Abstract
Cationic amino acids contribute to α-defensin bactericidal activity. Curiously, although Arg and Lys have equivalent electropositive charges at neutral pH, α-defensins contain an average of nine Arg residues per Lys residue. To investigate the role of high α-defensin Arg content, all Arg residues in mouse Paneth cell α-defensin cryptdin 4 (Crp4) and rhesus myeloid α-defensin 4 (RMAD-4) were replaced with Lys to prepare (R/K)-Crp4 and (R/K)-RMAD-4, respectively. Lys-for-Arg replacements in Crp4 attenuated bactericidal activity and slowed the kinetics of Escherichia coli ML35 cell permeabilization, and (R/K)-Crp4 required longer exposure times to reduce E. coli cell survival. In marked contrast, Lys substitutions in RMAD-4 improved microbicidal activity against certain bacteria and permeabilized E. coli more effectively. Therefore, Arg→Lys substitutions attenuated activity in Crp4 but not in RMAD-4, and the functional consequences of Arg→Lys replacements in α-defensins are dependent on the peptide primary structure. In addition, the bactericidal effects of (R/K)-Crp4 and (R/K)-RMAD-4 were more sensitive to inhibition by NaCl than those of the native peptides, suggesting that the high Arg content of α-defensins may be under selection to confer superior microbicidal function under physiologic conditions.
Mammalian α-defensins mediate innate immunity primarily in phagolysosomes of neutrophils and in the intestine following secretion by small intestinal Paneth cells (16, 30, 33). α-Defensins are ∼4-kDa cationic and amphipathic peptides with broad-spectrum bactericidal activities. Structurally, they consist of a three-stranded β-sheet structure that is established by three invariantly paired disulfide bonds (16, 32). In vitro, α-defensins are microbicidal against gram-positive and gram-negative bacteria, fungi, spirochetes, protozoa, and enveloped viruses (1, 4, 9, 16, 35, 44). Most α-defensins exert antibacterial effects by membrane disruption, inducing permeabilization of target cell membranes, as inferred from the formation of transient defects or stable pores in model phospholipid bilayers (12, 41). For example, the bactericidal activity of the mouse α-defensin cryptdin 4 (Crp4) is directly related to peptide binding and disruption of phospholipid bilayers (28). Crp4 exhibits strong interfacial binding to model membranes, inducing “graded” fluorophore leakage from model membrane vesicles (5, 6, 29). The role of Paneth cell α-defensins in enteric mucosal immunity is evident from the immunity of transgenic mice expressing human α-defensin 5 in Paneth cells to oral Salmonella enterica serovar Typhimurium infection (27). Also, mice lacking matrix metalloproteinase 7 (MMP-7), the mouse Paneth cell pro-α-defensin convertase, are more susceptible to enteric infection and defective in clearance of orally administered bacteria ( 40, 42).
Despite having highly diverse primary structures (22, 23), α-defensins retain conserved biochemical features that include an invariant disulfide array (32), a canonical Arg-Glu salt bridge, a conserved Gly residue at CysIII+8, and high Arg content relative to that of Lys (45). Studies have shown consistently that bactericidal activity is independent of these highly conserved features of the peptide family, with the exception of the relatively high Arg content (19, 20, 25, 26, 45). Arg residues contribute to Crp4 bactericidal activity, as shown by the loss of function induced in Crp4 by charge reversal or charge neutralization at varied Arg residue positions (36). Because Arg and Lys have equivalent electropositive charges, the unusually high prevalence of Arg over Lys is remarkable given the extensive gene duplication and sequence diversification that characterize the α-defensin gene family (24, 38). Knowing that α-defensin bactericidal activity depends in part on the electropositive surface charge (13, 36), we hypothesized that Arg may be positively selected relative to Lys to confer a functional advantage, such as improved bactericidal activity (see Fig. S1 in the supplemental material).
To test this hypothesis, Crp4 and rhesus myeloid α-defensin 4 (RMAD-4) variants with all Arg residues converted to Lys, termed (R/K)-Crp4 and (R/K)-RMAD-4, respectively, were assayed in vitro for possible attenuating effects of Lys substitutions (see Fig. 1; see also Fig. S2 in the supplemental material). The fact that these peptides are highly bactericidal but differ markedly in primary structure provided a rationale for their investigation in this study. Although the (R/K)-Crp4 molecule had modestly attenuated microbicidal activity and delayed kinetics of Escherichia coli cell permeabilization and bacterial cell killing, (R/K)-RMAD-4 generally had bactericidal activity greater than or equal to that of native RMAD-4. Thus, the notion that Arg is under positive selection in α-defensins to confer superior microbicidal activity is not supported as a general rule. Instead, the functional effects of changes in basic amino acids depend on their position in the context of individual peptide primary structures.
FIG. 1.
The primary structures of recombinant and synthetic α-defensins analyzed in these studies. Arg residue positions with Lys substitutions are in bold font.
MATERIALS AND METHODS
Mutagenizing PCR for generation of (R/K)-Crp4 and (R/K)-proCrp4 cDNA templates.
The Crp4, procryptdin 4 (proCrp4), and RMAD-4 cDNA templates were generated as cDNAs by PCR. (R/K)-Crp4 and (R/K)-proCrp4 were amplified from Crp4- and proCrp4-encoding plasmids by PCR using mutagenizing primers. Using Crp4 cDNA as a template, (R/K)-Crp4 [(R7K/R13K/R16K/R18K/R24K/R31K/R32K)-Crp461-92] was produced by PCR mutagenesis. Arg codons were converted to Lys codons by mutagenesis in three series of three-step PCRs, as follows.
In the first round of three-step reactions, the Arg7, Arg16, and Arg18 codons were converted to Lys in three steps: in step 1, the forward mutagenizing primer R7K-Crp4-f (5′-AT GAATTC ATGGG TTTGT TATGC TATTG TAAG-3′) was paired with the reverse mutagenizing primer Crp4-R16/18K-r (5′-ACGTC CCTTT AACTT TTTCT CCTCT-3′); in step 2, the forward primer Crp4-R16/18K-f (5′-AGAGG AGAAA AAGTT AAAGG GACT-3′), the reverse complement of Crp4-R16/18K-r, was paired with the reverse primer Crp4-stop-SalI-r (5′-TATGT CGACT CATCA GCGGC GGGGG CAGCA GT-3′); in step 3, 0.005 μl (each) from reaction mixtures 1 and 2 was combined with the primers R7K-Crp4-f and Crp4-stop-SalI-r to produce a full-length product containing the R7K, R16K, and R18K mutations.
In the second series of reactions, the Arg13, Arg31, and Arg32 codons of the (R7K/R16K/R18K)-Crp4 template were converted to Lys codons using a three-step procedure similar to that outlined above. In step 1, R7K-Crp4-f was paired with R13K-Crp4-r (5′-AACTT TTTCG CCCTT TTTGC AGTG-3′), the reverse mutagenizing primer; in step 2, R13K-Crp4-f (5′-CACTG CAAAA AGGGC GAAAA AGTT-3′), the reverse complement of R13K-Crp4-r, was paired with the reverse primer Crp4-R31/32K-r (5′-TATAT GTCGA CTCAC TTCTT TGGGC AGCAG TACAA AAATC G-3′); in step 3, the primer R7K-Crp4-f and the reverse primer Crp4-R31/32K-r were combined with 0.005 μl (each) of reaction mixtures 1 and 2 to generate a full-length (R7K/R13K/R16K/R18K/R31K/R32K)-Crp4 amplification product.
