Abstract
Hospital-acquired Enterococcus faecium isolates responsible for nosocomial outbreaks and invasive infections are enriched in the orf2351 and orf2430 genes, encoding the SgrA and EcbA LPXTG-like cell wall-anchored proteins, respectively. These two surface proteins were characterized to gain insight into their function, since they may have favored the rapid emergence of this nosocomial pathogen. We are the first to identify a surface adhesin among bacteria (SgrA) that binds to the extracellular matrix molecules nidogen 1 and nidogen 2, which are constituents of the basal lamina. EcbA is a novel E. faecium MSCRAMM (microbial surface component recognizing adhesive matrix molecules) that binds to collagen type V. In addition, both SgrA and EcbA bound to fibrinogen; however, SgrA targeted the alpha and beta chains, whereas EcbA bound to the gamma chain of fibrinogen. An E. faecium sgrA insertion mutant displayed reduced binding to both nidogens and fibrinogen. SgrA did not mediate binding of E. faecium cells to biotic materials, such as human intestinal epithelial cells, human bladder cells, and kidney cells, while this LPXTG surface adhesin is implicated in E. faecium biofilm formation. The acm and scm genes, encoding two other E. faecium MSCRAMMs, were expressed at the mRNA level together with sgrA during all phases of growth, whereas ecbA was expressed only in exponential and late exponential phase, suggesting orchestrated expression of these adhesins. Expression of these surface proteins, which bind to extracellular matrix proteins and are involved in biofilm formation (SgrA), may contribute to the pathogenesis of hospital-acquired E. faecium infections.
Enterococcus faecium has emerged as an important opportunistic gram-positive pathogen, responsible for nosocomial infections and hospital outbreaks worldwide (1, 30, 49). E. faecium can cause a variety of infections in immunocompromised patients, such as urinary tract infections, surgical site infections, bacteremia, and endocarditis. Hospital-acquired E. faecium isolates recovered from clinical sites and isolates associated with nosocomial outbreaks clearly differ from clinically nonrelevant E. faecium (25). These hospital-acquired E. faecium strains show high-level resistance to ampicillin and ciprofloxacin (24), are enriched in putative virulence genes, such as esp (23), which encodes the enterococcal surface protein and is implicated in biofilm formation (16, 47), genes putatively encoding the PilA and PilB pilus-like structures (17), and three cell wall-anchored LPXTG surface proteins, designated Orf903, Orf2351, and Orf2430 (18). These three surface proteins have in common that they contain an N-terminal signal peptide, a nonrepetitive A domain, and a C-terminal cell wall sorting signal (CWS), which is comprised of a highly conserved LPXTG-like (Leu-Pro-X-Thr-Gly, where X denotes any amino acid) sortase substrate motif and a hydrophobic domain followed by positively charged amino acids (27, 40). After translocation of the precursor protein, the LPXTG motif is cleaved by sortase, which subsequently anchors the surface protein to the cell wall peptidoglycan (27, 28).
A class of LPXTG surface proteins is comprised of the microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (37, 38), which contain, in addition to having characteristics of gram-positive cell wall-anchored surface proteins, at least one DE variant of the immunoglobulin G (IgG)-like fold in the N-terminal A domain (9). MSCRAMMs are often bifunctional proteins that adhere to one or more components of the host extracellular matrix (ECM). The ECM is a complex three-dimensional structure surrounding cells in mammalian tissue and a critical site for initial bacterial attachment and colonization. To date, two MSCRAMMs have been described for E. faecium that bind to components of the ECM, named Acm (adhesin of collagen from E. faecium) and Scm (second collagen adhesin of E. faecium). Acm interacts with collagen type I and to a lesser extent with collagen type IV (31, 35), and Scm binds to collagen type V and fibrinogen (42). These two collagen-binding MSCRAMMs were found ubiquitously among enterococcal isolates of clinical and nonclinical origin. However, while a functional acm gene is predominantly present in clinical isolates promoting adherence to collagen type I, it often occurs as an insertion element-interrupted pseudogene in isolates of nonclinical origin (33). These isolates did not bind collagen type I. In a rat endocarditis model in which equal amounts of an acm deletion mutant and the wild type were injected intraventricularly, the mutant strain was outnumbered by the wild type on the heart valve vegetations (32).
In this study, we functionally characterized two LPXTG surface proteins, Orf2351 and Orf2430, which we recently described (18) and which are now renamed SgrA and EcbA, respectively. We assessed whether these surface proteins can bind to protein ligands of the ECM. Furthermore, we investigated the function of an sgrA mutant in adhesion to biotic and abiotic surfaces.
(Part of this study was presented as a poster at the 108th General Meeting of the American Society for Microbiology, Boston, MA, 1 to 5 June 2008 [abstr. 08-GM-A-2548-ASM].)
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
The bacterial strains and plasmids used in this study are listed in Table 1. Rosetta-gami (DE3) pLysS Escherichia coli cells, containing the pRSETB vector (Invitrogen Corporation, Breda, The Netherlands), were used for expression of recombinant protein and were grown at 37°C in Luria-Bertani broth or agar, supplemented with carbenicillin (50 μg/ml) and chloramphenicol (34 μg/ml). E. faecium strains were grown aerobically at 37°C on Trypticase soy agar II plates supplemented with 5% sheep blood (Becton Dickinson, Alphen aan den Rijn, The Netherlands) or in brain heart infusion (BHI) broth, and when appropriate, the antibiotics chloramphenicol and gentamicin were used at concentrations of 10 μg/ml, and 125 μg/ml, respectively. Antibiotics were obtained from Sigma-Aldrich (St. Louis, MO). A temperature-sensitive vector (pTEX5500ts) was used to introduce an insertion-deletion mutation in the sgrA gene of E. faecium E1162, a clinical bloodstream isolate. Extracellular matrix molecules, collagen type I to type V, laminin (ultrapure), fibronectin, fibrinogen (plasminogen free), and vitronectin were all from human plasma (Sigma-Aldrich B.V., Zwijndrecht, The Netherlands). Bovine serum albumin (BSA) was purchased from Serva (Heidelberg, Germany). Human recombinant nidogen 1 and nidogen 2 containing an N-terminal His tag were purchased from R&D Systems (Abingdon, United Kingdom).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10F | E. coli host strain for routine cloning | Invitrogen |
| DH5α | E. coli host strain for routine cloning | 14 |
| Rosetta-Gami | DE3 pLysS for high-level recombinant protein expression | Novagen |
| E. faecium | ||
| E135 | Community surveillance feces isolate; esp, sgrA, and ecbA negative | 50, 51 |
| E155 | Hospital outbreak-associated isolate; esp, sgrA, and ecbA positive | 4, 51 |
| E300 | Hospital outbreak-associated isolate, from urine; esp, sgrA, and ecbA positive | 12, 51 |
| E470 | Hospital outbreak-associated isolate; esp, sgrA, and ecbA positive | 44, 51 |
| U0317 | Clinical isolate, from urine; esp, sgrA, and ecbA positive | This study |
| E1162 | Clinical blood isolate; Chls Gens; esp, sgrA, and ecbA positive | 16 |
| E1162ΔsgrA | E1162 with a disrupted sgrA gene; Chlr Gens; esp and ecbA positive | This study |
| TX0016 | Clinical endocarditis isolate; sgrA and ecbA positive | 2 |
| Plasmids | ||
| pRSETB | Expression vector | Novagen |
| pAH2351 | pRSETB derivative, to express rSgrA protein (residues 76W to 297N) | This study |
| pAH2430 | pRSETB derivative, to express rEcbA protein (residues 35E to 347T) | This study |
| pTEX5500ts | Shuttle plasmid; temp-sensitive in gram-positive hosts; Chlr Genr | 31 |
| pEF4 | pTEX5500ts:sgrA-Up, pTEX5500ts with a cloned sgrA-Up gene fragment; Chlr Genr | This study |
| pEF9 | pTEX5500ts:sgrA-Up-sgrA-Dn, pTEX5500ts with cloned sgrA gene fragments flanking the cat gene, for generating an sgrA insertion-deletion mutation; Chlr Genr | This study |
Chl, chloramphenicol; Gen, gentamicin; ts, temperature sensitive. Superscripts “s” and “r” indicate sensitivity and resistance, respectively.
