Abstract
Pseudomonas aeruginosa is responsible for potentially life-threatening infections in individuals with compromised defense mechanisms and those with cystic fibrosis. P. aeruginosa infection is notable for the appearance of a humoral response to some known antigens, such as flagellin C, elastase, alkaline protease, and others. Although a number of immunogenic proteins are known, no effective vaccine has been approved yet. Here, we report a comprehensive study of all 262 outer membrane and exported P. aeruginosa PAO1 proteins by a modified protein microarray methodology called the nucleic acid-programmable protein array. From this study, it was possible to identify 12 proteins that trigger an adaptive immune response in cystic fibrosis and acutely infected patients, providing valuable information about which bacterial proteins are actually recognized by the immune system in vivo during the natural course of infection. The differential detections of these proteins in patients and controls proved to be statistically significant (P < 0.01). The study provides a list of potential candidates for the improvement of serological diagnostics and the development of vaccines.
Pseudomonas aeruginosa is a gram-negative bacterium represented by different strains. PAO1 is the most representative one, and it contains one chromosome comprising 5,570 open reading frames (ORFs) (21). Ubiquitous in the environment, it rarely causes respiratory tract infections in healthy individuals but causes life-threatening lung infections in cystic fibrosis (CF) patients (24, 27), linking it directly to the disease's poor prognosis (38).
CF is a hereditary disease affecting 30,000 people in the United States alone, and it results from the impaired function of the cystic fibrosis transmembrane conductance regulator-encoded chloride channel. This defect leads to the accumulation of nutrient-rich mucus inside the respiratory tract, which is readily colonized by bacteria and difficult for immune cells and antibiotics to penetrate (13). Although these patients are generally colonized early by other pathogens, such as Staphylococcus aureus and Haemophilus influenzae, by the age of 18, 85% have chronic pseudomonal lung infections (23).
P. aeruginosa is also associated with nosocomial infections, especially in the immunocompromised (6, 10). Multidrug-resistant strains in intensive care units as well as outside the hospital setting have been described, making the development of alternate treatment strategies a necessity (9, 29). CF patients and other at-risk individuals would benefit from a functional vaccine, but none has been approved for use so far (8, 16).
In the late 1970s, evidence about proteins acting as important virulence factors in Pseudomonas aeruginosa infection called attention to exotoxin A (protein PA1148), alkaline protease (protein PA1248), and elastase (protein PA3724) (18, 43). Currently, diagnostic serology for P. aeruginosa is mostly based on these three proteins and has a sensitivity of 80% in chronic infections (36). This test is much less sensitive at the onset of infection, when serology alone does not detect pseudomonal infection in more than 43% of patients. The discovery of new antigens to complement the current panel of antigens would increase diagnostic sensitivity in all infection phases.
Pseudomonas presents different antigens on the surface depending on the microenvironment it colonizes, the mucoid phenotype being a hallmark for lung infections (5, 35). Which proteins are present on the pathogen's surface in each microenvironment has not yet been fully characterized; however, this information would be invaluable for diagnostics development and reverse vaccinology studies.
Early studies executed in the 1980s probed Western blots made from gel electrophoresis-fractionated P. aeruginosa extracts with serum to reveal reactive bands. Some of these reactive bands were identified to be specific outer membrane proteins (OMPs), such as OMPs E, H2, and I, as well as porin F, but due to technical limitations, not all bands could be identified (15, 20). Notably, all of these proteins, as well as the secreted proteins alkaline protease and elastase from the earlier studies, are externally displayed proteins, confirming the importance of this class of proteins in triggering an immune response, as they are readily seen by the immune system. Therefore, it would be useful to execute a systematic comprehensive study to analyze all predicted OMPs, both known and hypothetical, in an unbiased screening experiment.
These also represent good vaccine candidates based on data from phase I and II clinical trials using two P. aeruginosa OMPs in vaccines (8). A phase III trial of the external surface flagellin protein also showed promising results with previously uninfected CF patients (7). However, testing proteins as candidate antigens ordinarily requires antigen expression and purification, which are notoriously difficult for membrane proteins, making a comprehensive study of all P. aeruginosa OMPs with traditional methods impossible, particularly because the P. aeruginosa genome is very GC rich, which reduces the yield of protein in common expression systems.
Protein microarrays provide one mechanism for executing an unbiased screen. Recent studies demonstrate immune responses from plague patients against a Yersinia pestis protein microarray containing 144 virulence related factors, 14 of which were detected by all the patients tested (25). Similar protein microarray studies have been done for selected antigens from Francisella tularensis (39), Borrelia burgdorferi (44), Coxiella burnetti (2), severe acute respiratory syndrome coronavirus (45), and others. In some cases, difficulty in producing proteins has been encountered. In the C. burnetti study, ∼500 proteins could not be studied (∼25%) due to difficulties in producing some individual proteins for array spotting. The high-throughput nature of microarrays allows easy execution of comparative studies of the humoral response mounted against different pathogens. In a recent study, Keasey and collaborators demonstrated that by probing a Yersinia pestis-specific protein microarray with serum from animals infected with other gram-negative bacteria, it was possible to find shared cross-reactive proteins (common to several pathogens), fingerprint proteins (common to two or more bacteria), and signature proteins (specific to each pathogen) (17).
However, the traditional protein microarray printing method, which relies upon printing purified proteins, may encounter limitations. Chief among these is the difficulty in producing and purifying a broad range of proteins in order to obtain both adequate and consistent protein yields, resulting in an excessively large dynamic range of protein yields displayed on the array. This is particularly relevant in circumstances in which the targets are either hydrophobic or large.
To address these issues, we previously reported the development of a novel method for displaying proteins on protein microarrays, called the nucleic acid-programmable protein array (NAPPA), which is particularly advantageous for membrane proteins. In contrast to the traditional method for producing protein microarrays, NAPPA entails printing the cDNAs that encode the proteins that are “self-assembled” at the time of the experiment by transcribing and translating proteins in situ on the array surface (33). The cDNAs are situated in an expression vector such that they produce proteins with a carboxyl-terminal fusion tag that both allows protein capture at the surface and confirms full-length translation. This unique approach obviates the need to express and purify any protein, avoids protein aggregation by immediate capture on the slide surface, and ensures that the proteins displayed have been produced freshly at the time of assay.
NAPPAs display a uniform amount of protein in each feature (34) and work well for detecting humoral immune responses in a micro-enzyme-linked immunosorbent assay (micro-ELISA) format (1, 32). NAPPA has been demonstrated to express and display membrane proteins with an efficiency that exceeds 90% (34), perhaps by avoiding cell toxicity and protein aggregation. Based on the observation that NAPPA-immobilized proteins interact with other proteins as expected (33), we reasoned that many relevant epitopes should be displayed and that NAPPA could be used to test candidate membrane antigens in P. aeruginosa.
