Abstract
A characteristic feature of biofilm formation is the production of a protective extracellular polymeric matrix. In the gram-positive bacterium Bacillus subtilis, the biofilm matrix is synthesized by the products of the epsABCDEFGHIJKLMNO operon (hereafter called the eps operon) and yqxM-sipW-tasA loci. Transcription from these operons is repressed by two key regulators, AbrB and SinR. Relief of inhibition is necessary to allow biofilm formation to proceed. Here we present data indicating that Abh, a sequence and structural homologue of AbrB, regulates biofilm architecture by B. subtilis when colony morphology and pellicle formation are assessed. Data indicating that abh expression is dependent on the environmental signals that stimulate the activity of the extracytoplasmic function σ-factor σX are shown. We demonstrate that expression of slrR, the proposed activator of yqxM transcription, is positively controlled by Abh. Furthermore, Abh is shown to activate transcription from the promoter of the eps operon through its control of SlrR. These findings add to the increasingly complex transcriptional network that controls biofilm formation by B. subtilis.
Multicellular behaviors exhibited by microorganisms are survival strategies often associated with “stationary-phase” growth. The gram-positive soil-dwelling bacterium Bacillus subtilis is capable of many such behaviors, including cannibalism (16), genetic competence (19), exoprotease production (13), and biofilm formation (7, 20). If unfavorable conditions persist, B. subtilis is also capable of sporulation, a process that results in the formation of a dormant stress-resistant endospore (43). Both the processes of sporulation and biofilm formation are controlled in B. subtilis by the global regulator of multicellular behavior, Spo0A (7, 20, 43). Spo0A exhibits regulatory control when phosphorylated. Phosphorylation occurs through the action of a complex phosphorelay that is initiated in response to multiple environmental stimuli (9, 17). The promoter regions of Spo0A-regulated genes have been determined to possess different binding affinities for the activated regulator (15). Therefore, the impact of Spo0A∼P is determined by the extent to which Spo0A∼P accumulates within an individual cell. This simple but effective mechanism permits Spo0A∼P to control multiple incompatible cell states (15, 55). For example, transcription of the genes required for biofilm formation is induced before transcription of the genes required for spore formation (55). In this case, formation of a biofilm could perhaps allow scarce nutrients to be shared among the community in the hope that environmental conditions would improve so that cells do not have to instigate the irreversible and energetically expensive sporulation pathway.
Biofilm formation by B. subtilis occurs upon activation of two transcription factors, Spo0A and DegU (34, 40). DegU activates transcription of yvcA and yuaB, but how the products of these loci contribute to the formation of a mature biofilm remains unknown (30, 53, 54). As discussed above, Spo0A is required for biofilm formation and its activation is triggered by the lipopeptide surfactin (7, 20, 33). Downstream from Spo0A are two parallel pathways of repression and antirepression (2). Transcription of the operons required for the synthesis of the extracellular matrix is repressed by two key regulators, namely, AbrB and SinR (21, 28). Spo0A∼P-dependent derepression of both the AbrB and SinR regulons occurs by the induction of antagonist proteins that inhibit the ability of both repressors to bind DNA, namely, AbbA which interacts with AbrB, and SinI which interacts with SinR (1, 2). Spo0A∼P also directly inhibits abrB transcription (15, 46, 51). Thus, deletion of either abrB or sinR results in increased extracellular matrix production and a more rugose biofilm (34).
A key feature of biofilm formation is the synthesis of the extracellular matrix and the inhibition of motility (3, 34, 48). To date, two components of the biofilm matrix formed by B. subtilis strain NCIB3610 have been described, an exopolysaccharide and a protein called TasA. The chemical composition of the exopolysaccharide remains undefined, but it is known that the machinery required for its synthesis is encoded by the 15-gene epsABCDEFGHIJKLMNO operon (hereafter called the eps operon) (7, 34). The molecular function of all but one of the products of the eps operon is unknown, but EpsE interacts with the flagellar motor to render the cells immotile during biofilm formation (3). TasA, the major protein component of B. subtilis biofilm is the product of a three-gene operon, the yqxM-sipW-tasA operon (hereafter called the yqxM operon). The yqxM operon additionally encodes the proteins required for the correct localization of TasA within the matrix of the biofilm (6, 11).
Abh is a sequence and structural homologue of AbrB with 70% identity in the DNA binding domain (5). Despite this, the physiological role of Abh has remained relatively unknown. Most information concerning Abh function is derived from a study by Strauch et al. (49) who identified the first set of genes regulated by Abh. The genes identified regulate the production of antimicrobial compounds. The genes identified as directly regulated by Abh were also shown to be directly regulated by AbrB, thereby suggesting a significant Abh and AbrB regulatory overlap (35, 49). Production of Abh is regulated at the level of transcription (27, 49). Expression of abh is directly repressed by AbrB, and consequently, genes that are regulated by Abh are also indirectly controlled by Spo0A∼P (see above) (49). In addition, transcription of abh is activated by RNA polymerase in the presence of the extracytoplasmic function (ECF) σ-factors, σX (in vitro and in vivo data) and σW (in vitro data) (26, 27). More recently, by using a more stringent set of conditions, abh was also ascribed to the σM regulon (14). The B. subtilis genome encodes seven ECF σ-factors, six of which are “anchored” to the cytoplasmic membrane by their cotranscribed antagonist (32, 38, 58). Upon the sensing of a specific external stress, intramembrane proteolysis of the antagonist allows release of a specific σ-factor into the cytoplasm where it is free to interact with RNA polymerase and regulate their specific regulon (24).
Using an “undomesticated” isolate of B. subtilis, NCIB3610, we demonstrate that the AbrB homologue Abh regulates biofilm architecture. We show that the level of Abh synthesis is controlled by an unidentified environmental signal that stimulates the activity of the ECF σ-factor σX under biofilm formation conditions. Furthermore, we show that Abh regulates the activation of transcription from the eps operon which provides the extracellular polysaccharide component of the matrix and inhibits flagellum-based motility during biofilm formation (3, 7). It has previously been shown that biofilm formation requires the transcriptional activator SlrR (12, 31), and using single-cell analysis, we show that transcription of slrR is positively controlled by Abh. Consistent with this observation, the mutant biofilm generated in the absence of abh can be complemented by ectopic expression of slrR. Our data suggest that Abh indirectly activates biofilm formation via activation of slrR, which in turn increases the number of cells actively transcribing the genes required for synthesizing both the protein and polysaccharide components of the biofilm matrix.
MATERIALS AND METHODS
General strain construction and growth conditions.
