Abstract
Salmonella survives and replicates in host cells by using a type III secretion system to evade host immune defenses. The innate immune system plays an important role as a first line of defense against pathogens and is mediated in part by Toll-like receptors (TLRs); however, the infection dynamics of Salmonella enterica serovar Typhimurium within macrophages stimulated with TLR ligands is poorly understood. We studied the infection dynamics of Salmonella in murine macrophages previously exposed to TLR ligands and report that treatment of macrophages with four different TLR agonists resulted in their increased phagocytic capacity toward Salmonella but not fluorescent microspheres. Further analysis revealed that the intracellular replication of Salmonella was enhanced in TLR-stimulated macrophages in a manner requiring a functional type III secretion system and enhanced transcriptional activity of the sseA virulence gene operon. Studies of mice that normally resolve an acute primary infection with Salmonella revealed that pretreatment of animals with CpG DNA had a detrimental effect on disease outcome. CpG-treated mice infected with Salmonella all succumbed to infection and had higher bacterial loads in the spleen than did control animals. These data suggest that Salmonella can exploit macrophages activated via the innate immune system for increased intracellular survival.
Salmonella enterica is an intracellular gram-negative bacterium that infects multiple hosts and causes a variety of diseases (46). An important virulence strategy of S. enterica is the ability to survive and replicate within eukaryotic host cells, particularly epithelial cells and macrophages, inside a modified phagosome called the Salmonella-containing vacuole (SCV) (9). The type III secretion system (T3SS) encoded in Salmonella pathogenicity island 2 (SPI-2) plays an essential role in intracellular replication by translocating effector proteins into the host cell for SCV maintenance, modification of the vacuolar compartment, and subverting innate defenses such as reactive oxygen and nitrogen species (11, 24, 47, 49, 52).
The innate immune system is a first line of defense against pathogens such as Salmonella and is operationalized in part by signals from Toll-like receptors (TLRs) (1, 6) that recognize conserved microbial ligands at the surface of cells or within endosomes (2). To date, 13 mammalian TLRs have been identified along with numerous TLR ligands, signaling molecules, and regulatory molecules. There are several lines of evidence suggesting that TLR activation may have therapeutic potential. For example, administration to mice of oligodeoxynucleotides (ODN) that contain the CpG motif can provide protection against live pathogens including Listeria and Francisella (19), herpes simplex virus type 2 (HSV-2) (3, 25, 42), and Mycobacterium tuberculosis (30). Treatment with the TLR3 ligand, poly(I:C), can protect mice from infection with HSV-1 and HSV-2 (4, 27). Synthetic TLR ligands for use in cancer treatments, vaccines, infectious disease, and allergy are already in development, with some in the early phases of clinical testing (32).
Modulating the innate immune response through TLR agonist treatment is an area of research that has received increased attention. However, little is known about how TLR stimulation of macrophages impacts intracellular infection by S. enterica serovar Typhimurium, a pathogen uniquely adapted to live within macrophage cells. In this study we examined the impact of TLR ligand pretreatment of macrophages and mice on the outcome of infection following infection with S. enterica serovar Typhimurium. Our data suggest an adaptive survival mechanism in Salmonella that makes use of macrophages previously stimulated with TLR ligands.
MATERIALS AND METHODS
Strains and growth conditions.
S. enterica serovar Typhimurium strain SL1344 was used throughout (abbreviated S. Typhimurium) (54). Bacteria were cultured in Luria-Bertani (LB) broth and on LB agar containing streptomycin (50 μg/ml). An ssaR mutant (ΔssaR) with a deletion in ssaR encoding an essential protein in the SPI-2 T3SS has been described previously (10). The hha mutant (Δhha) has a deletion in the hha gene encoding a repressor of the SPI-2 genomic island and has been described previously (45). All mutants were unmarked, in-frame deletions constructed in the wild-type SL1344 background. RAW 264.7 murine macrophages and J774.1 murine macrophages were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (HyClone). Cells were cultured at 37°C in 5% CO2 without antibiotics.
TLR ligand treatment.
Macrophage cells were treated with TLR ligands for 16 to 20 h before infection with either 100 ng/ml lipopolysaccharide from Escherichia coli (Sigma), 10 μg/ml poly(I:C) (Sigma), 10 μg/ml of Staphylococcus aureus-derived TLR2-active substance (Sigma; supplied as peptidoglycan [PG] and defined as such here, but it may contain trace amounts of lipoprotein) (34), or 10 μg/ml CpG-ODN (5′-TCG TCG TTT TGT CGT TTT GTC GTT-3′) (Mobix; McMaster University, Canada). This time point was previously shown to elicit innate antiviral activity in RAW 264.7 cells against HSV (3). Control cells were grown identically but treated with phosphate-buffered saline (PBS).
Phagocytosis and intracellular replication experiments. (i) Standard experiment.
RAW 264.7 cells were seeded 24 h before infection at a density of 2.0 × 105 cells per well in 24-well culture dishes and treated with TLR ligands for the last 16 to 20 h before infection. An overnight culture of S. Typhimurium SL1344 was washed with PBS and then opsonized with 20% normal human serum in DMEM for 25 min at 37°C. Bacteria were diluted in DMEM, and macrophages were infected at a multiplicity of infection (MOI) of approximately 50:1. After 30 min of infection, the cells were washed three times with PBS and incubated in DMEM with 100 μg/ml gentamicin for 2 h, after which the drug concentration was reduced to 10 μg/ml. At 2 h and 20 h postinfection, cells were washed three times with PBS, lysed with 1% Triton X-100-0.1% sodium dodecyl sulfate, and bacteria in the cell lysates were plated to enumerate intracellular CFU. CFU data for treatment groups at 2 h were normalized by dividing the 2-h CFU values of each treatment group by the average 2-h CFU value for the PBS controls, and the results were plotted as the phagocytic index. The relative increase in the number of intracellular bacteria was calculated by dividing the CFU values at 20 h by the CFU values at 2 h postinfection for each treatment group. Intracellular replication was normalized by dividing by the relative increase for each treatment group by the average relative increase of the PBS control, and this value was plotted as the replication index. All treatment conditions were performed in triplicate for a given experiment, and data measuring bacterial uptake and intracellular replication were averaged from 4 to 12 independent experiments.
(ii) IFN-γ treatment.
In experiments where gamma interferon (IFN-γ) was used, macrophages were stimulated with 10 ng/ml IFN-γ in conjunction with TLR ligand treatment 16 to 20 h prior to infection.
(iii) Bacterial dosing experiments.
In some experiments the bacterial dose used to infect TLR-stimulated macrophages was reduced in order to obtain numbers of intracellular bacteria at 2 h postinfection similar to those of the PBS-treated control cells.
Phagocytosis of fluorescent microspheres.