PCR amplification and cloning of sgrA and ecbA.
Fragments of sgrA (encoding residues 76W to 297N) and ecbA (encoding residues 35E to 347T) were amplified by PCR using the primer sets SgrA_fw and SgrA_rv and EcbA_fw and EcbA_rv (Table 2). PCRs were performed at 25-μl volumes using HotStarTaq HiFidelity DNA polymerase (Qiagen Benelux B.V., Venlo, The Netherlands) with 10 pmol of each primer and 10 nmol of E. faecium TX0016 chromosomal DNA. DNA was isolated as described previously (3, 18). Restriction sites for the NdeI and BamHI enzymes (New England Biolabs, Ipswich, MA) were incorporated at the 5′ ends of the primers to facilitate directional cloning. In SgrA_fw and EcbA_fw, a 5′ ATG and 6× CAT were added for the in-frame expression of recombinant SgrA and EcbA. In SgrA_rv and EcbA_rv, a TGA stop codon was added. The two PCR fragments were purified by use of the PCR purification kit (Qiagen) and cloned into the pRSETB expression vector (Invitrogen), resulting in pAH2351 and pAH2430, respectively, and were subsequently used to transform One Shot TOP10F′ chemically competent E. coli (Invitrogen) according to the manufacturer's instructions. Recombinant plasmid was isolated with a plasmid purification kit (Qiagen) and analyzed by sequencing.
TABLE 2.
Oligonucleotides used in this study
| Primer namea | Oligonucleotide sequence (5′-3′) |
|---|---|
| ddl_fw | GAGACATTGAATATGCCTTATG |
| ddl_rv | AAAAAGAAATCGCACCG |
| Scm_fw | CTAACTGGTAACTATGGCTTGT |
| Scm_rv | GTCCGTGCTGTCACTTGT |
| Acm_fw | TCAGCAGTAATGTCACTTCGTTG |
| Acm_rv | GAATAGGCTGTTCATCTGCTCG |
| SgrA_fw | GGAATTCCATATGCGGGGTTCTCATCATCATCATCATCATGGTTGGGATGGTAATGGAAGTTCA |
| SgrA_rv | CGCGGATCCGCGTCAGTTCAAGGTTCTACTACCAGT |
| EcbA_fw | GGAATTCCATATGCGGGGTTCTCATCATCATCATCATCATGAAATTACTCATCCACAAACGGTA |
| EcbA_rv | CGCGGATCCGCGTCATGTAGTGTCAATCGTATAAGG |
| SgrA_fw2 | AATGAACGGGCAAATGAG |
| SgrA_rv2 | CTTTTGTTCCTTAGTTGGTATGA |
| EcbA_fw2 | GCAGTTTACAATGGTGTGAAGCAA |
| EcbA_rv2 | CGGCTAATGAGTATTTGTCGTTCC |
| SgrA-Up_fw | TTCCGCGGCCGCTATGGCCGACGTCGTCGACGCGTGGCTGAGTATAATTGCAG |
| SgrA-Up_rv | CGCGGATCCGCGCAAATTCTCCGTGATCGTCAT |
| SgrA-Dn_fw | AACTGCAGAACCAATGCATTGGGAGAAATGCAGGGAGCAAC |
| SgrA-Dn_rv | GCCTTAAGGCTTCACTCGATGGAAGAGAGAAC |
| SgrA_fw3 | ATGAAG AAAACAGCGACCGT |
| Orf2352_rv | TTATTCAATTTTAGATCTGT |
| SgrA_ser_F | AGTTGGACAGTTGTTGGACC |
| SgrA_ser_R | CTCGTGCTTCCTGTGCTACT |
| Cam_F | TTTAATAAAATTGATTTAGA |
| Cam_R | CCTGAATAGAGTTCATAAAC |
fw, forward; rv, reverse.
Expression and purification of recombinant SgrA and EcbA.
The plasmids pAH2351 and pAH2430 were used to transform Rosetta-Gami (DE3) pLysS chemically competent E. coli (Novagen, Gibbstown, NJ). These E. coli cells were grown to an optical density at 660 nm (OD660) of ∼0.7 in 200 ml Luria-Bertani broth supplemented with 0.02 M glucose. Expression of recombinant SgrA and EcbA containing a six-His tag at their N-terminal ends (rSgrA and rEcbA) was induced by 10 mM isopropyl-β-d-thiogalactopyranoside. The recombinant fusion proteins were purified by use of immobilized Ni2+ affinity chromatography (Probond; Invitrogen), all according to the manufacturer's instructions. Purified rSgrA and rEcbA were dialyzed overnight against 0.1× phosphate-buffered saline (PBS) at 4°C and concentrated by lyophilization, and purity was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE, Western blot, and ligand affinity blot analysis.
Protein samples were equally mixed with sample buffer (100 mM Tris-HCl, 5% dithiothreitol, 2% SDS, 0,004% bromophenol blue, and 20% glycerol) and boiled for 5 min. Equal amounts of protein were electrophoresed through a 10% SDS-polyacrylamide gel. Western blotting was carried out as described previously (18). Membranes were blocked with 4% skim milk (Campina Holland, Alkmaar, The Netherlands) in PBS-0.1% Tween 20 for 1 h at 37°C. Incubation with primary antibody was carried out for 1 h in 1% BSA in PBS-1% Tween 20 at 37°C, followed by two washes 10 min (each) in PBS-0.1% Tween 20 at 37°C. Subsequently, membranes were incubated for 1 h with goat anti-mouse IgG(H+L)-horseradish peroxidase (HRP) (Bio-Rad Laboratories, Veenendaal, The Netherlands) in 1% BSA in PBS-1% Tween 20 at 37°C. Membranes were washed twice with PBS-0,1% Tween 20, and proteins were visualized using the ECL Plus Western blotting detection system (GE Healthcare, Diegem, Belgium) and exposed to film (Hyperfilm ECL; GE Healthcare). For the ligand affinity blots, 20 μg of ECMs were separated by SDS-PAGE and blotted onto nitrocellulose, and after blocking, the membranes were incubated for 1 h at room temperature with 5.0 μg/ml of rSgrA or rEcbA, followed by 1 h of incubation with anti-His IgG-HRP antibodies (Qiagen) at 37°C. For the nonreducing ligand affinity blots, SDS was omitted from buffers.
Biotinylation of recombinant proteins.
Recombinant nidogen 1, nidogen 2, and rSgrA containing an N-terminal His tag were biotinylated using sulfo-N-hydroxysuccinimide-biotin according to the manufacturers instructions (Pierce Biotechnology, Rockford, IL). Biotinylation of the ECM proteins and rSgrA was analyzed by Western blotting using a streptavidin-HRP conjugate (Pierce).
mRNA expression analysis by reverse transcription (RT)-PCR.
For mRNA expression of the sgrA, ecbA, acm, and scm genes, E135 and E1162 cells were resuspended in PBS to an OD660 of 1.0 (∼1 × 109 CFU/ml) and pelleted by centrifugation (6,500 × g for 1 min). Total RNA was isolated and purified, and cDNA synthesis was done as described previously (6, 18). cDNA was used as a template for PCR using the primer pairs SgrA_fw2 and SgrA_rv2, EcbA_fw2 and EcbA_rv2, Acm_fw and Acm_rv, and Scm_fw and Scm_rv as depicted in Table 2. As an internal control, the housekeeping gene ddl (encoding d-alanine,d-alanine ligase) was amplified using primers ddl_fw and ddl_rv (Table 2). RNA samples not treated with reverse transcriptase were used as a control to detect DNA contamination in the total RNA preparations.