The B. subtilis strains used and constructed in this study are detailed in Table 1. Escherichia coli strain MC1061 [F′ lacIq lacZM15 Tn10 (tet)] was used for the construction and maintenance of plasmids. B. subtilis JH642 and 168 derivatives were generated by transformation of competent cells with plasmids or DNA using standard protocols (22). SPP1 phage transductions, for introduction of DNA into B. subtilis strain NCIB3610, were conducted as described previously (53). Both E. coli and B. subtilis strains were routinely grown in Luria-Bertani (LB) medium (10 g NaCl, 5 g yeast extract, and 10 g tryptone [all per liter]) or MSgg medium (5 mM potassium phosphate and 100 mM morpholinepropanesulfonic acid [MOPS] at pH 7.0 supplemented with 2 mM MgCl2, 700 μM CaCl2, 50 μM MnCl2, 50 μM FeCl3, 1 μM ZnCl2, 2 μM thiamine, 0.5% glycerol, 0.5% glutamate, and amino acids as appropriate at a final concentration of 50 μg ml−1) at 37°C (7, 11). When appropriate, the following antibiotics and concentrations were used: ampicillin at 100 μg ml−1), chloramphenicol at 5 μg ml−1, erythromycin at 5 μg ml−1, lincomycin at 25 μg ml−1, kanamycin at 25 μg ml−1, and spectinomycin at 100 μg ml−1.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype or descriptiona | Source or constructionb |
|---|---|---|
| 168 | trpC2 | BGSC |
| HB0801 | sigM::kan | 36 |
| HB0803 | sigW::mls | 36 |
| HB0804 | sigX::spc | 36 |
| JH642 | trpC2 pheA1 | 42 |
| NCIB3610 | Prototroph | BGSC |
| NRS1314 | degU::pBL204 (cat) | 53 |
| NRS1434 | trpC2 pheA1 amyE::Pabh-abh::spc | pBL154 → JH642 |
| NRS1893 | trpC2 pheA1 amyE::Phy-spank-abh-lacI (spc) | pNW400 → JH642 |
| NRS1894 | trpC2 pheA1 Δabh::cat | This study |
| NRS1900 | Δabh::cat | SPP1 NRS1894 → NCIB3610 |
| NRS1901 | Δabh::cat amyE::Phy-spank-abh-lacI (spc) | SPP1 NRS1893 → NRS1900 |
| NRS1904 | Δabh::cat amyE::Pabh-abh::spc | SPP1 NRS1434 → NRS1900 |
| NRS1916 | trpC2 pheA1 amyE::Pabh-gfp (cat) | pNW402 → JH642 |
| NRS1917 | amyE::Pabh-gfp (cat) | SPP1 NRS1916 → NCIB3610 |
| NRS1937 | trpC2 pheA1 amyE::Phy-spac-abh-lacI (cat) | pNW403 → JH642 |
| NRS1964 | trpC2 pheA1 amyE::Phy-spankslrR-lacI (spc) | pNW407 → JH642 |
| NRS1965 | amyE::Phy-spank-slrR-lacI (spc) | SPP1 NRS1964 → NCIB3610 |
| NRS1966 | Δabh::cat amyE::Phy-spankslrR-lacI (spc) | SPP1 NRS1964 → 1900 |
| NRS1968 | trpC2 sacA::PslrR-gfp-lacI (kan) | pNW408 → 168 |
| NRS2241 | trpC2 pheA1sacA::PepsA-gfp (kan) | pNW502 → JH642 |
| NRS2242 | sacA::PepsA-gfp (kan) | SPP1 NRS2241 → NCIB3610 |
| NRS2388 | trpC2 sacA::PyqxM-gfp (kan) | pNW602 → 168 |
| NRS2394 | sacA::PyqxM-gfp (kan) | SPP1 NRS2388 → NCIB3610 |
| NRS2547 | Δabh::cat sacA::PepsA-gfp (kan) | SPP1 NRS1894 → NRS2242 |
| NRS2549 | Δabh::cat sacA::PyqxM-gfp (kan) | SPP1 NRS1894 → NRS2394 |
| NRS2550 | sacA::PslrR-gfp-lacI (kan) | SPP1 NRS1968 → NCIB3610 |
| NRS2551 | Δabh::cat sacA::PslrR-gfp-lacI (kan) | SPP1 NRS1968 → NRS1900 |
| NRS2552 | Δabh::cat amyE::Phy-spank-abh-lacI (spc) sacA::PslrR-gfp-lacI (kan) | SPP1 NRS1968 → NRS1901 |
| NRS2553 | Δabh::cat amyE::Pabh-abh::spc sacA::PslrR-gfp-lacI (kan) | SPP1 NRS1968 → NRS1904 |
| NRS2556 | Δabh::cat amyE::Pabh-abh::spc sacA::PepsA-gfp (kan) | SPP1 NRS1434 → NRS2547 |
| NRS2557 | Δabh::cat amyE::Pabh-abh::spc sacA::PyqxM-gfp (kan) | SPP1 NRS1434 → NRS2549 |
| NRS2562 | amyE::Phy-spank-slrR-lacI sacA::PyqxM-gfp (kan) | SPP1 NRS1964 → NRS2394 |
| NRS2566 | amyE::Phy-spank-slrR-lacI sacA::PslrR-gfp (kan) | SPP1 NRS1964 → NRS2550 |
| NRS2571 | sigW::mls | SPP1 HB0803 → NCIB3610 |
| NRS2572 | sigX::spc | SPP1 HB0804 → NCIB3610 |
| NRS2574 | sigW::mls amyE::Phy-spac-abh-lacI (cat) | SPP1 NRS1937 → NRS2571 |
| NRS2575 | sigX::spc amyE::Phy-spac-abh-lacI (cat) | SPP1 NRS1937 → NRS2572 |
| NRS2576 | sigM::kan amyE::Pabh-gfp (cat) | SPP1 NRS1916 → NRS2570 |
| NRS2577 | sigX::spc amyE::Pabh-gfp (cat) | SPP1 NRS1916 → NRS2572 |
| NRS2578 | sigW::mls amyE::Pabh-gfp (cat) | SPP1 NRS1916 → NRS2571 |
| NRS2605 | Δabh::cat amyE::Phy-spank-slrR-lacI (spc)sacA::PepsA-gfp (kan) | SPP1 NRS1964 → NRS2547 |
| NRS2606 | Δabh::cat amyE::Phy-spank-slrR-lacI (spc)sacA::PyqxM-gfp(kan) | SPP1 NRS1964 → NRS2549 |
| NRS2716 | degU::pBL204 (cat) sacA::PslrR-gfp-lacI (kan) | SPP1 NRS1314 → NRS2550 |
Drug resistance cassettes are indicated as follows: cat, chloramphenicol resistance; kan, kanamycin resistance; mls, lincomycin-erythromycin resistance; spc, spectinomycin resistance.
The direction of strain construction is indicated with DNA or phage (SPP1) (→) recipient strain. BGSC is the Bacillus Genetic Stock Center.
Strain construction.
To delete abh, long-flanking homology PCR was used based on a method previously described (37). The region overlapping the translational start codon of abh was amplified using primers NSW504 (5′-CTGCGATCACGCCATCTTTCATCG-3′) and NSW505 (5′-GTTATCCG CTCACAATTCTGATTTCATAAAAACCCTTCTTCC-3′), and the region overlapping the termination stop codon of abh was amplified using primers NSW506 (5′-CGTCGTGACTGGGAAACAAAGAATAAAATTATGCTAAAAAAGGC-3′) and NSW510 (5′-CGATTTCCTGCAAATTATCCAATGATGCGG-3′). The translational start codon is indicated by a single underline, and the translational termination codon is indicated by double underline. Sequences in italics are homologous to the primer sequences used to amplify the chloramphenicol resistance gene. The chloramphenicol resistance gene was amplified from plasmid pCBB31 using primers NSW107 (5′-GTTTTCCCAGTCACGACG-3′) and NSW108 (5′-GAATTGTGAGCGGATAAC-3′). The purified fragments were combined in a PCR using primers NSW504 and NSW510 using LA Taq (TaKaRa). The fragment generated was transformed into B. subtilis JH642, and the resulting colonies were screened by PCR to ensure double recombination at the abh locus (NRS1894 Δabh::cat). The mutated abh gene was transferred to strain NCIB3610 by phage transduction, selecting for chloramphenicol resistance (strain NRS1900).
Plasmid construction.