RAW 264.7 cells were seeded onto 2.5-cm glass coverslips at a density of ∼105 cells/ml in 35- by 10-mm petri dishes and treated with TLR agonists as described above. Cells were incubated with 1.0-μm green-yellow opsonized fluorescent microspheres (Molecular Probes) at a density of 107 beads/ml. Phagocytosis was allowed to proceed for 3.5 h, and then cells were washed three times with PBS and transferred to Attofluor cell chambers (Invitrogen) in HEPES buffer for live-cell imaging. Cells were treated with 3 μM 5,6-carboxy SNARF-1 acetoxymethyl ester, acetate (Molecular Probes), to label the cytoplasm. Confocal microscopy was performed using a Leica DMI 600B microscope with a Leica TCS SP5 AOBS scanner (63× objective with a 1.3 numerical aperture) and Leica Application Suite Advanced Fluorescence Software. Internalized beads were identified using an in-house script for image processing in ImageJ (see supplemental material). Approximately 800 cells were scored for each treatment group. Images were imported into Adobe Photoshop, version 7.0, and assembled in Adobe Illustrator CS for labeling.
Immunofluorescence.
Macrophages were infected as described above onto 1.2-cm glass coverslips. At 2 h after infection, cells were washed and fixed with 2.5% paraformaldehyde-PBS solution (pH 7.4) for 30 min at 37°C. Samples were washed with PBS and blocked and permeabilized with 10% normal goat serum and 0.2% saponin in PBS for 30 min. Primary and secondary antibodies diluted in 0.2% saponin in PBS were applied to samples for 60 min and then mounted onto glass slides using mounting medium (Dako). Primary antibodies used were rabbit anti-Salmonella O antiserum group B factors 1, 4, 12, and 27 (Difco); rat anti-LAMP-1 (lysosomal-associated membrane protein 1) (clone 1D4B; Developmental Studies Hybridoma Bank, Iowa City, IA), and mouse anti-mono- and polyubiquitinated conjugates (monoclonal) (clone FK2; Biomol International). Secondary antibodies were Alexa Fluor 488-conjugated donkey anti-rat and goat anti-mouse antibodies and Alexa Fluor 568-conjugated donkey anti-rabbit and goat anti-rabbit antibodies, all used at 1:200 (Molecular Probes). DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) was used at 1:2,500 (Molecular Probes). Wide-field fluorescence microscopy was performed using a Leica DMI 600B (100× objective; 1.4 numerical aperture) and Velocity, version 4, software. Images were collected using a Hamamatsu Orca ER-AG camera.
Electron microscopy.
Macrophages were treated with TLR agonists or PBS and infected with S. Typhimurium as described above. At 2 h postinfection, the cells were fixed with 2% glutaraldehyde (Canemco) in 0.1 M sodium cacodylate (NaCac), pH 7.4, for 1 h at room temperature. Fixed samples were incubated in 0.2 M NaCac twice for 5 min each time and postfixed with 1% osmium tetroxide in 0.1 M NaCac for 30 min. Samples were dehydrated in a graded alcohol series and infiltrated with TAAB 812 (Canemco) in TAAB 812-100% ethanol at 1:1 for 1 h and at 3:1 for 1 h. Samples were incubated twice in TAAB 812 for 1 h. Excess TAAB 812 was aspirated off the samples, and small BEEM capsules were placed on the surface of the cell monolayer and polymerized at 65°C overnight. BEEM capsules were topped off with TAAB 812 and polymerized again. Capsules were snapped off, and ultrathin sections (70 nm; Ultracut T; Leica) were placed onto 200-mesh Cu/Pd grids. Sections were stained with saturated uranyl acetate for 5 min, rinsed with water, and stained with lead citrate for 2 min. Sections were viewed with a 1200 EX TEMScan instrument (JEOL, Tokyo).
Promoter activity for sseA.
Transcriptional activity of the sseA promoter within SPI-2 was examined using a transcriptional reporter fused to lacZ that was reported previously (8, 45). The reporter strain was cultured overnight in LB medium containing ampicillin (100 μg/ml) and then washed in PBS and diluted into DMEM for the macrophage infection experiments described above. Promoter activity was measured by quantifying β-galactosidase activity from host cell lysates prepared at 2 h and 10 h postinfection. In parallel, the number of intracellular bacteria giving rise to this enzyme activity was quantified as the number of CFU. β-Galactosidase activity (measured in relative light units) was normalized to the number of CFU to facilitate direct comparisons between technical and biological replicates.
Animal experiments.
Animal protocols were in accordance with the Canadian Council on the Use of Laboratory Animals and approved by the local Animal Ethics Committee. C57BL/6 mice and 129×1SvJ mice were bread in-house. Mice used for experimentation were female F1 offspring of 129×1SvJ female and C57BL/6 male parents. C57BL/6 mice have a nonfunctional Nramp1s (Slc11a1) allele and have defects in controlling intracellular Salmonella replication, whereas 129×1/SvImJ mice are homozygous for the Nrampr allele and are better able to limit intracellular replication of Salmonella. Mice were infected intravenously (i.v.) via the tail vein with 1 × 103 CFU of S. Typhimurium in a 0.1-ml volume. Groups of mice were simultaneously injected intraperitoneally (i.p.) with 50 μg of CpG (ODN 1826; Coley Pharmaceuticals, Ottawa, Ontario, Canada) or PBS. The health status of the animals was monitored following infection, and mice that lost 17% of their original body weight were deemed to have reached clinical endpoint and were euthanized.
To assess the systemic bacterial burden in mice, animals were euthanized, and single-cell suspensions were prepared from the spleens of infected mice in RPMI 1640 medium. An aliquot of the suspension was lysed with water for 30 s and then evaluated for the numbers of viable bacteria. Serial 10-fold dilutions of cell lysates were prepared in 0.9% saline, and 100-μl aliquots were plated on brain heart infusion agar. Bacterial colonies were enumerated following overnight incubation at 37°C.
RESULTS
Treatment with TLR agonists leads to increased uptake of S. Typhimurium.
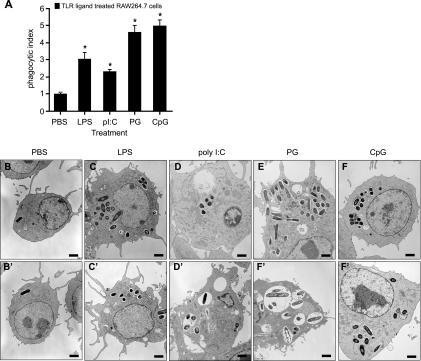
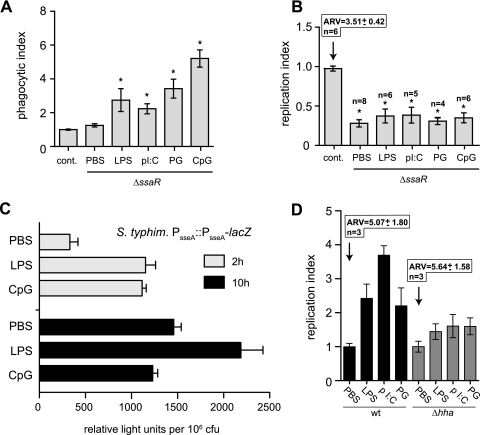
To understand how treatment of macrophages with TLR ligands influences the bacterial infection process by Salmonella, RAW 264.7 macrophages were treated prior to infection with TLR agonists including lipopolysaccharide ([LPS] TLR4), poly(I:C) (TLR3), PG/lipoprotein (TLR2), or CpG (TLR9) or with PBS as a control. Treatment with all TLR agonists significantly increased the number of intracellular bacteria at 2 h postinfection (Fig. 1A), a time prior to the onset of intracellular replication (12). At this time point, the number of intracellular bacteria reflects the number of bacteria phagocytosed during the initial 30-min infection. Specifically, treatment with LPS, poly(I:C), PG, or CpG increased bacterial uptake at 2 h postinfection by 3.0-, 2.3-, 4.6-, or 5.0-fold, respectively, compared to controls. We further verified these data by infecting TLR ligand-treated cells and examining infected cells by transmission electron microscopy at 2 h postinfection. In these experiments macrophages treated with PBS and then infected typically had zero to three visible bacteria, whereas cells treated with any of the TLR agonists had numerous (>10) intracellular S. Typhimurium bacteria, each enclosed within SCVs (Fig. 1B through F). These data demonstrate that treatment of macrophages with TLR agonists leads to greater phagocytic uptake of S. Typhimurium than in untreated cells.