Enzyme-linked immunosorbent assay (ELISA) and whole-cell ELISA.
Polystyrene microtiter plates (Greiner Microlon from Greiner Bio-one, Alphen aan den Rijn, The Netherlands) were coated overnight at 4°C with 10 μg/ml of ECMs in 50-μl volumes, and BSA was used as a negative control. After three washes with wash buffer (PBS-0.05% Tween 20), nonspecific sites were blocked with 100-μl blocking buffer (4% BSA in PBS-0.05% Tween 20) for 1 h at 37°C. Various concentrations of recombinant protein in 1% BSA in PBS-0.05% Tween 20 were added to the wells and incubated at 37°C. After 2 h, unbound protein was removed by washing three times. Bound proteins were detected by anti-His IgG-HRP antibodies (Qiagen) in 1% BSA in PBS-0.05% Tween 20 (1:5,000 dilution) for 1 h at 37°C.
Binding of E1162 and the sgrA mutant strain, E1162ΔsgrA, to immobilized nidogen 1, fibrinogen, laminin, and fibronectin (all 25 μg/ml) was assayed three times in duplicate by whole-cell ELISA. Plate-grown bacteria were resuspended in PBS to an OD660 of 0.2 (2 × 108 CFU/ml), and 100 μl was added to wells of a microtiter plate and allowed to bind overnight at 4°C. The wells were washed three times with wash buffer and subsequently blocked with blocking buffer for 1 h at 37°C. Binding of E1162 and E1162ΔsgrA E. faecium was assayed by incubation for 1 h at 37°C with rabbit antienterococcus serum (kindly provided by J. Huebner). Bound antibodies were detected by incubation with a peroxidase-conjugated goat anti-rabbit IgG (1:5,000; Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 37°C. Both antibodies were diluted in PBS with 1% BSA and 0.05% Tween 20. For both types of ELISAs, 50 μl of 0.11 M acetate buffer with 1.6% 3,3,5,5-tetramethylbenzidine and 0.8% ureumperoxide was added to each well, and the reaction was stopped after 10 min with 50-μl 0.5 M H2SO4. The absorbance was measured at 450 nm with an ELISA reader.
Mutagenesis of sgrA by insertional inactivation.
The sgrA gene of E. faecium E1162 was disrupted by construction of an insertion-deletion mutation as described previously (16, 31). In brief, a 373-bp sgrA fragment designated SgrA-Up was amplified from genomic E. faecium E1162 DNA by using the primers SgrA-Up_fw and SgrA-Up_rv, which contain SalI and BamHI restriction sites, respectively (Table 2). The PCR product was digested with SalI and BamHI and ligated to similarly digested pTEX5500ts, resulting in pEF4. Similarly, a 218-bp fragment designated SgrA-Dn was amplified by using the primers SgrA-Dn_fw and SgrA-Dn_rv, including NsiI and EcoRI restriction sites incorporated at the 5′ ends of the primers to facilitate directional cloning into pEF4, resulting in pEF9 (Table 1). The pEF9 plasmid was introduced into competent E. faecium E1162 by electroporation. After transformation, gentamicin-resistant colonies were picked and grown overnight at 42°C in BHI broth supplemented with gentamicin and subsequently plated on BHI agar plates with chloramphenicol and grown at 42°C. Single-crossover integration into SgrA-Up and SgrA-Dn complementary sequences of E. faecium E1162 was tested by PCR. Single-crossover mutants were grown overnight for eight serial passages in BHI culture supplemented with chloramphenicol at 42°C to cure pEF9. Selection for double-crossover recombination was performed by replica plating on BHI agar plates containing either chloramphenicol or gentamicin. The insertional inactivation of the sgrA gene in double-crossover mutants was analyzed by PCR with the primers SgrA_fw3 and Orf2352_rv, sequencing, and Southern hybridization.
Southern blot analysis.
Southern blot analysis was performed on chromosomal DNA isolated from E. faecium E1162 and E1162ΔsgrA to confirm disruption of the chromosomal sgrA gene. Chromosomal DNA was digested with EcoRI (Roche diagnostics, Almere, The Netherlands), and fragments were separated by agarose gel electrophoresis and blotted onto an Hybond N+ nylon membrane (GE Healthcare, Diegem, Belgium). DNA was fixed onto the membrane by incubation for 2 min in 0.4 M NaOH followed by a neutralization in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 1 min. The membrane was hybridized overnight at 42°C with 100 ng probe. Probes were generated by PCR in 50-μl volumes using the primer pairs Cam_F and Cam_R and SgrA_ser_F and SgrA_ser_R from pTEX5500ts and chromosomal DNA of E1162, respectively, as depicted in Table 2. Amplified DNA probes were purified with a PCR purification kit (Qiagen) and labeled according to the ECL nucleic acid labeling kit (GE Healthcare). The membranes were exposed to Hyperfilm ECL (GE Healthcare).
Cell surface hydrophobicity assay.
Cell surface hydrophobicity was measured using a hexadecane extraction of E1162 and E1162ΔsgrA cultures as previously described (39). Hydrophobicity is expressed as the percentage of cells that are extracted by the hexadecane as measured by OD.
Adherence to cell lines.
Human colorectal adenocarcinoma cells (Caco-2) and Madin-Darby canine kidney cells (MDCK) were cultured in Dulbecco's modified Eagle Medium (DMEM) (Gibco, Invitrogen, Paisley, United Kingdom) supplemented with 10% heat-inactivated fetal calf serum (Integro B.V., Zaandam, The Netherlands), 1% nonessential amino acids (Gibco), and 2 mM glutamine (Gibco) and incubated in a humidified, 37°C incubator with 5% CO2. Human bladder carcinoma cells (T24) were cultured in Eagle's minimal essential medium (EMEM) (BioWhittaker/Lonza, Breda, The Netherlands) supplemented with 50 μg/ml gentamicin (Gibco) and 10% heat-inactivated fetal calf serum (Integro B.V., Zaandam, The Netherlands). Cells were collected every seventh day by washing the monolayer twice with 0.022% disodium-EDTA; Acros Organics, Morris Plains, NJ) in PBS and trypsinizing the cells using 50 μg/ml trypsine (Gibco) in 0.022% disodium-EDTA in PBS. Differentiated Caco-2 cells were prepared by seeding cells from passages 25 to 45 in 12-well tissue culture plates (Costar) at 1.6 × 105 cells/ml in DMEM with all supplements and replacing the culture medium every second day. Differentiated Caco-2 cells were used 14 to 16 days after seeding. Twelve-well plates with MDCK or T-24 cells were prepared 3 days before use by seeding 1.6 × 105 cells/ml in DMEM or EMEM with the necessary supplements. The medium was replaced after 2 days.
Overnight-grown cultures of E135, E1162, and E1162ΔsgrA in BHI broth were diluted (1:50) and grown at 37°C to an OD660 of 0.4. Bacteria were harvested by centrifugation (6,500 × g; 3 min) and resuspended in DMEM or EMEM to 1 × 107 CFU/ml. For each strain, 1 ml bacterial suspension was added to the wells (100 bacteria to 1 cell). Plates were centrifuged (175 × g; 1 min) and incubated for 1 h at 37°C in 5% CO2 to allow bacterial adherence to the Caco-2, MDCK, and T24 cells. After incubation, monolayers were rinsed three times with DMEM/EMEM and cells were lysed with 1% Triton X-100 (Merck, Darmstadt, Germany) in PBS for approximately 5 min at room temperature. Adherent bacteria were quantified by plating serial dilutions on Trypticase soy agar II plates and counting CFU. The inoculum was also plated to determine viable counts. The assay was performed in duplicate and repeated three times.