Plasmid pBL154 was used to complement the Δabh strain by ectopic expression at the amyE locus using its native promoter. The abh coding region plus 172 bp of the upstream region was PCR amplified using primers BL189 (5′-GCGCGGATCCCAAGGAACTGTGTGTAAC-3′) and BL198 (5′-GCGCGAATTCTTATTCTTTTAAAGCGGC-3′) and ligated into pDG1730 (18) at the BamHI and EcoRI restriction sites. Ectopic expression of abh at amyE was placed under the control of two separate isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoters Phy-spank and Phy-spac using plasmid backbones pDR111 (8) and pPL82 (44), respectively. Plasmids pNW400 (pDR111 derived) and pNW403 (pPL82 derived) were generated by amplification of the abh coding region and the native ribosome binding site using primers NSW502 (5′-CGTAAAGCTTGGAAGAAGGGTTTTTATGAAATCAATAGGTGTT-3′) and NSW503 (5′-CGTAGCATGCTTATTCTTTTAAAGCGGCTTG-3′). PCR products were ligated into pDR111 and pPL82 at the HindIII and SphI restriction sites. Plasmid pNW402 was used to assay abh expression utilizing green fluorescent protein (GFP) production as a reporter that was driven by the upstream promoter region of abh (−500 bp relative to the A nucleotide of the abh translational start codon). The promoter region was amplified using primer pair NSW521 (5′-GCTAGAATTCCTGCGATCACGCCATCTTTCA-3′) and NSW522 (5′-CGTAAAGCTTAAAAACCCTTCTTCCTTTAAA-3′) and ligated into pBL165 using the EcoRI and HindIII sites (48). Plasmid pNW408 was constructed by PCR amplification of the upstream region of the slrR gene as defined by Chu et al. (12) using primer pair NSW710 (5′-GTCGAATTCCTAGACAATCGCATATAA TTCTTTG-3′) and NSW711 (5′-GTCAAGCTTCTAGAAATTCTCCTCTATTCCTGTCG-3′). The amplified fragment was cloned into vector pMF302 (15) at the EcoRI and HindIII restriction sites. The newly constructed plasmid was digested with EcoRI and BamHI to release the Pslr-gfp fragment that was ligated into pSac-Kan to allow integration at the sacA locus (39). Plasmid pNW602 was used to assay yqxM expression utilizing GFP production as a reporter that was driven by the upstream region of yqxM (−490 bp relative to the A nucleotide of the yqxM translational start codon). The promoter region was amplified using primers NSW620 (5′-GATGAATTCTCAAGTTAAATGGTATTGAT-3′) and NSW621 (5′-GATAAGCTTGTAAAACACTGTAACTTG-3′) and ligated into pBL165 using the EcoRI and HindIII sites (48) to generate pNW600. pNW600 was then digested with EcoRI and BamHI to release the PyqxM-gfp fragment that was ligated into pSac-Kan to allow integration at the sacA locus (39). Plasmid pNW502 was used to assay epsA expression utilizing GFP production as a reporter that was driven by the upstream region of epsA (−300 bp relative to the A nucleotide of the epsA translational start codon). The promoter region was amplified using primers NSW618 (5′-GTCGAATTCGAAATTCTCCTCTAATCCTG-3′) and NSW619 (5′-GATAAGCTTCATAGCCTTCAGCCTT-3′) and ligated into pBL165 using the EcoRI and HindIII sites (48) to generate pNW501. pNW501 was then digested with EcoRI and BamHI to release the PepsA-gfp fragment that was cloned into pSac-Kan to allow integration at the sacA locus (39). Plasmid pNW407 was used to induce slrR expression in an IPTG-dependent manner. Primers slrR-F (5′-CGTAAAGCTTAGAGGAGAATTTCATATTATGATTGGAAGAATTATCCG-3′) and slrR-R (5′-AAAAGCATGCTCATCTTCCCTTTGTTTTTAAAAAGGATTTG ACTTCATG-3′) were used to amplify the slrR gene and its native ribosome binding site. The PCR product was ligated into pDR111 (8) at the HindIII and SphI restriction sites. All plasmids generated were sequenced to ensure the absence of PCR-incorporated sequence errors.
Flow cytometry.
The fluorescence of strains harboring GFP promoter fusions was measured in biofilm-forming conditions after 18 h of incubation at 37°C using the previously described method (55). Single-cell fluorescence was directly measured on a BD FACSCalibur (BD Biosciences). Single cells were identified on the basis of forward and side scatter, while GFP fluorescence was analyzed using 488 nm excitation with detection at 530 ± 30 nm. Data were captured using Cell Quest Pro (BD Biosciences) and further analyzed using FlowJo software version 4.3. For data profiles that gave a normal distribution, the geometric mean fluorescence was used as a measure of gene expression. The final fluorescence value was generated by subtraction of the geometric mean generated for the autofluorescence of each strain's nonfluorescent parent. The number of GFP-positive cells was calculated as the number of cells that exhibited a fluorescence signal greater than that generated by their nonfluorescent parent.
Biofilm formation conditions and image analysis.
Biofilms were grown as described previously (7). After either 18 or 40 h of growth at 37°C, biofilm images were captured using a Leica MZ16 FA stereoscope using LAS software version 2.7.1. Scale bars depicted on each figure as a white or black bar represent the distance of 5 mm. Air-surface interface biofilms called pellicles were formed as described previously (7).
DNA footprinting analysis.
Purification of Abh and AbrB was conducted as previously described (49). The DNA fragment used in DNase I footprinting assays was the following: an EcoRI-HindIII fragment of approximately 350 bp (containing the slrR and epsA promoter regions) was amplified using primers NSW737 (5′-CGTGAATTCCAGCACGAATCTGTG-3′) and NSW739 (5′-CGTAAGCTTCAGCTGATTAATAGA-3′) from the chromosome of strain NCIB3610 and cloned into pUC19 to yield pNW420. The plasmid containing the fragment was linearized at a unique restriction enzyme site flanking the insert and labeled using [α-32P]dATP (Amersham) and the Klenow enzyme, followed by inactivation of the Klenow enzyme and the release of the singly end-labeled fragment via digestion at a unique site on its opposite flank. Labeled DNA fragments were purified using standard polyacrylamide gel electrophoresis and electroelution techniques. Protein binding buffer composition (1×) was 50 mM Tris (either pH 7 or pH 8), 10 mM MgCl2, 100 mM KCl, 10 mM 2-mercaptoethanol, and 100 μg/ml bovine serum albumin. We have previously determined that optimal Abh binding affinity occurs at pH 7 in vitro, whereas optimal AbrB affinity occurs in a broad plateau from pH 8 to above pH 9.5, with only a slight decrease (about twofold) seen for binding at pH 7 (5). Therefore, we performed the in vitro DNA binding reactions using AbrB at pH 8 and the Abh reactions at both pH 7 and pH 8. DNase I footprinting assays were performed at room temperature (22°C) and analyzed as described previously (50, 57).
Electrophoretic mobility shift assay (EMSA).
The binding conditions and target DNA were as described above for the footprinting analysis. Binding was allowed to proceed for 15 min at room temperature, and the reaction mixtures were loaded onto 6% polyacrylamide gels (1× Tris-borate-EDTA [TBE] buffer). After electrophoresis, the gels were dried, imaged using an Amersham Typhoon 9000 phosphorimager, and quantitated using ImageQuant software.
RESULTS AND DISCUSSION
Abh controls B. subtilis biofilm architecture.
We initiated this study with the aim of investigating whether the paralogue of the biofilm inhibitor AbrB (12, 21) Abh controlled biofilm formation based upon the knowledge that Abh and AbrB share an overlapping regulon (35, 49). We used rugose colony morphology and the formation of an air/surface interface biofilm called a pellicle as two independent indicators of biofilm formation (7). These types of biofilms have been shown to depend on the biosynthesis of the extracellular matrix (7, 55). All of our studies have been conducted using the undomesticated strain NCIB3610, as it forms more robust biofilms by comparison with isolates derived from the domesticated strain 168 (7). The morphologies of biofilms formed in the presence and absence of Abh are shown in Fig. 1. These images clearly show that the Δabh strain (NRS1900) displays reduced biofilm architecture by comparison to the wild-type strain. This can be complemented by ectopic expression of abh under the control of its own promoter (NRS1904). The alteration in biofilm formation is apparent in both the colony assay (Fig. 1A) and the pellicle assay (Fig. 1B). Additionally, deletion of abh from the domesticated B. subtilis strain, JH642, resulted in a threefold decrease in biofilm formation as measured using a crystal violet-based biofilm microtiter plate assay (data not shown). Further evidence to support our conclusion that Abh positively controls biofilm formation was generated as part of a systematic analysis of all known transcriptional regulators of B. subtilis strain ATCC 6051 (29). The impact of deletion on pellicle formation was assessed, and Abh was shown to promote pellicle formation by B. subtilis strain ATCC 6051 (29). In toto, we conclude that Abh is required for the formation of a wild-type biofilm by B. subtilis.
FIG. 1.
