Abstract
Here we show that the Salmonella enterica serovar Typhimurium PhoQ sensor kinase lessens the cytotoxicity of reactive nitrogen species (RNS) generated by inducible nitric oxide synthase (iNOS) in the innate response of mononuclear phagocytic cells. This observation is consistent with the expression patterns of PhoP-activated genes during moderate nitrosative stress in the innate host response. In contrast, RNS synthesized during high-NO fluxes of gamma interferon (IFN-γ)-activated macrophages repress PhoP-activated lpxO, pagP, and phoP gene transcription. Because PhoP-regulated Salmonella pathogenicity island 2 (SPI2) genes are also repressed by high-order RNS (39), we investigated whether the NO-mediated inhibition of PhoPQ underlies the repression of SPI2. Our studies indicate that a third of the expression of the SPI2 spiC gene recorded in nonactivated macrophages depends on PhoQ. Transcription of spiC is repressed in IFN-γ-primed macrophages in an iNOS-dependent manner, irrespective of the phoQ status of the bacteria. Transcription of spiC is restored in IFN-γ-treated, iNOS-deficient macrophages to levels sustained by a phoQ mutant in nonactivated phagocytes, suggesting that most NO-dependent repression of spiC is due to the inhibition of PhoPQ-independent targets. Comparison of the intracellular fitness of spiC, phoQ, and spiC phoQ mutants revealed that PhoPQ and SPI2 have codependent and independent effects on S. Typhimurium survival during innate nitrosative stress. However, the intracellular survival of most S. Typhimurium bacteria is conferred by the PhoPQ two-component regulator, and the SPI2 type III secretion system is repressed by high-order RNS of IFN-γ-activated macrophages.
The intracellular pathogen Salmonella enterica has evolved a network of signal transduction pathways to withstand the barrage of host defenses encountered in nonfusogenic phagosomes in the form of antimicrobial peptides and acid pH levels, as well as reactive oxygen species and reactive nitrogen species (RNS). The PhoPQ two-component signal transduction relay coordinates several aspects of the pathogenic lifestyle of S. Typhimurium (22). The PhoQ sensor kinase preferentially exerts phosphatase activity in resting Salmonella enterica serovar Typhimurium (7); however, exposure of the bacteria to antimicrobial peptides, low Mg2+ concentrations or acidic pH levels shifts phosphatase to autokinase activity, resulting in a net gain phosphorylation of PhoQ at His-277 (2, 3, 21, 51). The relay of this phosphate to PhoP Asp-55 unleashes the direct or indirect transcription of over 200 loci (45), many of which enhance the resistance of S. Typhimurium to acid pH levels, oxidative stress, metals, bile, and antimicrobial peptides (3, 8, 21, 27, 29, 59). Consequently, S. Typhimurium strains defective in PhoPQ signaling grow poorly in macrophages and are at least 10,000-fold attenuated when tested in several experimental murine models of salmonellosis (19, 43).
Nitric oxide (NO) is an integral component of host defense against the enteropathogen S. Typhimurium. The RNS emanating from inducible NO synthase (iNOS) enzymatic activity hasten the anti-Salmonella activities of human and murine macrophages while increasing the resistance of mice to systemic and gastrointestinal salmonellosis (1, 38, 56, 60, 63, 64). The mechanisms by which RNS add to the anti-Salmonella arsenal of phagocytes are not completely understood. NO and its congeners arrest DNA replication, damage DNA, and inhibit transcription of the Salmonella pathogenicity island 2 (SPI2) type III secretion system essential for S. Typhimurium virulence (39, 52, 53). In addition, RNS generated in acidic environments typical of gastric juice repress the PhoPQ-regulated acid tolerance response that protects rapidly growing S. Typhimurium bacteria against inorganic acid stress (5). S. Typhimurium is exposed to a similar repertoire of RNS within the gastric lumen and acidified phagosomes of gamma interferon (IFN-γ)-stimulated macrophages (36, 40). NO itself, as well as nitrogen dioxide (NO2) and dinitrogen trioxide (N2O3), arising from either the autooxidation of NO or the condensation of acidified nitrite (HNO2), are similarly generated in the lumen of phagosomes and in the stomach (36, 40). Because the PhoPQ signal transduction pathway is susceptible to inhibition by RNS produced under acidic conditions (5), we set out to investigate whether the PhoP regulon is functional in the context of the nitrosative stress engendered in the innate or IFN-γ-activated responses of professional phagocytes.
MATERIALS AND METHODS
Bacterial strains.
Salmonella enterica serovar Typhimurium strain ATCC 14028s was used as the wild type and as a background for the construction of mutations and lacZY-transcriptional fusions of both PhoP-activated genes and the SPI2 effector and translocator spiC (Table 1). The mutations were generated by following the λ Red-mediated gene replacement method (10). Primers encoding 60 nucleotides homologous to a target gene followed by 20 nucleotides homologous to the pKD13 template plasmid (Table 2) were used for the PCR amplification of the Flp recombinant target (FRT)-flanked kanamycin resistance cassette. The amplicons were DpnI digested and electroporated into S. Typhimurium strain TT22236 carrying the pTP2223 plasmid that expresses the λ Red recombinase under Ptac control. Mutations were moved into S. Typhimurium strain 14028s by P22-mediated transduction, and pseudolysogens were eliminated by streaking on Evans blue uranine agar plates. In-frame deletions were generated by recombining the two FRT sites flanking the kanamycin resistance cassette with the Flp recombinase encoded in the pCP20 plasmid (9). The mutations were confirmed by PCR analysis. Transcriptional lacZY fusions were constructed by pCP20-mediated integration of the pCE36 plasmid carrying a promoterless lacZY operon into unique FRT scars (14).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Salmonella Typhimurium strains | ||
| 14028s | Wild type | ATCC |
| TT22236 | LT2 carrying pTP2223 | 50 |
| AV0201 | ΔspiC::FRT | 39 |
| AV0212 | ΔspiC::lacZY | 39 |
| AV0475 | ΔphoQ::FRT | 5 |
| AV0609 | ΔpagP::lacZY | This study |
| AV0611 | ΔlpxO::lacZY | 5 |
| AV0612 | ΔphoQ::FRT ΔpagP::lacZY | This study |
| AV0614 | ΔphoQ::FRT ΔlpxO::lacZY | 5 |
| AV07181 | ΔphoQ::FRT ΔspiC::lacZY | This study |
| AV0830 | ΔphoQ::FRT ΔspiC::km | This study |
| Plasmids | ||
| pCP20 | bla cat cI857 PRflp pSC101 oriTS | 9 |
| pKD13 | bla FRT ahp FRT PS1 PS4 oriR6K | 10 |
| pCE36 | ahp FRT lacZY+ this oriR6K | 14 |
| pTP2223 | PLAClam bet exo Tetr | 49 |
TABLE 2.
