Abstract
The mechanisms controlling progression of conjugative DNA processing from a preinitiation stage of specific plasmid strand cleavage at the transfer origin to a stage competent for unwinding the DNA strand destined for transfer remain obscure. Linear heteroduplex substrates containing double-stranded DNA binding sites for plasmid R1 relaxosome proteins and various regions of open duplex for TraI helicase loading were constructed to model putative intermediate structures in the initiation pathway. The activity of TraI was compared in steady-state multiple turnover experiments that measured the net production of unwound DNA as well as transesterase-catalyzed cleavage at nic. Helicase efficiency was enhanced by the relaxosome components TraM and integration host factor. The magnitude of stimulation depended on the proximity of the specific protein binding sites to the position of open DNA. The cytoplasmic domain of the R1 coupling protein, TraDΔN130, stimulated helicase efficiency on all substrates in a manner consistent with cooperative interaction and sequence-independent DNA binding. Variation in the position of duplex opening also revealed an unsuspected autoinhibition of the unwinding reaction catalyzed by full-length TraI. The activity reduction was sequence dependent and was not observed with a truncated helicase, TraIΔN308, lacking the site-specific DNA binding transesterase domain. Given that transesterase and helicase domains are physically tethered in the wild-type protein, this observation suggests that an intramolecular switch controls helicase activation. The data support a model where protein-protein and DNA ligand interactions at the coupling protein interface coordinate the transition initiating production and uptake of the nucleoprotein secretion substrate.
Controlled duplex DNA unwinding is a crucial prerequisite for the expression and maintenance of genomes. Genome-manipulating and -regulating proteins are central to that biological function in recognizing appropriate DNA targets at initiation sequences and unwinding the complementary strands to provide single-stranded DNA (ssDNA) templates for nucleic acid synthesis and other processing reactions. The protein machineries involved include nucleic acid helicases. DNA helicases are powerful enzymes that convert the energy of nucleoside triphosphate hydrolysis to directional DNA strand translocation and separation of the double helix into its constituent single strands (for reviews, see references 13, 14, 16, 38, 55, and 64). By necessity, these enzymes interact with DNA strands via mechanisms independent of sequence recognition. At replication initiation helicases gain controlled access to the double-stranded genome at positions determined by the DNA binding properties of initiator proteins that comprise an origin recognition complex (1, 9, 17, 31, 45, 66). The mechanisms supporting localized unwinding within the complex include initiator-induced DNA looping, wrapping, and bending and feature regions of low thermodynamic stability. The exposed ssDNA mediates helicase binding followed by directional translocation along that strand until the enzyme engages the duplex for unwinding.
In the MOBF family of conjugation systems, the plasmid DNA strand destined for transfer (T strand) is unwound from its complement by a dedicated conjugative helicase, TraI of F-like plasmids or TrwC of the IncW paradigm. These enzymes are remarkable in that the same polypeptides additionally harbor in a distinct domain a DNA transesterase activity. That function is required to recognize and cleave the precise phosphodiester bond, nic, in the T strand where unwinding of the secretion substrate begins. In current models the conjugative helicases are thus targeted to the transfer origin (oriT) of their cognate plasmid by the high-affinity DNA sequence interactions of their N-terminal DNA transesterase domains. In the bacterial cell, recruitment and activation of the conjugative helicase occur not on naked DNA but within an initiator complex called the relaxosome (67). For the F-like plasmid R1, sequence-specific DNA binding properties of the plasmid proteins TraI, TraY, TraM, and the host integration factor (IHF) direct assembly of the relaxosome at oriT (10, 12, 29, 33, 51, 52). Integration of protein TraM confers recognition features to the relaxosome, which permit its selective docking to TraD, the coupling protein associated with the conjugative type IV secretion system (T4CP) (2, 15, 49). In current models, the T4CP forms a hexameric translocation pore at the cytoplasmic membrane that not only governs substrate entry to the envelope spanning type IV secretion machinery but also provides energy for macromolecular transport via ATP hydrolysis (36, 50). These models propose that T4CPs provide not only a physical bridge between the plasmid and the type IV transporter but also a unique control function in distinguishing one plasmid (relaxosome) from another (7, 8). Before the current study (see accompanying report [41]), evidence indicating that regulation of the initiation of conjugative DNA processing also takes place at this interface had not been reported.
F plasmid TraI protein, originally named Escherichia coli DNA helicase I, was initially characterized in the Hoffman-Berling laboratory (19). The purified enzyme exhibits properties in vitro consistent with its function in conjugative DNA strand transfer including a very high 1,100-bp/s rate of duplex unwinding, high processivity, and a 5′-to-3′ directional bias (relative to the strand to which it is bound) (34, 54). Together these features should readily support the observed rate of conjugative DNA translocation as well as concomitant replacement synthesis of the mobilized T strand from the 3′ OH product of nic cleavage.
Comparatively little is known about the mechanisms of initiating TraI helicase activity. The enzyme requires ssDNA 5′ to the duplex junction (32), and a minimum length of 30 nucleotides (nt) is necessary to promote efficient duplex unwinding on substrates lacking oriT (11, 54). To our knowledge, oriT is the only sequence where the helicase activity is naturally initiated, however. Moreover, the unique fusion of a helicase to the site- and strand-specific DNA transesterase domains within MOBF enzymes is expected to pose intriguing regulatory challenges during initiation. The combination within a single polypeptide of a site-specific DNA binding capacity with a helical motor activity would seem counterproductive. The extraordinary efficiency of these proteins in intercellular DNA strand transfer belies this prediction and instead hints strongly at a coordinated progression of the initiation pathway. Since relaxosome assembly is thus far insufficient to initiate helicase activity on supercoiled oriT substrates in vitro, we have developed a series of heteroduplex DNA substrates which support the unwinding reaction and model possible intermediate structures of R1 plasmid strand transfer initiation (10). In this system linear double-stranded DNA (dsDNA) substrates with a central region of sequence heterogeneity trap defined lengths of R1 oriT sequence in unwound conformation. Unexpectedly, efficient helicase activity initiated from a melted oriT duplex required ssDNA twice as long (60 nt) as that previously observed on substrates lacking this sequence (11).
In the current report, we describe an application of these models where variation in the position of duplex opening in the vicinity of nic, as well as the additional presence of auxiliary relaxosome proteins, has revealed novel insights into control of a conjugative helicase involving both DNA and protein interactions. Moreover, we observe a sequence-independent stimulation of the unwinding reaction in the presence of T4CP TraD. These results support a model where docking of the preinitiation relaxosome assembly to the T4CP alters the composition and architecture of the complex in a manner essential to the subsequent initiation of T-strand unwinding.
MATERIALS AND METHODS
Enzymes.
DNA-modifying reagents were used according to the manufacturers' recommendations. Lambda exonuclease was purchased from New England Biolabs. All restriction enzymes, S1 nuclease, T4 DNA ligase, and T4 polynucleotide kinase were provided by Fermentas. The thermally stable polymerase used for every application was Dynazyme II (Finnzymes). Pyruvate kinase and lactate dehydrogenase were provided by Roche. The R1 plasmid proteins TraM, TraY, and TraD; full-length TraI; the 34-kDa transesterase domain TraIN309; and E. coli IHF were purified as described previously (41). A 158-kDa truncated version of R1 TraI lacking the N-terminal DNA transesterase domain (TraIΔN308) was overexpressed from pAR45 (described below). Cultures (eight of 600 ml) of E. coli BL21 C41(DE3) (42) carrying pAR45 were grown at 37°C in LB medium with 100 μg/ml ampicillin. Isopropyl-1-thio-α-d-galactopyranoside was added to 1 mM at a culture density of an A600 of 0.5 to 0.6. After 5 h of shaking at 30°C, the cells were harvested by centrifugation. The pellet was resuspended in 100 ml of 20 mM spermidine, 2 mM EDTA, 200 mM NaCl, and frozen at −80°C. Prior to purification, the cells were thawed on ice for 2 h. Six milliliters of lysis buffer per 1 g of cells was added to this solution to achieve final concentrations of 50 mM Tris-HCl (pH 7.6), 30 mM NaCl, 5% sucrose, 0.15% (vol/vol) Brij 58, 0.1 mM phenylmethanesulfonyl fluoride (PMSF), and 0.5 mg/ml lysozyme. After 1 h at 0°C, the lysis mixture was centrifuged at 100,000 × g for 90 min at 4°C. The supernatant was collected and slowly precipitated with 30% solid ammonium sulfate with constant stirring on ice. After addition, the mixture was stirred at 0°C for 30 min and then centrifuged at 27,000 × g for 35 min at 4°C. The pellet was resuspended in 20 ml buffer I (20 mM Tris-HCl [pH 7.6], 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 50 mM NaCl, 0.1 mM PMSF, 10% [vol/vol] glycerol) and dialyzed overnight against a 100-fold volume of buffer I at 4°C. The dialyzed sample was loaded onto two 5-ml heparin HP columns connected in tandem (Amersham Bioscience) and equilibrated with buffer I. The column was developed with a 0.05 M to 1.2 M gradient of NaCl in buffer II (20 mM Tris-HCl [pH 7.6], 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 10% glycerol). Fractions containing TraIΔN308 eluted at 410 mM NaCl. Soluble ammonium sulfate was added to the pooled peak fractions to a 1 M final concentration. This solution was loaded onto two 5-ml phenyl Sepharose HP columns connected in tandem (Amersham Bioscience) and equilibrated with buffer III [20 mM Tris-HCl (pH 7.6), 0.1 mM EDTA, 1 M (NH4)2SO4, 1 mM DTT, 0.1 mM PMSF, 10% glycerol]. The protein was eluted with a decreasing 1.0 to 0.0 M gradient of (NH4)2SO4 in buffer IV (20 mM Tris-HCl [pH 7.6], 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 10% glycerol). TraIΔN308 eluted at 350 mM (NH4)2SO4. The peak fractions were dialyzed against buffer IV overnight at 4°C and concentrated with Amicon filter devices (Millipore). The concentrated protein was supplemented with glycerol to 40% (vol/vol) and stored at −80°C. The purity of the protein was 98%, and the apparent molecular mass of 158 kDa was confirmed by Coomassie blue staining following denaturing polyacrylamide gel electrophoresis. The protein concentration was estimated using the Bradford assay (Bio-Rad). The ATP hydrolysis activity of the purified fractions was compared to that of the full-length TraI protein using the pyruvate kinase-lactate dehydrogenase-coupled enzyme assay (22) as described previously (59).