FIG. 1.
Treatment of RAW 264.7 cells with TLR ligands increases phagocytic uptake of S. Typhimurium. (A) RAW 264.7 cells were treated with PBS, LPS, poly(I:C) (pI:C), PG, or CpG-ODN for 20 h before infection with wild-type S. Typhimurium. Levels of intracellular bacteria at 2 h postinfection are presented as the mean with standard errors from three technical replicates from 10 experiments, normalized to the PBS control. Asterisks represent significant differences from the PBS control (P < 0.05, one-way analysis of variance). (B to F) Transmission electron micrographs of RAW 264.7 cells treated with PBS or TLR agonists and then infected with S. Typhimurium for 2 h. Two representative images are shown for each treatment.
Treatment of RAW 264.7 cells with TLR agonists has little effect on phagocytosis of fluorescent microspheres.
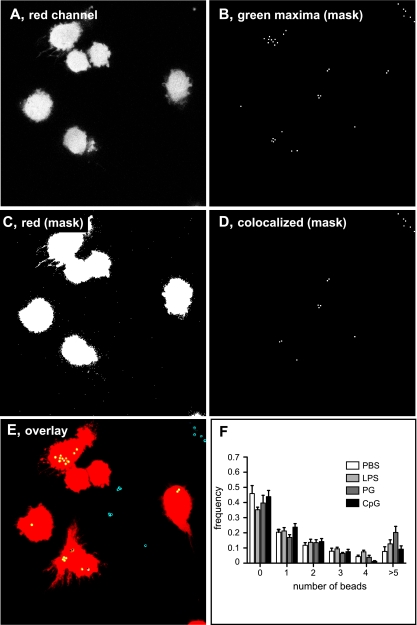
Treatment of macrophages with TLR ligands increased their uptake of S. Typhimurium. To see if this activity was specific to Salmonella or the result of a more general increase in phagocytic capacity, the effect of TLR ligands on the phagocytosis of fluorescent microspheres was examined using live-cell imaging. Although there was no significant increase in the number of internalized beads in macrophages treated with LPS, PG, or CpG compared to the PBS control (Fig. 2), there was a trend toward an increase in the proportion of cells with more than five fluorescent beads following LPS or PG treatment that did not reach statistical significance compared to the PBS treatment. Similarly, the proportion of macrophages with no intracellular microspheres was not different between the treatment groups (Fig. 2F). These data suggest that the enhanced phagocytic capacity of cells activated by TLR ligands is more specific for Salmonella than for inert microspheres, which is in keeping with other reports on preferential uptake by macrophages of E. coli and S. aureus but not latex beads following TLR ligand treatment (16).
FIG. 2.
Treatment with TLR agonists does not affect phagocytosis of fluorescent microspheres. Fluorescent microspheres were incubated with RAW 264.7 macrophages that had been pretreated with TLR agonists. Cells were treated with SNARF-1 dye for 30 min to stain the cytoplasm and then viewed with a confocal microscope for live-cell imaging. Maximum fluorescence intensity from the red channel (host cell cytoplasm; A) and green channel (beads; B) was collected and processed using an automated in-house script to create masks for beads (B) and host cell cytoplasm (C). Beads that colocalized with the host cell cytoplasm were identified (D), and then complete Z-stacks were overlaid (E). Internalized beads are show in yellow, external beads attached to the surface of cells are in yellow with a filled black dot, and external beads are in blue with a filled black dot. (F) The distribution of beads in the cell population (400 to 850 cells) was plotted as the means with standard deviations from three separate experiments.
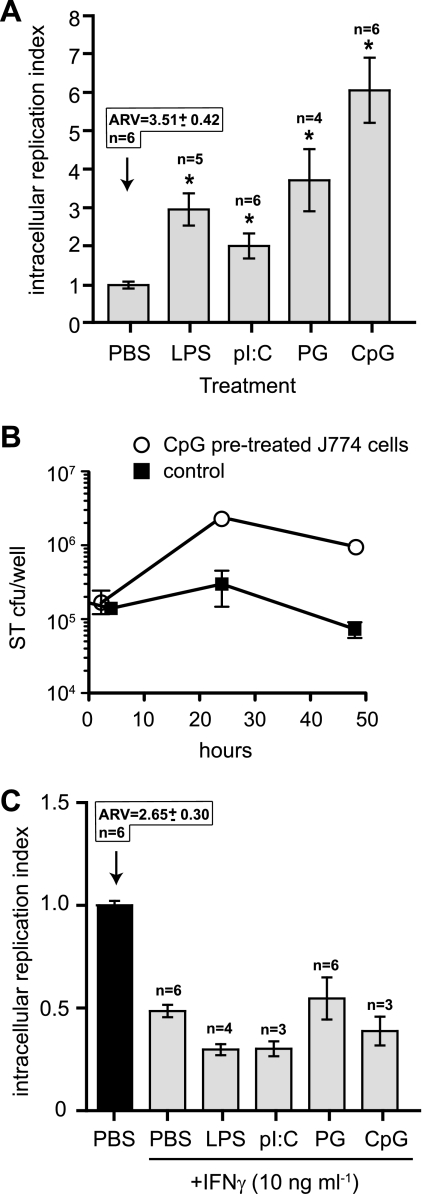
TLR ligands increase intracellular replication of S. Typhimurium that can be inhibited with IFN-γ.
TLR ligand treatment of macrophages resulted in increased phagocytic capacity for S. Typhimurium. However, because S. Typhimurium has evolved to replicate within macrophages by using a T3SS (26), we were interested in the fate of this intracellular population of bacteria following uptake into TLR-activated cells. Treatment of cells with any of the TLR agonists increased the intracellular replication of S. Typhimurium over an 18-h period over and above the bacterial replication seen in PBS-treated control cells (Fig. 3A). Treatment with LPS, poly(I:C), PG, and CpG increased bacterial replication by 3.0-, 2.0-, 3.7-, and 6.1-fold, respectively, compared to the PBS-treated control. To examine this further, we treated J774 macrophage cells with CpG DNA or PBS and followed the fate of intracellular Salmonella over time. In this macrophage cell line, Salmonella replicated to ∼10-fold higher numbers in TLR-stimulated cells than in unstimulated cells, which is similar to results seen in RAW264.7 cells (Fig. 3B). Thus, while treatment of cells with TLR agonists increased bacterial uptake, it did not result in increased S. Typhimurium killing activity.
FIG. 3.