Adherence to polystyrene.
Adherence of E135, E1162, and E1162ΔsgrA to abiotic material (polystyrene) was measured by crystal violet staining and assayed as described previously (20, 47).
Statistics.
The Student t test was used to assess statistical significant differences.
DNA sequencing.
PCR products were sequenced by use of the BigDye Terminator 3.1 reaction kit and an ABI PRISM 3100 capillary DNA sequencer (both from Applied Biosystems, Foster City, CA).
RESULTS
Structural organization of the SgrA and EcbA LPXTG surface proteins.
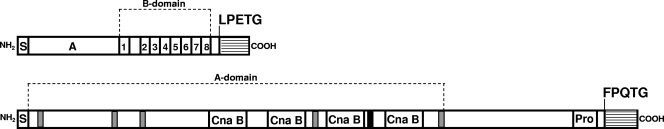
Recently we identified a 975-bp gene (orf2351; ZP_00602747) and a 3,228-bp gene (orf2430; ZP_00603098) encoding 325-amino-acid (aa) and 1,076-aa surface-exposed LPXTG proteins, which were specifically enriched in hospital-acquired E. faecium isolates (Fig. 1) (18). The orf2351 gene is renamed to sgrA, for the serine-glutamate repeat containing protein A, and orf2430 to ecbA for E. faecium collagen binding protein A (see below). In silico E. faecium TX0016 analysis revealed the absence of a sortase gene in the vicinity of the ecbA and sgrA genes, suggesting that these surface proteins may be anchored to the cell wall by the putative Orf2128 housekeeping class A sortase.
FIG. 1.
Schematic representation of the SgrA LPXTG surface protein and the EcbA MSCRAMM. Organization of the SgrA and EcbA proteins identified from the E. faecium TX0016 genome sequence as published at DDBJ/EMBL/GenBank is shown. Signal sequences are depicted by an S, a putative ligand binding domain is indicated by an A, and the Ser-Ser-Glu-Ser-Ser-Thr repeats are numbered. The cell wall sorting signal is depicted as a striped box, starting at the LPETG or FPQTG sortase substrate motif. The putative latching regions are depicted in gray, the Cna B domains are indicated by white boxes, the phenylalanine box is indicated in black, and the proline rich region is indicated by “Pro.”
The SgrA LPXTG surface protein contains a 28-aa N-terminal signal sequence (region S), followed by a 126-aa nonrepetitive region A, which may be a putative ligand-binding domain. Putative IgG-like folds were not present in this domain, suggesting that the SgrA protein cannot be classified as an MSCRAMM. Close to the C-terminal end, a 77-aa B-repeat domain was identified which contains eight Ser-Ser-Glu-Ser-Ser-Thr repeats and at the C terminus a typical CWS, containing an LPETG motif. Secondary structure prediction of SgrA using the 3D-PSSM/Phyre servers (21) predicted a 95-aa-long C-terminal coiled structure from amino acids 119-Pro to 213-Asp (relative to the signal peptide cleavage site), including the B domain, which is possibly involved to span the cell wall to expose the putative ligand-binding A domain from the surface to the extracellular environment (8, 15). A BLASTP homology search revealed significant similarity of the B domain of SgrA, with a putative LPXTG sequence containing a cell wall anchor family protein of E. faecalis V583 (EF0093; NP_813896), which contains nine SESST repeats (six SETSNT and three SESSST repeats) close to the C-terminal end, suggesting structural similarity with SgrA.
The EcbA LPXTG surface protein contains a 34-aa N-terminal signal sequence, followed by a large 696-aa nonrepetitive region A and a C-terminal CWS (Fig. 1) with a FPQTG sortase substrate motif. EcbA is predicted to be an MSCRAMM, since the A domain contains multiple MSCRAMM-like features, which are also found in the fibrinogen-binding MSCRAMM family of Staphylococcus aureus (13). It contains five low-consensus 9-amino-acid TYTFTDYVD-like sequences (TVTVELDLA, TVTDTNGLN, TYTIKIDVE, TVTLTLDEK, and TVTVPYEKL) which are implicated to be a latching cleft. In addition, EcbA harbors four putative Cna B domains which are predicted to adopt a variant of an IgG-like fold (8, 9). The C terminus contains a PxxxPxxPxxPxxPxxPxxPxxxxxxxP (where x denotes any amino acid) proline-rich region directly upstream of the putative FPQTG sortase substrate motif. Furthermore, a stretch of phenylalanine amino acids (FxFxFFxF) was identified in the central part of the protein, and the function, if any, remains to be elucidated. The EcbA protein displays similarity with putative cell wall-anchored proteins of E. faecalis (90% with EF1896 [GenBank accession no. NP_815578.1] and 84% with EF2347 [GenBank accession no. NP_816002.1]) and Listeria welshimeri serovar 6b strain SLCC5334 (30% with lwe0767 [accession no. YP_848968.1]). The EF1896 protein, also called Fss3, showed binding to the γ-chain of fibrinogen (41). E. faecium E1162 genome analysis (W. van Schaik et al., unpublished data) revealed that in addition to the ecbA gene, an ecbA-like gene was present on the E1162 chromosome. The EcbA and EcbA-like proteins share 90% amino acid identity, and the highest sequence heterogeneity occurs within the signal peptide sequence.
Heterogeneity in the sgrA gene.
To determine the level of sequence heterogeneity in the sgrA gene among E. faecium strains, the sgrA genes of five strains (E155, E300, E470, E1162, and U0317) were sequenced. The sgrA gene did not show any sequence variation among clinically relevant isolates at either the DNA or protein level, demonstrating that the sgrA gene is highly conserved. PCR amplification of sgrA in 64 isolates (25 clinical, 17 outbreak-associated, 10 hospital surveillance, 6 community surveillance, 3 environmental, and 3 animal E. faecium isolates) identified in 56 isolates the expected 671-bp fragment. However, in eight isolates from various sources, aberrant DNA fragments of ∼450 bp, ∼750 bp, and ∼1,500 bp were detected and were subsequently sequenced to determine the level of sequence heterogeneity. The occurrence of variant fragment sizes in these isolates was due to variation in the number of B repeats of sgrA, and four of the eight isolates also contained premature stop codons, suggesting that these sgrA genes are pseudogenes.
mRNA expression of ecbA, sgrA, acm, and scm in different stages of growth.