A third set of three-step PCR mutagenesis reactions was used to replace Arg24 with Lys, thus generating the final (R/K)-Crp4 amplification product. In step 1, the amplification product from the second set of mutagenesis reactions, (R7K/R13K/R16K/R18K/R31K/R32K)-Crp4, was a template for reaction mixtures containing the R7K-Crp4-f primer with the reverse mutagenizing primer R24K-Crp4-r (5′-GTACA AAAAC TTTAT TCCAC AAGTC CC-3′); in step 2, R24K-Crp4-f (5′-GGGAC TTGTG GAATA AAGTT TTTGT AC-3′) was paired with Crp4-PKK-r (5′-TAT GTCGAC TCACT TCTTG GGGCA GCAGT ACAAA AACTT T-3′); in step 3, as before, 0.005-μl samples from reaction mixtures 1 and 2 were combined with R7K-Crp4-f and Crp4-PKK-r to produce the (R/K)-Crp4 template DNA product for the final reactions for cloning into pET-28a.
The (R/K)-proCrp4 construct [(R67/73/76/78/84/91/92K)-proCrp420-92] was prepared by another three-step PCR amplification procedure. In step 1, (R/K)-Crp4 template DNA was amplified with the forward primer PC4Cod54/63-f (5′-CTTCA TGAAA AATCT TTGAG AGGTT TGTTA-3′) paired with the reverse primer Crp4-PKK-r; in step 2, native proCrp4 cDNA was amplified with the forward primer EcoRI-Met-PC4-f (5′-GCGC GAATTC ATGGA TCC TATCC AAAAC ACA-3′) with the reverse primer PC4Cod54/63-r (5′-TAACA AACCT CTCAA AGATT TTTCA TGAAG-3′), the reverse complement of PC4Cod54/63-f, to amplify the prosegment coding region (34, 36, 40); in step 3, 0.005-μl samples of reaction mixtures 1 and 2 were combined with the primer EcoRI-Met-PC4-f and the reverse primer Crp4-PKK-r to amplify the final (R/K)-proCrp4 coding sequence for cloning into pET-28a (36). (R/K)-RMAD-4 [(R1/2/5/7/10/13/14/26/33K)-RMAD-462-94] was custom synthesized by CPC Scientific, Inc. (San Jose, CA) and refolded as described previously (28).
Preparation of peptides.
Recombinant proteins were expressed and purified as His6-tagged fusion proteins as described previously (34, 37). Expression of recombinant fusion proteins was induced by adjusting exponentially growing E. coli BL21-CodonPlus (DE3)-RIL cells with 0.1 mM isopropyl-β-d-1-thiogalactopyranoside and incubating them at 37°C for 6 h in Terrific broth as described in earlier reports (14, 34, 37). Cells were lysed by sonication in 6 M guanidine-HCl-100 mM Tris (pH 8.0), and the suspension was clarified by centrifugation. His6-tagged fusion proteins purified by nickel-nitrilotriacetic acid resin affinity chromatography (Qiagen, Valencia, CA) were cleaved with cyanogen bromide and purified by analytical C18 reverse-phase high-pressure liquid chromatography (RP-HPLC). Peptide homogeneity was confirmed in analytical acid-urea polyacrylamide gel electrophoresis (AU-PAGE), a highly sensitive measure of defensin foldamers. Molecular masses were verified by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry, and peptides were quantified both by amino acid analysis and by using extinction coefficient calculations at 280 nm, performed at ExPASY (http://ca.expasy.org/tools).
To obtain homogeneous and properly folded peptides, (R/K)-Crp4, (R/K)-proCrp4, and (R/K)-RMAD-4 samples (0.5 to 2.0 mg/ml) were reduced with 1 mg/ml of dithiothreitol in 6 M guanidine-HCl, 0.2 M Trizma base, and 2 mM EDTA (pH 8.2) for 4 h at 50°C. Reduced peptides were adjusted to 5% acetic acid, and the reduced forms were purified by analytical C18 RP-HPLC (data not shown). Reduced peptides were refolded with stirring in 0.1 M NH4HCO3, 2.0 mM EDTA, 0.1 mg/ml cysteine, and 0.1 mg/ml cystine (pH 7.8) at concentrations of 100 to 300 μg/ml under air at 4°C. Peptide refolding was monitored by analytical C18 RP-HPLC at 24-h intervals, followed by a final purification performed by analytical C18 RP-HPLC.
Sensitivity to proteolysis by MMP-7, trypsin, and elastase.
Samples of proCrp4 and (R/K)-proCrp4 (11 μg) and of Crp4 and (R/K)-Crp4 (5 μg) were incubated with or without 0.5 molar equivalents of MMP-7 in 1 mM HEPES, 15 mM NaCl, and 0.5 mM CaCl2 (pH 7.4) for 18 h at 37°C. Similarly, samples of proCrp4 (12 μg) and Crp4, (R/K)-Crp4, RMAD-4, and (R/K)-RMAD-4 (6 μg) were incubated with human neutrophil elastase in 50 mM Tris-150 mM NaCl (pH 7.5) at 37°C for 2 h at a substrate:enzyme ratio of 40:1 (14). Twelve micrograms of proCrp4 and 6 μg of Crp4, (R/K)-Crp4, RMAD-4, and (R/K)-RMAD-4 were incubated with bovine pancreatic trypsin (Sigma-Aldrich, St. Louis, MO) in 50 mM ammonium bicarbonate at a 1:50 proteinase-to-protein ratio for 2 h. Peptide sensitivity to proteolysis was assayed by Coomassie blue staining of peptide digests following AU-PAGE separations (14, 20, 34).
Bactericidal peptide assays.
Cultures of Escherichia coli ML35, Listeria monocytogenes 10403S, Staphylococcus aureus 710a, and Vibrio cholerae 0395 were purchased from the American Type Culture Collection (Manassas, VA), and all Salmonella enterica serovar Typhimurium strains were provided by Samuel I. Miller (University of Washington). Bacteria growing exponentially in Trypticase soy broth were collected by centrifugation, washed, and resuspended in assay buffer containing 10 mM piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES), pH 7.4, supplemented with 0.01 volume (1% vol/vol) Trypticase soy broth (PIPES-TSB). Bacteria (5 × 106/ml) were exposed to various concentrations of peptide for 1 h at 37°C in 50 μl PIPES-TSB. Reaction mixtures were diluted 1:100 in 10 mM PIPES, pH 7.4, and plated on Trypticase soy agar plates using an Autoplate 4000 plating device (Spiral Biotech Inc., Bethesda, MD), and bacterial cell survival was determined by counting CFU after overnight growth at 37°C and expressed as a function of peptide concentration (28, 29, 34). For determinations of bactericidal activity as a function of peptide exposure time, log-phase bacteria (5 × 106/ml) were exposed to peptides for intervals of 0 to 2 h in PIPES-TSB. Samples were removed from the mixtures of bacteria and peptides at 30-min intervals and diluted 100-fold in 10 mM PIPES buffer, and bacterial cell survival was measured as described above.
Peptide-induced permeabilization of E. coli ML35.