Using bioinformatics tools based on amino acid composition, homology to known OMPs, and finding transmembrane ß-barrels, we identified 266 gene products among all of the 5,570 ORFs in PAO1 that were likely to reside in the outer membrane, are part of exposed surface organelles, or are secreted into the surrounding media. These proteins were represented on the surface of the NAPPAs, and patient sera were used to screen for their antigenicity.
MATERIALS AND METHODS
Selection of OMPs by using bioinformatics tools.
Several different bioinformatics tools, such as BOMP (3) and PROFtmb (4), were used for ß-barrel prediction; PROFtmb uses a profile based on a hidden Markov model, while BOMP uses other sequence-based algorithms to predict bacterial transmembrane β-barrels in the structure. PSORT (11) and Pence (26) were used for the prediction of subcellular localization. PSORT uses a combined algorithm to predict subcellular location by analyzing signal peptide, transmembrane helices, homology to annotated proteins, amino acid composition, and structural motifs. Pence uses database text annotations from homologs and machine learning to substantially improve the prediction of subcellular location. To achieve the most comprehensive OMP list, we combined the predicted results from all approaches and compared them to available annotation. A specialist in the field verified the results to eliminate obvious false positives and to add candidates of interest based on updated literature that might have been missed by the bioinformatics tools, defining the final set of 266 proteins to be studied.
Transfer of ORFs from the entry vector into the pANT7-GST expression vector.
Pseudomonas aeruginosa entry clones representing full-length ORFs in pDONR221 were previously made in our laboratory (21). The full-length sequences of all of the ORFs tested in this work were verified. Reactions for transfer from pDONR221 into pANT7-cGST were done with LR kits from Invitrogen, following the manufacturer's instructions and as previously described (28). Transfer reaction products were transformed into a T1 phage-resistant Escherichia coli DH5α host strain and plated onto LB agar-ampicillin 48-well grid plates for single-colony selection. After colony growth, a customized Megapix robot (Genetix) was used to count colonies, capture an image, isolate the indicated number of single colonies from each sector of the agar dish, and inoculate the colony into 1 ml growth media (LB-antibiotic). Cultures were grown overnight at 37°C, their growth was verified via measurement of optical densities at 600 nm, and aliquots were stored at −80°C as 15% glycerol stocks.
Plasmid DNA isolation.
An aliquot of the glycerol stocks was diluted in LB medium (1:300) and replated on LB agar (100 μg/ml ampicillin), and on the following day, isolated colonies were used to inoculate 1.5-ml Terrific Broth (100 μg/ml ampicillin) cultures in the 96-deep-well block format (34). Cultures were grown for 24 h at 37°C and pelleted by centrifugation at a relative centrifugal force of 5,000 for 15 min. Plasmid DNA was obtained by regular alkaline lysis, scaled up to a 96-well format, followed by anion exchange purification using the reagents and protocols provided by the resin manufacturer (Macherey-Nagel). Agar spotting and volume transfers from glycerol inoculation to plasmid DNA purification were done using robotics whenever possible (Biomek FX from Beckman-Coulter, Genmate from Tecan, and WellMate from Matrix).
Sample preparation for microarray printing.
In brief, purified DNA was precipitated using 10% sodium acetate (pH 5.5) and 60% isopropanol, washed with 200 μl of 80% ethanol, and dried. The dried DNA pellet was resuspended in a printing solution containing 50 ng/μl of capture antibody (Amersham), 3 mg/ml of bovine serum albumin (Sigma), and 2 mM bis(sulfosuccinimidyl) suberate (Pierce). The printing solution was transferred to a 384-well printing plate (Genetix) and arrayed onto aminosilane-coated glass slides. The printing was performed using a Genetix Q2 microarrayer with 300-μm solid tungsten pins, and slides were stored dry at room temperature.
NAPPA slide processing.
The slides were blocked in Superblock (Pierce) for 1 h at room temperature, rinsed with double deionized water, and dried using house air. An incubation chamber (Grace Bio) was applied to the array surface, and the cell-free transcription and translation mix (T7-TNT system; Promega) was prepared following the manufacturer's instructions and added to the slides. The slides were incubated in a chilling oven (Torrey Pines) at 30°C for 1.5 h and at 15°C for 0.5 h. Following activation with cell-free lysate, the slides were blocked with blocking buffer (5% milk in phosphate-buffered saline [PBS] supplemented with 0.2% Tween 20) for 1 h. For the detection of proteins on the array, the slides were incubated with 300 μl of primary antibody (10 μg/ml anti-glutathione S-transferase [GST] antibody; Cell Signaling Technologies) by using a LifterSlip (Erie) for 1 h at room temperature. The slides were rinsed with the blocking buffer and incubated with secondary antibody (10 μg/ml anti-mouse immunoglobulin G [IgG]; Amersham) by using the same conditions as for the primary antibody. The GST moiety is at the carboxyl terminus of each expressed protein; thus, detection confirms full-length translation. For the detection of serum signal, slides were placed in a hybridization chamber (Corning) and 2 ml of diluted serum (1:600 in blocking buffer) was added. The slides were incubated for 16 h at 4°C on a rotator. Following incubation with serum, slides were washed with blocking buffer and incubated with secondary antibody against human IgGs (1.6 μg/ml; Jackson ImmunoLabs) by using the same chamber previously described, for 1 h at room temperature. The slides for both protein signal and serum signal were developed using 600 μl of tyramide signal amplification solution (Perkin Elmer) and scanned using a ScanArray ProScanArray HT scanner (Perkin Elmer). Scanning conditions were 43% photomultiplier tube gain, 79% laser power, and 50-μm resolution for visualization and 33% photomultiplier tube gain, 80% laser power, and 20-μm resolution for quantification. False-color images were generated using ScanArray Express. DNA binding was measured by incubating the slides in 20 ml 1× PBS containing 34 μl PicoGreen reagent. The array images were quantified using MicroVigene software, version 2.501 (VigeneTech).
Serum collection.
A collection comprising 22 serum samples from patients attending the Cystic Fibrosis Clinic at the Children's Hospital in Boston, Massachusetts, was used. The ages of patients ranged from 5 to 54 years. Sputum samples were collected at the time of the outpatients' visit or admission, and they were all positive for P. aeruginosa. At this time, blood, from which the sera for the protein array were generated, was collected. The lung function of the subjects, as determined by spirometry, ranged from normal to 33%. Half of the patients experienced exacerbation as determined by the attending physicians.