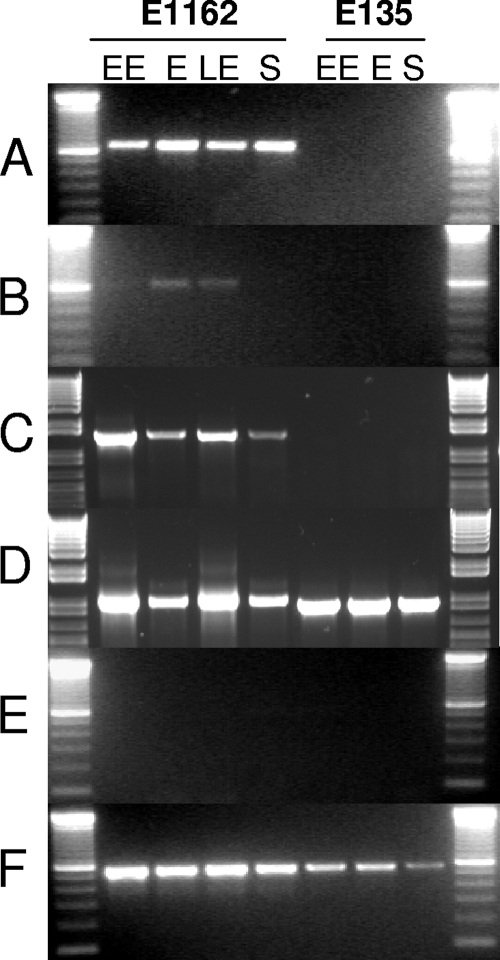
To analyze whether ecbA and sgrA and the collagen-binding MSCRAMMs acm and scm are expressed simultaneously in different stages of growth, E. faecium E1162 (positive for sgrA, ecbA, acm, and scm) and E135 (negative for sgrA and ecbA and positive for acm and scm) cells were grown in BHI broth. At four different time points, early exponential (OD660 = 0.30 after 4.5 h), exponential (OD660 = 0.750 after 5.5 h), late exponential (OD660 = 1.00 after 6.5 h), and stationary (OD660 = >1.00 after 8 h) phases of growth, cells were harvested and transcription of the five genes, ecbA, sgrA, acm, scm, and the ddl internal control, was analyzed by PCR on cDNA generated from total mRNA. In strain E1162, transcripts of sgrA, acm, and scm were detected in all phases of growth whereas transcripts of ecbA were detected only in the exponential and late exponential phases (Fig. 2). The E135 strain did not display expression of ecbA, sgrA, and acm, while scm was expressed. Notably, PCR revealed a larger amplification product for the acm gene in E135 (data not shown). Control PCRs on DNase-treated total mRNA samples in which the reverse transcriptase reaction was omitted were negative, and ddl was constitutively expressed in all samples.
FIG. 2.
mRNA expression of sgrA, ecbA, acm, and scm in different stages of growth. Panels A to D show mRNA expression of sgrA, ecbA, acm, and scm in E1162 (left part) or E135 (right part) cells isolated in early exponential (EE), exponential (E), late-exponential (LE), or stationary (S) phase of growth in BHI broth at 37°C. E135 lacks both the sgrA and ecbA genes. The acm and scm genes are expressed in all stages of growth in E1162 (panels C and D), while acm is not expressed in the E135 strain. Control ddl PCRs (E) with total mRNA preparations in which the RT reaction was omitted were all negative, demonstrating an absence of DNA contamination. Control ddl RT-PCRs (internal housekeeping control) (F) with E. faecium-specific ddl primers were all positive. The results are presented as amplified PCR products electrophoresed on an ethidium bromide-stained 1.5% agarose gel.
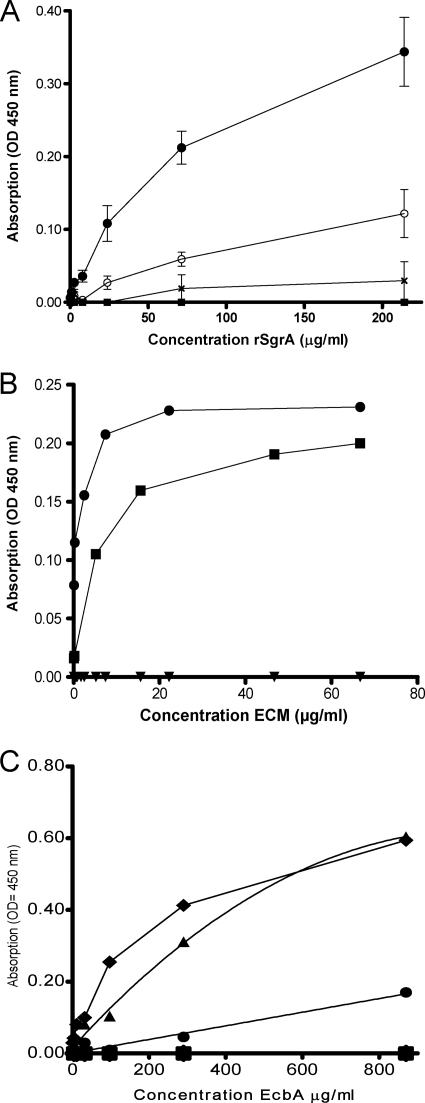
The rSgrA protein binds to human nidogen 1, its homolog nidogen 2, and the α- and β-chains of fibrinogen.
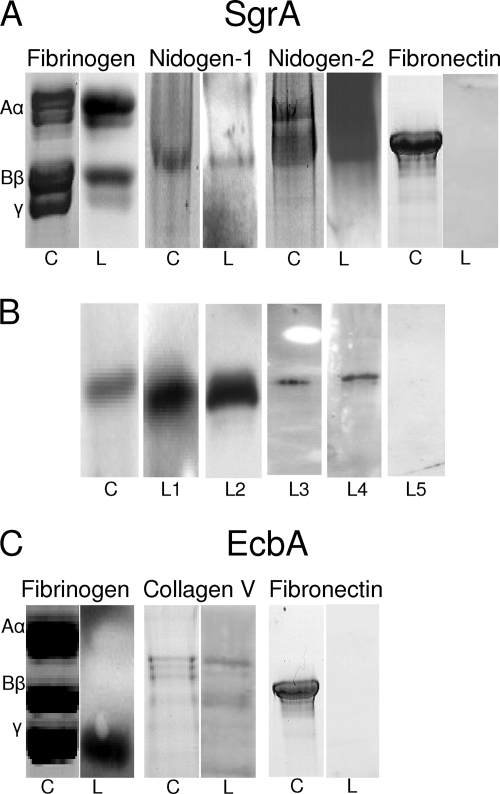
To assess whether rSgrA has the ability to bind to proteins of the ECM, fibronectin, fibrinogen, vitronectin, and BSA (negative control) were immobilized on a microtiter plate and the ability of rSgrA to bind to these components was assayed by ELISA (Fig. 3A). The rSgrA protein bound to immobilized fibrinogen in a dose-dependent manner (apparent Kd = 1.6 μM) and not significantly to fibronectin, vitronectin, or BSA. We performed ligand affinity blotting under reducing conditions to confirm binding to fibrinogen and not to fibronectin. Fibrinogen polypeptide chains separated through SDS-PAGE and stained with Coomassie blue appeared as three bands, designated Aα (63.5 kDa), Bβ (56 kDa), and γ (47 kDa), while fibronectin was displayed as one single 250-kDa band (Fig. 4). Using this technique, binding of rSgrA could be localized to the α- and β-chains of fibrinogen. The negative control, fibronectin, did not display binding of rSgrA.
FIG. 3.
Binding of rSgrA and rEcbA to ligands of the ECM as assessed by ELISA. Panel A shows concentration-dependent binding of rSgrA to immobilized fibrinogen (black circles) and not to fibronectin (black ×), vitronectin (open circles), or BSA (black squares). The OD450s were corrected for the response of six-His IgG-HRP antibodies with the respective ECM proteins. Data points represent the means of OD450s ± standard deviations for three independent experiments with two different purified rSgrA protein batches. Panel B demonstrates concentration-dependent binding of biotinylated nidogen 1 (black squares) and nidogen 2 (black circles) to immobilized rSgrA in a saturable manner and not to BSA (black inverse triangles). Bound proteins were detected using streptavidin peroxidase conjugate. The data points are representative values of three independent experiments with two different purified rSgrA protein batches. Panel C indicates concentration-dependent binding of rEcbA to immobilized collagen type V (black triangles) and fibrinogen (black squares) and not to collagen types I to IV, vitronectin, laminin, or BSA. The data points are representative values of four independent experiments with three different purified rEcbA protein batches.
FIG. 4.