Primers used for the construction of mutations and RT-PCR and real-time RT-PCR analysis
| Function and gene | Primer/probe sequencea |
|---|---|
| Mutation construction | |
| ΔpagP::FRT | F, 5′- ACGTGGCGACAGCCTGAGCATTATGATTTGTATGTCCCCGCCATTACCTGGCATGCGCGCTGGAGCTGCTTCGAAGGTT |
| R, 5′-AAGACTTTTTAATTCACAACTGAAGCATACCCTTCCCCATCAAAACTGGAAACGCATCCATTCCGGGGATCCGTCGACCT | |
| ΔlpxO::FRT | F, 5′-GCCGGAACGACGCGGGAGGATGTGATCAACAGATTTGAACTGCTCAGGACGCTCGCGTGCTGGAGCTGCTTCGAAGGTT |
| R, 5′-TTTGCGCTCCAGCACTCTGTGTAACGGCCCCCACTGCAGTATCCACTCCCTGAACTCGCTTTCCGGGGATCCGTCGACCT | |
| RT-PCR analysis | |
| phoP | F, 5′-CTTGGATCCGGATTCATGCTGGCAGTTTT |
| R, 5′-TGGAAGCTTTCCAGGTCATTTAAGAACAAAGAA | |
| Hyb, 5′-6-FAM-TGAAGGTTCAGCTCCAGGATTCAGG-BHQ-1 | |
| rpoD | F, 5′-GTGGCTTGCAATTCCTTGAT |
| R, 5′-AGCATCTGGCGAGAAATACG | |
| Hyb, 5′-6-FAM-ATAAGTTCGAATACCGTCGCGGCTACA-BHQ-1 | |
| mig-14 | F, 5′-CCCTATACCGGGAGGTGTTT |
| R, 5′-GGCAAGAAGCAGCGTAAATC |
Dually labeled oligonucleotide probes contain both the fluorescent dye 6-carboxyfluorescein (6-FAM) and Black Hole quencher 1 (BHQ-1). F, forward; R, reverse.
Effects of NO on in vitro PhoPQ and SPI2 gene transcription.
Transcription of PhoP-activated genes and the spiC SPI2 gene was induced in vitro by culturing S. Typhimurium in 8 μM MgCl2 N salts medium (12). Briefly, S. Typhimurium strains harboring lacZY transcriptional fusions grown overnight in Luria-Bertani (LB) medium were subcultured 1:100 in high-Mg2+ N salts medium [5 mM KCl, 7.5 mM (NH4)SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 38 mM glycerol, 0.1% Casamino Acids supplemented with 10 mM MgCl2 and 100 mM Tris-HCl, pH 7.6]. The bacteria were grown at 37°C in a shaker incubator until they reached an optical density at 600 nm (OD600) of 0.5. The cells were washed three times in 8 μM MgCl2 N salts medium, pH 6.9, and cultured for the indicated amounts of time in 8 μM MgCl2 N salts medium, pH 6.9, for the induction of PhoP-activated gene expression. Spermine NONOate (Cayman Chemical, Ann Arbor, MI) dissolved in 10 mM Tris-HCl, pH 7.4, was added to S. Typhimurium cultures at the beginning of growth in 8 μM MgCl2 N salts medium, pH 6.9. Alternatively, S. Typhimurium isolates grown overnight in LB broth were subcultured 1:50 in minimal EG medium [0.2 g/liter MgSO4, 2 g/liter C6H8O7-H2O, 10 g/liter K2HPO4, 3.5 g/liter Na(NH4)HPO4-4H2O, and 4 g/liter d-glucose], pH 7.0 (62). The cultures were grown to an OD600 of 0.4 (∼2 × 108 CFU/ml). Acid-inducible expression of spiC was initiated by subculturing S. Typhimurium for 2 h in fresh EG medium adjusted to pH 4.4 with HCl. Selected cultures were grown in the presence or absence of 500 μM spermine NONOate at 37°C with shaking. The expression of the lacZY transcriptional fusions was quantified spectrophotometrically as β-galactosidase enzymatic activity using the substrate o-nitrophenyl-β-d-galactopyranoside. β-Galactosidase activity is expressed in Miller units using the following equation: 1,000 × [(OD420 − 1.75 × OD550)/(T(min) × V(ml) × OD600)].
Macrophages.
Peritoneal macrophages were harvested from C57BL/6 and congenic B6.129S6-Cybbtm1din/J (gp91phox−/−) (48) or iNOS−/− (37) mice 4 days after intraperitoneal inoculation of 1 mg/ml sodium periodate (13). The peritoneal exudate cells were resuspended in RPMI+ medium (RPMI 1640 cell culture medium [Mediatech, Inc., Manassas, VA] supplemented with 10% heat-inactivated fetal bovine serum [BioWhittaker Inc., Walkersville, VA], 15 mM HEPES, 2 mM l-glutamine, 1 mM sodium pyruvate [Sigma-Aldrich, St. Louis, MO]), containing 100 U/ml penicillin and 100 μg/ml streptomycin (Mediatech). The peritoneal exudate cells were seeded at either 4 × 105 cells/ml in 24-well plates for phoP transcriptional studies or 2 × 105 cells/ml in 96-well plates (BD Biosciences, San Jose, CA) for macrophage-killing and β-galactosidase activity assays. The macrophages were enriched by adherence after 48 h of culture at 37°C in a 5% CO2 incubator. Where indicated, the macrophages were treated with 200 U/ml IFN-γ (Life Technologies, Gaithersburg, MD) for 18 h prior to S. Typhimurium infection.
Macrophage killing assays.
Macrophages were infected at a multiplicity of infection of 2 with S. Typhimurium previously opsonized with 10% normal mouse serum for 20 min. Extracellular bacteria were removed from the monolayers 25 min after infection by washing with prewarmed RPMI+ medium containing 12 μg/ml gentamicin (Sigma-Aldrich) (39). The number of bacteria remaining in the macrophages at various times of infection was determined after lysis of host cells with 0.25% deoxycholate and plating serial dilutions on LB agar plates. Percent survival was calculated as (CFU tn/CFU t0) × 100, where CFU tn and CFU t0 are the number of bacteria recovered after n hours and 25 min of infection, respectively.
Measurement of intracellular transcription of PhoP-activated genes and spiC.