DNA constructions.
Oligonucleotides used are listed in Table 1. To generate different heteroduplex DNA substrates containing distinct 60-nt stretches of sequence heterogeneity, templates for generating modified strands complementary to the R1 T strand were constructed by replacing each of the desired sequences of pDE100 with a nonhomologous region. Exchange of the oriT DNA required two independent PCRs. According to the strategy described previously (10), each reaction created one arm of the cDNA and half of the planned DNA heterogeneity including a (22-nt) region of terminal overlap with the product of the second PCR. All noncomplementary sequences were present in a 5′ extension of the primer; thus, the identity of the oriT sequences at the primer 3′ end determined the position of oriT replacement in the final constructions. In the first PCR 20 ng pDE100, 0.002 μM of pBST-std (10), and 0.2 μM of reverse (IR, NIC, and G2028) primers were used. The second PCR mixture contained 0.002 μM of pBSR-std (10) and 0.2 μM of forward primer IR, NIC, or G2028, respectively (Table 1). The reactions were started by adding 1 U of Dynazyme II. Conditions for amplifying pde160NIC and pde160G2028 were 94°C for 3 min; 30 cycles of 94°C for 30 s, 60°C for 40 s, and 72°C for 1.5 min; and 72°C for 5 min. A 50°C annealing temperature was used for pde160IR. The products were gel purified and eluted in distilled water, and the proper forward and reverse pairs were hybridized via their complementary extension region in the presence of 2 U Dynazyme II and 200 μM of each deoxynucleoside triphosphate at 94°C for 1.5 min, 94°C for 30 s, 64°C for 1 min, and 72°C for 2 min. The terminally overlapping fragments anneal in each cycle and were extended outward in both directions to fill in the remaining duplex arms. After eight cycles of hybridization, 0.5 μM of the primers pBST-st and pBSR-st was added to each reaction mixture and the newly created DNA was amplified at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 64°C for 40 s, and 72°C for 1.5 min, and finally at 72°C for 5 min. The final PCR products were digested with 5 U of EcoRI and PstI before ligation with pBluescript II KS(−) (Stratagene). Sequences of the resulting constructions pde160IR, pde160NIC, and pde160G2028 were verified.
TABLE 1.
Primers used during the DNA heteroduplex preparations
| Namea | Nonhomologous 5′-extension regionb | Homology to oriT sequencec |
|---|---|---|
| NIC fwd | 5′-CACAGCCGGATTTTGATAATGTCGCGAACACGCTGATC | GTTGAGCCTTTTTGTGGAGT-3′ (binds to 2009 to 1990) |
| NIC rev | 5′-ACATTATCAAAATCCGGCTGTGTCAGGCACTCCTTCCACAAATC | TTTGCTATTTGAATCATTAACTTAT-3′ (binds to 2070 to 2094) |
| IR fwd | 5′-CACAGCCGGATTTTGATAATGTCGCGAACACGCTGATC | CCACCAACCTGTTGAGCCT-3′ (binds to 2020 to 2002) |
| IR rev | 5′-ACATTATCAAAATCCGGCTGTGTCAGGCACTCCTTCCACAAATC | GAATCATTAACTTATGTTTTAAATAATGT-3′ (binds to 2081 to 2109) |
| G2028 fwd | 5′-CACAGCCGGATTTTGATAATGTCGCGAACACGCTGATC | GGAGTGGGTTAAATTATTTACG-3′ (binds to 1995 to 1974) |
| G2028 rev | 5′-ACATTATCAAAATCCGGCTGTGTCAGGCACTCCTTCCACAAATC | ACAAGTTTTTGCTGATTTGC-3′ (binds to 2056 to 2075) |
| UtraIHel fwd | 5′-ACGAATTCATATGCCAGCTTCACAGGACG-3′ | Binds to 922 to 938 in the traI gene (AY423546) |
| UtraIHel rev | 5′-TTCACACAGGAAACAGCTATGACC-3′ | Binds to 835 to 811 in pBluescript II SK(−) |
fwd, forward primer; rev, reverse primer.
Underlined sequences represent complementary termini where first and second PCR products hybridize.
See reference 19.
Expression construction pAR45 carries a 4.3-kb truncated traI gene of plasmid R1 (GenBank accession no. AY423546) under the control of the Ptac promoter. pAR45 was constructed in two steps: a 5.4-kb SphI fragment lacking the first 478 bp of traI coding sequence but containing the remaining traI gene plus the 5′ portion of the downstream gene traX was isolated from pHP2 (68) and ligated in pMS470Δ8 (18) linearized with SphI to generate pAR31 (10.7 kb). To introduce an ATG start codon immediately upstream of codon 309 of traI, pAR31 was cut with NdeI (within the pMS470Δ8 vector) and ClaI (removing the 5′ end of traI to codon 360). In step 2 the coding region for amino acids 309 to 360 was reconstructed by PCR amplification from a subclone carrying the first 1,082 bp of traI. The forward UtraIHel primer created an NdeI site and the start codon. The correct sequence of the PCR-derived traI region present in the resulting pAR45 construction was verified.
The heteroduplex substrates were generated by hybridization of a radiolabeled oriT-containing T strand and the (partially) complementary unlabeled R-strand DNA as described previously (10). In every case, a 1,059-bp AlfIII/PvuI fragment of pDE100 provided the T-strand template, and the R strand was synthesized from constructions where specific oriT segments were replaced with heterogeneous sequences (pde160NIC, pde160IR, and pde160G2028). Each of the desired strands was generated independently first as a dsDNA product using 0.2 μM of pBST-std and pBSR-std primers (Table 1) and conditions described previously (10). T strand for each substrate was radiolabeled with [α-32P]dATP during synthesis. Selective degradation of the unwanted complementary strand was achieved in the same reaction buffer by subsequent treatment with 10 U lambda exonuclease for 30 min at 37°C. Specificity of strand degradation was determined by phosphorylation of the priming oligonucleotide with T4 polynucleotide kinase prior to amplification. After digestion, DNA was purified using Qiagen PCR purification columns. Yield of ssDNA products was determined by agarose gel electrophoresis. The heteroduplex substrates were created as described previously (10) except that the radiolabeled T-strand products were combined with a twofold excess of unlabeled R strand in 10 mM Tris-HCl (pH 8.0) and 200 mM NaCl. Verification of the resulting open duplex structure was performed with S1 nuclease, NdeI, and SalI treatment.
T-strand cleavage and unwinding.
T-strand cleavage and unwinding were assayed as described previously (10). Briefly, the 20-μl reaction mixture contained 12.5 ng heteroduplex DNA and no or various concentrations of effector proteins TraM, TraD, TraY, and IHF independently or in combination, as indicated. When present, effector proteins were maintained in the mixture for 5 min at room temperature prior to initiation of the reaction by addition of either full-length TraI or TraIΔN308 protein. The reaction was terminated after 20 min at 37°C by addition of 0.9 mg/ml of proteinase K, 22.5 mM EDTA, and 0.1% sodium dodecyl sulfate (final concentrations) and further incubation at 37°C for 20 min. Reaction products were combined with 0.1 volume of loading dye (40% glycerol, 0.1% bromophenol blue) and resolved electrophoretically through 1.4% agarose in Tris-borate-EDTA buffer at 5 V/cm for 2.5 h. Radiolabeled DNA products were visualized with a Typhoon 9400 following autoradiography of the dried gel. Data were quantified using ImageQuant software (Molecular Dynamics). Statistical analysis was routinely performed using a one-sided Student t test (Systat), and significance is indicated in the legends to figures. Data describing total unwinding or cleaving and unwinding of input DNA as a function of variation in single protein concentrations were fitted to hyperbola or sigmoidal or peak curves using SigmaPlot 11.0 software to produce the plot lines shown graphically.