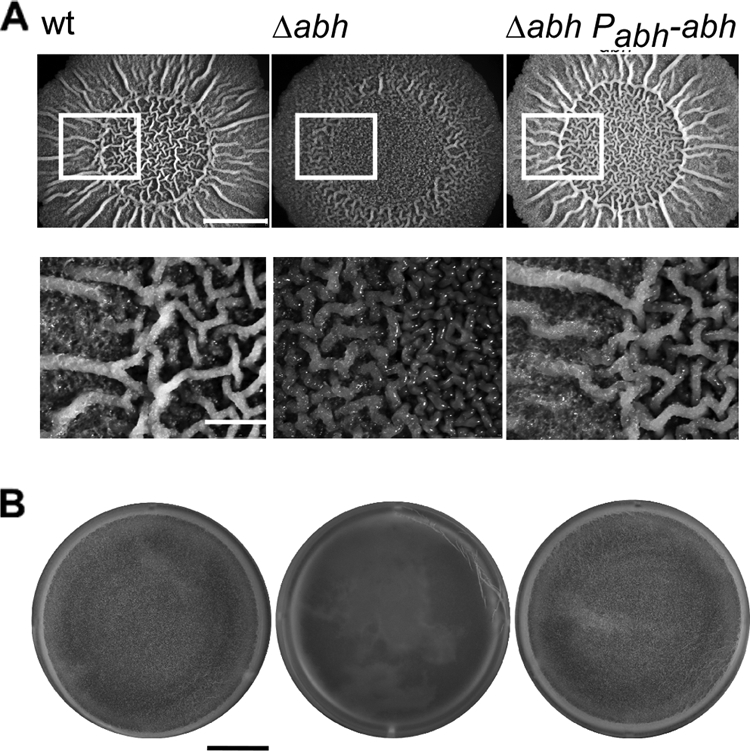
Abh controls biofilm architecture. (A) Representative images showing biofilm architecture after 40 h of growth at 37°C on MSgg medium. The wild-type (wt) strain (NCIB3610), Δabh (NRS1900), and Δabh+Pabh-abh (NRS1904) strains are shown. Bars, 5 mm (top images) and 1.25 mm (bottom images). (B) Pellicle morphology after 18 h growth at 37°C in MSgg medium. The wild-type (wt) strain (NCIB3610), Δabh (NRS1900), and Δabh+Pabh-abh (NRS1904) strains are shown. Bar, 8 mm.
σX activates abh expression.
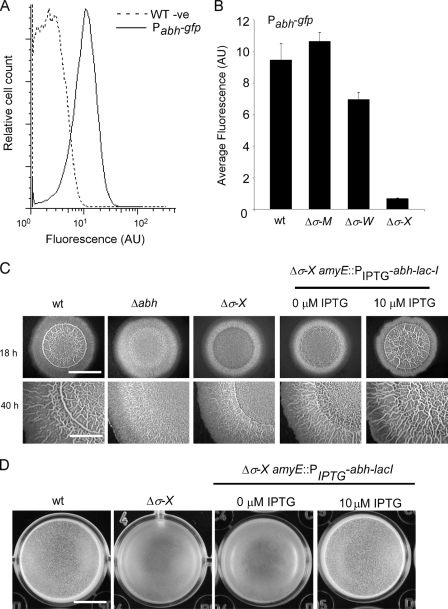
We were interested in establishing how transcription of abh was controlled under biofilm formation conditions. Using both in vitro and in vivo techniques, abh expression has previously been shown to be transcribed by the RNA polymerase core enzyme in the presence of the extracytoplasmic function sigma factor σX (26, 27). In vitro analysis also indicates that σW can activate abh transcription, although this has not been determined in vivo (26). More recently, it was proposed that abh transcription is activated by σM (14), raising the number of ECFs potentially responsible for abh transcription in vivo to three. We wanted to test which one of σX, σW, or σM had the dominant role in regulating abh transcription under biofilm formation conditions in the undomesticated strain NCIB3610. To do this, a Pabh-gfp reporter construct was generated (pNW402) and introduced into the wild-type NCIB3610 (NRS1917). The level of abh expression was measured in vivo during biofilm formation for each of the individual σ-factor mutants (σM, σW, and σX mutants) using flow cytometry. To compensate for any differences in background fluorescence, each strain carrying the Pabh-gfp construct was analyzed at the same time as a parent strain lacking the transcriptional fusion (see Materials and Methods). This ensured that the changes in fluorescence which were measured were not a result of changes in cell shape or some other phenotype. It was determined that Pabh-gfp displayed a narrow unimodal Gaussian distribution, indicative of transcription in all cells within the biofilm population (Fig. 2A). As Pabh-gfp expression followed a normal distribution, the geometric mean of the fluorescence value of the population of cells was used directly as a measure of gene expression. The expression of Pabh-gfp is σX dependent during biofilm formation, with ca. 18-fold reduction (P < 0.01) in Pabh-gfp expression being measured in the σX mutant by comparison with the wild type (Fig. 2B). The data also indicate that there is a small reduction in the level of abh transcription in the σW mutant while the σM mutant displays wild-type Pabh-gfp expression levels (Fig. 2B). These findings are slightly different from a recent study conducted using a laboratory isolate of B. subtilis where only upon deletion of both the σX and σM mutant genes was a large reduction in abh transcription observed (35). This highlights the strain-specific differences that exist between undomesticated and domesticated strains of B. subtilis and the value of analyzing different isolates of the same bacterial species.
FIG. 2.
σX activates abh expression. (A) Expression of Pabh-gfp in the wild-type strain (NRS1917) with the NCIB3610 wild-type strain as a nonfluorescent control. Average fluorescence is shown in arbitrary units (AU). WT -ve, wild-type negative control. (B) Expression of Pabh-gfp was measured in the wild type (wt) (NRS1917), σM mutant (Δσ-M) (NRS2576), σW mutant (Δσ-W) (NRS2578), and σX mutant (Δσ-X) (NRS2577). The values show the means plus standard errors of the means (error bars) from at least three independent experiments. In panels A and B, the cells were incubated for 18 h under biofilm formation conditions and analyzed using flow cytometry. (C) Representative images showing biofilm architecture for the wild-type (NCIB3610), Δabh (NRS1900), σX mutant (NRS2572), and σX mutant + PIPTG-abh-lacI (NRS2575) strains after 18 and 40 h of growth at 37°C. Bars, 5 mm. (D) Same as panel C) for the wild-type (NCIB3610), σX mutant (NRS2572), and σX mutant + PIPTG-abh-lacI (NRS2575) strains. Bar, 5 mm.
Artificial induction of abh transcription in the σX mutant restores wild-type biofilm architecture.
In good agreement with the observation that the σX mutant has a low level of abh transcription, the σX mutant displayed a reduction in the complexity of biofilm architecture that was comparable to that of the Δabh strain (Fig. 2C). Both the abh and σX mutant strains fail to develop the complex “raised bundles” and “veins” seen to develop in the wild type. Additionally, unlike the wild-type strain, the σX mutant was unable to from a robust and rugose pellicle (Fig. 2D). In contrast, the σM and σW mutants that had wild-type levels of abh transcription also displayed wild-type biofilm architecture and formed a rugose pellicle (data not shown). To confirm that the reduction in biofilm complexity observed in the absence of the σX mutant was a consequence of reduced abh expression, a copy of abh was placed under the control of a heterologous promoter and integrated into the amyE locus in the σX mutant strain (NRS2575). Induction of abh transcription with IPTG restored rugose wild-type biofilm architecture to the σX mutant and allowed robust pellicle formation (Fig. 2C and D). These results demonstrate that abh is the main downstream target of σX that is responsible for developing the mature structure of the biofilm formed by B. subtilis.