The 16 non-CF patient samples were collected at various clinics at the University of California San Francisco Medical Center. Samples were from intubated intensive care unit patients with newly acquired P. aeruginosa strains. To confirm that patients were infected with P. aeruginosa, bacteria were isolated from endotracheal aspirates. Isolates from airway secretions with high bacterial counts (mean CFU/ml = 6 × 107) were typically associated with patients diagnosed with ventilator-associated pneumonia; 50% of patients in this group died. Cultures from airway secretions with lower counts (mean CFU/ml = 3.4 × 105) were isolated from patients who were subsequently discharged or transferred to acute or long-term-care facilities. All isolates were cultured on Pseudomonas isolation agar, and the identities were confirmed by MicroScan Combo assay (Dade Behring, CA) according to the manufacturer's instructions. Sera from blood samples obtained from a third group of patients with newly acquired P. aeruginosa in their respiratory secretions and a preexisting diagnosis of chronic obstructive pulmonary disease (COPD) were also used for this study. One of the patients in this cohort developed clinical signs and symptoms of ventilator-associated pneumonia; this patient subsequently died.
Fifteen healthy, P. aeruginosa-free controls were used to screen NAPPA slides containing the 262 Pseudomonas aeruginosa ORFs.
All samples collected for this study were collected with institutional review board approval and handled as deidentified sera. The serum supply for this study was exhausted during the NAPPA screening and ELISA/Western blot validation studies.
Image analysis and quantification.
After the slides were scanned, spot intensities were measured using the MicroVigene software (VigeneTech) for microarray analysis. For each serum sample, the average of four spots (two slides per patient and two spots per antigen on each slide) was calculated for each antigen. The mean of all spots in each slide was obtained (disregarding controls), and the Z-score for each antigen in relation to the slide mean was generated. Slide images were also printed for visual confirmation by two independent researchers based on spot intensity and morphology.
Western blot confirmation.
Proteins of interest were produced in solution by using the in vitro transcription/translation (IVTT) system. Plasmid DNA (500 ng) purified by anion exchange and resuspended in diethyl pyrocarbonate-treated water was used to program protein production in IVTT reactions (20 μl). An aliquot of 3 μl of IVTT product was subjected to electrophoresis using 4 to 20% Criterion Precast gels (Bio-Rad), and the proteins were then transferred to a polyvinylidene difluoride membrane (GE Healthcare), which was blocked in SuperBlock (Thermo Scientific) overnight and incubated with patient serum (1:600 in 1× PBS containing 5% milk and 0.05% Tween 20) for 1 h at room temperature. Signal detection was performed using 1:1,000 anti-human IgG (Jackson ImmunoResearch) and SuperSignal West Femto (Thermo Scientific). Images were obtained using Chemi Doc (Bio-Rad).
ELISA for confirmation.
Proteins of interest were produced as described for Western blotting and bound to a preblocked anti-GST-coated plate (GE Healthcare). A standard ELISA protocol was followed using patient or control serum diluted at 1:300 in 1× PBS containing 5% milk and 0.05% Tween 20. Anti-human IgG (Jackson ImmunoResearch) and Super Signal ELISA Pico (Thermo Scientific) were used for detection. Luminescence was read on the SpectraFluor Plus reader (Tecan), and images were obtained using Chemi Doc (Bio-Rad).
Statistical analysis.
To examine reproducibility of the signals obtained in ELISAs, duplicate features of selected antigens were probed with serum from multiple patients. For each of the patients, we computed the coefficients of variation ([standard deviation/mean] × 100%) of the log-transformed signals of the duplicate spots. The coefficients of variation ranged from 0.03% to 1.28%. The ratio of the positive samples and controls for each antigen was computed as the average ratio of the antigen's raw signal to the average raw signal of negative internal controls across the assays. The signal intensities were log transformed, and positive samples were compared to the internal negative controls within each assay by using an analysis of variance adjusted for assay effect. The normality assumptions for each antigen were verified using the Shapiro-Wilks test. All ratios and P values adjusted for multiple comparisons for the tested antigens are listed in Table 1 and Table S2 in the supplemental material. The P values were adjusted for multiple comparisons by using the Benjamini-Hochberg procedure.
TABLE 1.
NAPPA Z-score results, visual confirmation, ELISA ratios, and statistical analysis results for the top 12 antigensa
| Protein | CF patients (n = 22) |
NCF patients (n = 16) |
Controls (n = 15) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Z-score hit no. of patients | Visual hit no. of patients | Max Z in hits | NAPPA P valueb | Hit/control antigen ELISA ratio (P value) | Z-score hit no. of patients | Visual hit no. of patients | Max Z in hits | NAPPA P valueb | Hit/control antigen ELISA ratio (P value) | Z-score hit no. of controls | Visual hit no. of controls | Max Z in hits | |
| PA0044 | 21 | 20 | 14.40 | 0.00528172 | 5.6 (<0.01) | 7 | 8 | 9.03 | 0.768344093 | 3.5 (<0.01) | 5 | 4 | 10.93 |
| FliC_A | 20 | 20 | 12.75 | 5.93484E−06 | 10.7 (<0.001) | 1 | 2 | 3.34 | 0.392514931 | 2.4 (<0.01) | 1 | 1 | 2.88 |
| PA5369 | 13 | 20 | 6.88 | 2.10671E−05 | 2.2 (<0.01) | 0 | 1 | 2.0 (0.44) | 0 | 0 | |||
| PA1080 | 10 | 14 | 4.79 | 0.021995242 | 11.2 (<0.001) | 1 | 1 | 1.72 | 1 | 4.2 (<0.01) | 0 | 0 | |
| PA3841 | 10 | 13 | 9.27 | 7.73051E−06 | 7.9 (<0.001) | 2 | 4 | 1.95 | 0.007746094 | 5.0 (<0.001) | 0 | 0 | |
| PA0973 | 12 | 13 | 5.33 | 1.61306E−05 | 8.2 (0.001) | 1 | 1 | 5.80 | 0.055059338 | 23.5 (<0.05) | 0 | 0 | |
| PA4922 | 12 | 12 | 9.69 | 0.018302992 | 8.6 (<0.001) | 1 | 1 | 2.69 | 0.008411395 | 6.0 | 0 | 0 | |
| PA0807 | 13 | 12 | 8.43 | 0.002835889 | 8.5 (0.07) | 0 | 0 | 0 | 0 | ||||
| PA3931 | 8 | 12 | 5.95 | 0.000122245 | 5.5 | 0 | 0 | 0 | 0 | ||||
| PA3407 | 10 | 10 | 3.71 | 0.078060019 | 13.6 (<0.001) | 15 | 10 | 7.77 | 1.80176E−05 | 15.1 (<0.01) | 2 | 0 | 1.83 |
| PA4110 | 9 | 10 | 10.28 | 1.20979E−05 | 13.0 (<0.001) | 1 | 2 | 3.84 | 0.001818193 | 9.4 (<0.01) | 0 | 0 | |
| PA2300 | 12 | 9 | 6.99 | 2.24864E−05 | 6.0 (<0.001) | 0 | 1 | 2.6 (0.01) | 0 | 0 | |||
The results for the 12 top antigens found are here shown, including the number of patients in which their Z-score was positive, the number of patients in which they were a visual hit, the maximum(Max) Z-score for that specific antigen among hits, the P value for the analysis of the intensity of the specific antigen in patients and control groups, and the ELISA results. Table S2 in the supplemental material shows the extended analysis, including results for all the 262 antigens tested.