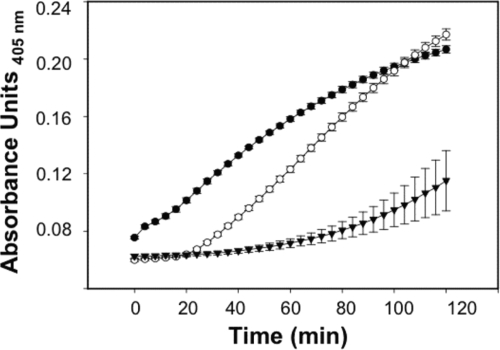
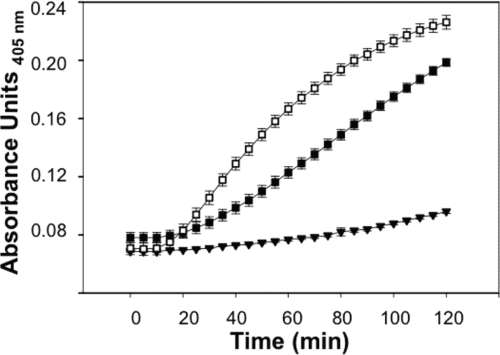
E. coli strain ML35 [lacZ(Con) ΔlacY] does not take up the lactose analogue 2-nitrophenyl β-d-galactopyranoside (ONPG) unless permeabilized by membrane-disruptive agents, including defensins (18). Upon membrane disruption, ONPG diffuses into the bacterial cell cytoplasm and is hydrolyzed by β-galactosidase (β-Gal) and converted to 2-nitrophenol (ONP), which can be measured by absorbance at 405 nm (18). Log-phase E. coli ML35 cells were washed and resuspended in 10 mM PIPES-TSB. In triplicate, bacteria (5 × 106 cells/ml) were exposed to peptides in the presence of 2.5 mM ONPG for 2 h at 37°C (8, 11, 40). The kinetics of ONPG hydrolysis was measured by determining the absorbance at 405 nm using a Spectra-Max plate spectrophotometer (Molecular Devices, Sunnyvale, CA). ProCrp4, Crp4, and (R/K)-Crp4 were assayed at 6 μM for data shown in Fig. 4. ProCrp4, RMAD-4, and (R/K)-RMAD-4 were assayed at 3 μM for data shown in Fig. 7. Error bars denote standard deviations of the means.
FIG. 4.
E. coli ML35 permeabilization by Crp4, proCrp4, and (R/K)-Crp4 at 6 μM. E. coli ML35 cells growing in log phase were exposed to peptides as shown at 37°C in 100 μl PIPES-TSB buffer with 2.5 mM ONPG. ONPG hydrolysis was measured by absorbance at 405 nm for 2 h. Symbols: proCrp4, ▾; Crp4; •; and (R/K)-Crp4, ○. (R/K)-Crp4 displayed delayed E. coli ML35 permeabilization kinetics compared to Crp4.
FIG. 7.
E. coli ML35 cell permeabilization by proCrp4, RMAD-4, and (R/K)-RMAD-4 at 3 μM. E. coli ML35 cells were exposed to peptides in 100 μl of PIPES-TSB buffer and 2.5 mM ONPG and assayed for ONPG hydrolysis as for Fig. 4. Symbols: RMAD-4, ▪; (R/K)-RMAD-4, □; proCrp4, ▾. (R/K)-RMAD-4 displayed increased permeabilization kinetics for E. coli ML35 compared to results for RMAD-4.
RESULTS
Arg-to-Lys mutagenesis attenuates Crp4 bactericidal activity.
Electropositive charge has been implicated as a determinant of Crp4 bactericidal activity (36). As an initial test of the hypothesis that Arg is prevalent in α-defensins because it provides greater microbicidal activity than Lys at the same position, (R/K)-Crp4 and (R/K)-proCrp4 peptide variants (Fig. 1) (see Materials and Methods) were purified for analyses of in vitro bactericidal peptide activity. Recombinant (R/K)-Crp4 and (R/K)-proCrp4 eluted as single peaks in C18 RP-HPLC, but AU-PAGE analysis of the HPLC-purified molecules revealed heterogeneity in the form of three differentially migrating peptide bands (data not shown). The peptide bands had identical molecular masses consistent with those predicted for (R/K)-Crp4 and (R/K)-proCrp4 monomers, respectively, suggesting that the peptide preparations contained misfolded variants. Accordingly, the proteins were reduced, refolded, and purified by analytical C18 RP-HPLC, and peptide homogeneity was verified by AU-PAGE analyses (see Fig. S2 in the supplemental material). In addition, the disulfide pairing assignments in (R/K)-Crp4 were verified by nuclear magnetic resonance spectrometry (data not shown).
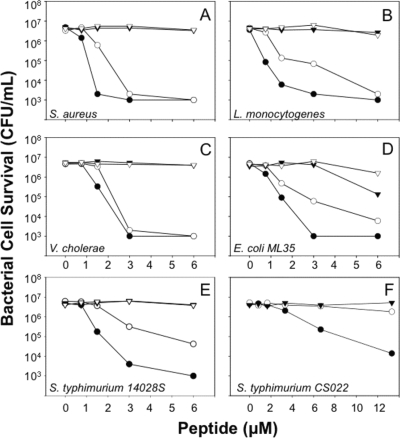
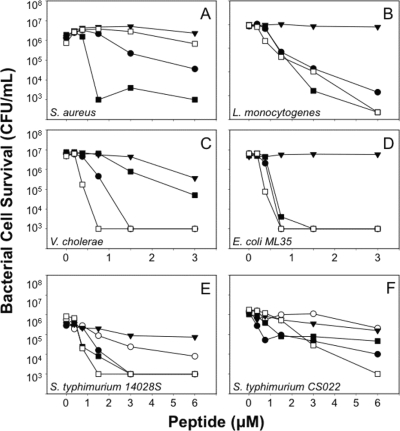
Lys replacements at Arg residue positions attenuated Crp4 bactericidal activity in in vitro bactericidal peptide assays. Against Staphylococcus aureus 502A, Listeria monocytogenes, and Vibrio cholerae, bacterial species that are sensitive to Crp4-mediated killing, (R/K)-Crp4 and Crp4 exhibited similar overall bactericidal activities (Fig. 2A to C, respectively). On the other hand, against less-sensitive bacterial species, such as E. coli ML35 and wild-type S. Typhimurium 14028S cells, (R/K)-Crp4 was markedly less bactericidal than native Crp4 (Fig. 2D and E). (R/K)-Crp4 attenuation was particularly evident against S. Typhimurium CS022, a phoP constitutive strain (Fig. 2F), the least α-defensin-sensitive strain tested (7, 10). The data shown in Fig. 2 are representative of a minimum of three independent assays against each organism performed on separate days. Previously, native Crp4 and a variant with two Lys-for-Arg substitutions, (R16K/R18K)-Crp4, were shown to have the same in vitro bactericidal activities (36), suggesting that complete Arg→Lys replacement may be needed to detect the observed attenuation of activity. Therefore, conversion of all Crp4 Arg residues to Lys diminished peptide activity, especially against bacterial cell targets with low inherent sensitivity to Crp4.
FIG. 2.