The β-galactosidase activity of S. Typhimurium strains harboring lpxO::lacZY, pagP::lacZY, or spiC::lacZY transcriptional fusions was studied in gp91phox−/− macrophages. Macrophages were challenged at a multiplicity of infection of 2 with lacZY-expressing S. Typhimurium strains opsonized with 10% normal mouse serum for 20 min as described previously (40). Extracellular bacteria were removed from the macrophage monolayers 25 min after challenge by washing with prewarmed RPMI+ medium containing 12 μg/ml gentamicin (Sigma-Aldrich). Selected groups of macrophages were treated with 250 μM of the competitive NOS inhibitor NG-monomethyl l-arginine (NMMA) acetate salt (Sigma-Aldrich). S. Typhimurium-infected macrophages were lysed with 0.25% deoxycholate 8 h after challenge. The number of viable bacteria was calculated after serial dilution and plating on LB agar plates. β-Galactosidase activity was measured in an Lmax luminometer (Molecular Devices, Sunnyvale, CA) with the Galacto-Light chemiluminescent reporter gene assay system according to the manufacturer's instructions (Applied Biosystems, Foster City, CA). Intracellular expression is represented as mean β-galactosidase activity ± standard deviation (SD)/106 bacteria.
Synthesis of cDNA from S. Typhimurium-infected macrophages.
Total RNA was isolated from S. Typhimurium-infected, wild-type, or iNOS-deficient macrophages (39). Monolayers of S. Typhimurium-infected macrophages were solubilized in TRIzol reagent (Invitrogen, Carlsbad, CA) containing silicon beads and processed in a bead-beater (Biospec Products, Inc., Bartlesville, OK). RNA was extracted with chloroform, precipitated with a 1:1 mixture of isopropyl alcohol-0.8 M sodium citrate solution, washed in ethanol, and dried in a speed vacuum. RNA samples were resuspended in RNase/DNase-free H2O and digested for 1 h with DNase (Promega, Madison, WI). RNA was further purified with an RNAeasy kit according to the manufacturer's instructions (Qiagen, Valencia, CA). Complementary cDNA was synthesized at 42°C for 30 min using Moloney murine leukemia virus reverse transcriptase (Promega), RNasin, deoxynucleoside triphosphates, and random hexanucleotides. The cDNA was used as a template for quantitative reverse transcription-PCR (RT-PCR) and standard PCR analysis.
PCR transcriptional profiling.
Quantitative RT-PCRs included cDNA, Takara Omnimix HS (Takara Bio, Inc., Japan), forward and reverse primers for the target genes, and fluorescently labeled DNA probes for the phoP gene or the housekeeping rpoD sigma factor (Table 2). Real-time PCRs consisted of a cycle at 94°C for 45 s; 25 to 30 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s; and a cycle at 59°C for 30 s. The resulting fluorescence was measured with a SmartCycler II thermocycler (Cepheid, Sunnyvale, CA). Standard PCRs contained S. Typhimurium cDNA, deoxynucleoside triphosphates, Taq DNA polymerase (Continental Lab Products, San Diego, CA), and forward and reverse primers for the PhoP-activated gene mig-14 or the housekeeping gene rpoD (Table 2).
Nitrite quantification.
The iNOS activity expressed by S. Typhimurium-infected macrophages was estimated by quantifying the NO oxidative end product NO2−. NO2− was measured spectrophotometrically at 550 nm after mixing supernatants of S. Typhimurium-infected macrophages with an equal volume of the Griess reagent (0.5% sulfanilamide and 0.05% N-1-naphthylethylenediamide hydrochloride in 2.5% acetic acid). The NO2− concentration in the supernatants was determined by regression analysis using an NaNO2 standard curve.
Statistical analysis.
Data are presented as means ± standard errors of the mean (SEM) or SD. The statistical significance between multiple comparisons was calculated with a two-way analysis of variance, followed by a Bonferroni posttest. Data were considered statistically significant when the P value was <0.05.
RESULTS
NO inhibits PhoPQ-dependent gene transcription stimulated by low levels of Mg2+.
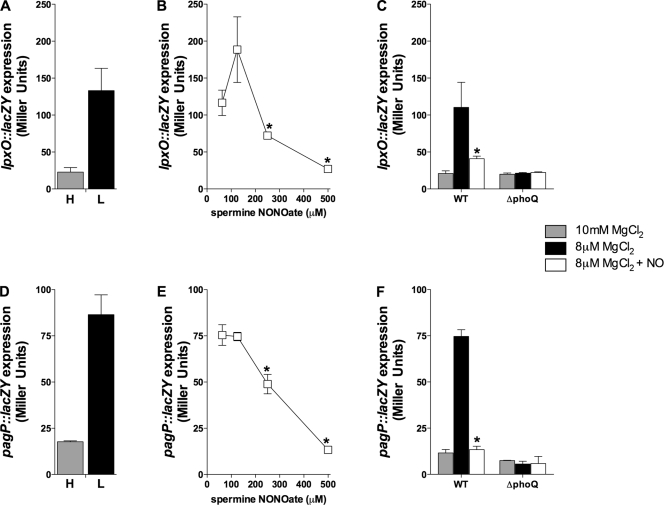
Previous work demonstrated that RNS produced under acidic conditions typically found in the stomach inhibit the PhoPQ-dependent acid tolerance response of rapidly growing S. Typhimurium (5). We therefore examined whether RNS produced at a neutral pH affect PhoPQ-dependent gene expression. As expected (21), the PhoP-activated genes lpxO and pagP were induced in S. Typhimurium grown in 8 μM MgCl2 N salts medium, pH 6.9 (Fig. 1A and D). Consistent with the RNS-mediated antagonism of PhoPQ signaling seen in S. Typhimurium cultured in EG medium, pH 4.4 (5), the NO donor spermine NONOate repressed in a dose-dependent manner the expression of lpxO and pagP sustained by S. Typhimurium grown in 8 μM MgCl2 N salts medium, pH 6.9 (Fig. 1B and E). Similar effects were seen with the PhoP-activated gene pmrD (data not shown). The inhibitory effects of spermine NONOate appear to be associated with its NO releasing capacity, since 500 μM of the spermine control did not reduce lpxO::lacZY or pagP::lacZY transcriptional activity (not shown). Moreover, the NO-mediated inhibition of transcription of PhoP-activated genes cannot be explained by the cytotoxicity of the drug, because the concentrations of spermine NONOate used here did not kill any of the S. Typhimurium strains tested (not shown). A total of 500 μM spermine NONOate reduced the transcription of these PhoP-activated genes to the basal level seen in controls grown in noninducing, 10 mM MgCl2 N salts medium, pH 7.6. The RNS-mediated repression of these loci appears to act on or upstream of the functional PhoPQ signal transduction pathway, because the marginal lpxO and pagP expression sustained in the absence of a functional PhoQ was unaltered upon NO treatment (Fig. 1C and F). Collectively, published data (5) and the findings described above demonstrate that RNS inhibit the PhoPQ-dependent gene expression stimulated by a variety of environmental conditions.
