during growth on methanol, respectively. Both microarray and proteomic studies revealed similar mtaCB3 expression patterns (15, 16). Conversely, we showed here that mtaCB1 could not support growth on methanol when driven by PmtaC3, despite the fact that growth was essentially at wild-type levels when mtaCB1 was expressed from stronger promoters [PmtaC1, PmtaC2, and PmcrB(tetO1)]. Less severe but similar results were obtained with mtaCB2 expressed from PmtaC3.
Our data also show that functional methanol-dependent MT1/MT2 activity can be obtained with any combination of mtaA, mtaB, and mtaC when the genes are expressed from a sufficiently strong promoter. We previously hypothesized that the various isozymes display differential regulation because they possess enzymatic activities tailored to different growth conditions (e.g., low substrate concentrations would favor an enzyme with a low Km and a low Vmax, while higher substrate concentrations would favor an enzyme with a high Km and a high Vmax) (6). Our goal in constructing strains that expressed each combination of synthetic operons that share conserved transcriptional and translational sequences was to provide biochemical support for this hypothesis. Our data clearly show that all combinations form active MT1/MT2 complexes. Unfortunately, we also showed that, despite our best efforts, the strains do not produce equivalent amounts of protein from the various synthetic constructs. Thus, although the qualitative conclusions drawn above are valid, quantitative comparisons of the biochemical and physiological data are inappropriate.
The surprising variation in amounts of the individual Mta proteins produced by essentially identical operons shows that the steady-state levels of the MT1/MT2 isozymes are regulated at the posttranscriptional level. Although clear differences between different synthetic operons expressed on the same growth substrate were observed, posttranscriptional regulation was especially pronounced when comparing the levels of proteins produced by the same operons under different growth conditions. Strikingly, the data show that the proteins are more abundant when they are required for growth than when they are not. Accordingly, when the cells are grown on methanol, for which the cells require MtaA, MtaB, and MtaC, the proteins are present at levels that are much higher than those observed when the cells are grown on TMA, for which these proteins are not required. Thus, the regulation is clearly physiologically relevant.
At least five potential mechanisms to account for the observed posttranscriptional regulation can be envisioned: (i) the mRNAs are produced in different amounts due to premature transcription termination on different substrates, (ii) the mRNAs have differential stabilities, (iii) the mRNAs are translated at different rates, (iv) the proteins display differential stabilities, and (v) the proteins display differential solubilities. The observation that mRNA levels of TMA- and methanol-grown cells are dramatically different strongly suggests that the effect is probably mediated by one of the first three mechanisms listed; however, distinguishing between these can be very difficult. Many studies have shown that translation efficiency is coupled to mRNA stability (17, 23, 26); thus, it is very difficult to disentangle poor translation from intrinsic mRNA stability. We can, however, gain insight into the molecular requirements by looking at sequences that are shared by the unstable mRNAs. Accordingly, it is likely that sequences within the mtaA, mtaC, and/or mtaB coding regions or within the mtaC2 RBS sequence mediate the effect. Additional experiments will be required to determine whether these effects are caused by substrate-specific effects on transcript elongation or translation efficiency, specific mRNA degradation in TMA-grown cells, or specific mRNA stabilization in methanol-grown cells. Regardless of which mechanism is responsible for the observed differences in mRNA abundance, these data suggest that control of transcription alone is not enough for these slow-growing, energy-limited microorganisms.
Supplementary Material
Acknowledgments
This work was supported in part by National Science Foundation grant MCB0517419. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. Support for R.B.O. was provided by the Philippine Agriculture-Fulbright Scholarship Program.
Footnotes
Published ahead of print on 18 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology, vol. 1 and 2. John Wiley and Sons, New York, NY.
- 2.Boccazzi, P., J. K. Zhang, and W. W. Metcalf. 2000. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri Fusaro. J. Bacteriol. 182:2611-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock, A. K., A. Kraft-Prieger, and P. Schonheit. 1994. Pyruvate—a novel substrate for growth and methane formation in Methanosarcina barkeri. Arch. Microbiol. 161:33-46. [Google Scholar]
- 4.Bose, A., and W. W. Metcalf. 2008. Distinct regulators control the expression of methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. Mol. Microbiol. 67:649-661. [DOI] [PubMed] [Google Scholar]
- 5.Bose, A., M. A. Pritchett, and W. W. Metcalf. 2008. Genetic analysis of the methanol- and methylamine-specific methyltransferase 2 genes of Methanosarcina acetivorans C2A. J. Bacteriol. 190:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bose, A., M. A. Pritchett, M. Rother, and W. W. Metcalf. 2006. Differential regulation of the three methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. J. Bacteriol. 188:7274-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deppenmeier, U. 2002. The unique biochemistry of methanogenesis. Prog. Nucleic Acid Res. Mol. Biol. 71:223-283. [DOI] [PubMed] [Google Scholar]
- 8.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 9.Ding, Y. H., S. P. Zhang, J. F. Tomb, and J. G. Ferry. 2002. Genomic and proteomic analyses reveal multiple homologs of genes encoding enzymes of the methanol:coenzyme M methyltransferase system that are differentially expressed in methanol- and acetate-grown Methanosarcina thermophila. FEMS Microbiol. Lett. 215:127-132. [DOI] [PubMed] [Google Scholar]
- 10.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guss, A. M., M. Rother, J. K. Zhang, G. Kulkarni, and W. W. Metcalf. 2008. New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2:193-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms, U., and R. K. Thauer. 1996. Methylcobalamin: coenzyme M methyltransferase isoenzymes MtaA and MtbA from Methanosarcina barkeri. Cloning, sequencing and differential transcription of the encoding genes, and functional overexpression of the mtaA gene in Escherichia coli. Eur. J. Biochem. 235:653-659. [DOI] [PubMed] [Google Scholar]
- 13.Hovey, R., S. Lentes, A. Ehrenreich, K. Salmon, K. Saba, G. Gottschalk, R. P. Gunsalus, and U. Deppenmeier. 2005. DNA microarray analysis of Methanosarcina mazei Gö1 reveals adaptation to different methanogenic substrates. Mol. Genet. Genomics 273:225-239. [DOI] [PubMed] [Google Scholar]
- 14.Keltjens, J. T., and G. D. Vogels. 1993. Conversion of methanol and methylamines to methane and carbon dioxide. In J. G. Ferry (ed.), Methanogenesis. Chapman & Hall, New York, NY.