P values are for comparison between the patient group and the control group.
For NAPPA, the reproducibility between duplicate NAPPA spots (serum signal) was determined by the coefficient of variation ([standard deviation/mean] × 100%) for the within-slide replicates. More than 70% of the antigens had coefficients of variation below 25%. For comparison among patients and controls, serum signal intensities for each particular antigen were computed as the average of four spots (two replicate spots from two duplicate slides), and a Z-score was computed for each antigen based on the average signal generated by that particular patient. The cutoff for positivity was defined as 1.6. A one-sided Wilcoxon test was used to compare the signals between controls and positive patients. The P values were adjusted for multiple comparisons by using the Benjamini-Hochberg procedure.
RESULTS
Selection of the proteins to be represented on the arrays.
Using the different tools described, it was possible to predict the proteins of interest as being those externally exposed by the pathogen. Table 2 was generated with the results from these different complementary tools plus the final hand curation from a specialist in the field (Stephen Lory). Many of these tools pointed to the same proteins, so a final nonredundant list of 266 proteins was assembled (see Table S1 in the supplemental material). This final list includes all the proteins mentioned in the introduction, which had been previously described as immunogenic, such as flagellin, exotoxin A, elastase, alkaline protease, OprI, OprH2, OprE, and OprF.
TABLE 2.
Tools used to predict OMPsa
| Source | No. of OMPs |
||
|---|---|---|---|
| Predicted | Confirmed by annotation | Passed hand curation | |
| BOMP | 120 | 61 | 112 |
| PROFtmb | 62 | 55 | 62 |
| PSORT | 165 | 108 | 154 |
| Pence | 142 | 112 | 137 |
| Genome annotation | 129 | NA | 123 |
| Experiment | 53 | NA | 53 |
| Total | 293 | 119 | 266 |
The chart displays the number of OMPs selected by each tool used, as well as the proportion of those that were confirmed by annotation and hand curation.
To be sure that the genes we tested were not unique to the PAO1 strain, we checked the Microbial Genome Database for Comparative Analysis (http://mbgd.genome.ad.jp/) to analyze the four P. aeruginosa strains whose genomic sequences were publicly available (PAO1, PA7, LESB58, and UCBPP-PA14) and found that more than 90% of the ORFs present in the PAO1 strain have nearly identical orthologs in at least one of the other strains. Specifically, for the 266 OMPs selected from PAO1, 230 have orthologs in all three of the other strains, 30 have orthologs in two other strains, and 3 have orthologs in one other strain. Thus, only two OMPs are PAO1 specific.
Generation of the slides containing P. aeruginosa proteins of interest.
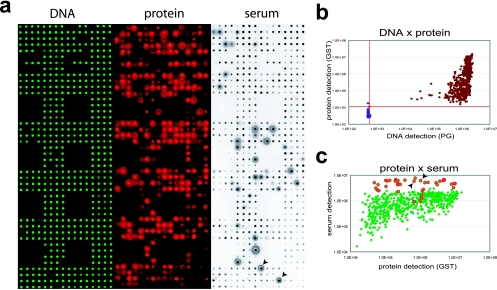
After PCR-based cloning of the full-length ORFs and full-length sequence verification (40) in the Gateway system (21), 262 of these 266 ORFs were successfully transferred to the in vitro expression vector pANT7-cGST and printed onto NAPPA slides in duplicate. NAPPA is a modified protein microarray in which expression clones (cDNAs) are initially printed and then transcribed/translated in situ to produce protein at the time of the experiment. As a first quality control step, we assessed binding of the plasmid DNA on the slides by staining with PicoGreen, which fluoresces when bound to DNA (Fig. 1a). The protein-encoding sequences are arranged such that they encode carboxyl-terminal GST tags for all proteins. Thus, to confirm protein expression and capture after the addition of the rabbit reticulocyte-based in vitro expression system, anti-GST antibody was used to detect the freshly produced proteins (Fig. 1a).
FIG. 1.
NAPPA quality control. (a) Expression clones encoding the target proteins fused to a C-terminal GST tag were printed along with a polyclonal anti-GST antibody in duplicate on the array surface. DNA capture was confirmed by PicoGreen (PG) staining (DNA), in situ protein expression and capture were assessed by GST detection using a monoclonal antibody (protein), and after incubation with serum, the response was measured with a horseradish peroxidase-linked anti-human IgG secondary antibody (serum). (b) DNA-versus-protein scatter plot showing background negative control spots (blue) and expression clone spots (brown) on a logarithmic plot. All clone spots showed protein and DNA levels at least three standard deviations higher than the mean background level (red lines). (c) Scatter plot comparing protein signal intensities (anti-GST antibody) and serum signal intensities (anti-human IgG antibody) for data from panel a. Red features correspond to visually confirmed hits. Duplicate signals for one example of the hits are indicated by the black arrows on both the serum panel and the graph.
The false-color image in Fig. 1a representing the protein array was adjusted to avoid saturating the bright pixels, and because of the narrow dynamic range of the print image, some spots appear to have no protein. However, as seen in the plotted quantitative values for the signal intensity in Fig. 1b, all of the array features where we printed an ORF had signals that were more than three standard deviations higher than the background signal from the control features (features with the same chemistry but which do not express protein).
To demonstrate that these arrays could be used to detect a human serum response and that the epitopes on the displayed proteins are accessible, a serum sample from a CF patient was used as a primary antibody to bind to the arrays (Fig. 1a). After we washed away the patient serum, a horseradish peroxidase-labeled secondary anti-human IgG was added and visualized with the tyramide signal amplification system, which generates fluorescent free radicals that attach to the slide surface at the site of antibody binding.