Bactericidal activity of recombinant Crp4, proCrp4, and mutants with Lys substitutions. Exponentially growing S. aureus (A), L. monocytogenes (B), V. cholerae (C), E. coli ML35 (D), S. Typhimurium 14028S (E), or S. Typhimurium CS022 (F) was exposed to peptides at 37°C in 50 μl of PIPES-TSB buffer for 1 h (see Materials and Methods). Following peptide exposure, the bacteria were plated on TSB-agar plates and incubated overnight at 37°C. Please note the higher peptide concentrations used in assays against S. Typhimurium CS022 (F). Surviving bacteria were counted as CFU/ml at each peptide concentration, and count values below 1 × 103 CFU/ml signify that no colonies were detected. Symbols: proCrp4, ▾; Crp4, •; (R/K)-proCrp4, ▿; and (R/K)-Crp4, ○. (R/K)-Crp4 exhibited reduced activity, particularly against S. Typhimurium strains.
In vitro activation of (R/K)-proCrp4 by MMP-7.
Native proCrp420-92 lacks both bactericidal and membrane-disruptive activities until cleaved by MMP-7 at Ser43 ↓ Ile44, which removes charge-neutralizing acidic amino acids present in the proCrp420-43 region (34, 40). To test whether the inhibitory effects of proregion Glu and Asp residues may be lost when Crp4 Arg positions are replaced with Lys, the in vitro bactericidal peptide activity of (R/K)-proCrp420-92 was assayed (Fig. 2). (R/K)-proCrp4 lacks bactericidal peptide activity, even against the Crp4-sensitive species V. cholerae and L. monocytogenes (Fig. 2A to E), similar to the absence of bactericidal activity in native proCrp420-92 (8, 34, 40). Because proCrp420-92 and (R/K)-proCrp420-92 both are inactive, we conclude that interactions between anionic proregion amino acids and the Crp4 moiety are equally inhibitory, regardless of whether the basic amino acids are Arg or Lys residues. Based on these findings, it seems unlikely that Arg is under selection to better maintain pro-α-defensins in an inactive state.
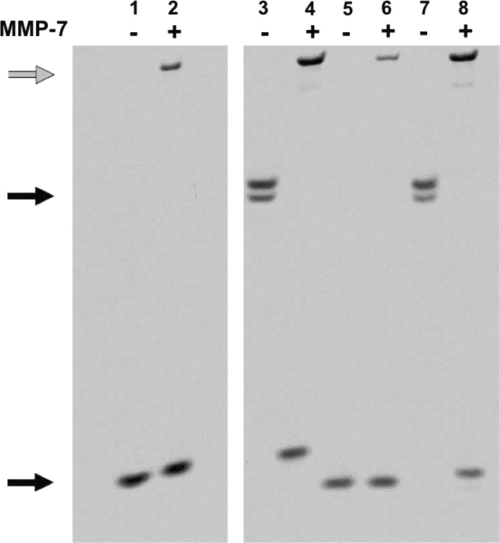
Mutagenesis of the canonical α-defensin salt bridge (Arg5-Glu13) in HNP-2 induced sensitivity to neutrophil elastase proteolysis (25). Also, because disulfide mutagenesis of Crp4 and RMAD-4 and their precursors results in extensive degradation by their respective convertases MMP-7 (20) and elastase (14), we tested whether Lys substitutions induce peptide susceptibility to proteolysis. (R/K)-proCrp4, (R/K)-Crp4, and corresponding native peptides were exposed to MMP-7, and their sensitivities to proteolysis were compared by AU-PAGE (Fig. 3). Consistent with Crp4 resistance to MMP-7 proteolysis (20), MMP-7 did not cleave (R/K)-Crp4 in vitro, and the major product of (R/K)-proCrp4 processing by MMP-7 was intact (R/K)-Crp4 (Fig. 3). Therefore, in vitro activation of the precursor with Lys substitutions was normal, evidence that Crp4 stability against MMP-7 proteolysis is not determined by electropositive amino acid side chain chemistry.
FIG. 3.
Products of MMP-7-mediated conversion of Crp4, proCrp4, and mutants with Lys substitutions. Eleven μg of recombinant proCrp4 and (R/K)-proCrp4 and 5 μg of Crp4 and (R/K)-Crp4 were incubated with or without 0.5 molar equivalents of MMP-7 in 0.10-strength HEPES buffer for 18 h at 37°C (see Materials and Methods). The digests were resolved by AU-PAGE and stained with Coomassie blue. The gray arrow denotes the MMP-7 band in lanes 2, 4, 6, and 8. Lanes: 1, Crp4 (lower black arrow); 2, Crp4 plus MMP-7; 3, proCrp4 (upper black arrow); 4, proCrp4 plus MMP-7; 5, (R/K)-Crp4; 6, (R/K)-Crp4 plus MMP-7; 7, (R/K)-proCrp4; 8, (R/K)-proCrp4 plus MMP-7.
Delayed permeabilization of E. coli ML35 by (R/K)-Crp4.
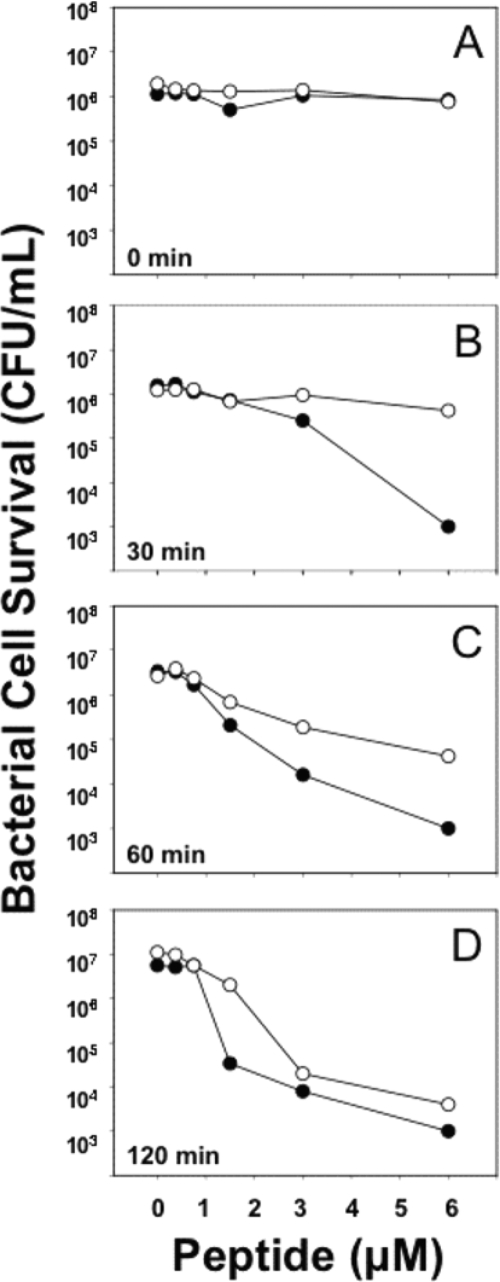
To test whether the bactericidal attenuation observed for (R/K)-Crp4 (Fig. 2) results from impaired peptide-induced bacterial cell permeabilization (18), Crp4 and (R/K)-Crp4 were compared in ONPG conversion assays performed with live E. coli ML35 cells (see Materials and Methods). α-Defensins generally induce bacterial cell death by target cell membrane disruption (11, 28, 29, 41), with eventual degradation of chemiosmotic gradients (18, 29). Membrane permeabilization of β-Gal-constitutive E. coli ML35 cells enables diffusion of the lactose analogue ONPG into the cells, where β-Gal converts colorless ONPG to ONP, which absorbs at 405 nm (18). In such assays, Crp4 induced rapid ONP production in E. coli ML35 cells (Fig. 4). In contrast, permeabilization of E. coli by 6 μM (R/K)-Crp4 was delayed, requiring a minimum of 20 min of (R/K)-Crp4 exposure before ONP was detected (Fig. 4). Furthermore, (R/K)-Crp4-induced permeabilization was defective at all concentrations (data not shown). In contrast to the lag observed for (R/K)-Crp4, proCrp4 induced baseline E. coli ML35 permeabilization throughout the assay, consistent with its well-documented lack of bactericidal activity (8, 20, 34, 40) (Fig. 2D). The delayed permeabilization of E. coli ML35 by (R/K)-Crp4 predicts that a prolonged period of exposure to the variant peptide would be required to affect bacterial cell survival.