RESULTS
Heteroduplex substrates model an intermediate stage of DNA transfer initiation.
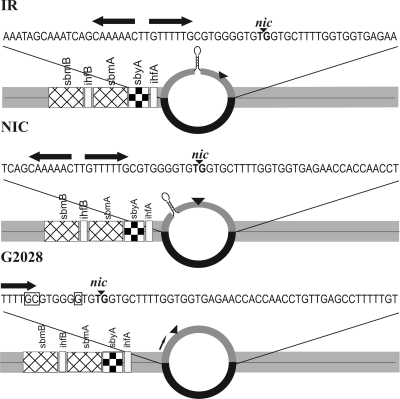
In earlier work we defined the ssDNA length requirements for TraI helicase loading at oriT by varying the extent of duplex opening at nic within heteroduplex DNA substrates (10). We found that the TraI helicase requires at least 60 nt in single-stranded form to enter and unwind duplex DNA efficiently. To date, the actual position of localized melting where helicase loading occurs during bacterial conjugation remains unknown (41). In this in vitro study, we explore whether the sequence composition of opened duplex affects TraI activities alone and in the additional presence of conjugation proteins. To this end, three heteroduplex substrates that maintained a constant length of R1 T-strand DNA in open conformation but shifted the position of unwinding were created. As a result, the segment of T-DNA exposed in single-stranded form and the distance separating that DNA from the binding sites for the additional relaxosome proteins varied. As illustrated in Fig. 1, the wild-type sequence of R1 T-strand DNA was present in all substrates but the central position of non-cDNA in the opposite strand of the heteroduplex was varied relative to nic (NIC), the inverted repeat (IR) spanning from G2050 to C2067, or the nic-distal nucleotide G2028 (numbering according to the work of Graus et al. [20]). This strategy was chosen to control the accessibility of bases within the inverted repeats that are key to ssDNA recognition by the TraI transesterase domain (65). In the IR substrate, the nic-proximal end of the inverted repeat is located at midpoint. Thus, the left arm of the repeat is 13 nt removed from the left duplex junction and nic lies 20 nt upstream from the right junction. In the NIC heteroduplex, the cleavage site assumes the central position such that the left arm of the inverted repeat adjoins the left duplex and 30 nt separate nic from the right duplex junction. The position of noncomplementarity in substrate G2028 begins in the middle of the right arm of the inverted repeat. Sequences proximal to this site are presumably able to base pair with their complementary strand in duplex DNA and may not or only transiently extrude a hairpin under our experimental conditions. This substrate thus exposes as ssDNA a minimum of bases 5′ to nic, which are known to mediate high-affinity binding by the TraI transesterase domain (65). The nick site of substrate G2028 is embedded in ssDNA 45 nt from the right duplex junction.
FIG. 1.
Heteroduplex substrates model intermediate DNA structures of transfer initiation. In each substrate 1,059 nt of the plasmid R1 T strand including oriT are hybridized to a complementary strand that contains additionally an identical 60-nt piece of non-cDNA (black). The extent of duplex opening in all substrates is kept constant while the composition of open sequence and its position relative to binding sites for relaxosome auxiliary proteins IHF (ihfA and ihfB), TraY (sbyA), and TraM (sbmA and sbmB) vary. The R1 T-strand sequence exposed as ssDNA is shown above each substrate. Functional features include the relaxase cleavage site, nic (black inverted triangle); inverted repeats (black arrows) upstream from nic; and a hairpin presumably formed when the inverted repeats are present in the open duplex (stem-loop structure on IR and NIC). Bases critical to sequence recognition by relaxase in the absence of the inverted repeat (65) are boxed (shown only for G2028).
Each substrate similarly harbors the specific sites of DNA binding for each auxiliary relaxosome protein in double-stranded form and in their native organization relative to each other and to nic, as illustrated in Fig. 1. The important variation between substrates, however, is the distance between the ssDNA available for TraI binding and the sites occupied by the auxiliary relaxosome components. For example, the 3′ border of the nearest auxiliary binding site, the consensus sequence ihfA, is positioned −1, −11, or −25 bp from the expected junction with unwound DNA for the IR, NIC, and G2028 substrates, respectively. Given that the reconstitution in vitro of IncF and IncW relaxosomes on supercoiled DNA has thus far not led to initiation of helicase activity (41), we still know little about localized melting of the transfer origin and loading of a conjugative helicase. The heteroduplex substrates described here aim to provide a plausible model for intermediates of R1 plasmid strand transfer initiation and reveal insights into control of a conjugative helicase on the level of both DNA and protein interactions.
DNA helicase activity varies with the position of duplex opening.
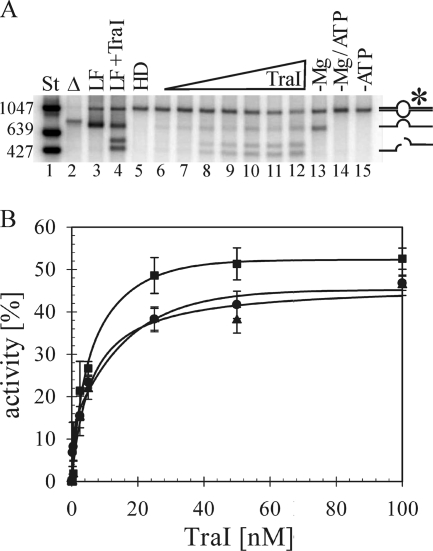
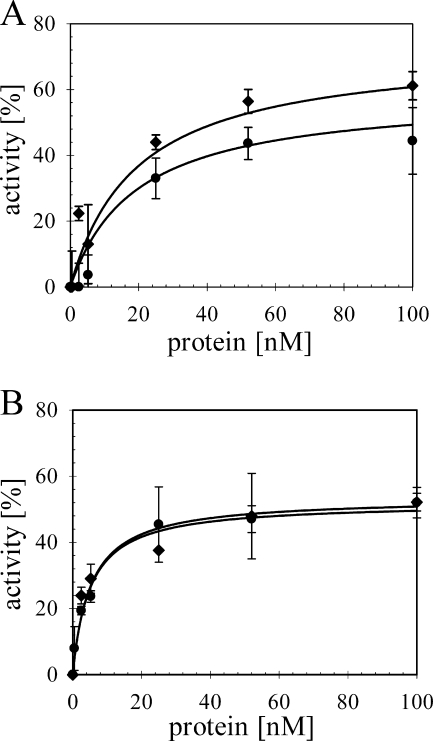
The relative efficiencies of TraI-catalyzed nic cleavage and DNA unwinding were compared on the series of heteroduplex substrates (Fig. 2). As expected for this assay (10), unwinding of the radiolabeled T strand was dependent on the presence of ATP (Fig. 2A, lane 15). Detection of the cleavage reaction at nic required a divalent cation (Fig. 2A, lanes 13 and 14). Given that the reaction products are resolved under nondenaturing conditions, visualization of the nicking reaction requires additionally the unwinding of the T strand; thus, no cleaved product was visible in the absence of ATP (Fig. 2A, lanes 14 and 15). The extent of the reaction is defined as the fraction of substrate unwound when the plateau of activity was reached. The ratio of cleaved and unwound T-DNA relative to unwound product was the same for each substrate. Maximal activity was observed on all heteroduplex substrates at 52 nM TraI (Fig. 2B), which corresponds to a ratio of 12 TraI monomers per duplex junction, also in good agreement with earlier work on substrates offering the same length of open duplex (10). Notably, at that concentration of TraI, 51% of substrate G2028 was unwound, whereas the yield of single-stranded product was significantly lower at 38% and 41% from IR and NIC, respectively. These results reflect the mean of three experiments using two independent preparations of each heteroduplex. Thus, a significant substrate-specific difference in the net production of unwound DNA was consistently observed with heteroduplex G2028 compared to NIC and IR.
FIG. 2.