FIG. 1.
NO inhibits PhoP-activated gene transcription. Expression of PhoP-activated genes was quantified as β-galactosidase activity of lpxO::lacZY (A to C) or pagP::lacZY (D to F) transcriptional fusions. A fraction of the bacteria grown in high (H) 10 mM MgCl2 N salts medium to an OD600 of 0.5 was cultured for 2 h at 37°C in low (L) 8 μM MgCl2 N salts medium. (B and E) The NO donor spermine NONOate was added to select cultures in 8 μM MgCl2 N salts medium. Cultures indicated in panels C and F were treated with 500 μM spermine NONOate. The data represent the means ± SD of six independent observations from three separate experiments. *, P value of <0.05 compared to untreated controls. WT, wild type.
PhoPQ-dependent gene transcription is repressed in NO-producing, IFN-γ-treated macrophages.
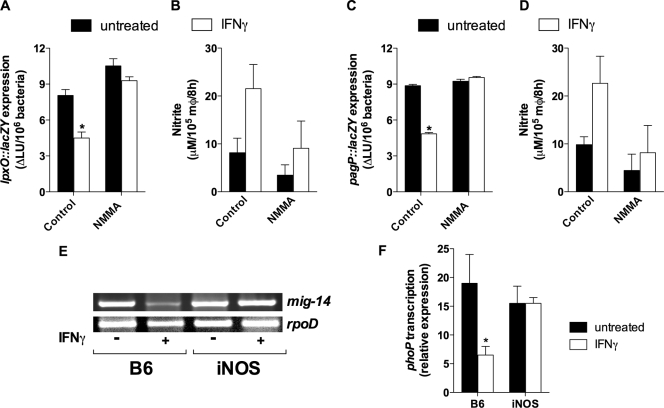
Next, we used gp91phox−/− macrophages to assess whether RNS generated by iNOS during innate or IFN-γ-primed responses of professional phagocytes affect PhoPQ-dependent gene transcription. This population of macrophages was chosen because it does not kill S. Typhimurium isolates, yet it sustains the SPI2-inhibitory nitrosative chemistry of wild-type phagocytes (40). The expression of lpxO and pagP was repressed (P < 0.001) in IFN-γ-stimulated, gp91phox-deficient macrophages (Fig. 2A and C). Addition of the NOS inhibitor NMMA not only reduced the NO output of IFN-γ-stimulated phagocytes (Fig. 2B and D) but restored lpxO and pagP transcription to the levels reported in unstimulated macrophages (Fig. 2A and C). The effect of host-derived RNS on PhoPQ-dependent gene transcription was independently studied by measuring the expression of mig-14. Consistent with the lpxO and pagP transcription patterns, mig-14 was repressed in an iNOS-dependent fashion inside IFN-γ-primed macrophages (Fig. 2E). Moreover, in accord with the expression patterns of the PhoP-activated genes pagP, lpxO, and mig-14, phoP transcripts were reduced about threefold in IFN-γ-treated macrophages compared to S. Typhimurium-infected, untreated phagocytes (Fig. 2F). These findings are in accord with the repression of phoP in S. Typhimurium exposed to NO in vitro (5). The levels of phoP mRNA remained unaltered in iNOS-deficient macrophages independently of their state of activation (Fig. 2F). These data suggest that the RNS generated by IFN-γ-primed macrophages inhibit the expression of PhoP-activated genes at the level of the PhoPQ two-component regulatory system.
FIG. 2.
Inhibition of PhoP-activated gene transcription by NO-producing, IFN-γ-treated macrophages. (A and C) Intracellular PhoP-activated gene expression was quantified in untreated or IFN-γ-primed gp91phox−/− macrophages by following the β-galactosidase activity of lpxO::lacZY and pagP::lacZY transcriptional fusions 8 h after infection. (B and D) A total of 250 μM of the competitive NOS inhibitor NMMA was added to selected groups of gp91phox−/− macrophages at the time of S. Typhimurium infection. Macrophage NO2− synthesis was quantified by the Griess reaction 8 h after infection. Data in panels A to D represent the mean ± SD of six to nine replicates from two to three separate experiments. *, a P value of <0.05 compared to untreated controls. (E) Transcription of the PhoP-activated gene mig-14 and the housekeeping gene rpoD was studied by RT-PCR of RNA samples isolated 20 h after S. Typhimurium infection from untreated (−) or IFN-γ-treated (+) peritoneal macrophages from C57BL/6 (B6) or iNOS−/− (iNOS) mice. (F) Transcription of phoP and the housekeeping sigma factor rpoD were quantified by real-time RT-PCR from RNA samples collected 20 h after S. Typhimurium infection of C57BL/6 (B6) or iNOS-deficient (iNOS) macrophages. The data represent the mean ± SEM of three to six independent experiments. Selected groups of macrophages were treated with 200 U/ml IFN-γ 18 h prior to infection. *, P value of <0.05 compared to untreated controls.
Contribution of RNS to the PhoQ-dependent intracellular survival of S. Typhimurium.
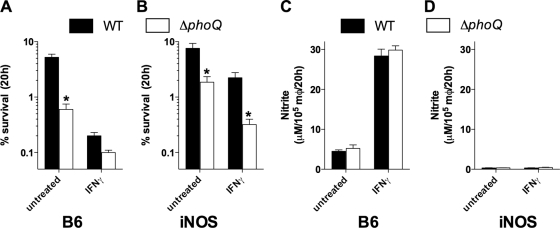
The PhoPQ two-component regulatory system is essential for the survival and proliferation of S. Typhimurium within professional phagocytes (18, 43). Because RNS produced enzymatically repress intracellular PhoP-dependent gene transcription (Fig. 2), the intracellular fitness of the S. Typhimurium phoQ mutant was investigated in untreated or IFN-γ-treated macrophages with differential nitrosative capacities (40). Compared to isogenic wild-type bacteria, 10-fold fewer S. Typhimurium phoQ mutant isolates were recovered from unstimulated macrophages isolated from C57BL/6 mice (Fig. 3A). IFN-γ increased both the iNOS-dependent, nitrosative capacity of the macrophages (Fig. 3C and D) and the intracellular killing of the wild type and the S. Typhimurium phoQ mutant (Fig. 3A). It should be noted that IFN-γ-primed macrophages contained twofold more wild-type than S. Typhimurium phoQ mutant isolates (Fig. 3A). Macrophages from iNOS-deficient mice were used to assess the contribution of RNS to the intracellular killing of the wild type and the S. Typhimurium phoQ mutant (Fig. 3B). Unstimulated, iNOS-deficient macrophages harbored about two- and fivefold more wild-type and S. Typhimurium phoQ mutant isolates, respectively (Fig. 3B), than iNOS-sufficient controls (Fig. 3A). Conversely, NO congeners generated by IFN-γ-primed macrophages appear to play a greater role controlling wild-type S. Typhimurium than the phoQ-deficient strain. IFN-γ-treated, iNOS-deficient macrophages harbored 10-fold more wild-type bacteria than IFN-γ-treated, iNOS-sufficient controls, whereas fourfold increases were seen in IFN-γ-treated, iNOS-deficient phagocytes infected with the S. Typhimurium phoQ mutant. Wild-type bacteria were still isolated in higher numbers than phoQ isogenic controls from IFN-γ-treated, iNOS-deficient macrophages. Together, these findings identify the PhoQ sensor kinase as a critical determinant of the antinitrosative defenses of the intracellular pathogen S. Typhimurium.