(R/K)-Crp4 bactericidal effects require extended peptide exposure.
As predicted by the delayed permeabilization of E. coli by (R/K)-Crp4, extensive peptide incubation was required to achieve bacterial cell killing. Bacterial cell survival was determined as a function of peptide exposure time by incubating E. coli ML35 cells with (R/K)-Crp4 or Crp4 and assessing bacterial cell survival by sampling at sequential 30-min intervals (Fig. 5). No loss of bacterial cell viability was evident in samples removed immediately after combining peptides with cells (Fig. 5A), and a 30-min exposure to 6 μM Crp4 reduced E. coli ML35 cell numbers by 3 to 4 orders of magnitude. In contrast, (R/K)-Crp4 had no measurable effect on bacterial cell survival under the same conditions (Fig. 5B). Exposure of E. coli to (R/K)-Crp4 for 60 min was associated with a loss of cell viability, but the microbicidal effect was less than that of Crp4 at all peptide concentrations tested (Fig. 5C and D). Thus, consistent with the delay in ONPG conversion and despite having an equivalent electropositive charge, (R/K)-Crp4 required longer exposure times than Crp4 to have bactericidal effects against E. coli. These findings are consistent with selection of Arg relative to Lys to optimize α-defensin bactericidal peptide activity, as has been reported for HNP-1 (45). However, comparable analyses of the primate α-defensin RMAD-4 do not support this generalization.
FIG. 5.
E. coli ML35 cell survival in response to Crp4 and (R/K)-Crp4 exposure time. E. coli ML35 cells were exposed to peptides at 37°C in 50 μl PIPES-TSB buffer (see Materials and Methods). Following peptide exposure for 0 (A), 30 (B), 60 (C), or 120 min (D), the peptide and bacterial mixtures were assayed for bacterial cell survival as for Fig. 2. Symbols: Crp4, •; (R/K)-Crp4, ○. (R/K)-Crp4 required longer periods of peptide exposure to affect bacterial cell survival.
RMAD-4 bactericidal activity is not attenuated by Arg-to-Lys mutagenesis.
To test whether Lys substitutions attenuate RMAD-4 activity in a manner consistent with results for (R/K)-Crp4, all Arg residue positions in RMAD-4 were replaced with Lys to prepare (R/K)-RMAD-4 by solid-phase synthesis (see Materials and Methods), and its biological activities were compared to those of native RMAD-4. Unlike the case of Crp4 with Lys substitutions (Fig. 2 and 5), corresponding mutagenesis in RMAD-4 did not attenuate peptide function against most bacterial species tested. Recombinant RMAD-4 and synthetic, refolded (R/K)-RMAD-4 were judged to be homogeneous by analytical C18 RP-HPLC and AU-PAGE analysis (14, 35, 37) (see Fig. S2 in the supplemental material). Additionally, the peptide masses were confirmed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (data not shown; see Materials and Methods).
Consistent with the mutagenesis of Crp4, (R/K)-RMAD-4 had less activity than native RMAD-4 against S. aureus (Fig. 6A). However, in marked contrast to the attenuation of (R/K)-Crp4 (Fig. 2 and 5), (R/K)-RMAD-4 was more bactericidal than native RMAD-4 against V. cholerae, E. coli ML35, and the phoP constitutive strain S. Typhimurium CS022 (Fig. 6C, D, and F), and the native and variant peptides were equally active against L. monocytogenes and S. Typhimurium 14028S (Fig. 6B and E). The data shown in Fig. 6 are representative of no fewer than three replicate dose-sensitivity assays for each target organism and were highly consistent (see Discussion). The reproducibility of these assays was illustrated by performing five replicate assays at three peptide concentrations against V. cholerae, an organism with greater sensitivity to (R/K)-RMAD-4 than to wild-type RMAD-4 (Fig. 6C; see also Fig. S4A in the supplemental material). Consistent with that dose-sensitivity curve, 0.75 μM (R/K)-RMAD-4 showed markedly greater potency than RMAD-4, and the difference in activity was the same as that shown in Fig. 6C, attesting to the reproducibility of these assays (see Fig. S4A in the supplemental material). Thus, Arg-to-Lys mutagenesis in (R/K)-RMAD-4 does not have the same general attenuating effect it does in Crp4.
FIG. 6.
Bactericidal peptide activity of (R/K)-RMAD-4. Peptide bactericidal peptide activity against exponentially growing S. aureus (A), L. monocytogenes (B), V. cholerae (C), E. coli ML35 (D), S. Typhimurium 14028S (E), or S. Typhimurium CS022 (F) was assayed as for Fig. 2. Symbols: RMAD-4, ▪; (R/K)-RMAD-4, □; proCrp4, ▾; Crp4, •; and (R/K)-Crp4, ○. (R/K)-RMAD-4 exhibited variable activity.
Increased permeabilization of E. coli ML35 by (R/K)-RMAD-4.
To test whether E. coli permeabilization kinetics were inherently delayed due to Lys substitutions or were consistent with bactericidal peptide activities, we measured (R/K)-RMAD-4-induced E. coli ML35 cell permeabilization in the ONPG conversion assay. Consistent with its enhanced bactericidal activity, (R/K)-RMAD-4 permeabilized E. coli more rapidly than RMAD-4, and higher levels of ONP were produced (Fig. 7). As expected, the proCrp4 negative control peptide lacked permeabilizing activity (Fig. 4 and 7). These outcomes, along with the (R/K)-Crp4 permeabilization data from Fig. 4, show that the consequences of Lys-for-Arg substitutions depend on their context within individual α-defensin primary structures and do not support the view that high Arg content evolved to optimize α-defensin bactericidal activity.
Proteolytic sensitivity of α-defensins with Lys substitutions to neutrophil elastase and trypsin.