TraI helicase activity varies when the sequence composition of the open duplex is altered. (A) An autoradiograph of a representative gel separating the TraI-catalyzed products of nic cleavage and unwinding of radiolabeled T strand (*) from a heteroduplex DNA substrate. For length comparison, specific dsDNA standards were generated via PCR with 5′-radiolabeled oligonucleotides (lane 1). Linear ssDNA not yet hybridized to heteroduplex was heat denatured (lane 2) or loaded directly without (lane 3) or following incubation with (lane 4) 100 nM TraI. Products generated from a heteroduplex substrate and no TraI (lane 5) or increasing concentrations of TraI—0.25 nM (lane 6), 0.5 nM (lane 7), 2.5 nM (lane 8), 5.2 nM (lane 9), 25 nM (lane 10), 52 nM (lane 11), and 100 nM (lane 12)—were resolved. Products of reaction mixtures containing 100 nM TraI but lacking magnesium (lane 13), ATP (lane 15), or both (lane 14) are shown. The positions of bands corresponding to starting material, as well as products of unwinding and both unwinding and cleavage, are shown schematically on the right. (B) The net production levels of unwound and cleaved T strand from three heteroduplex substrates, IR (•), NIC (▴), and G2028 (▪), with increasing concentrations of TraI were compared. The efficiency of TraI activity is expressed as the percentage of input DNA converted to the sum of unwound and unwound and cleaved products in the reaction. Values represent the means of four experiments using two independent preparations of each substrate. Standard deviations are shown.
Removing the TraI DNA transesterase domain equalizes helicase efficiency.
In accordance with their function, DNA helicases do not interact with DNA in a sequence-specific manner. Yet fusion of the TraI helicase to an N-terminal DNA transesterase domain enables high-affinity binding of TraI to specific single-stranded sequences in the vicinity of nic (25, 26, 35, 56, 65). In this assay, we analyze TraI activity in steady-state multiple turnover experiments that measure the net production of separated DNA strands as well as cleavage of the T-strand nick site. Importantly, the multiple turnover cycle is expected to comprise successive steps including enzyme binding, dissociation, rebinding, rates of translocation, and duplex unwinding. Variation at any step could alter the net production of unwound DNA. It is reasonable to hypothesize, therefore, that the reduced activity exhibited by full-length TraI on heteroduplex IR and NIC relative to G2028 may result from high-affinity binding by the DNA transesterase domain to T-strand sequences uniquely accessible in those substrates. In this model it follows that tight binding by the TraI N terminus would affect a subsequent step delaying activation of helical motor activity.
A second plausible explanation would be independent of specific sequence recognition by the protein and instead reflects the potential for the nic-proximal inverted repeat, which is presumably exposed as ssDNA in both IR and NIC, to form a stem-loop structure under these conditions (Fig. 1). If so, the length of single-stranded T DNA available to load the helicase would be less in the case of IR (29 nt) or NIC (39 nt) than in the case of G2028 (60 nt). The observed difference in overall unwinding efficiency might then reflect poor helicase loading.
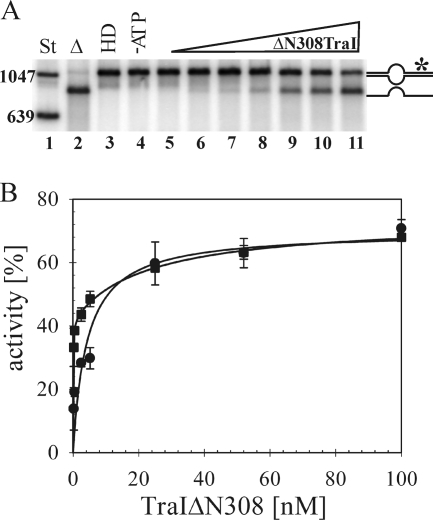
To differentiate between these possible explanations, we compared the relative efficiencies of the TraI helicase domain on the heteroduplex substrates in the absence of the DNA transesterase domain. The catalytic domains of the IncF TraI protein and the related TrwC protein from plasmid R388 can be separated physically and still exhibit biochemical activities in vitro equivalent to those of their full-length counterparts (6, 37, 57, 60). If the observed difference in helicase efficiency observed with these substrates reflects limited ssDNA for efficient loading, then a truncated form of TraI that maintains helicase activity but lacks the DNA transesterase domain should exhibit the same lowered activity on these DNAs as that of the full-length TraI. A 158-kDa form of TraI lacking the first 308 amino acids (TraIΔN308) was overexpressed and purified to homogeneity. The DNA-dependent nucleoside triphosphate-hydrolyzing activities of the helicase domain and full-length TraI were in reasonable agreement with each other (Km = 0.27 mM versus 0.22 mM; Vmax = 10,000 versus 13,736 mol ATP mol−1 TraI min−1) and prior results (32). Moreover, on partial (639-bp) duplex fragments that provide a 5′ loading strand but lack the R1 oriT sequence, the two forms of TraI exhibited equivalent helicase activities (not shown). The effects of increasing concentrations of the TraIΔN308 helicase domain on heteroduplexes G2028 and IR were then compared (Fig. 3). As expected, no cleaved product was observed in the absence of the TraI transesterase domain (Fig. 3A). In contrast to the performance of the full-length protein, however, no significant differences in helicase activity were detected for these heteroduplex DNAs using the independent helicase domain. Equivalent activity maxima were obtained with both heteroduplex DNAs at a concentration of 52 nM protein (Fig. 3B). A modest shift in the activity curve plotted for heteroduplex G2028 is apparent in the data summarized in Fig. 3 (n = 3 experiments), and yet this difference was not statistically significant when additional experiments were analyzed (n = 6). In summary, the TraI helicase domain does not exhibit a substrate-specific difference in efficiency similar to that of the full-length protein. Moreover, the potential for hairpin extrusion, which distinguishes these DNAs, does not appear to limit the efficiency of helicase loading under these conditions.
FIG. 3.
Unwinding efficiency of the helicase domain TraIΔN308 is substrate independent. (A) A representative autoradiogram detects products of TraIΔN308-catalyzed strand separation. Radiolabeled dsDNA fragments of known length were used as a standard (lane 1). DNA substrate was heat denatured (lane 2), loaded directly (lane 3), or incubated with 100 nM TraIΔN308 in the absence of ATP (lane 4) or with cofactor and increasing concentrations of enzyme: 0.25 nM (lane 5), 0.5 nM (lane 6), 2.5 nM (lane 7), 5.2 nM (lane 8), 25 nM (lane 9), 52 nM (lane 10), and 100 nM (lane 11). The positions of bands corresponding to heteroduplex (*) as well as unwound T strand are shown schematically on the right. (B) The extents of the reactions catalyzed by various concentrations of TraIΔN308 on substrates IR (•) and G2028 (▪) are compared. Activity is expressed as percentage of total substrate unwound in the reaction. Values represent the averages of three experiments, with two different substrate preparations.
Given that the sequence-specific inhibition of TraI helicase activity was lost in the absence of the transesterase domain, another possible explanation for the low activity may be a spatial limitation on loading imposed by the physical presence of the TraI N-terminal domain bound to recognition sequences within the open duplex. To investigate this possibility, the IR and G2028 substrates were preincubated with a 200-fold molar excess of the 34-kDa transesterase domain (TraIN309) (41) before the addition of increasing concentrations of the helicase domain (TraIΔN308). Again, for every concentration of protein tested, no variation in unwinding efficiency was observed with IR compared to the G2028 substrate despite the fact that IR presents the preferred transesterase binding sequence in open conformation while G2028 does not (Fig. 4).
FIG. 4.
Excess of isolated TraI transesterase domain did not alter TraIΔN308-catalyzed unwinding activity. T-strand unwinding and nic cleavage were reconstituted on heteroduplex substrates with separately purified TraI helicase domain and a 200:1 stoichiometry of transesterase domain TraIN309 to substrate. Reaction mixtures containing 400 nM of TraIN309 were incubated for 5 min at room temperature prior to addition of increasing concentrations of helicase TraIΔN308. The cleavage and unwinding efficiencies observed with IR (•) and G2028 (▪) are shown. Values represent the means of four experiments, with two different substrate preparations.
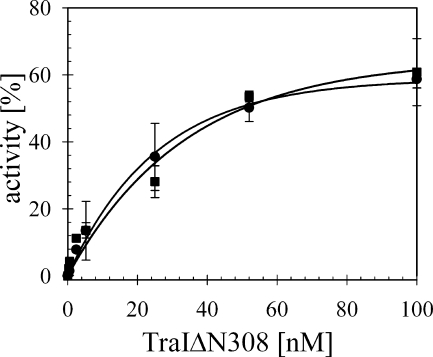
In summary, the sequence-dependent inhibition of helicase activity of the full-length TraI could not be reconstituted with the isolated helicase domain or a mixture of the separately purified transesterase and helicase domains. Fully consistent with the prior data, a direct comparison of the extent of the reaction supported by the full-length TraI compared to the helicase domain on all substrates indicated that, while full-length TraI exhibited lower activity on the NIC (Fig. 5A) and IR (not shown) substrates than did the helicase domain alone (Fig. 5A), this difference was again eliminated on substrate G2028 (Fig. 5B). As the simplest explanation, we propose that the substrate-specific reduction of TraI activity reflects the physical linkage of the DNA transesterase and conjugative helicase domains in one bifunctional protein. The high affinity of the transesterase domain for substrates containing its signature recognition features may impose a conformation within the helicase domain that is less effective for loading and translocation on the adjacent ssDNA, ultimately delaying the initiation of duplex unwinding. This observation implies that coordinate control of these concerted initiation steps is likely to apply to relaxosome regulation during conjugative transfer initiation.