Consistent with our findings, B. subtilis biofilm formation has previously been shown to depend on the combined efforts of σX, σW, and σM which have overlapping regulons (36). However, in contrast to our results, the single σX mutants were reported to exhibit wild-type biofilm architecture (36). σX is known to be required for survival of B. subtilis at high incubation temperatures (25); therefore, we tested whether σX, and therefore Abh, control of biofilm architecture, was temperature dependent and grew biofilms at 22°C (room temperature) and 37°C. (Mascher and coworkers [36] used 22°C, while we used 37°C.) The impact of mutating either σX or abh was reduced with a reduction in the incubation temperature (data not shown). These results indicate that at higher incubation temperatures, σX, and therefore Abh, play a more crucial role in controlling the architecture of the biofilm. They also suggest that at lower temperatures, some other unknown factor can compensate for abh under these particular environmental conditions.
It is still unclear what stimulates σX activity, but activation of σX has been shown to be upregulated when genes predicted to be responsible for drug efflux, peptide uptake, sugar metabolism, and antimicrobial production are inactivated (52). These observations suggest that the signal(s) which stimulates σX activation may include the export of toxic molecules, presence of cell density signals, and production of antimicrobial compounds. This led to the proposal that σX was required for maintaining cell envelope homeostasis (52). Therefore, B. subtilis biofilm formation can be activated by Spo0A in response to surfactin (33) but additionally can influenced by an unknown signal controlling σX function which may be perturbations of cell envelope homeostasis. The roles of both of these regulatory pathways in controlling biofilm formation in ecological settings remain to be evaluated.
Biofilm matrix production is controlled by Abh.
To understand why biofilm architecture was impacted by the absence of Abh, we considered what effect deletion of abh would have on the expression of the yqxM and eps operons. The yqxM and eps operons are two loci in B. subtilis that are critical for the biosynthesis of the extracellular matrix (6). To determine the impact of deleting abh on expression, the upstream promoter region for both the yqxM and eps operons was fused independently to gfp and the level of fluorescence was measured using flow cytometry in the presence and absence of abh. The eps and yqxM promoters are bistable and therefore active in only a subpopulation of cells (10, 55). We found that deletion of abh did not affect the bistable profile from being established, but we saw that the number of GFP-positive cells was reduced in the absence of abh for both promoters (Fig. 3A and B). For the PyqxM-gfp construct, the number of GFP-positive cells decreased from 50% of total population after 18 h of incubation to 35% (P < 0.01), and for the Peps-gfp construct, it went from 68% to 47% (P < 0.01) (Fig. 3C). When considering only the active population of cells that produce each matrix component, these data indicate that in a Δabh biofilm, the number of cells initiating transcription of the loci required for matrix production is reduced ca. 33%. We chose to monitor expression after 18 h of incubation, as this time point corresponds to the peak in expression from these promoters under static incubation conditions (55; our unpublished findings). Ectopic expression of abh under the control of its native promoter in the Δabh strain negated the impact of deleting abh and increased the number of cells expressing the PyqxM-gfp and Peps-gfp fusion (Fig. 3C). Consistent with these findings, it has been shown in strain ATCC 6051 that deletion of abh inhibits the formation of cell clusters which have been shown to depend on the biosynthesis of the extracellular matrix (29). Therefore, we conclude that the altered biofilm architecture produced by the Δabh strain is a result of a reduction in the number of cells producing the biofilm matrix components, namely, TasA and the exopolysaccharide.
FIG. 3.
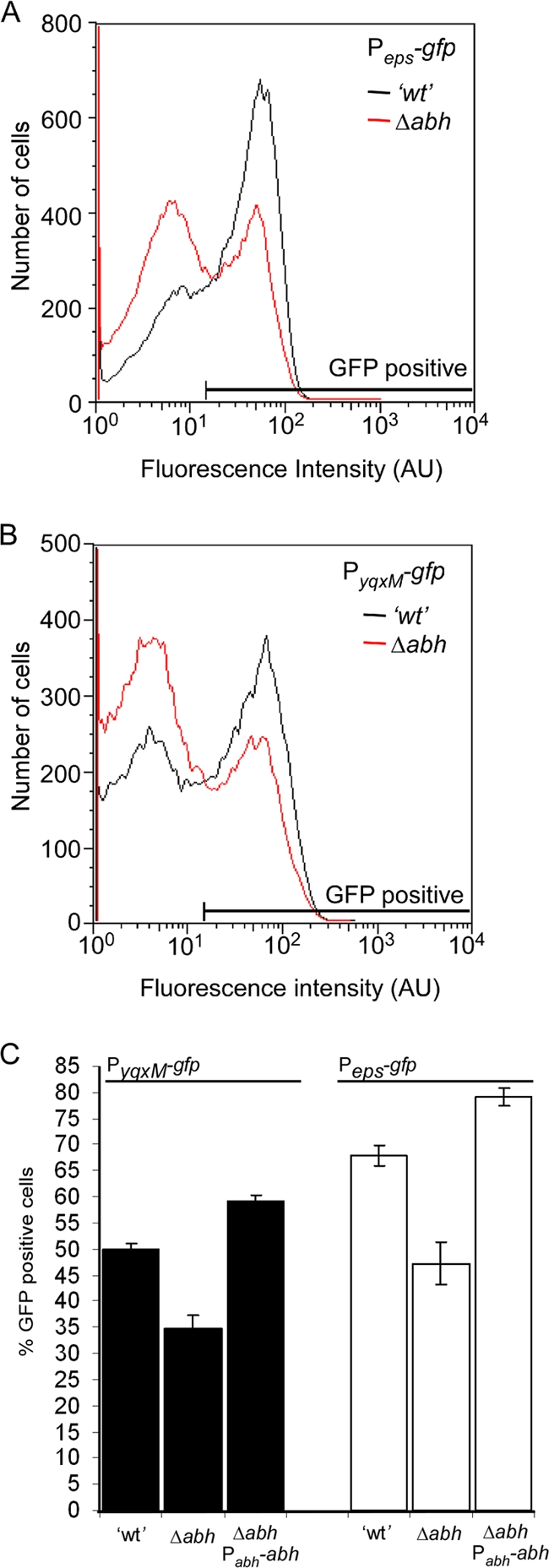
Abh activates transcription of the loci required for the synthesis of the biofilm matrix. PepsA-gfp (A) and PyqxM-gfp (B) expression was measured after 18 h of incubation under biofilm formation conditions using flow cytometry. Representative graphs of wild-type (′wt') (NRS2242 [Peps-gfp] and NRS2394 [PyqxM-gfp]) and Δabh (NRS2547 [Peps-gfp] and NRS2549 [PyqxM-gfp]) are shown. Fluorescence intensity is shown in arbitrary units (AU). (C) The average number of wild-type (NRS2242 [Peps-gfp] and NRS2394 [PyqxM-gfp]), Δabh (NRS2547 [Peps-gfp] and NRS2549 [PyqxM-gfp]) and Δabh + Pabh-abh (NRS2556 [Peps-gfp] and NRS2557 [PyqxM-gfp]) cells that were determined to be GFP positive are plotted. The values show the means and standard errors of the means (error bars) from at least three independent experiments.
Abh activates slrR transcription.