To determine whether Lys substitutions induce α-defensin susceptibility to proteolysis in vitro, proCrp4, Crp4, (R/K)-Crp4, RMAD-4, and (R/K)-RMAD-4 were exposed to neutrophil elastase and trypsin, and proteolysis was assayed by AU-PAGE analyses of peptide digests (see Materials and Methods). As expected (14, 20), both Crp4 and RMAD-4 withstood proteolytic digestion by both elastase and trypsin, and trypsin cleaved proCrp4 to generate a peptide that comigrated with Crp4 in AU-PAGE. The (R/K)-Crp and (R/K)-RMAD-4 peptides both were insensitive to elastase, but apparent cleavage was evident in trypsin digests of (R/K)-RMAD-4 (see Fig. S3 in the supplemental material). In addition, proCrp4 was cleaved by neutrophil elastase into fragments, some of which did not comigrate with Crp4 (see Fig. S3 in the supplemental material). Because (R/K)-Crp4 was resistant to both enzymes, the generalization that Lys substitutions increase inherent proteolytic sensitivity is not supported. On the other hand, the increased sensitivity of (R/K)-RMAD-4 to trypsin suggests that Arg-to-Lys replacements in certain contexts may induce greater trypsin susceptibility, depending on the positions of particular residues on the peptide surface.
Arg-to-Lys mutagenesis induces peptide inhibition by NaCl.
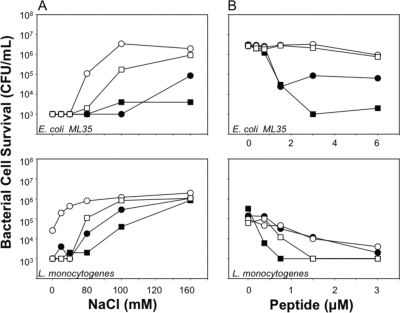
Because electrostatic conditions influence α-defensin bactericidal activity (17, 30, 31, 39), bactericidal peptide assays were performed in the presence of varied NaCl concentrations to determine whether Lys-for-Arg substitutions modulate sensitivity to inhibition by NaCl. To test this notion, bactericidal peptide assays were performed with E. coli ML35 and L. monocytogenes in the presence of RMAD-4 (3 μM), (R/K)-RMAD-4 (3 μM), Crp4 (6 μM), or (R/K)-Crp4 (6 μM) in solutions of increasing NaCl concentrations (Fig. 8A). In a series of consistently reproducible assays, variants with Lys substitutions were more sensitive to NaCl inhibition than corresponding native α-defensins. For example, although RMAD-4 was insensitive to 160 mM NaCl, (R/K)-RMAD-4 was highly attenuated at 80 mM NaCl (Fig. 8A), and when the NaCl concentration was fixed at 80 mM, both variants with Lys substitutions were inactive against E. coli ML35 (Fig. 8B, upper panel) and more extensively inhibited against L. monocytogenes. The data in Fig. 8 are representative of three independent determinations for both organisms. The reproducibility of the salt sensitivity findings was illustrated by performing five replicate assays with 1.5 μM RMAD-4 or (R/K)-RMAD-4 against L. monocytogenes at different NaCl concentrations (see Fig. S4B in the supplemental material). That experiment showed that (R/K)-RMAD-4 bactericidal activity decreased more than 10-fold at 20 mM NaCl while RMAD-4 retained potent killing activity against L. monocytogenes (see Fig. S4B in the supplemental material).
FIG. 8.
NaCl attenuates bactericidal peptide activities of variants with Lys substitutions. (A) E. coli ML35 (upper panel) or L. monocytogenes (lower panel) was exposed to 6 μM Crp4 or (R/K)-Crp4 or to 3 μM RMAD-4 or (R/K)-RMAD-4 at 37°C for 1 h in 50 μl PIPES-TSB buffer supplemented with NaCl as shown. Bacterial cell survival was quantitated as for Fig. 2. Variant peptides with Lys substitutions are more sensitive to inhibition by NaCl. (B) E. coli ML35 cells were exposed to the peptide concentrations shown for 1 h at 37°C in 50 μl of PIPES-TSB buffer containing 80 mM NaCl, and cell survival was assayed as described previously (see the legend to Fig. 2). Symbols: RMAD-4, ▪; (R/K)-RMAD-4, □; Crp4, •; (R/K)-Crp4, ○. Against E. coli, the α-defensins with Lys substitutions were less active in 80 mM NaCl than native α-defensins.
DISCUSSION
Electropositive charge is a determinant of α-defensin bactericidal peptide activity (36), and cationic charge in the peptide family is contributed disproportionately by a molar ratio of nine Arg residues for each Lys (Fig. 1; see Fig. S1 in the supplemental material). However, despite the equivalent electropositive charges of (R/K)-Crp4 and Crp4, the (R/K)-Crp4 molecule had diminished bactericidal peptide activities against diverse bacterial species (Fig. 2), delayed kinetics of E. coli cell permeabilization (Fig. 4), and required prolonged exposure times to have bactericidal effects on E. coli (Fig. 5). The Crp4 studies, therefore, supported the initial view that the conserved high Arg-to-Lys molar composition of the α-defensin family evolved to optimize microbicidal activities with short peptide exposure times. This conclusion is in general agreement with the outcome of Lys-for-Arg substitutions in HNP-1 (45). However, comparable mutagenesis and analysis of RMAD-4 showed that this conclusion does not apply across the peptide family in general. Thus, as is apparent from these findings, selecting one particular α-defensin molecule as representing the general properties of the peptide family is not a useful concept.
The α-defensins share four canonical structural features. Numbering residue positions in accordance with the Crp4 and RMAD-4 mature peptide N termini, the conserved features are the following: (i) the disulfide array, (ii) a salt bridge formed by Arg7-Glu15 hydrogen bonds, (iii) a conserved Gly at residue position 19, and (iv) the high Arg:Lys molar ratio investigated in this report. Most efforts to identify the function(s) of these canonical α-defensin features have focused on their roles as potential determinants of bactericidal activity. In most instances, mutagenesis at these canonical residue positions did not alter in vitro bactericidal activity (11, 14, 20, 26). For example, in the case of the disulfide array, the disulfide bonds confer protection against proteolysis by the activating convertases (14, 20). These collective findings suggest that the salt bridge and the Gly19 position may have evolved to provide a conserved function(s) in the peptide family that is independent of bactericidal activity. Possibly, these structural features are conserved to enable or facilitate efficient chaperone interactions, peptide folding, trafficking in the endoplasmic reticulum, protection against proteolysis, or receptor-mediated events. Thus, it is possible, perhaps likely, that the high Arg content contributes to aspects of α-defensin biology, such as those noted, that we have not investigated here.
Complete Lys-for-Arg replacements in RMAD-4 did not diminish bactericidal peptide activity (Fig. 6) or reduce the kinetics of E. coli cell permeabilization (Fig. 7). To the contrary, Lys substitutions in RMAD-4 resulted in equivalent or improved peptide activities against most bacterial cell targets, with the exception of S. aureus. In contrast to comparisons of HNP-1 and HNP-1 with Lys substitutions (45), in vitro bactericidal peptide assays of (R/K)-Crp4 and (R/K)-RMAD-4 in the presence of NaCl showed that both peptides were more sensitive to salt inhibition than the native molecules (Fig. 8). Perhaps Arg abundance contributes to optimization of microbicidal activities under higher NaCl concentrations. Thus, the consequences of Arg-to-Lys replacements in these two α-defensins do not support general conclusions applicable to the peptide family but instead depend on the context of their positions within specific peptide primary structures.