S. Typhimurium has enhanced replication in cells stimulated with TLR ligands. (A) RAW 264.7 cells were treated with PBS, LPS, poly(I:C), PG, or CpG-ODN for 20 h before infection with wild-type S. Typhimurium. The increase in intracellular bacteria between 2 to 18 h postinfection is shown as the mean with standard errors from three technical replicates from 4 to 10 experiments normalized to the PBS control. The absolute replication value (ARV) for the PBS control is given above the control bar, and the number of experiments used to produce the data is given above each treatment group. Asterisks represent significant differences from the PBS control (P < 0.05; one-way analysis of variance). (B) Intracellular replication of S. Typhimurium (ST) in J774 cells pretreated with CpG DNA prior to infection. Data are the means with standard errors. (C) The enhanced replication of S. Typhimurium in TLR ligand-treated cells is blocked by IFN-γ. RAW 264.7 cells were treated with IFN-γ in addition to PBS, LPS, poly(I:C), PG, or CpG-ODN or with PBS alone for 20 h before infection. Intracellular bacterial replication for the treatment groups was normalized to the PBS control and is shown as the mean with standard errors from three technical replicates from three to six experiments. The absolute replication value (ARV) for the PBS control is given above the control bar, and the number of experiments used to produce the data is given above each treatment group. Asterisks represent significant differences (P < 0.05, one-way analysis of variance) compared to the PBS control not treated with IFN-γ.
IFN-γ is a key activator of macrophages both in vivo and in vitro and plays an important role in resistance against intracellular pathogens such as Salmonella (44) and Mycobacterium tuberculosis (14, 22). To examine the effect of IFN-γ on the bactericidal ability of TLR-stimulated cells, RAW 264.7 macrophages were incubated with TLR ligands, IFN-γ, or TLR ligands in conjunction with IFN-γ before infection with S. Typhimurium. Unlike treatment with PBS or TLR ligands alone, treatment with IFN-γ or IFN-γ in conjunction with TLR ligands resulted in a net killing of internalized bacteria (Fig. 3C), indicating that the addition of IFN-γ can overcome the TLR signals that enhance intracellular survival and replication of S. Typhimurium.
Increased Salmonella replication following TLR stimulation is not due to initially high intracellular bacterial numbers.
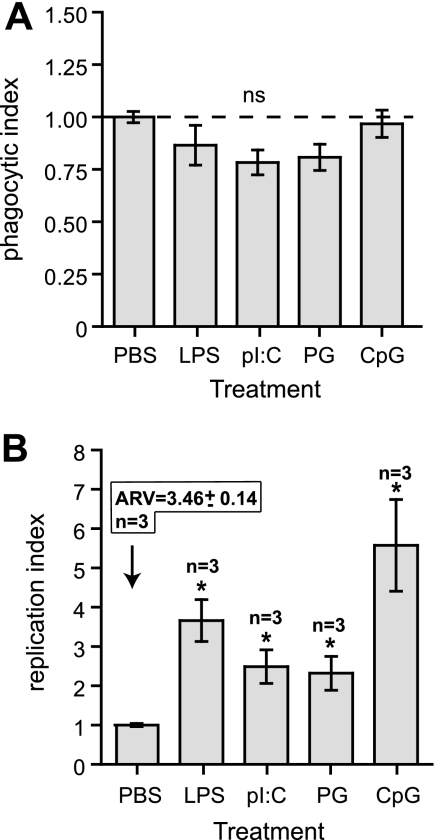
We considered that the enhanced intracellular replication of S. Typhimurium in TLR-activated macrophages was due to the initially higher level of intracellular bacteria in treated cells. To address this, we reduced the MOI in TLR-primed cells from 50 to as low as 5 in order to achieve equivalent numbers of intracellular bacteria at the 2-h time point in treated cells as well as controls (Fig. 4A). Once these conditions were reproducibly established, we followed the fate of intracellular S. Typhimurium under these conditions. Despite the initially reduced and normalized bacterial load at 2 h postinfection (phagocytic index of ∼1) (Fig. 4A), intracellular replication of S. Typhimurium remained significantly greater in cells primed with TLR agonists than in the PBS-treated controls (Fig. 4B), indicating that the enhanced bacterial replication following TLR ligand treatment is due not solely to the high bacterial burden following phagocytic uptake.
FIG. 4.
Enhanced replication of S. Typhimurium in cells treated with TLR ligands is not due to an initially high intracellular bacterial load. (A) RAW 264.7 cells were treated with PBS, LPS, poly(I:C), PG, or CpG-ODN for 20 h before infection with wild-type S. Typhimurium in which the MOI was adjusted to achieve similar numbers of intracellular bacteria between the groups at 2 h postinfection. Data are the means with standard errors from three separate experiments with three technical replicates per experiment. (B) Intracellular replication of S. Typhimurium was quantified and expressed as the mean with standard errors from three separate experiments, normalized to the PBS control. The absolute replication value (ARV) for the PBS control is given above the control bar. Asterisks represent significant differences from the PBS control (P < 0.05, one-way analysis of variance). ns, not significant.
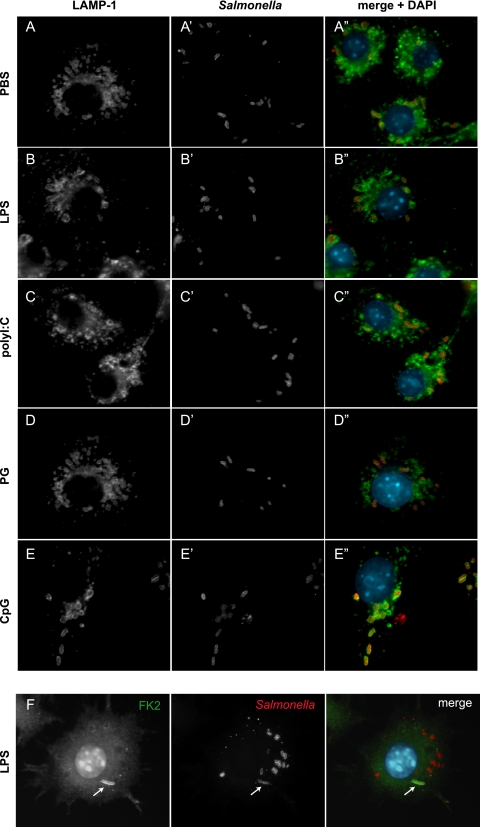
Salmonella growing in macrophages primed with TLR agonists is within LAMP-1-positive endosomes, and cytosolic escape leads to ubiquitination.
S. Typhimurium growing in mammalian cells occupies a membrane-bound endosome termed the SCV, for which LAMP-1 is required for vacuole maintenance (9). In addition, S. Typhimurium that escapes into the macrophage cytosol is targeted for degradation by the ubiquitin/proteasome system (40). To determine whether TLR agonist treatment of cells prior to infection changed either the recruitment of LAMP-1 to SCVs or cytosolic recognition by the ubiquitin system, we infected TLR-stimulated and control-treated macrophages and used immunofluorescence microscopy to quantify the distribution of intracellular S. Typhimurium. In all conditions tested, the numbers of LAMP-1-positive SCVs in infected cells were similar and not significantly different from one another (Fig. 5A to E). The fraction of LAMP-1-positive SCVs in cells treated with PBS, LPS, poly(I:C), PG, and CpG was 93.5%, 92.4%, 93.3%, 94.0%, and 93.4%, respectively, indicating that ∼7% of the intracellular bacterial population was not within LAMP-1-positive vacuoles. To determine the intracellular location of this population of bacteria, we costained infected cells with an anti-Salmonella antiserum and an antibody that recognizes mono- and polyubiquitinylated proteins (FK2) but not free ubiquitin (Fig. 5F). These data indicated that in all treatment groups the proportion of S. Typhimurium bacteria colocalizing with FK2 was ∼10% which, in conjunction with the LAMP-1 staining, fully accounted for the intracellular bacterial population. These LAMP-1 and FK2 distribution data are similar to data reported previously at a similar time point (5, 40). These data also indicated that TLR stimulation did not change the distribution of vacuole-associated versus cytosolic bacteria and did not affect cytosolic recognition of S. Typhimurium by the ubiquitin system.