The resulting images were entered into MicroVigene microarray quantification software and printed for visual analysis. Quantification showed that all the clones of interest (Fig. 1b) have sufficient immobilized DNA and display protein above the detection threshold (red lines) set three standard deviations higher than the no-DNA controls (blue spots). Moreover, the pilot serum screening revealed that even weak protein spots were detected by antibodies in patients' sera (Fig. 1c).
In addition to the quantitative analysis, the slides were examined for responses that were visually obvious (Fig. 1c). An example of a pair of duplicate features is indicated in the figure. Based on the range of proteins that were detected, all of the protein display levels in this experiment were adequate for serum detection.
Screening patients and controls.
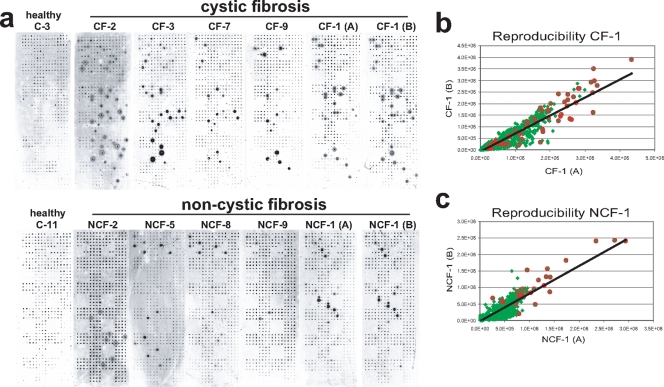
Replicate slides containing duplicate features were screened with independently prepared serum samples from each of 38 patients with documented P. aeruginosa infections (22 CF and 16 non-CF patients with various acute P. aeruginosa infections), as well as 15 healthy controls (Fig. 2a). Responses to the P. aeruginosa antigens were generally stronger among the CF patients than among non-CF patients in antigen number, in signal intensity, and in the fraction of responders. The mean signal intensities for the four features for each antigen (duplicate spots from replicate slides) were calculated for each patient, and a Z-score was computed for each antigen in each patient relative to the mean signal intensity of the slide [Z = (signal − mean)/standard deviation]. An initial cutoff for positives was set at a Z-score of ≥1.6, based on internal controls. The reproducibility for this quantification method is represented by the scatter plots of the same serum samples tested on different arrays seen in Fig. 2b and c. Samples are tightly grouped around the 45°-angle line that represents excellent reproducibility between replicate slides.
FIG. 2.
Serum screening. (a) A representative group of serum slides for controls and CF and non-CF (NCF) patients. (b and c) Reproducibility scatter plots of the serum responses for independently processed samples (samples A and B) from patients CF-1 and NCF-1, as seen in panel a. All detected spots are represented in green, and the visually confirmed hits are highlighted with red circles. The R2 values for CF-1 and NCF-1 were 0.85 and 0.80, respectively.
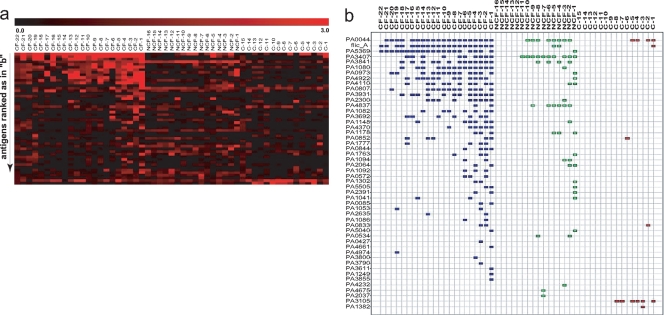
After this objective quantitative evaluation, slides were submitted to visual review and a list of visual hits was obtained. A heat map based on the Z-scores for this overlapping subset (Fig. 3a) shows the more intense response of CF patients. As seen on the map and in Table S2 in the supplemental material, responses in healthy individuals were rare and weak. A map of visually confirmed hits is shown in Fig. 3b.
FIG. 3.
Microarray analysis. (a) Z-scores were computed from the averages of all four features for each antigen (duplicate features from duplicate slides). Here, the Z-scores of the 48 visually confirmed positives were used to create a heat map. (b) A map displaying the visually confirmed hits from panel a. The order of antigens in the heat map and the order in the chart are the same.
A total of 48 antigens were found to be positive in at least one patient. Of these, 12 antigens were detected in 10 or more patients, comprising a group of more promising candidates for further studies.
Confirmation of microarray results.
All antigens that were positive by both Z-score and visual review as well as subsets of antigens positive by visual review only and Z-score only were tested by ELISA for confirmation, using sera from at least two different patients per antigen. A large percentage of the hits for antigens positive by both Z-score and visual review (70%) was also positive by ELISA, whereas none of the antigens positive by Z-score only or antigens negative by both methods demonstrated a positive ELISA response (Fig. S1 in the supplemental material). Interestingly, 87% of the hits corresponding to antigens positive only by visual review were also positive by ELISA, emphasizing the value of the visual review.
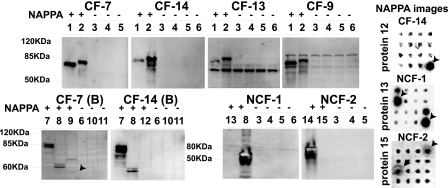
We also tested some of the antigens for response by immunoblotting. As expected, the proteins that were negative by NAPPA were also negative by immunoblotting. Moreover, six of the nine NAPPA-positive antigens tested were also positive by immunoblotting. The remaining three proteins were negative by immunoblotting but were unmistakably positive by NAPPA (Fig. 4), two of them being confirmed as positive by ELISA (PA1082 and PA4837). PA1302 was detected only by NAPPA. It is possible that their microarray signal arises from conformational epitopes displayed on the array but absent from the denatured immunoblots.
FIG. 4.
Western blot confirmation. Proteins of interest were generated by IVTT and analyzed by Western blotting using patient serum as the primary antibody. The numbers above the lanes correspond to antigens tested, and their NAPPA results for that specific patient are represented by a plus or minus sign. NAPPA images for some hits that were negative by Western blotting are shown. Proteins tested are numbered as follows: 1, PA1080; 2, PA0044; 3, PA2373; 4, PA2130; 5, PA2760; 6, no DNA control; 7, PA3841; 8, PA0973; 9, PA0807; 10, PA2685; 11, PA4179; 12, PA1082; 13, PA1302; 14, PA1094; 15, PA4837. Molecular masses are given to the left of the gels.
In silico analysis of the hits.
P. aeruginosa genome annotation was used to determine if the 48 positive hits showed any tendency to cluster based on their structure, presence of specific domains, protein function, or pathway involvement. After a thorough evaluation, it was not possible to find any common feature that drove subclustering of the hits (data not shown). Of course, these were all proteins preselected on the basis of common features, which might make it difficult to subcluster them.