Ligand affinity blotting demonstrated binding of rSgrA to nidogen and fibrinogen. (A) Human fibrinogen and fibronectin were separated through SDS-PAGE, while nidogen 1 and nidogen 2 were separated through native PAGE followed by Coomassie blue staining (indicated by “C”). The ligand affinity blots were probed with rSgrA (fibrinogen; left part) or biotinylated rSgrA (nidogen 1 and nidogen 2) and are indicated by an “L.” (B) Reciprocal ligand affinity binding assays. rSgrA was separated through SDS-PAGE and stained with Coomassie (lane C). Anti-His monoclonal antibodies (L1) or biotinylated ligands including fibrinogen (L2), nidogen 1 (L3), nidogen 2 (L4), and a negative control (biotinylated fibronectin; L5) were allowed to bind to rSgrA and were detected using a streptavidin conjugate. (C) Ligand affinity blotting under reducing conditions demonstrated binding of rEcbA to collagen type V and fibrinogen and not to fibronectin.
Interestingly, in an unpure laminin preparation, we detected binding of rSgrA to an unknown ligand as determined by ligand affinity blotting (data not shown). The glycoproteins designated nidogen 1 (also known as entactin) and nidogen 2 are tightly associated with the γ-chain of laminin and are often coisolated from basal membrane extracts (5, 22, 45). We therefore hypothesized that rSgrA, in addition to the binding activity for fibrinogen, may also display binding activity for either human nidogen. Nidogen 1, its homolog nidogen 2, and BSA as a negative control were biotinylated, since the recombinant nidogens contain, in addition to rSgrA, an N-terminal His tag, and the ability of these two ECM proteins to bind to rSgrA was assessed by ELISA. Levels of biotinylation of the proteins used in this assay were comparable as determined by Western blotting using a streptavidin peroxidase conjugate (data not shown). Biotinylated nidogen 1 and nidogen 2 bound to the rSgrA protein in a concentration-dependent and saturable manner, while no binding to BSA was observed (Fig. 3B). To confirm the interaction of rSgrA with both nidogens, ligand affinity Western blotting was carried out. The nidogen 1, nidogen 2, and fibronectin proteins were separated through PAGE under nonreducing conditions, blotted on nitrocellulose, and subsequently incubated with biotinylated rSgrA. The rSgrA protein bound to both nidogens and not to the negative control, fibronectin (Fig. 4A). As a control, reciprocal ligand affinity binding assays were performed to confirm our initial findings. In these assays, recombinant SgrA was separated through SDS-PAGE and either stained with Coomassie (Fig. 4B, lane C) or detected using anti-His antibodies (lane L1) or biotinylated ligands, including fibrinogen (lane L2), nidogen 1 (lane L3), nidogen 2 (lane L4), and a negative control (biotinylated fibronectin; lane L5), which were allowed to bind to rSgrA. All ligands showed binding to rSgrA except the negative control, fibronectin.
The rEcbA protein binds to collagen type V and the γ-chain of fibrinogen.
To analyze whether rEcbA has the ability to bind to proteins of the ECM, collagen types I to V, fibrinogen and ultrapure laminin, vitronectin, and BSA as a negative control were immobilized on a microtiter plate, and the ability of rEcbA to bind to these components was assayed by ELISA (Fig. 3C). The rEcbA protein bound to immobilized collagen type V and fibrinogen in a dose-dependent manner and not to the other ECM proteins or BSA. Ligand affinity blotting confirmed binding of rEcbA to these ligands of the ECM (Fig. 4B). The binding site for rEcbA could be localized to the α-chain of collagen type V and the γ-chain of fibrinogen. The negative control, fibronectin, did not display binding of rEcbA.
SgrA promotes interaction of E. faecium cells with nidogen 1 and 2 and fibrinogen.
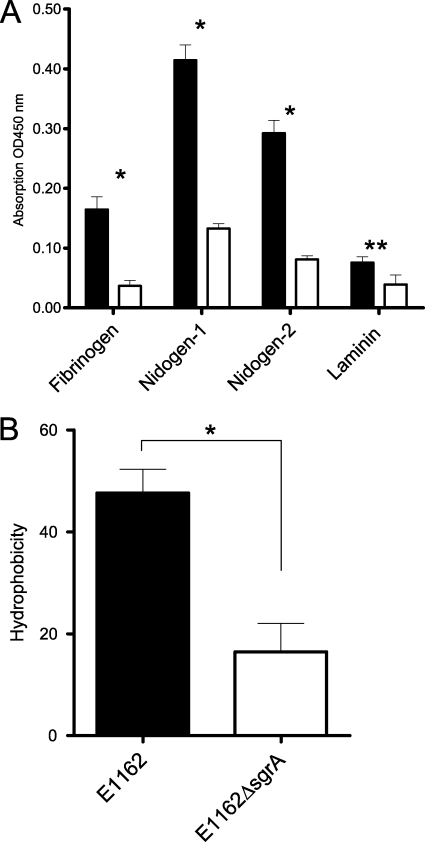
To study the role of SgrA in the interaction of E. faecium cells with nidogen 1, nidogen 2, and fibrinogen, the sgrA gene was inactivated by an insertion-deletion mutation in the clinical E. faecium E1162 isolate. Correct insertion-deletion mutation in the sgrA gene was confirmed by PCR, sequencing, and Southern hybdridization. In addition, Southern blotting using a probe directed against the serine-glutamic acid repeat region and the chloramphenicol (cat) cassette demonstrated that in the double-crossover E1162ΔsgrA mutant, the serine-glutamic acid repeat region was replaced by a cat cassette (data not shown). In vitro growth analyses in BHI broth and on solid medium did not reveal any growth defects between the mutant and the wild-type E1162 strain (data not shown). Subsequent whole-cell ELISA analysis in which nidogen 1, nidogen 2, fibrinogen, and laminin (negative control) were immobilized showed that the E1162ΔsgrA cells displayed significantly reduced binding to nidogen 1 (3.1-fold), nidogen 2 (3.6-fold), and fibrinogen (4.5-fold) and a minor though unexpected reduced binding to ultrapure laminin (1.9-fold) (Fig. 5A). We therefore determined the cell surface hydrophobicities of E1162 and E1162ΔsgrA cells using a hexadecane extraction, which revealed a significantly lower (2.9-fold) surface hydrophobicity of the E1162ΔsgrA strain than of the wild type, E1162 (Fig. 5B).
FIG. 5.
Adherence of E1162 and the isogenic mutant E1162ΔsgrA to ECM molecules by whole-cell ELISA. (A) Fibrinogen, nidogen 1, nidogen 2, and laminin (negative control) were immobilized on a microtiter plate, and E1162 and E1162ΔsgrA E. faecium cells were added to the wells and allowed to bind to these components. Adherent bacteria were detected using an antienterococcus serum followed by goat anti-rabbit IgG-HRP antibodies. Black bars represent wild-type E. faecium E1162, and white bars indicate the E1162ΔsgrA mutant. *, P < 0.005; **, P < 0.05. (B) Cell surface hydrophobicities of wild-type E1162 and an sgrA isogenic mutant (E1162ΔsgrA). The experiments were performed three times with similar results, and values represent means ± standard deviations of triplicate measurements. *, P = 0.0169.
Adhesion to biotic and abiotic surfaces.
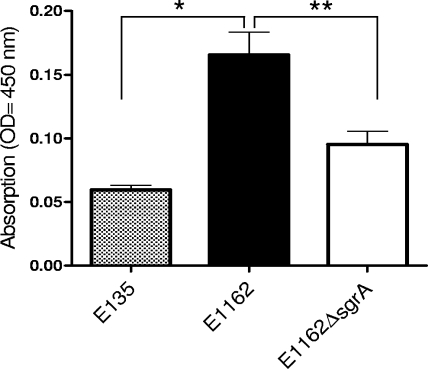
To further characterize the function of SgrA, adherence to biotic surfaces was assessed. The wild-type E. faecium strain E1162, its isogenic E1162ΔsgrA mutant, and an sgrA-negative E. faecium strain (E135) were assayed for their ability to adhere to intestinal epithelial cells (Caco-2), human bladder carcinoma cells (T24), and MDCK cells. Strain E1162 exhibited adherence to Caco-2, T24, and MDCK cells, while the E135 strain showed only low-level binding to these cell lines (data not shown). The E1162 and E1162ΔsgrA strains displayed comparable binding to Caco-2, MDCK, and T24 cells. To analyze whether SgrA is involved in adherence to abiotic material, strains E1162, E1162ΔsgrA, and E135 were analyzed for the ability to form a 24-hr biofilm on a polystyrene surface. The E1162 wild-type strain displayed a high level of biofilm formation on polystyrene, whereas the E135 strain showed a low level. The E1162ΔsgrA strain was significantly impaired in biofilm formation (Fig. 6) compared to the wild-type strain, E1162 (1.6-fold).