FIG. 5.
Salmonella cells growing in TLR-stimulated macrophages are in LAMP-1-positive endosomes, and cytosolic escape leads to ubiquitination. RAW 264.7 cells treated with PBS (A) or TLR agonists (B to E) and infected with S. Typhimurium for 2 h were stained with antibodies against Salmonella (red) and LAMP-1 (green). Representative images from three separate experiments are shown. (F) Cytosolic bacteria are ubiquitinylated. RAW 264.7 cells treated with PBS or TLR agonists were infected and then stained with antibodies directed against Salmonella and mono- and polyubiquitin. Shown are immunofluorescent images from LPS-treated cells representative of three independent experiments. Ubiquitinylation of cytosolic bacteria was observed with similar frequencies in PBS-treated cells and cells treated with the other TLR agonists (data not shown).
Increased S. Typhimurium replication in TLR-stimulated macrophages requires a functional SPI-2 T3SS.
The T3SS encoded within SPI-2 is well known to be important for intracellular survival and replication of S. Typhimurium within host macrophages. We first examined whether TLR stimulation of macrophages furnished an intracellular environment that obviated the need for SPI-2 type III secretion activity for replication. Macrophages treated with TLR ligands did not support intracellular replication of a Salmonella ssaR mutant lacking a major structural component of the SPI-2 T3SS (41) even though these mutant cells were phagocytosed to similar levels as wild-type cells (Fig. 6A and B). These data suggested that SPI-2 T3SS activity was important for the enhanced replication phenotype.
FIG. 6.
Enhanced intracellular replication of Salmonella in TLR-stimulated cells is dependent on functional SPI-2 type 3 secretion. RAW 264.7 cells were treated with PBS, LPS, poly(I:C) (pI:C), PG, or CpG-ODN for 20 h before infection with wild-type S. Typhimurium (control) or a mutant with a defective T3SS, ΔssaR. The intracellular bacterial population was quantified at 2 h postinfection (A), and the relative change in the intracellular bacterial population between 2 to 18 h postinfection was calculated (B). The absolute replication value (ARV) for the control is given above the control bar, and the number of experiments used to produce the data is given above each treatment group. Bars represent the mean, with standard errors normalized to control cells infected with wild-type bacteria. Asterisks denote significant differences from the wild-type control (P < 0.05, one-way analysis of variance). (C) Intracellular transcriptional activity of the sseA virulence operon promoter. RAW 264.7 cells treated with PBS, LPS, or CpG ODN were infected with S. Typhimurium (S. typhim) carrying a chromosomal sseA transcriptional reporter fused to lacZ. Intracellular β-galactosidase activity was measured at 2 h and 10 h postinfection and normalized to the number of intracellular bacteria to facilitate group comparisons. Data are the means with standard errors from triplicate determinations from three separate experiments (P = 0.042 and 0.005 [Mann Whitney test]for LPS and CpG treatments at 2 h compared to the PBS control; P = 0.026 and 0.117 [Mann Whitney test] for LPS and CpG treatments at 10 h compared to the PBS control). (D) The replication advantage of wild-type Salmonella in TLR-primed cells is lost in a genetic mutant with augmented basal activity of SPI-2 transcription. Δhha mutant cells lacking a SPI-2 repressor and with high basal activity of SPI-2 transcription were used to infect TLR-stimulated cells. The replication index was calculated for mutant cells and compared to wild-type cells in which SPI-2 transcription activity is induced inside cells following infection. Data are the means with standard errors from three experiments. The absolute replication value (ARV) for the PBS control is given above the control bars. wt, wild type; cont, control.
It is known that expression of Salmonella virulence factors encoded within SPI-2 is influenced by the innate immune functionality of the macrophage host cell. That is, S. Typhimurium responds at a genetic level to a more resistant host environment by augmenting gene expression in SPI-2 to facilitate intracellular survival and replication (55). To test whether the intracellular environment of TLR-primed macrophages produced quantitative changes in SPI-2 gene expression compared to control cells, we used the prototypical sseA transcriptional reporter that reports on global SPI-2 gene expression (13, 18). Reporter cells in which the transcriptional reporter was integrated in single copy on the bacterial chromosome were used to infect macrophages that were pretreated with either LPS or CpG-containing DNA, two of the TLR agonists giving rise to the greatest enhancement of intracellular replication. At 2 h and 10 h postinfection, times when SPI-2 gene expression activity in macrophages is highest (20), the number of intracellular bacteria was enumerated, and the β-galactosidase activity in host cell lysates arising from this population was quantified. At 2 h after infection, the sseA promoter activity was approximately threefold higher in cells primed with LPS (P = 0.042) or CpG DNA (P = 0.005) than in control cells (Fig. 6C). Enhanced activity of the sseA transcriptional reporter was maintained at 10 h postinfection in cells primed with LPS (P = 0.026) but returned to that of control cells by 10 h in cells pretreated with CpG DNA (P = 0.117) (Fig. 6C). The correlation between bacterial replication and patterns of SPI-2 transcriptional activity after infection suggested that transient enhanced transcriptional activity of the SPI-2 T3SS at 2 h, followed by normalization by 10 h, was involved in the enhanced bacterial replication seen in TLR-stimulated cells. To gain further insight into this possibility, we infected TLR-stimulated macrophages with a Salmonella mutant lacking hha, a gene coding for a SPI-2 repressor, that was previously shown to have high sustained SPI-2 expression activity due to derepression of the system (45, 50). In these experiments, the enhanced intracellular replication seen in TLR-stimulated cells was lost in the Salmonella hha mutant (Fig. 6D), further supporting the linkage between early enhanced SPI-2 expression activity and intracellular replication in TLR-stimulated cells. Together, this series of experiments demonstrated that enhanced intracellular replication of S. Typhimurium in cells treated with TLR ligands required a functional SPI-2 T3SS and that TLR ligands affected the magnitude of transcriptional activity from the sseA virulence gene operon.
CpG DNA impairs host defense against S. Typhimurium infection.