FIG. 3.
The PhoQ-mediated intracellular survival of S. Typhimurium is lost in NO-producing, IFN-γ-primed macrophages. The antimicrobial activity of untreated or IFN-γ-treated macrophages from C57BL/6 (B6) (A) and iNOS−/− (iNOS) (B) mice was recorded 20 h after infection with the wild type (WT) or the S. Typhimurium phoQ mutant. The nitrite-producing capacity of untreated and IFN-γ-treated macrophages from B6 (C) or iNOS-deficient (D) mice was determined 20 h after infection. The data represent the mean percent survival ± SEM of 14 independent observations from four separate experiments. *, P value of <0.001 compared to WT controls.
RNS repress intracellular SPI2 gene transcription by targeting PhoQ-independent signaling pathways.
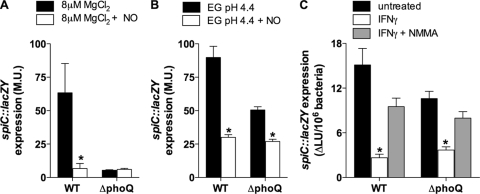
The iNOS-dependent inhibition of the PhoP regulon sustained by IFN-γ-primed macrophages (Fig. 2) closely resembles the NO-mediated inhibition of SPI2 transcription seen in activated macrophages (39). Since PhoPQ signaling can control SPI2 transcription (4, 12, 65), it is possible that the repression of SPI2 by NO congeners is a manifestation of adverse effects on PhoPQ. To test this hypothesis, spiC::lacZY transcriptional activity was investigated in the wild type and the S. Typhimurium phoQ mutant exposed to NO congeners. As reported for a variety of SPI2 loci (5, 39, 40), treatment of S. Typhimurium with 500 μM spermine NONOate repressed the spiC::lacZY expression induced in 8 μM MgCl2 N salts medium (Fig. 4A). In the absence of the sensor kinase PhoQ, not only was spiC::lacZY unresponsive to low MgCl2 levels, but its basal level of expression was unaltered by NO. In contrast to the predominant role that PhoPQ signaling exerts on SPI2 expression in S. Typhimurium grown in medium with low levels of Mg2+, less than 50% of the acid-inducible spiC::lacZY transcription appears to depend on PhoQ (Fig. 4B). Treatment of S. Typhimurium with 500 μM spermine NONOate completely inhibited the phoQ-dependent and -independent spiC::lacZY expression induced at low pH levels (Fig. 4B). As we have described for phoP-activated genes (5), a shift to pH 4.4 not only stimulated spiC::lacZY transcription (Fig. 4B) but also stopped bacterial growth (not shown). To determine if the iNOS-dependent inhibition of PhoP-activated gene transcription (Fig. 2) contributes to the intracellular repression of SPI2 (39), spiC::lacZY transcriptional activity was tested in the wild type and the phoQ-deficient S. Typhimurium mutant in macrophages from gp91phox−/− mice. This population of macrophages was selected because it does not kill S. Typhimurium, irrespective of the spiC status of the bacterium (39). To minimize further the effect that small variations in the intracellular bacterial burden may have on the estimation of spiC::lacZY expression, β-galactosidase activity was standardized to 106 CFU by counting the surviving bacteria capable of forming a colony on LB agar plates. spiC::lacZY expression was significantly (P < 0.01) reduced in the absence of a functional phoQ (Fig. 4C). In fact, a third of the intracellular spiC transcription sustained in unstimulated macrophages is dependent on a functional phoQ allele. This observation appears to support the idea that the intracellular expression of spiC relies on both PhoQ-dependent and -independent signals. The spiC::lacZY expression sustained by the wild type and the S. Typhimurium phoQ mutant was nonetheless repressed to similar levels in IFN-γ-primed macrophages (Fig. 4C). The NOS inhibitor NMMA restored spiC::lacZY expression of both the wild type and the S. Typhimurium phoQ mutant in IFN-γ-primed macrophages to levels sustained by the S. Typhimurium mutant in unstimulated phagocytes (Fig. 4C). Collectively, these data indicate that the RNS of IFN-γ-primed macrophages repress PhoQ-dependent and -independent signaling. Our data also indicate that the NO-dependent repression of intracellular spiC transcription seems to occur through the inhibition of PhoQ-independent signaling.
FIG. 4.
NO inhibits intracellular SPI2 transcription in PhoQ-independent ways. SPI2 expression was measured in the wild type (WT) and in the S. Typhimurium phoQ mutant strain harboring a spiC::lacZY transcriptional fusion. SPI2 expression was induced in 8 μM MgCl2 N salts medium (A) or in EG medium, pH 4.4 (B). A total of 500 μM spermine NONOate was added to selected cultures. Intracellular spiC::lacZY expression by the wild type or the S. Typhimurium phoQ mutant was quantified in untreated or IFN-γ-primed gp91phox−/− macrophages 8 h after infection. (C) Selected groups of IFN-γ-primed macrophages were treated with 250 μM NMMA at the time of infection. Data in panels A to C represent the means ± SD of six replicates from two separate experiments. *, P value of <0.05 compared with untreated controls.
RNS produced by IFN-γ-primed macrophages abrogate PhoQ- and SPI2-dependent intracellular survival of S. Typhimurium.
The contributions of the PhoPQ two-component regulatory and SPI2 type III secretion systems to the intracellular survival of S. Typhimurium were next investigated. Compared to wild-type controls, S. Typhimurium isolates harboring mutations in either phoQ or spiC were hypersusceptible to the antimicrobial activities of unstimulated, immunocompetent macrophages (Fig. 5A). It should be noted that twofold fewer S. Typhimurium phoQ mutant isolates were consistently (P < 0.05) recovered from the macrophages than spiC isogenic controls, indicating that PhoPQ plays a role in the intracellular survival of S. Typhimurium independent of its control of SPI2. Likewise, some of the contributions of SPI2 to the intracellular fitness of S. Typhimurium appear to be independent of PhoQ function, as suggested by the fact that a phoQ spiC double mutant was significantly (P < 0.05) more attenuated than S. Typhimurium deficient for only phoQ or spiC (Fig. 5A). The number of phoQ spiC S. Typhimurium bacteria isolated from unstimulated macrophages was similar to the number of wild-type bacteria isolated from IFN-γ-primed macrophages (Fig. 5A). The survival advantage associated with PhoQ and SPI2 was still manifested in untreated or IFN-γ-treated macrophages lacking iNOS (Fig. 5B). Collectively, these data suggest that NO congeners produced by IFN-γ-primed macrophages abrogate codependent and independent PhoPQ and SPI2 functions critical to the intracellular fitness of S. Typhimurium.