FIG. 6.
Biofilm formation on polystyrene. The ability of an sgrA-negative strain, E135 (gray bar), wild-type E1162 (black bar), or an sgrA isogenic mutant, E1162ΔsgrA (white bar) to form a 24-h biofilm on a polystyrene surface is shown. The experiments were performed twice with similar results, and values represent means ± standard deviations of 10 measurements. *, P < 0.002; **, P = 0.03.
DISCUSSION
Here we describe the function of the SgrA LPXTG protein and the EcbA MSCRAMM, two novel surface-exposed adhesins that are specific markers for hospital-acquired E. faecium isolates and that may play a role in the pathogenesis of E. faecium infections. We demonstrated that SgrA and EcbA are implicated in adhesion to components of the ECM, that SgrA mediates binding of E. faecium cells to its ligands of the ECM, and that SgrA is implicated in biofilm formation.
SgrA is the first characterized cell wall-anchored LPXTG surface adhesin of E. faecium that binds to components of the ECM. It is a highly conserved protein among multiresistant hospital-acquired E. faecium strains and binds to fibrinogen and two homologues proteins, designated nidogen 1 and nidogen 2. To our knowledge, we are the first to identify a bacterial cell surface adhesin that has nidogen as its cognate ligand. Nidogen 1 and nidogen 2 are sulfated monomeric glycoproteins of 150 and 200 kDa, respectively (5, 22). Nidogens are a major component of the basal lamina, a specialized membrane of the extracellular matrix. The basal membrane is a well-organized network and is also comprised of laminin, perlecan (a heparan sulfate proteoglycan), and collagen type IV, underlying epithelia, peripheral nerve axons, and muscle and fat cells. Fibrinogen is a large (340-kDa) plasma protein, composed of six polypeptide chains (two Aα, two Bβ, and two γ), and plays an important role in hemostasis and coagulation (19). Wild-type E. faecium E1162 cells bound to fibrinogen, nidogen 1, and nidogen 2, whereas the E1162ΔsgrA strain showed reduced binding to these components of the ECM. Peculiarly, the E1162ΔsgrA strain also showed reduced binding to ultrapure laminin. This finding may be explained by the fact that E1162ΔsgrA cells had a significantly reduced cell surface hydrophobicity, thereby potentially influencing efficiencies or mechanisms of binding to other ligands.
The SgrA LPXTG surface adhesin is not involved in binding to biotic surfaces, such as human intestinal epithelial cells, human bladder cells, and kidney cells. Instead, SgrA mediates adherence of E. faecium cells to a polystyrene abiotic surface, since the E1162ΔsgrA strain produced significantly less biofilm. Biofilm formation is considered to be an important pathogenic property of enterococci and has been demonstrated for a variety of infections (7, 11, 26, 29). Biofilm formation by the E1162ΔsgrA strain was not completely abolished, which is likely due to expression of Esp at the surface (16, 47). Alternatively, a different cell surface charge (46), glycolipids (43), or possibly other surface components, such as pili (17, 34), which are known to be implicated in biofilm formation of E. faecalis, may contribute to E. faecium biofilm formation.
In silico analysis revealed that the EcbA protein, a marker for hospital-acquired E. faecium, has all structural and typical features of an MSCRAMM. E. faecium E1162 genome analysis (van Schaik et al., unpublished) revealed, in addition to the ecbA gene, the presence of an ecbA-like gene which is highly similar to ecbA (90%). Due to the presence of two highly homologues genes and the current limited genetic tools for E. faecium, we were unable to construct a double ecbA and ecbA-like isogenic mutant of E. faecium E1162. Therefore, we assessed whether rEcbA bound to components of the ECM. Indeed, rEcbA bound to collagen type V and fibrinogen in a concentration-dependent manner, making EcbA the third MSCRAMM of E. faecium. Recently the Scm MSCRAMM was identified and characterized and also bound to collagen type V and fibrinogen (42). In addition, SgrA bound to fibrinogen as well. The presence of multiple LPXTG surface proteins with similar functions has also been described for S. aureus, where MSCRAMMs such as clumping factors A and B and the FnbpA and FnbpB proteins bind to fibrinogen, thereby targeting different fibrinogen chains (36, 48). The ligand affinity blot data suggest that SgrA and EcbA target different parts of the fibrinogen molecule. The rSgrA protein bound to the α and β subunits, whereas rEcbA bound only to the γ subunit, of fibrinogen. Future experiments including ligand affinity blotting using recombinant fibrinogen subunits isolated and purified from Lactococcus lactis, pepscan analysis, and the construction of amino acid substitution mutants will reveal the binding sites of rEcbA and rSgrA for fibrinogen and their other ligands.
Apparently, E. faecium expresses three (SgrA, EcbA, and Scm) fibrinogen-binding adhesins at its surface, and for SgrA and EcbA, it has been demonstrated that these proteins recognize different parts of the ligand. Possibly, these two different interactions may act synergistically to allow tight attachment to this ligand. Alternatively, the presence of multiple structurally distinct fibrinogen-binding surface proteins may allow E. faecium to attach to the ligand in the presence of anti-SgrA, -EcbA, or -Scm antibodies. Constitutive mRNA expression of sgrA, acm, and scm in different stages of growth and controlled ecbA expression from exponential to late exponential phase suggest that expression of these adhesins in E. faecium is fine-tuned to allow adherence to or trophism for a particular tissue. Given the aberrant size of the acm amplicon, an absence of acm expression in the E135 strain is likely due to insertion of a transposon in the acm gene (33).
Fibrinogen and fibrin are major components of blood clots during wound healing and are the major plasma proteins deposited on implanted foreign devices. This means that for patients with indwelling medical devices, SgrA may have a dual role in the infective process. In these patients, SgrA may facilitate attachment to polymer surfaces with subsequent growth on the device. Subsequently, SgrA and EcbA may mediate binding to ECM, deposited on these abiotic devices (10). As such, these surface adhesins may play a role in the pathogenesis of intravascular catheter-related infections.
Hospital-acquired E. faecium has multiple intrinsic and acquired antibiotic resistance traits, which seriously hamper treatment of infected patients. Novel alternative treatment and prevention strategies are therefore urgently required. In that respect, the development of combinatory vaccines that target LPXTG surface proteins, such as SgrA and EcbA, could be a promising approach.
Acknowledgments
We thank Willem van Schaik for critical reading of the manuscript. We thank Johannes Huebner for kindly providing rabbit antienterococcus serum.
This work was supported by ZonMW research grant 6100.008 from The Netherlands Organization for Health, Research and Development and by a grant from the European Union Sixth Framework Programme under contract LSHE-CT-2007-037410.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 8 September 2009.