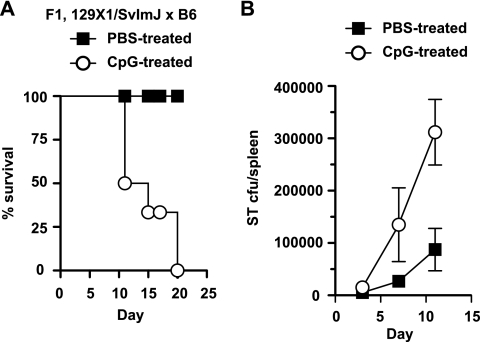
To gain further insight into the outcome of infection with S. Typhimurium following TLR stimulation, we delivered CpG DNA or PBS i.p. to Nramps/r heterozygote mice at the time of i.v. challenge with S. Typhimurium and monitored survival of the mice for 20 days. CpG DNA was used here because of the reproducible phenotype it elicited in cells in the previous experiments. All infected control mice that received PBS survived the infection. However, all mice given CpG DNA at the time of infection died by day 20 after inoculation (Fig. 7A). Because the spleen is a key site of Salmonella residence in vivo (17, 35), we examined the splenic bacterial load in infected mice at different days after infection in the presence or absence of concurrent CpG DNA treatment. In these experiments, CpG-treated mice had higher splenic bacterial loads beginning at about day 5 after inoculation, which continued at day 10 when the first CpG-treated mice succumbed to the primary infection. These data suggest that CpG DNA can impair the host defense against S. Typhimurium infection, correlating with greater bacterial burden in the spleen following infection.
FIG. 7.
CpG DNA impairs host defense against S. Typhimurium infection. (A) Concurrent with i.v. infection with S. Typhimurium, mice were given CpG DNA (open circles) or PBS (filled squares) delivered i.p. Mice were monitored after infection and scored for body weight loss and survival for 20 days. (B) Splenic bacterial load from mice treated with CpG DNA (open circles) or PBS (filled squares). Data are from a representative experiment with six mice per group.
DISCUSSION
Phagocytosis is a highly regulated process in macrophages, and a growing body of evidence links TLR stimulation with phagocytosis (7, 16, 31, 38, 48). Flow cytometry of murine macrophages treated with CpG showed increased phagocytosis of Burkholderia pseudomallei, S. enterica serovar Typhi, and E. coli in a dose-dependent manner (48). Doyle and coworkers expanded on this by showing that treatment of macrophages with CpG ODN, lipid A, PG, or poly(I:C) increased phagocytic uptake of E. coli and S. aureus but not of latex beads (16). Recently, others found that stimulation of RAW 264.7 cells with LPS, PG, CpG, and poly(I:C) increased phagocytosis of E. coli expressing green fluorescent protein (31). However, in the latter study, the degree of phagocytic enhancement did not differ with various TLR agonists, as we along with others (16) have observed, where a TLR9 agonist provided the strongest increase in phagocytosis and a TLR3 agonist provided the weakest. Analogously, phagocytosis of E. coli is impaired in macrophages lacking the adapter molecule MyD88, suggesting a connection between TLR signaling and phagocytosis (7). Although multiple TLR ligands can upregulate a phagocytic program in macrophages involving complement receptors, FcγR1 and scavenger receptors such as SR-A and MARCO (16), the spectrum of receptors associated with Salmonella uptake are not known at this point. Elucidating the surface phenotype of TLR-primed cells followed by functional studies might give insight into this question.
In regard to the effector functions of macrophages pretreated with TLR ligands prior to S. Typhimurium infection, input from a single TLR created an intracellular environment more conducive for S. Typhimurium survival and replication. This was true of all the TLR agonists tested here, which signal through TLR2, TLR3, TLR4, and TLR9. This enhanced bacterial replication was insensitive to the initial bacterial load's gaining entry during phagocytosis because in experiments with reduced inocula that normalized the initial intracellular bacterial population, Salmonella infecting TLR-stimulated cells still out-competed bacteria replicating in PBS-treated cells. Interestingly, coadministration of IFN-γ restricted the bacterial replication permitted in TLR-stimulated macrophages, which is likely mediated by enhanced production of reactive oxygen (44) and nitrogen species (36) following IFN-γ treatment. While we also performed similar experiments with freshly isolated mouse peritoneal macrophages, the degree of intracellular Salmonella replication following TLR ligand treatment was variable. One interpretation of the work with primary cells may relate to differences in their cytokine repertoire. For example, RAW264.7 cells do not produce IFN-γ in response to TLR ligands, whereas primary macrophages do (4, 15, 37). Likewise, although J774 cells respond to exogenous IFN-γ, they are a weak source of IFN-γ during infection (33). Given that IFN-γ is a key mediator of Salmonella control in vitro, it is likely that differences in IFN-γ competence underlie the difference between macrophage cell lines and primary cells under our in vitro conditions. These results are similar to those reported for the bovine pathogen S. enterica serovar Dublin, in which IFN-γ completely inhibited S. Dublin replication in macrophages (51). In the same study, CpG treatment partially inhibited the growth of M. tuberculosis but not S. Dublin, suggesting that TLR agonist stimulation has different effects on the recognition, uptake, and intracellular survival abilities of different bacteria. These observations are germane to current discourse on the use of immune enhancement as a novel alternative to antibiotics (21) as different intracellular pathogens have evolved different strategies for intracellular parasitism and for overcoming, or even exploiting, the innate immune system in host cells. The system we have presented here should be useful to probe the macrophage effector functions downstream of TLR activation that can be targets of professional intracellular pathogens.
In the case of Salmonella, activation of virulence genes in SPI-2 is required for evasion of both reactive oxygen and nitrogen species following phagocytosis (11, 49), which can be partially blocked in the presence of IFN-γ (36). This cytokine is critical for control of Salmonella since genetic mutations that perturb this cytokine pathway render individuals highly susceptible to disseminated nontyphoidal Salmonella infections (39). Together with data showing that S. Typhimurium upregulates SPI-2 gene expression in response to a more resistant host cell genotype (55), these observations suggest that virulence gene expression is tunable in magnitude, based on the activation state of the host cell. This also suggests that the interplay between the host immune system and bacterial adaptive mechanisms is under surveillance and acclimation by both bacteria and the host cell. The enhanced activity of the SPI-2 transcriptional reporter in TLR agonist-treated macrophages supports this notion. Although the mechanism by which Salmonella vets this intracellular environment is not yet known, it will be an important area for further study.
Although the protective role of TLRs in bacterial infections is well known, there are also data to suggest that stimulation of TLR signaling can exacerbate S. Typhimurium infection in certain in vivo contexts. For example, mice deficient in TIRAP (Toll/interleukin-1 receptor domain-containing adaptor protein), involved in signaling downstream of certain TLRs (28), show accelerated clearance of Salmonella infection, implying a mild deleterious effect of TLR/TIRAP signaling on resolution of primary infection (29). Similarly, TRIF and MyD88 signaling exacerbates infection by virulent S. Typhimurium following administration of a defective Salmonella vaccine strain (23), implying that the interface between Salmonella and even well-studied aspects of the innate immune system are not completely understood. Our data showed that administering CpG DNA to mice at the time of S. Typhimurium infection exacerbates the infectious process, leading to death. The increased bacterial load in the spleen of CpG-treated animals may mediate this outcome, in part, but we also suspect that additional immune-mediated influences are involved that will be the subject of follow-up work ongoing in our laboratory. It is also of interest that the outcome of infection in mice with another intracellular pathogen, B. pseudomallei, is improved following deletion of TLR2, the TLR involved in responding to B. pseudomallei LPS (53). Compared with TLR4−/− or wild-type mice, TLR2−/− mice had a survival advantage that correlated with lower bacterial organ load and reduced lung inflammation, suggesting that TLR2 stimulation enhanced the growth of B. pseudomallei and dissemination to the blood and spleen from the site of primary infection. In this model, loss of type 2 cytokines such as interleukin-10 could not explain the protection of TLR2−/− animals during B. pseudomallei infection, suggesting that other mechanisms, perhaps an enhanced ability to replicate intracellularly in the presence of TLR2 signaling, are involved. The ability of TLR ligands to enhance replication of wild-type S. Typhimurium but not T3SS mutants or other nonpathogenic organisms (43) suggests an adaptive mechanism in Salmonella that can exploit cells stimulated by the innate immune system.