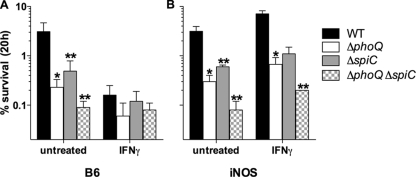
FIG. 5.
Abrogation of PhoPQ- and SPI2-dependent intracellular survival of S. Typhimurium in NO-producing, IFN-γ-primed macrophages. The percentage of wild-type (WT) S. Typhimurium and its isogenic phoQ, spiC, or phoQ spiC mutant control recovered from untreated or IFN-γ-treated macrophages from immunocompetent C57BL/6 (B6) (A) or iNOS−/− (iNOS) (B) mice was recorded 20 h after infection. The data represent the mean percent survival ± SEM of three to six independent observations from two separate experiments. *, P value of <0.001 compared to WT controls; **, P value of <0.05 compared to phoQ controls.
DISCUSSION
Because PhoPQ-dependent gene transcription is subject to repression by RNS generated under acidic conditions typically found in the stomach (5), we reasoned that PhoPQ signaling may also be inhibited by the nitrosative and oxidative stress associated with RNS of IFN-γ-stimulated macrophages (36, 40). The repression of the PhoP-activated genes lpxO, pagP, and mig-14 by NO congeners of IFN-γ-stimulated macrophages lends support to this hypothesis. NO generated in vitro in low-Mg2+ or acidic environments, which may recapitulate some aspects of the intraphagosomal environment, also repressed PhoPQ signaling. The mechanism for the NO-mediated inhibition of PhoPQ signaling is unknown at this time. NO inhibits the expression of the PhoP regulon in a low-Mg2+ environment known to directly activate the PhoQ sensor kinase. It is therefore possible that the RNS-dependent repression of PhoP-activated genes occurs at the level of the PhoPQ two-component regulatory system. Alternatively, the repression of the PhoP regulon may reflect the overall decrease of transcription and translation seen in S. Typhimurium undergoing nitrosative stress (5). It should be noted, however, that not all gene transcripts are affected equally in response to NO. For instance, NO downregulates about 10% of the S. Typhimurium transcriptome, of which SPI2 and PhoP-regulated genes are overwhelmingly represented, while RNS upregulate the transcription of approximately 10% of the genome (5). On the other hand, the fact that RNS inhibit the expression of spiC and PhoP-activated genes triggered at pH 4.4 in nondividing S. Typhimurium bacteria (5; see above) argues against the idea that the NO-mediated repression of transcription is an indirect manifestation of its effects on bacterial growth. Regardless of the mechanism of action, and given that S. Typhimurium bacteria harboring mutations in the PhoPQ two-component regulatory system grow poorly in macrophages and are highly attenuated in murine models of salmonellosis (18, 43), the RNS-mediated inhibition of the PhoP regulon represents a novel mechanism by which IFN-γ may contribute to host defense against this intracellular pathogen.
As expected (18, 43), the S. Typhimurium phoQ mutant was attenuated in macrophages. Defects in the iNOS hemoprotein increased the virulence of the S. Typhimurium phoQ mutant during the innate and IFN-γ-primed responses of macrophages. These data demonstrate a hitherto unknown role for the PhoPQ system in resistance to NO congeners of professional phagocytes. Despite growing better in iNOS-deficient macrophages, the S. Typhimurium phoQ mutant bacteria were still more readily killed by phagocytes unable to produce NO than isogenic wild-type bacteria. The known role of PhoPQ in remodeling the cell envelope may help explain the hypersusceptibility of the S. Typhimurium phoQ mutant to the killing activity of iNOS-deficient macrophages. PhoP-activated genes mediate resistance to a broad spectrum of defensins and α-helical peptides (28, 29, 35, 41, 44, 54, 55). Specifically, the outer membrane protease encoded by the PhoP-activated gene pgtE cleaves the α-helical peptide C18G (30), while the PhoP-regulated gene products of lpxO, pagP, pmrC, ugd, and ugtL reduce the permeability of the outer membranes of S. Typhimurium cells (46). LpxO-mediated hydroxylations stabilize the outer membrane by promoting hydrogen bonding between lipid A molecules, while the PagP-dependent palmitoylation increases hydrophobic interactions between the acyl chains of lipid A (25, 26, 32, 46). The negative charge of lipid A is reduced either by the PmrC- and Ugd-mediated addition of phosphoethanolamine and 4-aminoarabinose or by the UgtL-catalyzed reduction of phosphorylation (28, 31, 35). Finally, PhoP-mediated activation of mig-14 confers resistance to biologically active antimicrobial peptides by a yet unknown mechanism (6, 47). Consequently, by repressing the PhoP-activated lpxO, pagP, and mig-14 genes, RNS generated by IFN-γ-primed macrophages may increase the sensitivity of S. Typhimurium to host antimicrobial peptides.
Decreased antioxidant defenses may have also contributed to the enhanced susceptibility of the S. Typhimurium mutant to the antibacterial activity of iNOS-deficient macrophages. The PhoPQ regulatory system coordinates various antioxidant defenses. On one hand, the PhoP-activated gene product IraP protects S. Typhimurium from H2O2 cytotoxicity by stabilizing σS levels (58). On the other hand, PhoP activates the expression of the Cu/Zn superoxide dismutase CI (27), which is indispensable for full S. Typhimurium virulence in mice and survival within macrophages (11). Lastly, as discussed below, PhoP optimizes intracellular expression of the SPI2 type III secretion system associated with S. Typhimurium evasion of the NADPH phagocyte oxidase (20, 57, 61). The inhibition of PhoPQ-dependent signaling by RNS generated in IFN-γ-stimulated macrophages may therefore enhance the exposure of S. Typhimurium biomolecules to intracellular oxidative stress.