Antigen prediction tools were also used to determine if the list of hits would have been predicted in silico. Without structural data on the OMPs, we were limited to using tools that predicted linear epitopes from the primary structure. “Antigenic” is one such tool in EMBOSS, predicting potentially antigenic regions of a protein sequence by using the method of Kolaskar and Tongaonkar (19, 30, 37). “BepiPred” is another tool using a hidden Markov model and propensity scale method (22). Both of them can analyze protein sequences in batch mode. The Antigenic tool predicted that all the OMPs would be antigenic, whereas BepiPred predicted 27 OMPs to be immunogenic, only 2 of which were actually detected in these experiments. A list of 117 annotated B-cell epitope sequences for P. aeruginosa from the Immune Epitope Database was also assembled (31). The protein sequences of the 262 OMPs were scanned, and 65 OMPs containing one or more of these epitopes were found; two of these OMPs overlapped with NAPPA hits. In conclusion, current antigen prediction tools could not predict the observed NAPPA results.
DISCUSSION
The number of CF patients and the growing number of immunodeficient patients, especially due to human immunodeficiency virus infection, draw attention to the importance of opportunistic pathogens such as Pseudomonas aeruginosa. Because the abundance of P. aeruginosa proteins changes according to the microenvironment of the infection (5, 35), empirical studies that provide a better understanding of the human humoral response against this pathogen can contribute to improved diagnostics and specific vaccine development by highlighting the P. aeruginosa proteins that are both expressed and exposed during infection in vivo.
Our objective was to select the proteins predicted to reside on the outer surface of the pathogen by using bioinformatics tools. A list of 266 candidate proteins was defined, 262 of which were successfully subcloned in the in vitro expression vector of interest, for NAPPA microarray expression. In addition to offering a tool for unbiased representation for parallel study, the use of NAPPA circumvented some of the common problems associated with the study of membrane proteins, namely, the difficulty in producing and purifying them, as well as their instability. As soon as the protein chains are synthesized, they are captured and immobilized by the capture antibody, which might avoid aggregation. Moreover, the cell-free expression system also avoids the host toxicity problems associated with overloading the protein transportation machinery (14, 41). Altogether, this approach generated a uniform display of membrane proteins on the slides.
By displaying the proteins of interest in duplicate on a single slide, experimental artifacts are minimized, as all features are exposed to the same serum sample at the same time.
Among the 262 OMPs and the controls tested, 48 antigens were detected in at least one patient. Importantly, 12 antigens were positive in 10 or more patients (PA0044, PA5369, PA3407, PA3841, PA1080, PA0973, PA4922, PA4110, PA0807, PA3931, PA2300, and FliC_A), three of which were frequently observed for both CF and non-CF patient groups (PA0044, PA3407, and PA3841). We found several protein classes known to be immunogenic, such as exoenzymes and flagellum-related proteins. We also found three hypothetical proteins (PA5369, PA0807, and PA3931) that were among the top 12 of our list. The strong differential responses for these antigens in patients and controls support the notion that the protein microarray can be used to detect immune responses to membrane proteins, as can be seen by the significant P values (P < 0.01) in Table 1, representing the significance of NAPPA differential intensity detection between patients and controls for the top 12 hits. Moreover, the serum responses demonstrate that at least 48 of these antigens are clearly presented by the pathogen during infection. These antigens can be used for further studies targeting vaccine development and diagnostics, particularly the promising top 12, which were detected by a large number of patients.
There was a clear difference in the response patterns of CF and non-CF patients, considering both the number of antigens and the intensity of response. We attribute the stronger response of CF patients to the duration of infection, as these patients were chronically colonized since adolescence, whereas non-CF patients presented more-recent infections. This idea is corroborated by the observation that among the 11 non-CF patients who responded to at least one of these antigens, 7 either had COPD or were hospitalized in a long-term-care facility (see Table S3 in the supplemental material). The chronic nature of COPD can make the lung environment similar to that of a CF patient; however, a larger number of COPD patients would be needed to confirm this apparent tendency. Both children and adults mounted an immune response to the pathogen, and the evaluation of data on disease severity (forced expiratory volume in 1 s, disease outcome, and bacterial count), when available, showed no obvious correlation with the humoral response pattern. It is interesting to notice that some patients from both the CF group and the non-CF group showed no response to any of the antigens on the array. It is not clear if the absence of responses is due to host or pathogen factors, and the limited clinical data available for these deidentified samples make it impossible to address this here. Comparisons among the clinical and molecular data showed no correlation connecting specific antigens to prognosis.
Using NAPPA, we observed responses to most of the previously known antigens, such as exotoxin A, flagellin, OprH2, and porin F, as well as the more recently described OprL (PA0973) and ExoS (PA3841). We also observed responses to a considerable list of new P. aeruginosa antigens, including hypothetical proteins; however, we did not observe responses to alkaline protease or elastase. Although CF patients are generally described as chronically infected, they undergo phases of relative clearance, promoted by antibiotic treatment, during which the antibody titers to some pseudomonas antigens decrease significantly. This could explain some of the variation in responses among different patients and why we may have missed responses to some proteins. Also, it is known that responses to elastase and alkaline protease are weaker early in infection, ranging from 20% prevalence to 38% prevalence. Thus, given that these were all closely monitored patients, the samples may have been obtained prior to the development of strong humoral response to these late-responding proteins. Finally, as with any immunological method, NAPPA may not be the ideal presentation method for all antigens. Nevertheless, it successfully found antigens that had not been detected by other methods.
Of the three OMPs initially described to be immunogenic in the 1980s (OprI, OprH2, and OprE), only responses to OprH2 were found among our patients, but it is interesting to notice that no other recent studies report immunogenicity for OprI or OprE after natural Pseudomonas infection. It is possible that the use of current refined-protein identification techniques would identify what was called OprE and OprI as different OMPs. More-recent studies use OprI as an innate immunity trigger for its ability to act on Toll-like receptors (12).
The success of a protein-based vaccine for P. aeruginosa will rely on demonstrating protective immunity from the selected antigens. We regard this work as a first step toward identifying antigens that may prove useful in vaccines, that is, the identification of antigens that induce a humoral response in multiple patients. In these deidentified samples, we do not know whether these humoral responses correlated with bacterial clearance or not. Both sets of patients were monitored closely and were likely to have received antibiotics at the earliest sign of infection. The approaches by which vaccines have proved useful depend upon, among other factors, how the antigens are prepared, whether the antigens are engineered to enhance responses, which adjuvants are used, what vaccination schedule is used, and when the patients are vaccinated relative to their encounter with the pathogen. The hope here is that a vaccine would prepare individuals to mount a much more prompt response during early colonization, when the bacterial load is still controllable.