Supplementary Material
Acknowledgments
We thank members of the Coombes, Brumell, and Ashkar laboratories for helpful comments on this work, Tony Collins at the McMaster Biophotonics Facility for assistance with live-cell imaging, and Rick Mackenzie at the McMaster Integrated Microscopy Facility for processing samples for transmission electron microscopy.
This work was funded by an operating grant to B.K.C. from the Canadian Institutes of Health Research (CIHR; MOP 82704). B.K.C. is a CIHR New Investigator and the recipient of the ASM-Merck Young Investigator Award from the American Society of Microbiology and the Early Researcher Award from the Ontario Ministry of Research and Innovation.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 31 August 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 3.Ashkar, A. A., S. Bauer, W. J. Mitchell, J. Vieira, and K. L. Rosenthal. 2003. Local delivery of CpG oligodeoxynucleotides induces rapid changes in the genital mucosa and inhibits replication, but not entry, of herpes simplex virus type 2. J. Virol. 77:8948-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashkar, A. A., X. D. Yao, N. Gill, D. Sajic, A. J. Patrick, and K. L. Rosenthal. 2004. Toll-like receptor (TLR)-3, but not TLR4, agonist protects against genital herpes infection in the absence of inflammation seen with CpG DNA. J. Infect. Dis. 190:1841-1849. [DOI] [PubMed] [Google Scholar]
- 5.Birmingham, C. L., A. C. Smith, M. A. Bakowski, T. Yoshimori, and J. H. Brumell. 2006. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281:11374-11383. [DOI] [PubMed] [Google Scholar]
- 6.Blander, J. M., and R. Medzhitov. 2006. On regulation of phagosome maturation and antigen presentation. Nat. Immunol. 7:1029-1035. [DOI] [PubMed] [Google Scholar]
- 7.Blander, J. M., and R. Medzhitov. 2004. Regulation of phagosome maturation by signals from Toll-like receptors. Science 304:1014-1018. [DOI] [PubMed] [Google Scholar]
- 8.Brown, N. F., B. A. Vallance, B. K. Coombes, Y. Valdez, B. A. Coburn, and B. B. Finlay. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog. 1:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brumell, J. H., and S. Grinstein. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7:78-84. [DOI] [PubMed] [Google Scholar]
- 10.Brumell, J. H., C. M. Rosenberger, G. T. Gotto, S. L. Marcus, and B. B. Finlay. 2001. SifA permits survival and replication of Salmonella typhimurium in murine macrophages. Cell Microbiol. 3:75-84. [DOI] [PubMed] [Google Scholar]
- 11.Chakravortty, D., I. Hansen-Wester, and M. Hensel. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 195:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coombes, B. K., N. F. Brown, S. Kujat-Choy, B. A. Vallance, and B. B. Finlay. 2003. SseA is required for translocation of Salmonella pathogenicity island-2 effectors into host cells. Microbes Infect. 5:561-570. [DOI] [PubMed] [Google Scholar]
- 13.Coombes, B. K., N. F. Brown, Y. Valdez, J. H. Brumell, and B. B. Finlay. 2004. Expression and secretion of Salmonella pathogenicity island-2 virulence genes in response to acidification exhibit differential requirements of a functional type III secretion apparatus and SsaL. J. Biol. Chem. 279:49804-49815. [DOI] [PubMed] [Google Scholar]
- 14.Cooper, A. M., D. K. Dalton, T. A. Stewart, J. P. Griffin, D. G. Russell, and I. M. Orme. 1993. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J. Exp. Med. 178:2243-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Marzio, P., P. Puddu, L. Conti, F. Belardelli, and S. Gessani. 1994. Interferon gamma upregulates its own gene expression in mouse peritoneal macrophages. J. Exp. Med. 179:1731-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle, S. E., R. M. O'Connell, G. A. Miranda, S. A. Vaidya, E. K. Chow, P. T. Liu, S. Suzuki, N. Suzuki, R. L. Modlin, W. C. Yeh, T. F. Lane, and G. Cheng. 2004. Toll-like receptors induce a phagocytic gene program through p38. J. Exp. Med. 199:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlap, N. E., W. H. Benjamin, Jr., R. D. McCall, Jr., A. B. Tilden, and D. E. Briles. 1991. A “safe-site” for Salmonella typhimurium is within splenic cells during the early phase of infection in mice. Microb. Pathog. 10:297-310. [DOI] [PubMed] [Google Scholar]
- 18.Duong, N., S. Osborne, V. H. Bustamante, A. M. Tomljenovic, J. L. Puente, and B. K. Coombes. 2007. Thermosensing coordinates a cis-regulatory module for transcriptional activation of the intracellular virulence system in Salmonella enterica serovar Typhimurium. J. Biol. Chem. 282:34077-34084. [DOI] [PubMed] [Google Scholar]
- 19.Elkins, K. L., T. R. Rhinehart-Jones, S. Stibitz, J. S. Conover, and D. M. Klinman. 1999. Bacterial DNA containing CpG motifs stimulates lymphocyte-dependent protection of mice against lethal infection with intracellular bacteria. J. Immunol. 162:2291-2298. [PubMed] [Google Scholar]
- 20.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 21.Finlay, B. B., and R. E. Hancock. 2004. Can innate immunity be enhanced to treat microbial infections? Nat. Rev. Microbiol. 2:497-504. [DOI] [PubMed] [Google Scholar]
- 22.Flynn, J. L., M. M. Goldstein, K. J. Triebold, and B. R. Bloom. 1993. Major histocompatibility complex class I-restricted T cells are necessary for protection against M. tuberculosis in mice. Infect. Agents Dis. 2:259-262. [PubMed] [Google Scholar]
- 23.Foster, G. L., T. A. Barr, A. J. Grant, T. J. McKinley, C. E. Bryant, A. MacDonald, D. Gray, M. Yamamoto, S. Akira, D. J. Maskell, and P. Mastroeni. 2008. Virulent Salmonella enterica infections can be exacerbated by concomitant infection of the host with a live attenuated S. enterica vaccine via Toll-like receptor 4-dependent interleukin-10 production with the involvement of both TRIF and MyD88. Immunology 124:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallois, A., J. R. Klein, L. A. Allen, B. D. Jones, and W. M. Nauseef. 2001. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J. Immunol. 166:5741-5748. [DOI] [PubMed] [Google Scholar]
- 25.Harandi, A. M., K. Eriksson, and J. Holmgren. 2003. A protective role of locally administered immunostimulatory CpG oligodeoxynucleotide in a mouse model of genital herpes infection. J. Virol. 77:953-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 27.Herbst-Kralovetz, M. M., and R. B. Pyles. 2006. Quantification of poly(I:C)-mediated protection against genital herpes simplex virus type 2 infection. J. Virol. 80:9988-9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horng, T., G. M. Barton, R. A. Flavell, and R. Medzhitov. 2002. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature 420:329-333. [DOI] [PubMed] [Google Scholar]