Although it is clear that Abh promotes the formation of an architecturally complex biofilm (Fig. 1) by controlling transcription of the eps and yqxM operons (Fig. 3), the mechanism by which expression of the eps and yqxM operons was controlled by Abh remained unknown. As part of an independent study, we conducted microarray analysis to identify genes regulated by Abh (to be published separately). During data analysis, it was noted that transcription of slrR was reduced in the absence of abh (1.8-fold ± 0.03-fold reduction [mean ± standard error of the mean]; n = 3; P = 0.05). Past data demonstrated that SlrR is an activator of biofilm formation which positively activates transcription from the promoter of the yqxM operon in B. subtilis strain NCIB3610 and positively influences transcription from the promoters of the yqxM and eps operons in B. subtilis strain ATCC 6051 (12, 31). Therefore, we proposed that Abh activation of slrR transcription could provide the link between Abh and its role in controlling biofilm architecture. To confirm whether Abh was an activator of slrR expression, the slrR promoter region (as specified by Chu et al. [12]) was fused to gfp and introduced into the sacA locus on the chromosome. Fluorescence generated by the PslrR-gfp construct was measured using flow cytometry. PslrR-gfp expression during biofilm formation was found to be very low in the wild-type strain, which is consistent with previous reports (12). We determined that slrR expression exhibited a normal Gaussian distribution pattern, indicating that slrR is transcribed uniformly in all cells during biofilm formation (data not shown). It was previously shown that deletion of sinR resulted in a ca. 25-fold increase in slrR expression (12); therefore, for a positive control, the PslrR-gfp expression level was measured during biofilm formation in a ΔsinIR strain (NRS2554). Deletion of both sinI and sinR in combination resulted in an approximately eightfold increase in PslrR-gfp expression (data not shown). Additionally, DegU, an activator of biofilm formation (40), has recently been shown not to influence slrR transcription (56); therefore, for a negative control, we deleted degU and measured the level of PslrR-gfp expression. Consistent with previous findings (56), no difference in the level of expression in the absence of degU was observed (P = 0.25) (Fig. 4). In contrast, and consistent with our DNA microarray analysis, when the PslrR-gfp fluorescence was measured in the Δabh strain (NRS2551), a statistically significant reduction in the level of slrR transcription was observed (1.5-fold reduction; P < 0.01) (Fig. 4). Given that the wild-type levels of slrR transcription are very low (12), this is a significant finding. Ectopic expression of abh using either the native or inducible promoter returned PslrR-gfp expression back to a level that was comparable to that of the wild-type strain, confirming that the reduction in fluorescence was specifically due to the lack of abh (Fig. 4). These data indicate that in addition to SinR and AbrB inhibiting slrR transcription (12), Abh functions to enhance slrR expression.
FIG. 4.
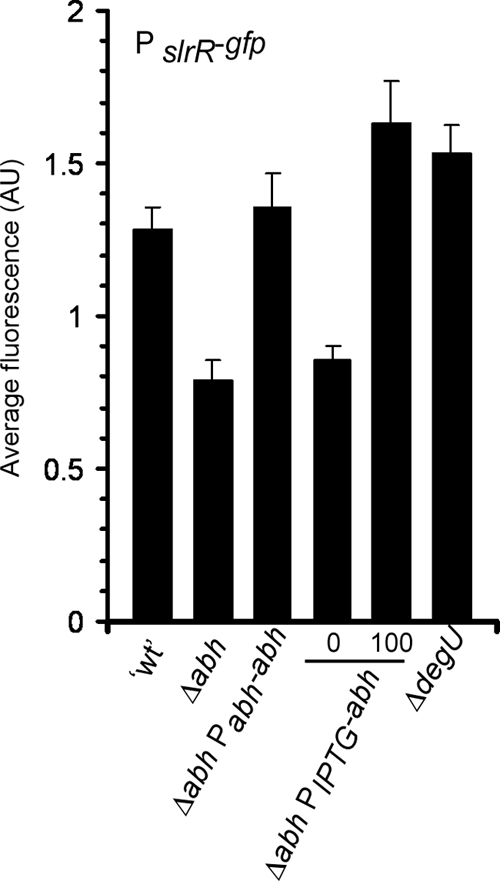
Abh activates slrR expression. Expression of PslrR-gfp was measured after 18 h of incubation under biofilm formation conditions at 37°C for the “wild type” (′wt') (NRS2550), Δabh mutant (NRS2551), Δabh Pabh-abh mutant (NRS2553), Δabh PIPTG-abh mutant (NRS2552), and degU mutant (NRS2716). Average fluorescence is shown in arbitrary units (AU). The level of IPTG added is shown below the bars (0 for 0 μM IPTG or 100 for 100 μM IPTG). The values show the means plus standard errors of the means (error bars) from at least three independent experiments.
Abh indirectly regulates slrR transcription.
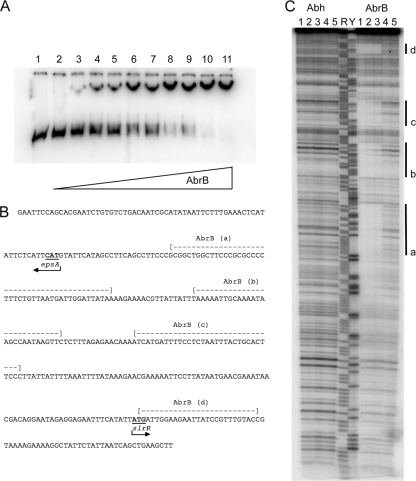
The data presented so far indicated that Abh influences expression of three loci required for biofilm formation (namely, the slrR gene and eps and yqxM operons) (Fig. 3 and 4). It should be noted that epsA and slrR are divergently transcribed, and therefore, the promoter regions are contained within the same fragment of intergenic DNA (Fig. 5B). It has been proposed that during control of sublancin production, Abh functions to relieve AbrB-mediated repression of transcription by outcompeting AbrB for shared regulatory binding sites in the promoter region (35). This “molecular displacement” model is unlikely to occur at the yqxM promoter, as Abh has not been found to bind to the yqxM promoter DNA in vitro (49). However, we wondered whether the molecular displacement model may account for the opposing influence of Abh and AbrB on transcription at the slrR and eps promoters. An inbuilt requirement for this model was the assumption that AbrB was able to bind to the DNA. While it is recognized that transcription of eps and slrR is repressed by AbrB, EMSAs using the region between the eps operon and the slrR gene and purified His-tagged AbrB did not demonstrate any binding interaction in vitro (12). It is our experience that when AbrB is epitope tagged, it is not as efficient at binding DNA as when it is purified unmodified (data not shown). Therefore, we chose to readdress the question of whether AbrB bound to the region between the eps operon and the slrR gene. To test this, we used EMSA analysis (Fig. 5A). The apparent dissociation constant under the conditions used (room temperature, pH 8, 100 mM KCl) was determined to be 0.65 μM ± 0.1 μM. Binding of AbrB to the region between the eps operon and the slrR gene was established as cooperative in nature, as less than 20% of AbrB was bound at 0.5 μM but greater than 80% of AbrB was bound at 0.9 μM (Fig. 5A). Having concluded that AbrB interacted directly with the region between the eps operon and the slrR gene, we used DNA footprinting analysis to test whether Abh could also bind to the DNA region between the eps operon and the slrR gene, and if so, whether the binding sites overlapped with those for AbrB.
FIG. 5.
Abh and AbrB regulate matrix production indirectly and directly, respectively. (A) Electrophoretic mobility shift assays demonstrate that AbrB binds to the region of DNA between the slrR gene and the eps operon. The concentration of AbrB is as follows: lane 1, 0 μM; lane 2, 0.1 μM; lane 3, 0.3 μM; lane 4, 0.38 μM; lane 5, 0.5 μM; lane 6, 0.6 μM; lane 7, 0.75 μM; lane 8, 0.9 μM; lane 9, 1.0 μM; lane 10, 2.0 μM; lane 11, 3.0 μM. (B) The DNA sequence used in the EMSA and DNA footprinting analysis is provided. The AbrB binding regions are indicated above the nucleotide sequence. The arrowheads indicate the translational start codon for both epsA and slrR. (C) DNA footprinting analysis of the region between the slrR gene and the eps operon. The concentrations of Abh and AbrB are as follows: lanes 1 and 2, 0 μM; lane 3, 0.3 μM; lane 4, 3 μM; lane 5, 30 μM. The four AbrB binding regions are labeled a to d and are highlighted by the thick black line. The nucleotides covered by the binding are shown in panel B. Maxam-Gilbert purine and pyrimidine sequence ladders are shown in lane R and lane Y, respectively, for reference.