- 15.Li, L., Q. Li, L. Rohlin, U. Kim, K. Salmon, T. Rejtar, R. P. Gunsalus, B. L. Karger, and J. G. Ferry. 2007. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Q., L. Li, T. Rejtar, B. L. Karger, and J. G. Ferry. 2005. Proteome of Methanosarcina acetivorans. Part II: comparison of protein levels in acetate- and methanol-grown cells. J. Proteome Res. 4:129-135. [DOI] [PubMed] [Google Scholar]
- 17.Lu, P., C. Vogel, R. Wang, X. Yao, and E. M. Marcotte. 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25:117-124. [DOI] [PubMed] [Google Scholar]
- 18.Maeder, D. L., I. Anderson, T. S. Brettin, D. C. Bruce, P. Gilna, C. S. Han, A. Lapidus, W. W. Metcalf, E. Saunders, R. Tapia, and K. R. Sowers. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188:7922-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf, W. W., J. K. Zhang, and R. S. Wolfe. 1998. An anaerobic, intrachamber incubator for growth of Methanosarcina spp. on methanol-containing solid media. Appl. Environ. Microbiol. 64:768-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Brien, J. M., R. H. Wolkin, T. T. Moench, J. B. Morgan, and J. G. Zeikus. 1984. Association of hydrogen metabolism with unitrophic or mixotrophic growth of Methanosarcina barkeri on carbon monoxide. J. Bacteriol. 158:373-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira, C. C., and J. E. McCarthy. 1995. The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270:8936-8943. [DOI] [PubMed] [Google Scholar]
- 24.Pritchett, M. A., and W. W. Metcalf. 2005. Genetic, physiological and biochemical characterization of multiple methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. Mol. Microbiol. 56:1183-1194. [DOI] [PubMed] [Google Scholar]
- 25.Pritchett, M. A., J. K. Zhang, and W. W. Metcalf. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapaport, L. R., and G. A. Mackie. 1994. Influence of translational efficiency on the stability of the mRNA for ribosomal protein S20 in Escherichia coli. J. Bacteriol. 176:992-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer, K., U. Harms, and R. K. Thauer. 1997. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Purification, properties and encoding genes of the corrinoid protein MT1. Eur. J. Biochem. 243:670-677. [DOI] [PubMed] [Google Scholar]
- 28.Sauer, K., and R. K. Thauer. 1999. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri—substitution of the corrinoid harbouring subunit MtaC by free cob(I)alamin. Eur. J. Biochem. 261:674-681. [DOI] [PubMed] [Google Scholar]
- 29.Sauer, K., and R. K. Thauer. 1998. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri—identification of the active-site histidine in the corrinoid-harboring subunit MtaC by site-directed mutagenesis. Eur. J. Biochem. 253:698-705. [DOI] [PubMed] [Google Scholar]
- 30.Sowers, K. R., J. E. Boone, and R. P. Gunsalus. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 59:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Meijden, P., H. J. Heythuysen, A. Pouwels, F. Houwen, C. van der Drift, and G. D. Vogels. 1983. Methyltransferases involved in methanol conversion by Methanosarcina barkeri. Arch. Microbiol. 134:238-242. [DOI] [PubMed] [Google Scholar]
- 32.van der Meijden, P., H. J. Heythuysen, H. T. Sliepenbeek, F. P. Houwen, C. van der Drift, and G. D. Vogels. 1983. Activation and inactivation of methanol: 2-mercaptoethanesulfonic acid methyltransferase from Methanosarcina barkeri. J. Bacteriol. 153:6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Meijden, P., L. P. J. M. Jansen, C. van der Drift, and G. D. Vogels. 1983. Involvement of corrinoids in the methylation M (2-mercaptoethanesulfonic acid) by methanol and enzymes from Methanosarcina barkeri. FEMS Microbiol. Lett. 19:247-251. [Google Scholar]
- 34.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 35.Woese, C. R., L. J. Magrum, and G. E. Fox. 1978. Archaebacteria. J. Mol. Evol. 11:245-251. [DOI] [PubMed] [Google Scholar]
- 36.Zehnder, A. J., and K. Wuhrmann. 1976. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 194:1165-1166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.