- 29.Jerke, S., A. Srinivasan, and S. J. McSorley. 2008. Expression of Toll/IL-1R domain-containing adaptor protein (TIRAP) is detrimental to primary clearance of Salmonella and is not required for the generation of protective immunity. Immunol. Lett. 116:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Juffermans, N. P., J. C. Leemans, S. Florquin, A. Verbon, A. H. Kolk, P. Speelman, S. J. van Deventer, and T. van der Poll. 2002. CpG oligodeoxynucleotides enhance host defense during murine tuberculosis. Infect. Immun. 70:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong, L., and B. X. Ge. 2008. MyD88-independent activation of a novel actin-Cdc42/Rac pathway is required for Toll-like receptor-stimulated phagocytosis. Cell Res. 18:745-755. [DOI] [PubMed] [Google Scholar]
- 32.Krieg, A. M. 2006. Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug Discov. 5:471-484. [DOI] [PubMed] [Google Scholar]
- 33.Lampe, M. F., C. B. Wilson, M. J. Bevan, and M. N. Starnbach. 1998. Gamma interferon production by cytotoxic T lymphocytes is required for resolution of Chlamydia trachomatis infection. Infect. Immun. 66:5457-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, H., M. M. Nooh, M. Kotb, and F. Re. 2008. Commercial peptidoglycan preparations are contaminated with superantigen-like activity that stimulates IL-17 production. J. Leukoc. Biol. 83:409-418. [DOI] [PubMed] [Google Scholar]
- 35.Mastroeni, P., A. Grant, O. Restif, and D. Maskell. 2009. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat. Rev. Microbiol. 7:73-80. [DOI] [PubMed] [Google Scholar]
- 36.McCollister, B. D., T. J. Bourret, R. Gill, J. Jones-Carson, and A. Vazquez-Torres. 2005. Repression of SPI2 transcription by nitric oxide-producing, IFNγ-activated macrophages promotes maturation of Salmonella phagosomes. J. Exp. Med. 202:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Means, T. K., B. W. Jones, A. B. Schromm, B. A. Shurtleff, J. A. Smith, J. Keane, D. T. Golenbock, S. N. Vogel, and M. J. Fenton. 2001. Differential effects of a Toll-like receptor antagonist on Mycobacterium tuberculosis-induced macrophage responses. J. Immunol. 166:4074-4082. [DOI] [PubMed] [Google Scholar]
- 38.Neal, M. D., C. Leaphart, R. Levy, J. Prince, T. R. Billiar, S. Watkins, J. Li, S. Cetin, H. Ford, A. Schreiber, and D. J. Hackam. 2006. Enterocyte TLR4 mediates phagocytosis and translocation of bacteria across the intestinal barrier. J. Immunol. 176:3070-3079. [DOI] [PubMed] [Google Scholar]
- 39.Ottenhoff, T. H., F. A. Verreck, E. G. Lichtenauer-Kaligis, M. A. Hoeve, O. Sanal, and J. T. van Dissel. 2002. Genetics, cytokines and human infectious disease: lessons from weakly pathogenic mycobacteria and salmonellae. Nat. Genet. 32:97-105. [DOI] [PubMed] [Google Scholar]
- 40.Perrin, A. J., X. Jiang, C. L. Birmingham, N. S. So, and J. H. Brumell. 2004. Recognition of bacteria in the cytosol of mammalian cells by the ubiquitin system. Curr. Biol. 14:806-811. [DOI] [PubMed] [Google Scholar]
- 41.Pfeifer, C. G., S. L. Marcus, O. Steele-Mortimer, L. A. Knodler, and B. B. Finlay. 1999. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect. Immun. 67:5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pyles, R. B., D. Higgins, C. Chalk, A. Zalar, J. Eiden, C. Brown, G. Van Nest, and L. R. Stanberry. 2002. Use of immunostimulatory sequence-containing oligonucleotides as topical therapy for genital herpes simplex virus type 2 infection. J. Virol. 76:11387-11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribes, S., S. Ebert, D. Czesnik, T. Regen, A. Zeug, S. Bukowski, A. Mildner, H. Eiffert, U. K. Hanisch, S. Hammerschmidt, and R. Nau. 2009. Toll-like receptor prestimulation increases phagocytosis of Escherichia coli DH5α and Escherichia coli K1 strains by murine microglial cells. Infect. Immun. 77:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenberger, C. M., and B. B. Finlay. 2002. Macrophages inhibit Salmonella typhimurium replication through MEK/ERK kinase and phagocyte NADPH oxidase activities. J. Biol. Chem. 277:18753-18762. [DOI] [PubMed] [Google Scholar]
- 45.Silphaduang, U., M. Mascarenhas, M. Karmali, and B. K. Coombes. 2007. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J. Bacteriol. 189:3669-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsolis, R. M., R. A. Kingsley, S. M. Townsend, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Of mice, calves, and men. Comparison of the mouse typhoid model with other Salmonella infections. Adv. Exp. Med. Biol. 473:261-274. [PubMed] [Google Scholar]
- 47.Uchiya, K.-I., M. A. Barbieri, K. Funato, A. H. Shar, P. D. Stahl, and E. A. Groisman. 1999. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18:3924-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Utaisincharoen, P., W. Kespichayawattana, N. Anuntagool, P. Chaisuriya, S. Pichyangkul, A. M. Krieg, and S. Sirisinha. 2003. CpG ODN enhances uptake of bacteria by mouse macrophages. Clin. Exp. Immunol. 132:70-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vazquez-Torres, A., Y. Xu, J. Jones-Carson, D. W. Holden, S. M. Lucia, M. C. Dinauer, P. Mastroeni, and F. C. Fang. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655-1658. [DOI] [PubMed] [Google Scholar]
- 50.Vivero, A., R. C. Banos, J. F. Mariscotti, J. C. Oliveros, F. Garcia-del Portillo, A. Juarez, and C. Madrid. 2008. Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, J. P., T. Hayashi, S. K. Datta, R. S. Kornbluth, E. Raz, and D. G. Guiney. 2005. CpG oligonucleotides partially inhibit growth of Mycobacterium tuberculosis, but not Salmonella or Listeria, in human monocyte-derived macrophages. FEMS Immunol. Med. Microbiol. 45:303-310. [DOI] [PubMed] [Google Scholar]
- 52.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 53.Wiersinga, W. J., C. W. Wieland, M. C. Dessing, N. Chantratita, A. C. Cheng, D. Limmathurotsakul, W. Chierakul, M. Leendertse, S. Florquin, A. F. de Vos, N. White, A. M. Dondorp, N. P. Day, S. J. Peacock, and T. van der Poll. 2007. Toll-like receptor 2 impairs host defense in gram-negative sepsis caused by Burkholderia pseudomallei (melioidosis). PLoS Med. 4:e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 55.Zaharik, M. L., B. A. Vallance, J. L. Puente, P. Gros, and B. B. Finlay. 2002. Host-pathogen interactions: host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc. Natl. Acad. Sci. USA 99:15705-15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.