It is still unclear whether and to what extent PhoPQ regulates SPI2 transcription (4, 12, 16, 17, 23, 33, 34, 42, 65, 66). Under our experimental conditions, it appears that about one third of the intracellular expression of the SPI2 spiC gene depends on an intact PhoPQ signaling system. This observation is consistent with the partial contribution of PhoQ to the regulation of spiC under acidic conditions, but it contrasts with the tight control that PhoPQ exerts on spiC in Mg2+ poor environments. Inhibition of NOS activity restored the intracellular expression of spiC::lacZY in both the wild type and the S. Typhimurium phoQ mutant to levels sustained by the S. Typhimurium phoQ mutant in unstimulated phagocytes, suggesting that the NO-dependent repression of intracellular spiC transcription occurs mostly through the inhibition of PhoQ-independent signaling. Therefore, NO must inhibit presently unknown targets. The two-component regulatory systems SsrA/SsrB and EnvZ/OmpR or the MarR-like regulator SlyA, all known regulators of SPI2 transcription (15, 66), are among these possible targets. Cysteine and tyrosine residues are preferred amino acid targets of RNS. With the exception of EnvZ and OmpR, all these regulatory proteins have one or more cysteine, while all but SlyA contain tyrosines. Future investigations will be needed to determine whether SsrA, SsrB, EnvZ, OmpR, or SlyA contains redox active residues that impart regulatory functions to these proteins.
Our data indicate that PhoPQ and SPI2 are primary targets of the RNS-dependent anti-S. Typhimurium activity of IFN-γ-activated macrophages, as shown by the fact that a phoQ spiC double mutant was killed as efficiently by untreated phagocytes as by IFN-γ-treated macrophages, irrespective of their nitrosative capacities. The data presented herein also show that the functions of PhoPQ and SPI2 overlap to a great extent. This could be explained by the aforementioned regulation of SPI2 by PhoP. However, PhoPQ and SPI2 may also act independently to establish a nonfusogenic phagosome that avoids contact with known oxygen-dependent and -independent host defenses (24). Although the protective mechanisms of PhoPQ and SPI2 appear to antagonize common host defenses, our investigations suggest some specialized functions as well. For example, the S. Typhimurium phoQ mutant is consistently less fit for survival in the intracellular environment than spiC-deficient isogenic controls. Moreover, a strain of S. Typhimurium lacking both phoQ and spiC is significantly more attenuated than controls bearing single mutations.
In summary, we have shown that the PhoPQ two-component regulatory system plays an important role in defending S. Typhimurium against NO congeners generated in the innate response, while it becomes a target of RNS engendered by activated phagocytes. The RNS-dependent inhibition of PhoPQ signaling and SPI2 type III secretion may expose S. Typhimurium to a deluge of antimicrobial effectors as varied as oxyradicals generated by the NADPH phagocyte oxidase or hydrolytic enzymes and antimicrobial peptides of the degradative pathway, thereby contributing to the cytotoxicity of the NO congeners of activated macrophages.
Acknowledgments
Support of this work was provided by the National Institutes of Health (AI054959 and T32 AI52066), the Burroughs Wellcome Fund, and the Schweppe Foundation.
We are grateful to James Laughlin and Bruce McCollister for providing assistance with the macrophage assays and Jessica Jones-Carson for editing the manuscript.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 8 September 2009.
REFERENCES
- 1.Alam, M. S., T. Akaike, S. Okamoto, T. Kubota, J. Yoshitake, T. Sawa, Y. Miyamoto, F. Tamura, and H. Maeda. 2002. Role of nitric oxide in host defense in murine salmonellosis as a function of its antibacterial and antiapoptotic activities. Infect. Immun. 70:3130-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. [DOI] [PubMed] [Google Scholar]
- 3.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlsma, J. J., and E. A. Groisman. 2005. The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57:85-96. [DOI] [PubMed] [Google Scholar]
- 5.Bourret, T. J., S. Porwollik, M. McClelland, R. Zhao, T. Greco, H. Ischiropoulos, and A. Vazquez-Torres. 2008. Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One 3:e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodsky, I. E., R. K. Ernst, S. I. Miller, and S. Falkow. 2002. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J. Bacteriol. 184:3203-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castelli, M. E., E. Garcia Vescovi, and F. C. Soncini. 2000. The phosphatase activity is the target for Mg2+ regulation of the sensor protein PhoQ in Salmonella. J. Biol. Chem. 275:22948-22954. [DOI] [PubMed] [Google Scholar]
- 8.Chamnongpol, S., and E. A. Groisman. 2002. Mg2+ homeostasis and avoidance of metal toxicity. Mol. Microbiol. 44:561-571. [DOI] [PubMed] [Google Scholar]
- 9.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, A. Vazquez-Torres, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deiwick, J., T. Nikolaus, S. Erdogan, and M. Hensel. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759-1773. [DOI] [PubMed] [Google Scholar]
- 13.Ding, A. H., C. F. Nathan, and D. J. Stuehr. 1988. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J. Immunol. 141:2407-2412. [PubMed] [Google Scholar]
- 14.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 15.Fass, E., and E. A. Groisman. 2009. Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, X., R. Oropeza, and L. J. Kenney. 2003. Dual regulation by phospho-OmpR of ssrA/B gene expression in Salmonella pathogenicity island 2. Mol. Microbiol. 48:1131-1143. [DOI] [PubMed] [Google Scholar]
- 17.Feng, X., D. Walthers, R. Oropeza, and L. J. Kenney. 2004. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol. Microbiol. 54:823-835. [DOI] [PubMed] [Google Scholar]
- 18.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 19.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallois, A., J. R. Klein, L. A. Allen, B. D. Jones, and W. M. Nauseef. 2001. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J. Immunol. 166:5741-5748. [DOI] [PubMed] [Google Scholar]
- 21.García Véscovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 22.García Véscovi, E., F. C. Soncini, and E. A. Groisman. 1994. The role of the PhoP/PhoQ regulon in Salmonella virulence. Res. Microbiol. 145:473-480. [DOI] [PubMed] [Google Scholar]
- 23.Garmendia, J., C. R. Beuzon, J. Ruiz-Albert, and D. W. Holden. 2003. The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella typhimurium SPI-2 type III secretion system. Microbiology 149:2385-2396. [DOI] [PubMed] [Google Scholar]
- 24.Garvis, S. G., C. R. Beuzon, and D. W. Holden. 2001. A role for the PhoP/Q regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell. Microbiol. 3:731-744. [DOI] [PubMed] [Google Scholar]
- 25.Gibbons, H. S., S. Lin, R. J. Cotter, and C. R. Raetz. 2000. Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, a new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. J. Biol. Chem. 275:32940-32949. [DOI] [PubMed] [Google Scholar]