REFERENCES
- 1.Ammerlaan, H. S., A. Troelstra, C. L. Kruitwagen, J. A. Kluytmans, and M. J. Bonten. 2009. Quantifying changes in incidences of nosocomial bacteraemia caused by antibiotic-susceptible and antibiotic-resistant pathogens. J. Antimicrob. Chemother. 63:1064-1070. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, R. C., B. E. Murray, and R. M. Rakita. 1994. Roles of antibodies and complement in phagocytic killing of enterococci. Infect. Immun. 62:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M. (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 4.Bonten, M. J., M. K. Hayden, C. Nathan, J. van Voorhis, M. Matushek, S. Slaughter, T. Rice, and R. A. Weinstein. 1996. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet 348:1615-1619. [DOI] [PubMed] [Google Scholar]
- 5.Carlin, B., R. Jaffe, B. Bender, and A. E. Chung. 1981. Entactin, a novel basal lamina-associated sulfated glycoprotein. J. Biol. Chem. 256:5209-5214. [PubMed] [Google Scholar]
- 6.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 7.Dautle, M. P., R. L. Ulrich, and T. A. Hughes. 2002. Typing and subtyping of 83 clinical isolates purified from surgically implanted silicone feeding tubes by random amplified polymorphic DNA amplification. J. Clin. Microbiol. 40:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deivanayagam, C. C., R. L. Rich, M. Carson, R. T. Owens, S. Danthuluri, T. Bice, M. Hook, and S. V. Narayana. 2000. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 8:67-78. [DOI] [PubMed] [Google Scholar]
- 9.Deivanayagam, C. C., E. R. Wann, W. Chen, M. Carson, K. R. Rajashankar, M. Hook, and S. V. Narayana. 2002. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 21:6660-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlan, R. M. 2001. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 33:1387-1392. [DOI] [PubMed] [Google Scholar]
- 11.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunne, W. M., Jr., and W. Wang. 1997. Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J. Clin. Microbiol. 35:388-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Hartford, O., P. Francois, P. Vaudaux, and T. J. Foster. 1997. The dipeptide repeat region of the fibrinogen-binding protein (clumping factor) is required for functional expression of the fibrinogen-binding domain on the Staphylococcus aureus cell surface. Mol. Microbiol. 25:1065-1076. [DOI] [PubMed] [Google Scholar]
- 16.Heikens, E., M. J. Bonten, and R. J. Willems. 2007. Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J. Bacteriol. 189:8233-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrickx, A. P., M. J. Bonten, M. van Luit-Asbroek, C. M. Schapendonk, A. H. Kragten, and R. J. Willems. 2008. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology 154:3212-3223. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickx, A. P., W. J. van Wamel, G. Posthuma, M. J. Bonten, and R. J. Willems. 2007. Five genes encoding surface-exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium clonal complex 17 isolates. J. Bacteriol. 189:8321-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrick, S., O. Blanc-Brude, A. Gray, and G. Laurent. 1999. Fibrinogen. Int. J. Biochem. Cell Biol. 31:741-746. [DOI] [PubMed] [Google Scholar]
- 20.Hufnagel, M., S. Koch, R. Creti, L. Baldassarri, and J. Huebner. 2004. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J. Infect. Dis. 189:420-430. [DOI] [PubMed] [Google Scholar]
- 21.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 22.Kohfeldt, E., T. Sasaki, W. Gohring, and R. Timpl. 1998. Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 282:99-109. [DOI] [PubMed] [Google Scholar]
- 23.Leavis, H., J. Top, N. Shankar, K. Borgen, M. Bonten, E. J. van, and R. J. Willems. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leavis, H. L., R. J. Willems, J. Top, and M. J. Bonten. 2006. High-level ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J. Clin. Microbiol. 44:1059-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leavis, H. L., R. J. Willems, W. J. van Wamel, F. H. Schuren, M. P. Caspers, and M. J. Bonten. 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS. Pathog. 3:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung, J. W., Y. L. Liu, T. D. Desta, E. D. Libby, J. F. Inciardi, and K. Lam. 2000. In vitro evaluation of antibiotic prophylaxis in the prevention of biliary stent blockage. Gastrointest. Endosc. 51:296-303. [DOI] [PubMed] [Google Scholar]
- 27.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 28.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72:3658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 31.Nallapareddy, S. R., K. V. Singh, and B. E. Murray. 2006. Construction of improved temperature-sensitive and mobilizable vectors and their use for constructing mutations in the adhesin-encoding acm gene of poorly transformable clinical Enterococcus faecium strains. Appl. Environ. Microbiol. 72:334-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallapareddy, S. R., K. V. Singh, and B. E. Murray. 2008. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect. Immun. 76:4120-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nallapareddy, S. R., K. V. Singh, P. C. Okhuysen, and B. E. Murray. 2008. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect. Immun. 76:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nallapareddy, S. R., K. V. Singh, J. Sillanpaa, D. A. Garsin, M. Hook, S. L. Erlandsen, and B. E. Murray. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 116:2799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nallapareddy, S. R., G. M. Weinstock, and B. E. Murray. 2003. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 47:1733-1747. [DOI] [PubMed] [Google Scholar]
- 36.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 37.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 38.Patti, J. M., and M. Hook. 1994. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 6:752-758. [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg, M., D. Gutnick, and E. Rosenberg. 1980. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9:29-33. [Google Scholar]
- 40.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sillanpaa, J., S. R. Nallapareddy, J. Houston, V. K. Ganesh, A. Bourgogne, K. V. Singh, B. E. Murray, and M. Hook. 2009. A family of fibrinogen-binding MSCRAMMs from Enterococcus faecalis. Microbiology 155:2390-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sillanpaa, J., S. R. Nallapareddy, V. P. Prakash, X. Qin, M. Hook, G. M. Weinstock, and B. E. Murray. 2008. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology 154:3199-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theilacker, C., P. Sanchez-Carballo, I. Toma, F. Fabretti, I. Sava, A. Kropec, O. Holst, and J. Huebner. 2009. Glycolipids are involved in biofilm accumulation and prolonged bacteremia in Enterococcus faecalis. Mol. Microbiol. 71:1055-1069. [DOI] [PubMed] [Google Scholar]
- 44.Timmers, G. J., W. C. van der Zwet, I. M. Simoons-Smit, P. H. Savelkoul, H. H. Meester, C. M. Vandenbroucke-Grauls, and P. C. Huijgens. 2002. Outbreak of vancomycin-resistant Enterococcus faecium in a haematology unit: risk factor assessment and successful control of the epidemic. Br. J. Haematol. 116:826-833. [DOI] [PubMed] [Google Scholar]
- 45.Timpl, R., M. Dziadek, S. Fujiwara, H. Nowack, and G. Wick. 1983. Nidogen: a new, self-aggregating basement membrane protein. Eur. J. Biochem. 137:455-465. [DOI] [PubMed] [Google Scholar]
- 46.van Merode, A. E., H. C. van der Mei, H. J. Busscher, K. Waar, and B. P. Krom. 2006. Enterococcus faecalis strains show culture heterogeneity in cell surface charge. Microbiology 152:807-814. [DOI] [PubMed] [Google Scholar]
- 47.Van Wamel, W. J., A. P. Hendrickx, M. J. Bonten, J. Top, G. Posthuma, and R. J. Willems. 2007. Growth condition-dependent Esp expression by Enterococcus faecium affects initial adherence and biofilm formation. Infect. Immun. 75:924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wann, E. R., S. Gurusiddappa, and M. Hook. 2000. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275:13863-13871. [DOI] [PubMed] [Google Scholar]
- 49.Werner, G., T. M. Coque, A. M. Hammerum, R. Hope, W. Hryniewicz, A. Johnson, I. Klare, K. G. Kristinsson, R. Leclercq, C. H. Lester, M. Lillie, C. Novais, B. Olsson-Liljequist, L. V. Peixe, E. Sadowy, G. S. Simonsen, J. Top, J. Vuopio-Varkila, R. J. Willems, W. Witte, and N. Woodford. 20 November 2008. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 13:pii=19046. [PubMed] [Google Scholar]
- 50.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, K. E. van, J. D. Van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
- 51.Willems, R. J., J. Top, M. van Santen, D. A. Robinson, T. M. Coque, F. Baquero, H. Grundmann, and M. J. Bonten. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]