Given the multiple strains of P. aeruginosa and the variation in antigen presentation by the pathogen in different infection types, a multivalent vaccine seems a likely solution. The 12 most frequently detected antigens here are good candidates for further evaluation, although we would not necessarily discard the other hits. Some of the other hits may have corresponded to early responses that we missed but which nevertheless contribute to a protective effect.
All the top 12 antigens plus several of the others were confirmed visually and by ELISA. Subsets of these may prove useful for diagnostic applications. A test analyzing the responses to PA5369, PA1080, PA3841, PA3407, and PA4837 displays a sensitivity of 87%, specificity of 100%, positive predictive value of 100%, and negative predictive value of 72% in this limited group of patients. These values improve when CF is established as a prior condition, suggesting possible applications for the early detection of P. aeruginosa infection in individuals with CF (42). Although further validation studies with independent samples would be needed to confirm these findings, these early results are promising.
Bioinformatics analysis of all positive antigens revealed no structural or functional commonalities among the proteins that acted as strong immunogens, and the antigen prediction tools did not predict which ones would show a strong humoral response. Thus, at the present, the experimental determination of the humoral response by using high-throughput tools like NAPPA may remain the only reliable method for assessing the humoral response to a large repertoire of proteins produced by a bacterial pathogen.
These promising antigens should now be submitted to animal studies to evaluate protective properties, both individually and in groups, and also tested for diagnostic purposes in a larger population of patients and controls.
Here, we have presented a list of 48 antigens from Pseudomonas aeruginosa which are clearly expressed during pulmonary infections and are potential candidates for diagnostics and vaccine development. As well, we demonstrated the applicability of a novel method for rapidly and comprehensively testing full-length bacterial OMPs for their ability to induce a humoral immune response in infected individuals. This method exploits the ability of NAPPAs to efficiently produce and display membrane proteins. Because of the ease of displaying proteins in the NAPPA format and the minute volumes of serum needed (∼10 μl), this method can be readily applied to many other pathogens. It clearly demonstrates the usefulness of the methodology for large-scale epidemiological surveys of human humoral responses, correlating clinical conditions of patients with the presence or lack of antibodies to specific bacterial products, thus identifying potentially protective antigens.
Supplementary Material
Acknowledgments
We thank E. A. Mendoza and J. Raphael (Harvard Institute of Proteomics) for excellent technical support.
This work was supported by NIAID contract DHHSN266200400053C and by a grant from the Cystic Fibrosis Foundation.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 8 September 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anderson, K. S., N. Ramachandran, J. Wong, J. V. Raphael, E. Hainsworth, G. Demirkan, D. Cramer, D. Aronzon, F. S. Hodi, L. Harris, T. Logvinenko, and J. Labaer. 2008. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J. Proteome Res. 7:1490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beare, P. A., C. Chen, T. Bouman, J. Pablo, B. Unal, D. C. Cockrell, W. C. Brown, K. D. Barbian, S. F. Porcella, J. E. Samuel, P. L. Felgner, and R. A. Heinzen. 2008. Candidate antigens for Q fever serodiagnosis revealed by immunoscreening of a Coxiella burnetii protein microarray. Clin. Vaccine Immunol. 15:1771-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berven, F. S., K. Flikka, H. B. Jensen, and I. Eidhammer. 2004. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 32:W394-W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bigelow, H. R., D. S. Petrey, J. Liu, D. Przybylski, and B. Rost. 2004. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Res. 32:2566-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bragonzi, A., D. Worlitzsch, G. B. Pier, P. Timpert, M. Ulrich, M. Hentzer, J. B. Andersen, M. Givskov, M. Conese, and G. Doring. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J. Infect. Dis. 192:410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz, E., E. Munoz, K. Agbaht, and J. Rello. 2007. Management of ventilator-associated pneumonia caused by multiresistant bacteria. Curr. Opin. Crit. Care 13:45-50. [DOI] [PubMed] [Google Scholar]
- 7.Döring, G., C. Meisner, and M. Stern. 2007. A double-blind randomized placebo-controlled phase III study of a Pseudomonas aeruginosa flagella vaccine in cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 104:11020-11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Döring, G., and G. B. Pier. 2008. Vaccines and immunotherapy against Pseudomonas aeruginosa. Vaccine 26:1011-1024. [DOI] [PubMed] [Google Scholar]
- 9.Driscoll, J. A., S. L. Brody, and M. H. Kollef. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351-368. [DOI] [PubMed] [Google Scholar]
- 10.Elting, L. S., E. B. Rubenstein, K. V. Rolston, and G. P. Bodey. 1997. Outcomes of bacteremia in patients with cancer and neutropenia: observations from two decades of epidemiological and clinical trials. Clin. Infect. Dis. 25:247-259. [DOI] [PubMed] [Google Scholar]
- 11.Gardy, J. L., M. R. Laird, F. Chen, S. Rey, C. J. Walsh, M. Ester, and F. S. Brinkman. 2005. PSORTb v. 2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21:617-623. [DOI] [PubMed] [Google Scholar]
- 12.Gartner, T., M. Baeten, S. Otieno, H. Revets, P. De Baetselier, and K. Huygen. 2007. Mucosal prime-boost vaccination for tuberculosis based on TLR triggering OprI lipoprotein from Pseudomonas aeruginosa fused to mycolyl-transferase Ag85A. Immunol. Lett. 111:26-35. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918-951. [DOI] [PubMed] [Google Scholar]
- 14.Grisshammer, R. 2006. Understanding recombinant expression of membrane proteins. Curr. Opin. Biotechnol. 17:337-340. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E., E. C. Mouat, and D. P. Speert. 1984. Quantitation and identification of antibodies to outer-membrane proteins of Pseudomonas aeruginosa in sera of patients with cystic fibrosis. J. Infect. Dis. 149:220-226. [DOI] [PubMed] [Google Scholar]