FIG. 5.
Sequence-specific inhibition of oriT unwinding is unique to full-length TraI. The capacity of isolated helicase domain TraIΔN308 (⧫) to unwind T-DNA was compared directly with that of wild-type TraI (•). Increasing concentrations of each enzyme were incubated with NIC substrate carrying the inverted repeat in open conformation (A) or G2028, lacking the inverted repeat (B). The average extent of strand separation is shown for three independent experiments.
Auxiliary proteins TraM and IHF enhance helicase action in a sequence-dependent manner.
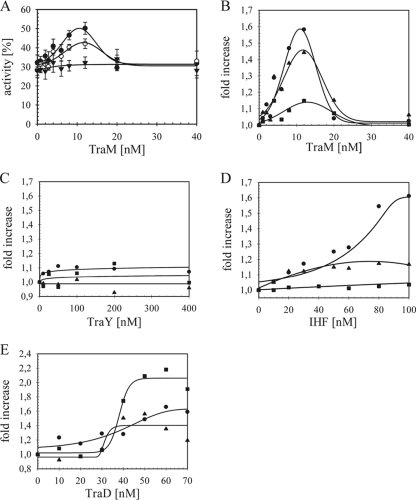
During bacterial conjugation, recruitment and activation of the conjugative helicase occur not on naked DNA but instead on a loading platform provided by the relaxosome. At the outset of this study, nothing was known about the capacity of auxiliary relaxosome components to contribute to the initiation of TraI unwinding activity at oriT in vivo or in vitro. To test each potential effector of the initiation pathway, conditions were selected under which the full-length TraI was combined with each substrate individually at a concentration sufficient to unwind approximately 50% of the input DNA. The effect of incubating increasing concentrations of each auxiliary protein with the substrate prior to starting the reaction was then tested. Under these conditions, a twofold stimulation of the reaction above the basal activity of TraI would represent 100% conversion of the starting material to unwound product. Prior incubation of substrates with comparable concentrations of bovine serum albumin as a negative control had no effect on helicase efficiency (not shown). By comparison, variation in TraI activity as a function of TraM concentration is illustrated in Fig. 6A, including the standard deviations obtained from multiple experiments. For clarity, the stimulatory effects of TraM derived from the same data are expressed as enhancement of the starting TraI activity (Fig. 6B). TraM increased TraI activity significantly on substrates IR (1.6-fold) and NIC (1.4-fold) but not with G2028 (Fig. 6B). A maximum effect of TraM on helicase activity was observed at a concentration of 12 nM for both substrates, while higher concentrations of this effector failed to stimulate activity. Increased TraI activity was thus achieved with a range of 1 to 1.5 tetramers of TraM per substrate. Given that multiple tetramers of TraM are expected to bind to the specific recognition sequences sbmA and sbmB (61), stimulation occurred when the sites were not fully occupied. The degree of enhancement caused by TraM varied with the substrate used. TraM enhanced helicase activity most effectively on the IR substrate, where the distance between the closest TraM binding site, sbmA, and the position of duplex opening is only 55 bp. With heteroduplex NIC, somewhat less stimulation was observed and the relevant sites are separated by 65 bp. No statistically significant variation in helicase activity was observed with TraM and G2028. In this substrate sbmA is the furthest removed (79 bp) from the border to open duplex. Consistent with these findings, equally strong enhancement due to TraM was observed at a lower concentration of TraI (25 nM), again specifically on substrate IR but not on G2028 (not shown). At higher concentrations of TraI, nearly 100% of substrate was already unwound, and so increased activity was not detectable (not shown). The ratio of cleaved and unwound T-DNA relative to unwound product did not vary due to the presence of TraM or any of the effectors tested. For clarity, error bars were omitted from increase values presented for all other effectors with each substrate in the remaining figure (Fig. 6B to E).
FIG. 6.
Protein effectors TraM, IHF, and TraD enhance oriT unwinding. The effect of additional proteins on TraI cleavage and unwinding activities was evaluated with the IR (•), NIC (○), and G2028 (▾) substrates. Reaction mixtures held constant concentrations of TraI (52 nM) and DNA (2 nM) and various concentrations of TraM (0 to 40 nM; monomer) (A and B), TraY (0 to 400 nM; monomer) (C), IHF (0 to 100 nM; heterodimer) (D), or TraD (0 to 70 nM; monomer) (E). With the use of data from three independent experiments, the average yield of product from each DNA in the presence of effector was compared to the activity of TraI alone. The identical data for TraM effects are expressed as increased unwinding activities (%) including standard deviations in panel A or expressed as increase relative to TraI initial activity (value set to 1.0) in panel B. The enhancement in TraI activities is similarly shown as a function of increasing effector concentration for panels C to E. Standard deviations were omitted for clarity. Statistically significant stimulation was observed for TraM with IR (P = 0.018) and NIC (P = 0.019); for IHF with IR (P < 0.001) and NIC (P = 0.039); and for TraD with IR (P = 0.022), NIC (P = 0.02), and G2028 (P = 0.036).
In a previous study employing the NIC substrate only, we attempted to detect effects for TraY protein and the E. coli IHF on helicase activity in this system (10). The results for both were negative. Consistent with those data, the presence of TraY had no impact on helicase efficiency on all substrates under our current conditions (Fig. 6C). The identical preparation of purified TraY did, however, stimulate nic cleavage on supercoiled oriT DNA catalyzed by either full-length TraI or the transesterase domain alone (41); thus, the possible explanation of an inactive TraY fraction could be eliminated in both studies.
In contrast to our earlier attempt, an enhancement of oriT unwinding mediated by IHF was now detected on both the IR and NIC substrates (Fig. 6D). Similarly to the observations with TraM, no significant stimulation of the reaction was observed using G2028, which presents the open duplex at the furthest position (+25 bp) upstream of the nearest IHF binding site, ihfA. As the concentration of IHF was further increased, the auxiliary effect on DNA unwinding was reduced. Concentrations exceeding 200 nM IHF markedly inhibited helicase activity on all substrates (not shown). This negative effect is likely due to a sequence-nonspecific interaction with DNA. In support of this hypothesis, we measured significant inhibition of the ssDNA-dependent ATPase activity of TraI due to IHF at similar protein-to-DNA ratios in the coupled enzyme assay (not shown).
T4CP TraD stimulates TraI in oriT unwinding.
TraD and its functional counterparts in other systems (50) are known to bind to ssDNA without sequence preference and with lower affinity to dsDNA. In the accompanying report, we show that the purified soluble domain of TraD, TraDΔN130, facilitates TraI-catalyzed nic cleavage in a manner that may require interactions between the proteins (41). The capacities of TraDΔN130 to additionally affect the extent of the unwinding reaction catalyzed by the full-length TraI were compared on all substrates (Fig. 6E). TraDΔN130 increased unwinding in each case. In good agreement with our previous observations, full-length TraI exhibited higher activity on substrate G2028 than on IR and NIC (not shown). The magnitude of TraDΔN130-mediated enhancement of the unwinding reaction was also most pronounced on substrate G2028. In contrast to results obtained with TraM and IHF, a marked dependence on TraD concentration was observed for helicase stimulation. The sigmoidal mode of enhancement may reflect the requirement for a cooperative interaction of TraD with protein and/or DNA ligand(s). The inflection points of these curves occurred between 30 and 40 nM concentrations of TraDΔN130, i.e., 15 to 20 monomers of protein per DNA substrate. Consistent with this proposal, the soluble domain of the related TrwB protein of plasmid R388, TrwBΔN70, forms active hexamers in vitro in a highly concentration-dependent manner (58).
Initiator proteins stimulate oriT unwinding by the truncated TraIΔN308 helicase domain.