Why Crp4 but not RMAD-4 bacterial cell killing activity is attenuated by Arg-for-Lys replacements remains unexplained. For example, mutagenesis of Arg to Lys in Crp4 and RMAD-4 does not modify the overall cationic charge of the peptides, but their activities are measurably altered in standard in vitro assays and by the presence of NaCl. Although Arg and Lys carry equivalent cationic charges at physiological pH, the charge is distributed differently by the two side chains. As previous authors have noted (45), the Lys side chain is more hydrophobic than that of Arg owing to its five methylene groups. Also, Arg interacts mainly via its guanidinium group, which enables Arg to form stronger electrostatic hydrogen bonding interactions, including salt bridges and additional contacts. Possibly, the dispersion of cationic charge over the guanidinium group may allow for a greater area of interaction or more stable hydrogen bonding interactions (21), and those stronger interactions may improve affinity for electronegative microbial cell envelopes and subsequent membrane disruption in media of higher ionic strength. On the other hand, the cationicity of the Lys ɛ-amino group is less dispersed, presenting a more-localized positive charge that also may favor interactions with bacterial cell surfaces under certain conditions. An analysis of atom densities in proteins (3) also reveals distinct properties of Arg and Lys in protein structures. For example, as noted by Karlin and colleagues (3, 15), Asp and Glu exhibit stronger attractions for Arg than Lys, Arg is a stronger base (higher pKa), and Arg is more hydrophobic in that it forms salt bridges with acidic residues in a buried state, in contrast to Lys, which has its ɛ-amino group exposed on protein surfaces (15). Whether the distinctive general biochemical features of these amino acids determine their roles in peptides as small as α-defensins is somewhat speculative but is open to investigation. In this respect, the acquisition and comparison of solution structures for the native and R/K α-defensins may be very useful in disclosing how these different side chain chemistries modulate activity. Structural determinations performed in the presence of model membranes may be particularly valuable.
Modeling studies show that the distribution of charge on the Crp4 and RMAD-4 peptide surfaces differs. For example, solution structures of Crp4 show that the cationic charge is distributed uniformly on one face of the peptide surface (13, 26), but the basic residues in RMAD-4 are positioned mainly in that region of the triple-stranded β-sheet most distal to the N and C termini (M. J. Cocco et al., unpublished data). Perhaps this more polarized distribution of Arg residues in RMAD-4 concentrates electropositive charge, which may become more localized upon their replacement with Lys. One tentative prediction of these studies is that when α-defensin cationicity is more broadly distributed, Arg-to-Lys substitutions are attenuating, but when Arg residues are clustered, as in RMAD-4, such replacements are neutral or even augment activity by further charge localization. This possibility may be open to analysis by study of a range of Arg/Lys replacement combinations rather than substitutions at all positions to provide additional data for structural interpretation of these differential effects. We also speculate that the more dispersed charge of Arg may reduce the favorability of interactions with Na+ and Cl− ions relative to that of Lys to diminish the inhibitory effects of salts on bactericidal action.
The increased sensitivity of peptides with Lys substitutions to inhibition by NaCl suggests that the prevalence of Arg as the basic amino acid in α-defensins confers improved bactericidal activity at higher ionic strength. However, in comparisons of human HNP-1 with a variant containing three Lys-for-Arg substitutions, native HNP-1 assayed against E. coli was more sensitive to salt inhibition than the variant peptide. Although this finding may be due to the low cationicity of HNP-1 relative to those of Crp4 and RMAD-4, it may be further evidence that effects of α-defensin mutagenesis must be interpreted in the context of individual peptide primary structures. Given these findings and the diversity of α-defensin primary structures, the effects of Lys-for-Arg substitutions in other α-defensins are likely to induce a spectrum of effects ranging from attenuation to gain of function, depending on the distribution of Arg residues on peptide surfaces (see Fig. S1 in the supplemental material). Such studies could be used to develop an α-defensin topology map capable of predicting the consequences of Arg-to-Lys replacements as well as substitutions to improve peptide action.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants DK044632 and AI059346 and the Human Frontiers Science Program (to A.J.O.).
We thank Michael E. Selsted for helpful discussions. Xiaoqing Qu, Steven Young, and Hector Salazar provided excellent technical assistance and advice.
Editor: F. C. Fang
Footnotes
Published ahead of print on 8 September 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aley, S. B., M. Zimmerman, M. Hetsko, M. E. Selsted, and F. D. Gillin. 1994. Killing of Giardia lamblia by cryptdins and cationic neutrophil peptides. Infect. Immun. 62:5397-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Baud, F., and S. Karlin. 1999. Measures of residue density in protein structures. Proc. Natl. Acad. Sci. USA 96:12494-12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borenstein, L. A., M. E. Selsted, R. I. Lehrer, and J. N. Miller. 1991. Antimicrobial activity of rabbit leukocyte defensins against Treponema pallidum subsp. pallidum. Infect. Immun. 59:1359-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummings, J. E., D. P. Satchell, Y. Shirafuji, A. J. Ouellette, and T. K. Vanderlick. 2003. Electrostatically controlled interactions of mouse Paneth cell alpha-defensins with phospholipid membranes. Austr. J. Chem. 56:1031-1034. [Google Scholar]
- 6.Cummings, J. E., and T. K. Vanderlick. 2007. Kinetics of cryptdin-4 translocation coupled with peptide-induced vesicle leakage. Biochemistry 46:11882-11891. [DOI] [PubMed] [Google Scholar]
- 7.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327-1334. [DOI] [PubMed] [Google Scholar]
- 8.Figueredo, S. M., C. S. Weeks, S. K. Young, and A. J. Ouellette. 2009. Anionic amino acids near the pro-alpha-defensin N terminus mediate inhibition of bactericidal activity in mouse pro-cryptdin-4. J. Biol. Chem. 284:6826-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganz, T., M. E. Selsted, D. Szklarek, S. S. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadjicharalambous, C., T. Sheynis, R. Jelinek, M. T. Shanahan, A. J. Ouellette, and E. Gizeli. 2008. Mechanisms of alpha-defensin bactericidal action: comparative membrane disruption by cryptdin-4 and its disulfide-null analogue. Biochemistry 47:12626-12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hristova, K., M. E. Selsted, and S. H. White. 1996. Interactions of monomeric rabbit neutrophil defensins with bilayers: comparison with dimeric human defensin HNP-2. Biochemistry 35:11888-11894. [DOI] [PubMed] [Google Scholar]