- 16.Holder, I. A. 2004. Pseudomonas immunotherapy: a historical overview. Vaccine 22:831-839. [DOI] [PubMed] [Google Scholar]
- 17.Keasey, S. L., K. E. Schmid, M. S. Lee, J. Meegan, P. Tomas, M. Minto, A. P. Tikhonov, B. Schweitzer, and R. G. Ulrich. 2009. Extensive antibody cross-reactivity among infectious gram-negative bacteria revealed by proteome microarray analysis. Mol. Cell. Proteomics 8:924-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klinger, J. D., D. C. Straus, C. B. Hilton, and J. A. Bass. 1978. Antibodies to proteases and exotoxin A of Pseudomonas aeruginosa in patients with cystic fibrosis: demonstration by radioimmunoassay. J. Infect. Dis. 138:49-58. [DOI] [PubMed] [Google Scholar]
- 19.Kolaskar, A. S., and P. C. Tongaonkar. 1990. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 276:172-174. [DOI] [PubMed] [Google Scholar]
- 20.Kubesch, P., B. U. von Specht, and B. Tummler. 1988. Immune response in cystic fibrosis to outer membrane proteins of Pseudomonas aeruginosa. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 269:395-410. [DOI] [PubMed] [Google Scholar]
- 21.Labaer, J., Q. Qiu, A. Anumanthan, W. Mar, D. Zuo, T. V. Murthy, H. Taycher, A. Halleck, E. Hainsworth, S. Lory, and L. Brizuela. 2004. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 14:2190-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen, J. E., O. Lund, and M. Nielsen. 2006. Improved method for predicting linear B-cell epitopes. Immunome Res. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurans, M., A. Arion, M. Fines-Guyon, A. Regeasse, J. Brouard, R. Leclercq, and J. F. Duhamel. 2006. Pseudomonas aeruginosa and cystic fibrosis: first colonization to chronic infection. Arch. Pediatr. 13(Suppl. 1):S22-S29. (In French.) [PubMed] [Google Scholar]
- 24.Leonard, A., T. Leal, and P. Lebecque. 2007. Cystic fibrosis and pseudomonas aeruginosa current and future strategies. J. Pharm. Belg. 62:25-28. (In French.) [PubMed] [Google Scholar]
- 25.Li, B., D. Zhou, Z. Wang, Z. Song, H. Wang, M. Li, X. Dong, M. Wu, Z. Guo, and R. Yang. 2008. Antibody profiling in plague patients by protein microarray. Microbes Infect. 10:45-51. [DOI] [PubMed] [Google Scholar]
- 26.Lu, Z., D. Szafron, R. Greiner, P. Lu, D. S. Wishart, B. Poulin, J. Anvik, C. Macdonell, and R. Eisner. 2004. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics 20:547-556. [DOI] [PubMed] [Google Scholar]
- 27.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murthy, T., A. Rolfs, Y. Hu, Z. Shi, J. Raphael, D. Moreira, F. Kelley, S. McCarron, D. Jepson, E. Taycher, D. Zuo, S. E. Mohr, M. Fernandez, L. Brizuela, and J. Labaer. 2007. A full-genomic sequence-verified protein-coding gene collection for Francisella tularensis. PLoS ONE 2:e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obritsch, M. D., D. N. Fish, R. MacLaren, and R. Jung. 2005. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: epidemiology and treatment options. Pharmacotherapy 25:1353-1364. [DOI] [PubMed] [Google Scholar]
- 30.Parker, J. M., D. Guo, and R. S. Hodges. 1986. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry 25:5425-5432. [DOI] [PubMed] [Google Scholar]
- 31.Peters, B., J. Sidney, P. Bourne, H. H. Bui, S. Buus, G. Doh, W. Fleri, M. Kronenberg, R. Kubo, O. Lund, D. Nemazee, J. V. Ponomarenko, M. Sathiamurthy, S. Schoenberger, S. Stewart, P. Surko, S. Way, S. Wilson, and A. Sette. 2005. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 3:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandran, N., K. S. Anderson, J. Raphael, E. Hainsworth, S. Sibani, W. R. Montor, M. Pacek, J. Wong, M. Eljanne, M. G. Sanda, Y. Hu, T. Logvinenko, and J. Labaer. 2008. Tracking humoral responses using self assembling protein microarrays. Proteomics Clin. Appl. 2:1518-1527. [DOI] [PMC free article] [PubMed]
- 33.Ramachandran, N., E. Hainsworth, B. Bhullar, S. Eisenstein, B. Rosen, A. Y. Lau, J. C. Walter, and J. LaBaer. 2004. Self-assembling protein microarrays. Science 305:86-90. [DOI] [PubMed] [Google Scholar]
- 34.Ramachandran, N., J. V. Raphael, E. Hainsworth, G. Demirkan, M. G. Fuentes, A. Rolfs, Y. Hu, and J. Labaer. 2008. Next-generation high-density self-assembling functional protein arrays. Nat. Methods 5:535-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309-322. [DOI] [PubMed] [Google Scholar]
- 36.Ratjen, F., H. Walter, M. Haug, C. Meisner, H. Grasemann, and G. Doring. 2007. Diagnostic value of serum antibodies in early Pseudomonas aeruginosa infection in cystic fibrosis patients. Pediatr. Pulmonol. 42:249-255. [DOI] [PubMed] [Google Scholar]
- 37.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 38.Saiman, L. 2004. Microbiology of early CF lung disease. Paediatr. Respir. Rev. 5(Suppl. A):S367-S369. [DOI] [PubMed] [Google Scholar]
- 39.Sundaresh, S., A. Randall, B. Unal, J. M. Petersen, J. T. Belisle, M. G. Hartley, M. Duffield, R. W. Titball, D. H. Davies, P. L. Felgner, and P. Baldi. 2007. From protein microarrays to diagnostic antigen discovery: a study of the pathogen Francisella tularensis. Bioinformatics 23:i508-i518. [DOI] [PubMed] [Google Scholar]
- 40.Taycher, E., A. Rolfs, Y. Hu, D. Zuo, S. E. Mohr, J. Williamson, and J. Labaer. 2007. A novel approach to sequence validating protein expression clones with automated decision making. BMC Bioinformatics 8:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, S., M. L. Bader, D. Drew, and J. W. de Gier. 2006. Rationalizing membrane protein overexpression. Trends Biotechnol. 24:364-371. [DOI] [PubMed] [Google Scholar]
- 42.West, S. E., L. Zeng, B. L. Lee, M. R. Kosorok, A. Laxova, M. J. Rock, M. J. Splaingard, and P. M. Farrell. 2002. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA 287:2958-2967. [DOI] [PubMed] [Google Scholar]
- 43.Woods, D. E., S. J. Cryz, R. L. Friedman, and B. H. Iglewski. 1982. Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect. Immun. 36:1223-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, Y., J. F. Bruno, and B. J. Luft. 2008. Profiling the humoral immune response to Borrelia burgdorferi infection with protein microarrays. Microb. Pathog. 45:403-407. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, H., S. Hu, G. Jona, X. Zhu, N. Kreiswirth, B. M. Willey, T. Mazzulli, G. Liu, Q. Song, P. Chen, M. Cameron, A. Tyler, J. Wang, J. Wen, W. Chen, S. Compton, and M. Snyder. 2006. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. USA 103:4011-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.