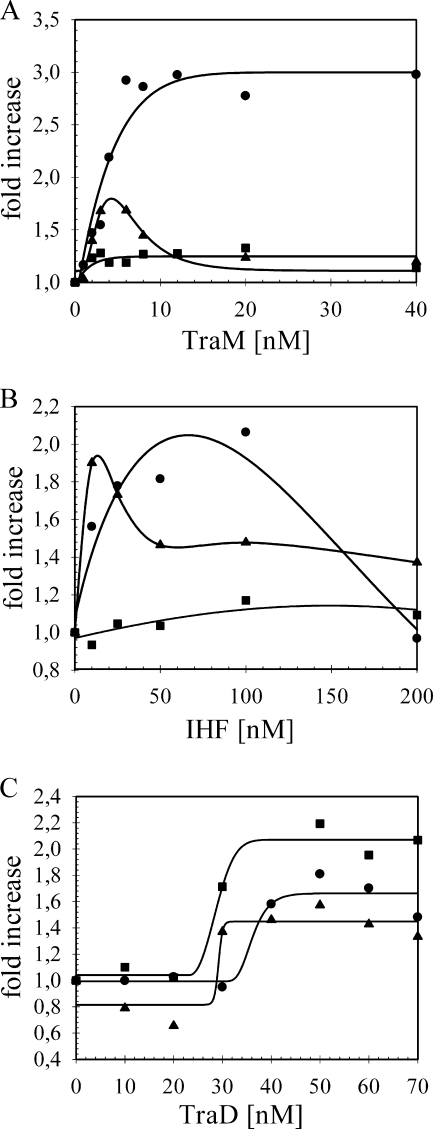
The ability of relaxosome components to facilitate T-strand unwinding may result from interactions between proteins, protein and DNA, or both. To explore these alternatives, we first confirmed that the effector proteins present in the assays were able to bind to oriT under the conditions used. Reconstitution of the effector ensemble on a heteroduplex substrate at concentrations sufficient to support the helicase activity maximum led to a detectable shift in the electrophoretic mobility of the substrate compared to DNA alone (not shown). We next compared the impact of effectors on the reaction catalyzed by the truncated TraIΔN308. If the nature of effector stimulation was exclusively due to an interaction between proteins, we reasoned that the absence of the 34-kDa N-terminal domain, which is dispensable for helicase activity, might shift the activity profiles from that observed with full-length TraI. No loss of enhancement was seen for any effector in combination with the helicase domain (Fig. 7). Generally, we observed for each effector the same substrate-specific dependencies for the magnitude of stimulation obtained with TraI. For the data set shown here (Fig. 7A), the extent of reaction supported by 52 nM of the isolated helicase domain was relatively low (19%). The additional presence of TraM stimulated the unwinding reaction and, given the unusually low initial TraIΔN308 activity, was able to support a threefold enhancement with substrate IR. A notable deviation from the observations with full-length protein was the absence of inhibition of the extent of unwinding at higher concentrations of TraM on the same substrate. Moreover, we detected a shift to lower concentrations of TraM, IHF, and TraD for these enhancement curves, relative to the TraI data, on all substrates where stimulation of helicase activity was significant (Fig. 7). These findings indicate that the relaxosome components' promotion of T-DNA unwinding by TraI occurs equally well with the truncated form TraIΔN308. If a direct interaction(s) between helicase and initiator protein is involved in the stimulation, the N-terminal 34-kDa domain of TraI is dispensable.
FIG. 7.
TraM, IHF, and TraD also stimulate TraIΔN308 activity. The effect of additional proteins on oriT unwinding catalyzed by the truncated TraIΔN308 form of the protein was compared using conditions equivalent to those of previous assays with wild-type TraI. Reactions were initiated by addition of TraIΔN308 (52 nM). The increase in TraIΔN308 unwinding activity on the IR (•), NIC (▴), and G2028 (▪) substrates is presented as a function of auxiliary protein concentration. The relative enhancement due to addition of TraM (0 to 40 nM; as monomer) (A), IHF (0 to 200 nM; as heterodimer) (B), or TraD (0 to 70 nM; as monomer) (C) is shown. The data represent the means of three experiments using two different substrate preparations. Standard deviations were omitted for clarity.
The presence of TraY in combination with TraIΔN308 had no significant influence on the reaction, as was the case for TraI (not shown). In a final set of experiments, we compared the effects of reconstituting the entire relaxosome plus TraDΔN130 on each substrate in combination with either full-length TraI or TraIΔN308 with that of stimulation supported by the individual initiator proteins. For the combined reaction mixtures, each effector was present at a concentration equivalent to that where the activity maximum was reached on the substrate exhibiting the strongest effect, e.g., 12 nM TraM, 100 nM IHF, 50 nM TraDΔN130, and 200 nM TraY were combined with 52 nM TraI. The same protein-to-substrate ratio was selected for TraY, which supported maximum stimulation of TraI in the transesterase reaction on supercoiled oriT DNA (41). In each case the extent of stimulation measured for the combined proteins was not significantly higher than that observed in the reaction mixtures containing the individual effector protein most active on a given substrate (not shown). The series of experiments was repeated six times using two independent preparations of each substrate.
In conclusion, exploitation of the current in vitro system did not provide additional insights into potential synergy or concerted effects of reconstitution of known components of the R1 relaxosome. Nonetheless, this system identified independent enhancer functions for the T4CP TraDΔN130 as well as for the auxiliary relaxosome proteins TraM and IHF in generating unwound T-DNA from oriT. Evidence presented here thus provides novel insights into levels of regulation imposed on the TraI protein within the strand transfer initiation cascade.
DISCUSSION
Conjugative DNA processing proceeds with remarkable efficiency; thus, facilitating mechanisms appear to streamline the initiation pathway. Details of that pathway remain obscure, and the ensemble reactions analyzed in this study reconstitute multiple steps that ultimately result in the unwinding of T-DNA from the R1 oriT. Known conjugative helicases do not initiate duplex unwinding on supercoiled oriT plasmids except under cellular conditions of bacterial conjugation. Heteroduplex substrates were thus constructed to model possible initiation intermediates presenting loading platforms of oriT DNA in open conformation. A permutation of the position of duplex opening relative to nic in the set of heteroduplex substrates revealed that the full-length TraI protein is subject to a sequence-dependent control not previously recognized. The mechanism is apparently autoregulatory, as separating the helicase from its N-terminal transesterase domain eliminated the difference in activity. Reconstitution of the unwinding reaction catalyzed by the truncated helicase domain in the presence of excessive amounts of the separately purified N-terminal transesterase domain failed to limit unwinding on any DNA substrate. Thus, the sequence dependence of the TraI inhibition was not linked to a reduced loading efficiency. Williams and Schildbach (65) showed that the transesterase domain of F plasmid TraI binds to oligonucleotides containing the inverted repeat sequence and nic in two different modes, which appear to involve the same or overlapping regions of the protein. Substrate G2028 should present sufficient DNA in single-stranded form (comparable to the 17-nt wild-type oligonucleotide used by these authors) to support one TraI binding mode but not the second competing mode involving the hairpin. Given that T-strand unwinding from G2028 was more efficient than that on substrates exposing also the inverted repeat sequences, we suggest that these distinct modes of TraI interaction with nic-proximal sequences may govern the regulation that we observe on the level of helicase initiation. Studies from the Schildbach laboratory have provided substantial detail concerning the contributions of specific oriT bases and amino acids of the transesterase domains to the remarkable specificity of the ssDNA binding interactions exhibited by the IncF family of proteins (24-26, 35, 56, 65). In ongoing work, comparison of a series of defined mutations within oriT combined with the corresponding mutant derivatives of purified TraI will enable us to dissect the DNA binding component of this novel regulatory interaction.
Physical tethering of the two TraI functional domains has persisted in evolution; thus, it would appear advantageous to localize the conjugative helicase to oriT via sequence recognition properties of the transesterase domain and yet simultaneously repress a concerted unwinding event. It follows that a commitment to unwinding from nic requires activation of the molecular control switch linking the two domains. The combined properties of site-specific DNA binding and helical motor activity are shared by initiators of DNA replication in eukaryotic DNA tumor viruses (3, 47, 53). Details of the mechanisms of intramolecular regulation of helicase activity mediated by modular accessory domains or interactions between subdomains are emerging for nucleic acid helicases and translocases (38, 55) including the well-characterized E. coli Rep protein (4). The molecular nature of autoregulation imposed on TraI in combination with oriT substrates is currently unknown. Under conditions conducive to plasmid strand transfer, signals transduced via the T4CP may reverse the repression, allowing the helicase to commit to translocation or duplex entry. The switch makes sense since replication systems do not risk unwinding unaccompanied by DNA synthesis. In the case of conjugative replication, a direct handoff to the T4CP and replication factors may be the preferred solution. The capacity to detect the repression in vitro provides an approach to elucidate the nature of this molecular switch. Simple addition of purified TraD alone or in combination with all effector proteins did not restore the overall unwinding efficiency of TraI or TraIΔN308 on the NIC or IR substrates to the level observed with G2028 (not shown).
Several regulatory effects on the conjugative helicase were observed for additional proteins in the model, however. We provide the first evidence for a stimulatory effect of relaxosome auxiliary factors as well as the T4CP TraD on T-strand unwinding from oriT. Effector-enhanced unwinding was observed with both full-length TraI and the helicase domain alone. The heteroduplex substrates vary in the distance between the site of helicase loading and the array of binding sites for other relaxosome proteins; thus, it was possible to confirm that the magnitude of stimulation induced by TraM and IHF varied directly with the proximity of their recognition sequences. The steady-state multiple turnover experiments performed in this study were sufficient to reveal both positive and negative effects on helicase efficiency but cannot provide information about what aspect of activity is affected. The data imply the involvement of protein interactions. Stimulation of helicase activity was observed at concentrations of IHF and TraM where their respective sites of binding would not be saturated. Given that the open conformation of the model substrates is constitutively present, stimulation of the helicase detected under these conditions may reflect artificially low effector protein concentrations compared to requirements for initiation on supercoiled plasmid. Recognition of the role that these proteins play in activating the helicase paves the way for detailed biochemical investigation of the underlying mechanisms. Importantly, those data will provide a rational basis for subsequent genetics approaches essential to understanding the process of T-strand unwinding during bacterial conjugation.