- 13.Jing, W., H. N. Hunter, H. Tanabe, A. J. Ouellette, and H. J. Vogel. 2004. Solution structure of cryptdin-4, a mouse Paneth cell alpha-defensin. Biochemistry 43:15759-15766. [DOI] [PubMed] [Google Scholar]
- 14.Kamdar, K., A. Maemoto, X. Qu, S. K. Young, and A. J. Ouellette. 2008. In vitro activation of the rhesus macaque myeloid alpha-defensin precursor proRMAD-4 by neutrophil serine proteinases. J. Biol. Chem. 283:32361-32368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlin, S., Z. Y. Zhu, and F. Baud. 1999. Atom density in protein structures. Proc. Natl. Acad. Sci. USA 96:12500-12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehrer, R. I. 2007. Multispecific myeloid defensins. Curr. Opin. Hematol. 14:16-21. [DOI] [PubMed] [Google Scholar]
- 17.Lehrer, R. I., A. Barton, K. A. Daher, S. S. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehrer, R. I., A. Barton, and T. Ganz. 1988. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J. Immunol. Methods 108:153-158. [DOI] [PubMed] [Google Scholar]
- 19.Lehrer, R. I., T. Ganz, and M. E. Selsted. 1988. Oxygen-independent bactericidal systems. Mechanisms and disorders. Hematol. Oncol. Clin. N. Am. 2:159-169. [PubMed] [Google Scholar]
- 20.Maemoto, A., X. Qu, K. J. Rosengren, H. Tanabe, A. Henschen-Edman, D. J. Craik, and A. J. Ouellette. 2004. Functional analysis of the α-defensin disulfide array in mouse cryptdin-4. J. Biol. Chem. 279:44188-44196. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, J. B., J. M. Thornton, J. Singh, and S. L. Price. 1992. Towards an understanding of the arginine-aspartate interaction. J. Mol. Biol. 226:251-262. [DOI] [PubMed] [Google Scholar]
- 22.Ouellette, A. J. 2006. Paneth cell alpha-defensin synthesis and function. Curr. Top. Microbiol. Immunol. 306:1-25. [DOI] [PubMed] [Google Scholar]
- 23.Ouellette, A. J., and C. L. Bevins. 2001. Paneth cell defensins and innate immunity of the small bowel. Inflamm. Bowel Dis. 7:43-50. [DOI] [PubMed] [Google Scholar]
- 24.Patil, A., A. L. Hughes, and G. Zhang. 2004. Rapid evolution and diversification of mammalian α-defensins as revealed by comparative analysis of rodent and primate genes. Physiol. Genomics 20:1-11. [DOI] [PubMed] [Google Scholar]
- 25.Rajabi, M., E. de Leeuw, M. Pazgier, J. Li, J. Lubkowski, and W. Lu. 2008. The conserved salt bridge in human alpha-defensin 5 is required for its precursor processing and proteolytic stability. J. Biol. Chem. 283:21509-21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosengren, K. J., N. L. Daly, L. M. Fornander, L. M. Jonsson, Y. Shirafuji, X. Qu, H. J. Vogel, A. J. Ouellette, and D. J. Craik. 2006. Structural and functional characterization of the conserved salt bridge in mammalian Paneth cell alpha-defensins: solution structures of mouse cryptidin-4 and (E15D)-cryptidin-4. J. Biol. Chem. 281:28068-28078. [DOI] [PubMed] [Google Scholar]
- 27.Salzman, N. H., M. M. Chou, H. de Jong, L. Liu, E. M. Porter, and Y. Paterson. 2003. Enteric salmonella infection inhibits Paneth cell antimicrobial peptide expression. Infect. Immun. 71:1109-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satchell, D. P., T. Sheynis, S. Kolusheva, J. E. Cummings, T. K. Vanderlick, R. Jelinek, M. E. Selsted, and A. J. Ouellette. 2003. Quantitative interactions between cryptdin-4 amino terminal variants and membranes. Peptides 24:1793-1803. [DOI] [PubMed] [Google Scholar]
- 29.Satchell, D. P., T. Sheynis, Y. Shirafuji, S. Kolusheva, A. J. Ouellette, and R. Jelinek. 2003. Interactions of mouse Paneth cell alpha-defensins and alpha-defensin precursors with membranes: prosegment inhibition of peptide association with biomimetic membranes. J. Biol. Chem. 278:13838-13846. [DOI] [PubMed] [Google Scholar]
- 30.Selsted, M. E. 2007. A pocket guide to explorations of the defensin field. Curr. Pharm. Des. 13:3061-3064. [DOI] [PubMed] [Google Scholar]
- 31.Selsted, M. E. 1993. Investigational approaches for studying the structures and biological functions of myeloid antimicrobial peptides. Genet. Eng. (N.Y.) 15:131-147. [DOI] [PubMed] [Google Scholar]
- 32.Selsted, M. E., and S. S. Harwig. 1989. Determination of the disulfide array in the human defensin HNP-2. A covalently cyclized peptide. J. Biol. Chem. 264:4003-4007. [PubMed] [Google Scholar]
- 33.Selsted, M. E., and A. J. Ouellette. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551-557. [DOI] [PubMed] [Google Scholar]
- 34.Shirafuji, Y., H. Tanabe, D. P. Satchell, A. Henschen-Edman, C. L. Wilson, and A. J. Ouellette. 2003. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J. Biol. Chem. 278:7910-7919. [DOI] [PubMed] [Google Scholar]
- 35.Tanabe, H., A. J. Ouellette, M. J. Cocco, and W. E. Robinson, Jr. 2004. Differential effects on human immunodeficiency virus type 1 replication by α-defensins with comparable bactericidal activities. J. Virol. 78:11622-11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe, H., X. Qu, C. S. Weeks, J. E. Cummings, S. Kolusheva, K. B. Walsh, R. Jelinek, T. K. Vanderlick, M. E. Selsted, and A. J. Ouellette. 2004. Structure-activity determinants in Paneth cell alpha-defensins: loss-of-function in mouse cryptdin-4 by charge-reversal at arginine residue positions. J. Biol. Chem. 279:11976-11983. [DOI] [PubMed] [Google Scholar]
- 37.Tanabe, H., J. Yuan, M. M. Zaragoza, S. Dandekar, A. Henschen-Edman, M. E. Selsted, and A. J. Ouellette. 2004. Paneth cell alpha-defensins from rhesus macaque small intestine. Infect. Immun. 72:1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taudien, S., P. Galgoczy, K. Huse, K. Reichwald, M. Schilhabel, K. Szafranski, A. Shimizu, S. Asakawa, A. Frankish, I. F. Loncarevic, N. Shimizu, R. Siddiqui, and M. Platzer. 2004. Polymorphic segmental duplications at 8p23.1 challenge the determination of individual defensin gene repertoires and the assembly of a contiguous human reference sequence. BMC Genomics 5:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran, D., P. Tran, K. Roberts, G. Osapay, J. Schaal, A. Ouellette, and M. E. Selsted. 2008. Microbicidal properties and cytocidal selectivity of rhesus macaque theta defensins. Antimicrob. Agents Chemother. 52:944-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weeks, C. S., H. Tanabe, J. E. Cummings, S. P. Crampton, T. Sheynis, R. Jelinek, T. K. Vanderlick, M. J. Cocco, and A. J. Ouellette. 2006. Matrix metalloproteinase-7 activation of mouse Paneth cell pro-alpha-defensins: Ser43 ↓ Ile44 proteolysis enables membrane-disruptive activity. J. Biol. Chem. 281:28932-28942. [DOI] [PubMed] [Google Scholar]
- 41.White, S. H., W. C. Wimley, and M. E. Selsted. 1995. Structure, function, and membrane integration of defensins. Curr. Opin. Struct. Biol. 5:521-527. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, C. L., A. J. Ouellette, D. P. Satchell, T. Ayabe, Y. S. Lopez-Boado, J. L. Stratman, S. J. Hultgren, L. M. Matrisian, and W. C. Parks. 1999. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286:113-117. [DOI] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Zhu, S. 2008. Discovery of six families of fungal defensin-like peptides provides insights into origin and evolution of the CSαβ defensins. Mol. Immunol. 45:828-838. [DOI] [PubMed] [Google Scholar]
- 45.Zou, G., E. de Leeuw, C. Li, M. Pazgier, P. Zeng, W. Y. Lu, J. Lubkowski, and W. Lu. 2007. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J. Biol. Chem. 282:19653-19665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.