The results of our analysis are shown in Fig. 5C and indicate that Abh does not bind to the region between the eps operon and the slrR gene. No regions of DNase protection were visible with increasing concentrations of Abh (Fig. 5C, lanes 3, 4 and 5). The purified Abh used in the experiments was confirmed to be active using a DNA target (C47) previously shown to bind purified Abh in vitro (data not shown) (4). In contrast, the DNA footprinting analysis supports the EMSA analysis and indicates that AbrB binds to the region between the eps operon and the slrR gene at four distinct regions (Fig. 5B and C, lanes 3, 4, and 5). These data clearly demonstrate that AbrB repression of eps and slrR transcription is directly mediated. The fact that Abh was found not to bind to either the region between the eps operon and the slrR gene (Fig. 5C) or to the promoter of the yqxM operon (49) indicates that Abh activation of matrix production is achieved through an indirect mechanism and therefore is different from that proposed for the regulation of sublancin production (35, 49).
Abh controls matrix production through regulation of slrR transcription.
There is conflicting evidence in the literature as to whether SlrR activates transcription from only the promoter of the yqxM operon or whether SlrR also activates expression from the promoter of the eps operon (compare reference 12 to reference 31). Our data demonstrate that deletion of abh results in a reduction in the number of cells that activate transcription from the operons required for matrix production (eps and yqxM) (Fig. 3), as well as demonstrating a reduction in the level of slrR transcription (Fig. 4). The simplest model that could be proposed to explain our findings in the absence of any direct interaction is that Abh indirectly influences slrR transcription (Fig. 4 and 5) and SlrR subsequently controls eps and yqxM transcription. Integral to this model is SlrR activation of eps expression. Therefore, it was necessary to clarify whether or not SlrR activated transcription from the eps promoter.
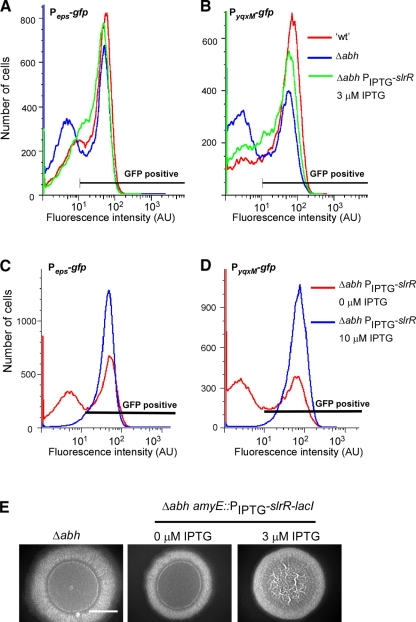
To determine whether SlrR influenced transcription of the eps operon, we chose to use flow cytometry with strains containing the Peps-gfp reporter fusion. This allowed us to examine expression at the single-cell level under biofilm formation conditions. We used strains that contained the PyqxM-gfp fusion as a positive control (12, 31). Strains were constructed that contained the Δabh mutation so that we could simultaneously determine whether the decrease in slrR transcription seen in the Δabh strain (Fig. 4) accounted for the reduced number of cells that expressed the operons required for matrix production (Fig. 3). The level of slrR transcription was artificially modulated by placing slrR under the control of the IPTG-inducible promoter at the amyE locus in strains NRS2605 (Δabh PIPTG-slrR-lacI Peps-gfp) and NRS2606 (Δabh PIPTG-slrR-lacI PyqxM-gfp) (Table 1). The number of cells expressing the matrix-encoding operons was calculated using flow cytometry after growth in the presence of 0 μM, 3 μM, and 10 μM IPTG for 18 h. The wild-type and Δabh strains served as controls (Fig. 6A and B). The data show that for both reporter fusions, the addition of 3 μM IPTG resulted in an increase in the number of cells expressing the operons required for matrix production. This was concluded as the profile observed by flow cytometry was comparable to that seen for the wild type (Fig. 6A and B). In contrast, at higher concentrations of IPTG (e.g., 10 μM and 100 μM IPTG [data not shown]), it was found that all of the cells in the biofilm activated transcription from the promoters of the eps and yqxM operons (Fig. 6C and D). These findings demonstrate that SlrR either directly or indirectly controls transcription from the eps operon and that small changes in the level of SlrR have a large influence on transcription.
FIG. 6.
Abh activates matrix production by activation of slrR. PepsA-gfp (A and C) and PyqxM-gfp (B and D) expression was measured after 18 h of incubation under biofilm formation conditions using flow cytometry. Representative graphs of wild type (′wt') (NRS2242 [Peps-gfp] and NRS2394 [PyqxM-gfp]) and Δabh mutants (NRS2547 [Peps-gfp] and NRS2549 [PyqxM-gfp]) and Δabh amyE::PIPTG-slrR mutants (NRS2605 [Peps-gfp] and NRS2606 [PyqxM-gfp]) are shown. The IPTG concentrations are indicated in the panels. Fluorescence intensity is shown in arbitrary units (AU). (E) Representative images showing biofilm architecture for the Δabh (NRS1900) and Δabh + PIPTG-slrR-lacI (NRS1966) strains after 18 h at 37°C. The IPTG concentrations are indicated. Bar, 5 mm.
Induction of PIPTG-slrR-lacI with 3 μM IPTG in the Δabh mutant restored the transcription profile with respect to both the Peps and PyqxM promoters back to the wild-type profile (Fig. 6A and B). Consistent with this, a low level of slrR induction was found to compensate for the Δabh mutation with respect to biofilm architecture. This was evident when slrR was induced with 3 μM IPTG, which restored the complex “vein”-like structures to the colony formed by the Δabh strain after 18 h of growth (Fig. 6E). (This was the time point at which matrix expression was assayed.) These findings confirm that matrix production is activated by Abh-mediated regulation of slrR transcription. Additionally, the data demonstrate that under biofilm-forming conditions, SlrR activates expression of both of the operons required for the biosynthesis of the protein (TasA) and polysaccharide components of the biofilm matrix.
Concluding remarks.
We have shown that Abh positively regulates biofilm architecture and that abh transcription is controlled in a σX-dependent manner under biofilm formation conditions in response to an unidentified signal. This suggests that upon release of σX into the cytoplasm by RsiX, its membrane-bound antagonist (25), σX can associate with the RNA polymerase core enzyme to activate abh transcription. We predict that Abh is the sole (or at least the major) downstream target of σX responsible for controlling biofilm architecture, since ectopic expression of abh in the σX mutant strain fully complements the σX mutant biofilm defect (Fig. 2C). The intramembrane-cleaving protease RasP has recently been shown to activate biofilm formation by B. subtilis (23). RasP is one of a number of proteases that is required to degrade RsiW, the σW anti-sigma factor. Degradation of RsiW results in the release of σW from the membrane and allows the genes in the σW regulon to be activated (47). It remains to be tested whether RasP also controls induction of σX, but if it does, this would be consistent with the biofilm phenotype of both the rasP and σX mutant strains (23).
The role for Abh in controlling biofilm architecture is the opposite of its paralogue AbrB which inhibits biofilm formation by binding to the promoter regions and blocking transcription from multiple promoters (34, 41, 54) (Fig. 5). We demonstrate that Abh controls the expression of three AbrB-repressed targets, the slrR gene and the eps and yqxM operons that are required for biofilm formation (Fig. 4 and 5A). We eliminate the possibility that Abh positively regulates biofilm architecture by counteracting AbrB-mediated repression exerted on the operons required for biosynthesis of the extracellular matrix since DNA footprinting analysis revealed that Abh does not bind directly to the region between the eps operon and the slrR gene (Fig. 5). This is consistent with previous findings indicating that Abh regulation of expression of the yqxM operon is indirectly mediated (49). Taken together with our findings that low levels of induction of slrR in the Δabh strain can complement matrix production and biofilm architecture, the data suggest that Abh exerts its regulatory control solely through indirect activation of slrR transcription.