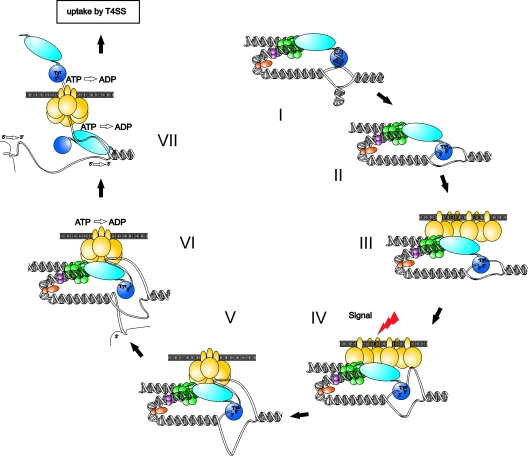
Addition of the data presented here and in the accompanying report (41) to the prior information obtained in vivo or extrapolated from in vitro data including the available evidence concerning T4CP control supports the following model for regulation of T-strand production and delivery to the translocation pathway of the MOBF type IV secretion system (Fig. 8).
FIG. 8.
Model for regulation of T-strand production and delivery to the translocation pathway of the MOBF type IV secretion system (T4SS). Steps in the initiation reaction cascade (I to VII) are described in the text. The subsequent translocation process per se and the steps terminating plasmid strand transfer are not shown. Protein components are illustrated as follows: the bifunctional TraI transesterase (blue) with active site tyrosine (Tyr) and TraI helicase domains (light blue), IHF (orange), TraY (purple), TraM (green), and the T4CP TraD (yellow).
Preinitiation.
The relaxosome assembles on supercoiled oriT DNA and assumes a complex (but yet-undefined) higher-order structure determined by the sum of topological distortions induced by each effector interacting with the DNA. As reported earlier (65), the TraI transesterase domain interacts with nic-proximal sequences containing the inverted repeat in two distinct binding modes (Fig. 8, steps I and II). Within the relaxosome, TraI transesterase in the step II binding modus is proficient for cleaving-joining activity at nic and is stimulated most substantially by IHF but also independently by the presence of TraY and TraM (10, 27, 33, 41, 44, 48). The relaxosome is anchored to the T4CP TraD at the cytoplasmic membrane via interactions with TraM (2, 15, 39, 40, 49) and TraI (step III). TraD stimulates DNA cleavage at nic in combination with full-length TraI via mechanisms not supported by the isolated transesterase domain (41). TraD requires an additional F plasmid protein(s) to form stable multimers in the membrane (23). We propose that TraD is not yet assembled in the hexameric form required for ATP hydrolysis (58, 59). Localized melting of oriT at this stage may or may not be sufficient to load the TraI helicase, but duplex unwinding does not occur.
Activation of the transporter/substrate interface.
Translocation on ssDNA and duplex entry require modulation of a molecular switch in the full-length TraI protein and probably an extension of duplex melting at nic (Fig. 2 to 4) (41). Conditions that would induce these changes presumably emerge from intercellular contacts between bacteria and are communicated over conjugative pili to the T4 secretion apparatus (signal, step IV). In that case docking of the preinitiation complex to the type IV secretion system via TraD would confer competence for T-strand production. Interactions between TraD and relaxosome components thus complete the signal transduction pathway to protein and nucleoprotein targets. TraI switches to translocation mode, altering the N-terminal domain's high-affinity interactions with the inverted repeat and surrounding sequences (step V). Both 3′ and 5′ ends of nic remain sequestered by TraI (5). The altered protein contacts or increased exposure of ssDNA changes TraD conformation and ligand interaction; TraD's latent nucleoside triphosphatase is activated (58, 59) (step VI).
Commitment to T-strand mobilization.
Uptake of the nucleoprotein adduct into the TraD translocation channel is mediated by recognition of translocation signals on the TraI protein (46, 62, 63) (step VII). A second TraI protein is loaded to the exposed T strand to facilitate replacement-strand synthesis of the secreted DNA. Simultaneous handoff of the 3′ end of nic to DNA polymerase III (69) and commitment of the 5′ nucleoprotein complex to the translocation pathway mark the end of the initiation reaction.
In this model, we argue that if replacement-strand synthesis occurs at a rate equivalent to that observed for mobilization of the E. coli chromosome (800 bp/s), TraI-catalyzed activity at the junction of plasmid strands would be necessary to work in concert with DNA polymerase III (30, 43). It follows that, if the monomer of TraI covalently linked to the 5′ end of the T strand is actively secreted in the initial step of translocation, then the TraI monomer positioned at the duplex junction is by necessity distinct from that piloting the T-strand uptake and secretion through the T4 machinery. The speed of T-strand unwinding may in turn be increased by the mechanical force (21, 28) generated by an ATPase-dependent propulsion of the T strand through the central channel of the T4CP conjugative pore (36).
Acknowledgments
We are grateful to L. Frost, B. Mayer, R. Zechner, K. Zangger, F. de la Cruz, and W. Keller for helpful discussions.
This work was financed by the Austrian FWF grants P18607 and W901-B05 (DK: Molecular Enzymology) and the EU grant FP6 PL 019023.
Footnotes
Published ahead of print on 18 September 2009.
REFERENCES
- 1.Barry, E. R., and S. D. Bell. 2006. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 70:876-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beranek, A., M. Zettl, K. Lorenzoni, A. Schauer, M. Manhart, and G. Koraimann. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 186:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. [DOI] [PubMed] [Google Scholar]
- 4.Brendza, K. M., W. Cheng, C. J. Fischer, M. A. Chesnik, A. Niedziela-Majka, and T. M. Lohman. 2005. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl. Acad. Sci. USA 102:10076-10081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 6.Byrd, D. R., J. K. Sampson, H. M. Ragonese, and S. W. Matson. 2002. Structure-function analysis of Escherichia coli DNA helicase I reveals non-overlapping transesterase and helicase domains. J. Biol. Chem. 277:42645-42653. [DOI] [PubMed] [Google Scholar]
- 7.Cabezón, E., E. Lanka, and F. de la Cruz. 1994. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 176:4455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabezón, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 9.Costa, A., and S. Onesti. 2008. The MCM complex: (just) a replicative helicase? Biochem. Soc. Trans. 36:136-140. [DOI] [PubMed] [Google Scholar]
- 10.Csitkovits, V. C., D. Dermic, and E. L. Zechner. 2004. Concomitant reconstitution of TraI-catalyzed DNA transesterase and DNA helicase activity in vitro. J. Biol. Chem. 279:45477-45484. [DOI] [PubMed] [Google Scholar]
- 11.Csitkovits, V. C., and E. L. Zechner. 2003. Extent of single-stranded DNA required for efficient TraI helicase activity in vitro. J. Biol. Chem. 278:48696-48703. [DOI] [PubMed] [Google Scholar]
- 12.de la Cruz, F., L. S. Frost, R. J. Meyer, and E. L. Zechner. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev., in press. [DOI] [PubMed]
- 13.Delagoutte, E., and P. H. von Hippel. 2002. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: structures and properties of isolated helicases. Q. Rev. Biophys. 35:431-478. [DOI] [PubMed] [Google Scholar]
- 14.Delagoutte, E., and P. H. von Hippel. 2003. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Q. Rev. Biophys. 36:1-69. [DOI] [PubMed] [Google Scholar]
- 15.Disqué-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enemark, E. J., and L. Joshua-Tor. 2008. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 18:243-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsburg, S. L. 2004. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 68:109-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fürste, J. P., W. Pansegrau, R. Frank, H. Blöcker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 19.Geider, K., and H. Hoffmann-Berling. 1981. Proteins controlling the helical structure of DNA. Annu. Rev. Biochem. 50:233-260. [DOI] [PubMed] [Google Scholar]
- 20.Graus, H., A. Hödl, P. Wallner, and G. Högenauer. 1990. The sequence of the leading region of the resistance plasmid R1. Nucleic Acids Res. 18:1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha, T. 2007. Need for speed: mechanical regulation of a replicative helicase. Cell 129:1249-1250. [DOI] [PubMed] [Google Scholar]
- 22.Hackney, D. D., and P. K. Clark. 1985. Steady state kinetics at high enzyme concentration. The myosin MgATPase. J. Biol. Chem. 260:5505-5510. [PubMed] [Google Scholar]