- 26.Gibbons, H. S., C. M. Reynolds, Z. Guan, and C. R. Raetz. 2008. An inner membrane dioxygenase that generates the 2-hydroxymyristate moiety of Salmonella lipid A. Biochemistry 47:2814-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Golubeva, Y. A., and J. M. Slauch. 2006. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutase SodCI is a member of the PhoPQ regulon and is induced in macrophages. J. Bacteriol. 188:7853-7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groisman, E. A., J. Kayser, and F. C. Soncini. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. USA 89:11939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guina, T., E. C. Yi, H. Wang, M. Hackett, and S. I. Miller. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar Typhimurium promotes resistance to alpha-helical antimicrobial peptides. J. Bacteriol. 182:4077-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 32.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 33.Kim, C. C., and S. Falkow. 2004. Delineation of upstream signaling events in the Salmonella pathogenicity island 2 transcriptional activation pathway. J. Bacteriol. 186:4694-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, A. K., C. S. Detweiler, and S. Falkow. 2000. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J. Bacteriol. 182:771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, H., F. F. Hsu, J. Turk, and E. A. Groisman. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lundberg, J. O., E. Weitzberg, J. A. Cole, and N. Benjamin. 2004. Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2:593-602. [DOI] [PubMed] [Google Scholar]
- 37.MacMicking, J. D., C. Nathan, G. Hom, N. Chartrain, D. S. Fletcher, M. Trumbauer, K. Stevens, Q. W. Xie, K. Sokol, N. Hutchinson, et al. 1995. Altered responses to bacterial infection and endotoxic shock in mice lacking inducible nitric oxide synthase. Cell 81:641-650. [DOI] [PubMed] [Google Scholar]
- 38.Mastroeni, P., A. Vasquez-Torrez, F. C. Fang, Y. Xu, S. Khan, C. E. Hormeache, and G. Dougan. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCollister, B. D., T. J. Bourret, R. Gill, J. Jones-Carson, and A. Vazquez-Torres. 2005. Repression of SPI2 transcription by nitric oxide-producing, IFN-γ-activated macrophages promotes maturation of Salmonella phagosomes. J. Exp. Med. 202:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCollister, B. D., J. T. Myers, J. Jones-Carson, M. Husain, T. J. Bourret, and A. Vazquez-Torres. 2007. N2O3 enhances the nitrosative potential of IFN-γ-primed macrophages in response to Salmonella. Immunobiology 212:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLeod, G. I., and M. P. Spector. 1996. Starvation- and stationary-phase-induced resistance to the antimicrobial peptide polymyxin B in Salmonella typhimurium is RpoS (σS) independent and occurs through both phoP-dependent and -independent pathways. J. Bacteriol. 178:3683-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miao, E. A., J. A. Freeman, and S. I. Miller. 2002. Transcription of the SsrAB regulon is repressed by alkaline pH and is independent of PhoPQ and magnesium concentration. J. Bacteriol. 184:1493-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. USA 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, S. I., W. S. Pulkkinen, M. E. Selsted, and J. J. Mekalanos. 1990. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium. Infect. Immun. 58:3706-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monsieurs, P., S. De Keersmaecker, W. W. Navarre, M. W. Bader, F. De Smet, M. McClelland, F. C. Fang, B. De Moor, J. Vanderleyden, and K. Marchal. 2005. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J. Mol. Evol. 60:462-474. [DOI] [PubMed] [Google Scholar]
- 46.Murata, T., W. Tseng, T. Guina, S. I. Miller, and H. Nikaido. 2007. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:7213-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarre, W. W., T. A. Halsey, D. Walthers, J. Frye, M. McClelland, J. L. Potter, L. J. Kenney, J. S. Gunn, F. C. Fang, and S. J. Libby. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56:492-508. [DOI] [PubMed] [Google Scholar]
- 48.Pollock, J. D., D. A. Williams, M. A. Gifford, L. L. Li, X. Du, J. Fisherman, S. H. Orkin, C. M. Doerschuk, and M. C. Dinauer. 1995. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9:202-209. [DOI] [PubMed] [Google Scholar]
- 49.Poteete, A. R., and A. C. Fenton. 1984. Lambda red-dependent growth and recombination of phage P22. Virology 134:161-167. [DOI] [PubMed] [Google Scholar]
- 50.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prost, L. R., M. E. Daley, V. Le Sage, M. W. Bader, H. Le Moual, R. E. Klevit, and S. I. Miller. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26:165-174. [DOI] [PubMed] [Google Scholar]
- 52.Richardson, A. R., K. C. Soliven, M. E. Castor, P. D. Barnes, S. J. Libby, and F. C. Fang. 2009. The base excision repair system of Salmonella enterica serovar Typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog. 5:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schapiro, J. M., S. J. Libby, and F. C. Fang. 2003. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc. Natl. Acad. Sci. USA 100:8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi, Y., M. J. Cromie, F. F. Hsu, J. Turk, and E. A. Groisman. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53:229-241. [DOI] [PubMed] [Google Scholar]
- 55.Shi, Y., T. Latifi, M. J. Cromie, and E. A. Groisman. 2004. Transcriptional control of the antimicrobial peptide resistance ugtL gene by the Salmonella PhoP and SlyA regulatory proteins. J. Biol. Chem. 279:38618-38625. [DOI] [PubMed] [Google Scholar]
- 56.Shiloh, M. U., J. D. MacMicking, S. Nicholson, J. E. Brause, S. Potter, M. Marino, F. Fang, M. Dinauer, and C. Nathan. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10:29-38. [DOI] [PubMed] [Google Scholar]
- 57.Suvarnapunya, A. E., and M. A. Stein. 2005. DNA base excision repair potentiates the protective effect of Salmonella pathogenicity island 2 within macrophages. Microbiology 151:557-567. [DOI] [PubMed] [Google Scholar]
- 58.Tu, X., T. Latifi, A. Bougdour, S. Gottesman, and E. A. Groisman. 2006. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. USA 103:13503-13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Velkinburgh, J. C., and J. S. Gunn. 1999. PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vazquez-Torres, A., J. Jones-Carson, P. Mastroeni, H. Ishiropoulus, and F. C. Fang. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]
- 62.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 63.Webb, J. L., M. W. Harvey, D. W. Holden, and T. J. Evans. 2001. Macrophage nitric oxide synthase associates with cortical actin but is not recruited to phagosomes. Infect. Immun. 69:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White, J. K., P. Mastroeni, J. F. Popoff, C. A. Evans, and J. M. Blackwell. 2005. Slc11a1-mediated resistance to Salmonella enterica serovar Typhimurium and Leishmania donovani infections does not require functional inducible nitric oxide synthase or phagocyte oxidase activity. J. Leukoc. Biol. 77:311-320. [DOI] [PubMed] [Google Scholar]
- 65.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 66.Yoon, H., J. E. McDermott, S. Porwollik, M. McClelland, and F. Heffron. 2009. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 5:e1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]