There are several open-ended questions that arise as a consequence of this work. Starting at the top of the regulatory cascade, the environmental signal that activates σX has not been identified. Our data indicate that under specific environmental conditions the σX pathway will provide a route to stimulate biofilm formation. Downstream of σX, it remains to be determined how Abh influences slrR transcription as we know from DNA footprinting analysis that this is unlikely to be directly mediated. With respect to SlrR, it is unknown how SlrR activates transcription from the promoter of the eps operon, as the eps promoter region lacks the proposed SlrR binding site (12). Additionally, it is unknown why overexpression of slrR results in activation of eps and yqxM transcription in all of the cells in the biofilm. These findings indicate that SlrR is somehow capable of overriding the influence of SinR- and AbrB-mediated repression. It is also interesting to note that the regulatory network that controls slrR expression described by Kobayashi (31) is repressed by YwcC, a member of the TetR-like repressor family of transcriptional regulators (45). The TetR family of regulators are known to respond to antimicrobial agents that could also trigger σX activity (45). Thus, slrR transcription may be activated by two separate pathways which are activated upon sensing of antimicrobial agents that disrupt cell envelope homeostasis. In summary, our findings indicate that the production of the biofilm matrix is influenced by an as yet unidentified environmental stimulus that regulates the activity of the ECF σ-factor, σX. These findings highlight the extensive ways by which B. subtilis regulates the transcription of the operons required for biofilm formation.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (grant numbers BB/C520404/1 and BB/E001572/1).
We acknowledge Adel Ibrahim from the College of Life Sciences cloning service for constructing pNW407. We thank Kazuo Kobayashi and Beth Lazazzera for helpful discussions, Adam Ostrowski for pNW602, and John Helmann for generously sharing strains with us.
Footnotes
Published ahead of print on 18 September 2009.
REFERENCES
- 1.Bai, U., I. Mandic-Mulec, and I. Smith. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 7:139-148. [DOI] [PubMed] [Google Scholar]
- 2.Banse, A. V., A. Chastanet, L. Rahn-Lee, E. C. Hobbs, and R. Losick. 2008. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 105:15547-15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair, K. M., L. Turner, J. T. Winkelman, H. C. Berg, and D. B. Kearns. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636-1638. [DOI] [PubMed] [Google Scholar]
- 4.Bobay, B. G., A. Andreeva, G. A. Mueller, J. Cavanagh, and A. G. Murzin. 2005. Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins. FEBS Lett. 579:5669-5674. [DOI] [PubMed] [Google Scholar]
- 5.Bobay, B. G., G. A. Mueller, R. J. Thompson, A. G. Murzin, R. A. Venters, M. A. Strauch, and J. Cavanagh. 2006. NMR structure of AbhN and comparison with AbrBN: first insights into the DNA binding promiscuity and specificity of AbrB-like transition state regulator proteins. J. Biol. Chem. 281:21399-21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branda, S. S., F. Chu, D. B. Kearns, R. Losick, and R. Kolter. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59:1229-1238. [DOI] [PubMed] [Google Scholar]
- 7.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 10.Chai, Y., F. Chu, R. Kolter, and R. Losick. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 67:254-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu, F., D. B. Kearns, S. S. Branda, R. Kolter, and R. Losick. 2006. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 59:1216-1228. [DOI] [PubMed] [Google Scholar]
- 12.Chu, F., D. B. Kearns, A. McLoon, Y. Chai, R. Kolter, and R. Losick. 2008. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol. Microbiol. 68:1117-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl, M. K., T. Msadek, F. Kunst, and G. Rapoport. 1992. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 267:14509-14514. [PubMed] [Google Scholar]
- 14.Eiamphungporn, W., and J. D. Helmann. 2008. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita, M., J. E. Gonzalez-Pastor, and R. Losick. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 17.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 18.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 19.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9-17. [DOI] [PubMed] [Google Scholar]
- 20.Hamon, M. A., and B. A. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199-1209. [DOI] [PubMed] [Google Scholar]
- 21.Hamon, M. A., N. R. Stanley, R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2004. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol. Microbiol. 52:847-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 23.Heinrich, J., T. Lunden, V. P. Kontinen, and T. Wiegert. 2008. The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiology 154:1989-1997. [DOI] [PubMed] [Google Scholar]
- 24.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 25.Huang, X., A. Decatur, A. Sorokin, and J. D. Helmann. 1997. The Bacillus subtilis σX protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J. Bacteriol. 179:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, X., K. L. Fredrick, and J. D. Helmann. 1998. Promoter recognition by Bacillus subtilis σW: autoregulation and partial overlap with the σX regulon. J. Bacteriol. 180:3765-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, X., and J. D. Helmann. 1998. Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search. J. Mol. Biol. 279:165-173. [DOI] [PubMed] [Google Scholar]
- 28.Kearns, D. B., F. Chu, S. S. Branda, R. Kolter, and R. Losick. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 55:739-749. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi, K. 2007. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J. Bacteriol. 189:4920-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi, K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66:395-409. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, K. 2008. SlrR/SlrA control the initiation of biofilm formation in Bacillus subtilis. Mol. Microbiol. 69:1399-1410. [DOI] [PubMed] [Google Scholar]
- 32.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 33.Lopez, D., M. A. Fischbach, F. Chu, R. Losick, and R. Kolter. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 106:280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez, D., H. Vlamakis, and R. Kolter. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33:152-163. [DOI] [PubMed] [Google Scholar]
- 35.Luo, Y., and J. D. Helmann. 2009. Extracytoplasmic function σ factors with overlapping promoter specificity regulate sublancin production in Bacillus subtilis. J. Bacteriol. 191:4951-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mascher, T., A. B. Hachmann, and J. D. Helmann. 2007. Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function σ factors. J. Bacteriol. 189:6919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto, T., K. Nakanishi, K. Asai, and Y. Sadaie. 2005. Transcriptional analysis of the ylaABCD operon of Bacillus subtilis encoding a sigma factor of extracytoplasmic function family. Genes Genet. Syst. 80:385-393. [DOI] [PubMed] [Google Scholar]
- 39.Middleton, R., and A. Hofmeister. 2004. New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid 51:238-245. [DOI] [PubMed] [Google Scholar]
- 40.Murray, E. J., T. B. Kiley, and N. R. Stanley-Wall. 2009. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology 155:1-8. [DOI] [PubMed] [Google Scholar]
- 41.Nagórska, K., K. Hinc, M. A. Strauch, and M. Obuchowski. 2008. Influence of the σB stress factor and yxaB, the gene for a putative exopolysaccharide synthase under σB control, on biofilm formation. J. Bacteriol. 190:3546-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 43.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 44.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramos, J. L., M. Martinez-Bueno, A. J. Molina-Henares, W. Teran, K. Watanabe, X. Zhang, M. T. Gallegos, R. Brennan, and R. Tobes. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robertson, J. B., M. Gocht, M. A. Marahiel, and P. Zuber. 1989. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc. Natl. Acad. Sci. USA 86:8457-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schobel, S., S. Zellmeier, W. Schumann, and T. Wiegert. 2004. The Bacillus subtilis sigmaW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 52:1091-1105. [DOI] [PubMed] [Google Scholar]
- 48.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strauch, M. A., B. G. Bobay, J. Cavanagh, F. Yao, A. Wilson, and Y. Le Breton. 2007. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J. Bacteriol. 189:7720-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauch, M. A., V. Webb, G. B. Spiegelman, and J. A. Hoch. 1990. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. USA 87:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner, M. S., and J. D. Helmann. 2000. Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the σX and σW factors in Bacillus subtilis. J. Bacteriol. 182:5202-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verhamme, D. T., T. B. Kiley, and N. R. Stanley-Wall. 2007. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol. Microbiol. 65:554-568. [DOI] [PubMed] [Google Scholar]
- 54.Verhamme, D. T., E. J. Murray, and N. R. Stanley-Wall. 2009. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J. Bacteriol. 191:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vlamakis, H., C. Aguilar, R. Losick, and R. Kolter. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 22:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winkelman, J. T., K. M. Blair, and D. B. Kearns. 2009. RemA (YlzA) and RemB (YaaB) regulate extracellular matrix operon expression and biofilm formation in Bacillus subtilis. J. Bacteriol. 191:3981-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, K., and M. A. Strauch. 2001. DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein. J. Bacteriol. 183:4094-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshimura, M., K. Asai, Y. Sadaie, and H. Yoshikawa. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150:591-599. [DOI] [PubMed] [Google Scholar]