- 23.Haft, R. J., E. G. Gachelet, T. Nguyen, L. Toussaint, D. Chivian, and B. Traxler. 2007. In vivo oligomerization of the F conjugative coupling protein TraD. J. Bacteriol. 189:6626-6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harley, M. J., and J. F. Schildbach. 2003. Swapping single-stranded DNA sequence specificities of relaxases from conjugative plasmids F and R100. Proc. Natl. Acad. Sci. USA 100:11243-11248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harley, M. J., D. Toptygin, T. Troxler, and J. F. Schildbach. 2002. R150A mutant of F TraI relaxase domain: reduced affinity and specificity for single-stranded DNA and altered fluorescence anisotropy of a bound labeled oligonucleotide. Biochemistry 41:6460-6468. [DOI] [PubMed] [Google Scholar]
- 26.Hekman, K., K. Guja, C. Larkin, and J. F. Schildbach. 2008. An intrastrand three-DNA-base interaction is a key specificity determinant of F transfer initiation and of F TraI relaxase DNA recognition and cleavage. Nucleic Acids Res. 36:4565-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard, M. T., W. C. Nelson, and S. W. Matson. 1995. Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J. Biol. Chem. 270:28381-28386. [PubMed] [Google Scholar]
- 28.Johnson, D. S., L. Bai, B. Y. Smith, S. S. Patel, and M. D. Wang. 2007. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell 129:1299-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karl, W., M. Bamberger, and E. L. Zechner. 2001. Transfer protein TraY of plasmid R1 stimulates TraI-catalyzed oriT cleavage in vivo. J. Bacteriol. 183:909-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, S., H. G. Dallmann, C. S. McHenry, and K. J. Marians. 1996. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell 84:643-650. [DOI] [PubMed] [Google Scholar]
- 31.Konieczny, I. 2003. Strategies for helicase recruitment and loading in bacteria. EMBO Rep. 4:37-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn, B., M. Abdel-Monem, H. Krell, and H. Hoffmann-Berling. 1979. Evidence for two mechanisms for DNA unwinding catalyzed by DNA helicases. J. Biol. Chem. 254:11343-11350. [PubMed] [Google Scholar]
- 33.Kupelwieser, G., M. Schwab, G. Högenauer, G. Koraimann, and E. L. Zechner. 1998. Transfer protein TraM stimulates TraI-catalyzed cleavage of the transfer origin of plasmid R1 in vivo. J. Mol. Biol. 275:81-94. [DOI] [PubMed] [Google Scholar]
- 34.Lahue, E. E., and S. W. Matson. 1988. Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J. Biol. Chem. 263:3208-3215. [PubMed] [Google Scholar]
- 35.Larkin, C., S. Datta, M. J. Harley, B. J. Anderson, A. Ebie, V. Hargreaves, and J. F. Schildbach. 2005. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure 13:1533-1544. [DOI] [PubMed] [Google Scholar]
- 36.Llosa, M., F. X. Gomis-Rüth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 37.Llosa, M., G. Grandoso, M. A. Hernando, and F. de la Cruz. 1996. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264:56-67. [DOI] [PubMed] [Google Scholar]
- 38.Lohman, T. M., E. J. Tomko, and C. G. Wu. 2008. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 9:391-401. [DOI] [PubMed] [Google Scholar]
- 39.Lu, J., and L. S. Frost. 2005. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J. Bacteriol. 187:4767-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu, J., J. J. Wong, R. A. Edwards, J. Manchak, L. S. Frost, and J. N. Glover. 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. 70:89-99. [DOI] [PubMed] [Google Scholar]
- 41.Mihajlovic, S., S. Lang, M. V. Sut, H. Strohmaier, C. J. Gruber, G. Koraimann, E. Cabezón, G. Moncalián, F. de la Cruz, and E. L. Zechner. 2009. Plasmid R1 conjugative DNA processing is regulated at the coupling protein interface. J. Bacteriol. 191:6877-6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 43.Mok, M., and K. J. Marians. 1987. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J. Biol. Chem. 262:16644-16654. [PubMed] [Google Scholar]
- 44.Nelson, W. C., M. T. Howard, J. A. Sherman, and S. W. Matson. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J. Biol. Chem. 270:28374-28380. [PubMed] [Google Scholar]
- 45.Nielsen, O., and A. Lobner-Olesen. 2008. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep. 9:151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker, C., and R. J. Meyer. 2007. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol. Microbiol. 66:252-261. [DOI] [PubMed] [Google Scholar]
- 47.Parsons, R., M. E. Anderson, and P. Tegtmeyer. 1990. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J. Virol. 64:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ragonese, H., D. Haisch, E. Villareal, J. H. Choi, and S. W. Matson. 2007. The F plasmid-encoded TraM protein stimulates relaxosome-mediated cleavage at oriT through an interaction with TraI. Mol. Microbiol. 63:1173-1184. [DOI] [PubMed] [Google Scholar]
- 49.Sastre, J. I., E. Cabezón, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schröder, G., and E. Lanka. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 185:4371-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwab, M., H. Gruber, and G. Högenauer. 1991. The TraM protein of plasmid R1 is a DNA-binding protein. Mol. Microbiol. 5:439-446. [DOI] [PubMed] [Google Scholar]
- 52.Schwab, M., H. Reisenzein, and G. Högenauer. 1993. TraM of plasmid R1 regulates its own expression. Mol. Microbiol. 7:795-803. [DOI] [PubMed] [Google Scholar]
- 53.Seo, Y. S., F. Muller, M. Lusky, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc. Natl. Acad. Sci. USA 90:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sikora, B., R. L. Eoff, S. W. Matson, and K. D. Raney. 2006. DNA unwinding by Escherichia coli DNA helicase I (TraI) provides evidence for a processive monomeric molecular motor. J. Biol. Chem. 281:36110-36116. [DOI] [PubMed] [Google Scholar]
- 55.Singleton, M. R., M. S. Dillingham, and D. B. Wigley. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76:23-50. [DOI] [PubMed] [Google Scholar]
- 56.Stern, J. C., and J. F. Schildbach. 2001. DNA recognition by F factor TraI36: highly sequence-specific binding of single-stranded DNA. Biochemistry 40:11586-11595. [DOI] [PubMed] [Google Scholar]
- 57.Street, L. M., M. J. Harley, J. C. Stern, C. Larkin, S. L. Williams, D. L. Miller, J. A. Dohm, M. E. Rodgers, and J. F. Schildbach. 2003. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim. Biophys. Acta 1646:86-99. [DOI] [PubMed] [Google Scholar]
- 58.Tato, I., I. Matilla, I. Arechaga, S. Zunzunegui, F. de la Cruz, and E. Cabezón. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 282:25569-25576. [DOI] [PubMed] [Google Scholar]
- 59.Tato, I., S. Zunzunegui, F. de la Cruz, and E. Cabezón. 2005. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. USA 102:8156-8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Traxler, B. A., and E. G. Minkley, Jr. 1988. Evidence that DNA helicase I and oriT site-specific nicking are both functions of the F TraI protein. J. Mol. Biol. 204:205-209. [DOI] [PubMed] [Google Scholar]
- 61.Verdino, P., W. Keller, H. Strohmaier, K. Bischof, H. Lindner, and G. Koraimann. 1999. The essential transfer protein TraM binds to DNA as a tetramer. J. Biol. Chem. 274:37421-37428. [DOI] [PubMed] [Google Scholar]
- 62.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 63.Vergunst, A. C., M. C. van Lier, A. den Dulk-Ras, T. A. Stuve, A. Ouwehand, and P. J. Hooykaas. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA 102:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Hippel, P. H., and E. Delagoutte. 2003. Macromolecular complexes that unwind nucleic acids. Bioessays 25:1168-1177. [DOI] [PubMed] [Google Scholar]
- 65.Williams, S. L., and J. F. Schildbach. 2006. Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Res. 34:426-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zakrzewska-Czerwinska, J., D. Jakimowicz, A. Zawilak-Pawlik, and W. Messer. 2007. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 31:378-387. [DOI] [PubMed] [Google Scholar]
- 67.Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann, E. Lanka, G. Muth, W. Pansegrau, C. M. Thomas, B. M. Wilkins, and M. Zatyka. 2000. Conjugative DNA transfer processes, p. 87-174. In C. M. Thomas (ed.), The horizontal gene pool. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 68.Zechner, E. L., H. Pruger, E. Grohmann, M. Espinosa, and G. Högenauer. 1997. Specific cleavage of chromosomal and plasmid DNA strands in gram-positive and gram-negative bacteria can be detected with nucleotide resolution. Proc. Natl. Acad. Sci. USA 94:7435-7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zechner, E. L., C. A. Wu, and K. J. Marians. 1992. Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. III. A polymerase-primase interaction governs primer size. J. Biol. Chem. 267:4054-4063. [PubMed] [Google Scholar]