Abstract
Porphyromonas gingivalis forms communities with antecedent oral biofilm constituent streptococci. P. gingivalis major fimbriae bind to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) present on the streptococcal surface, and this interaction plays an important role in P. gingivalis colonization. This study identified the binding domain of Streptococcus oralis GAPDH for P. gingivalis fimbriae. S. oralis recombinant GAPDH (rGAPDH) was digested with lysyl endopeptidase. Cleaved fragments of rGAPDH were applied to a reverse-phase high-pressure liquid chromatograph equipped with a C18 column. Each peak was collected; the binding activity toward P. gingivalis recombinant fimbrillin (rFimA) was analyzed with a biomolecular interaction analysis system. The fragment displaying the strongest binding activity was further digested with various proteinases, after which the binding activity of each fragment was measured. The amino acid sequence of each fragment was determined by direct sequencing, mass spectrometric analysis, and amino acid analysis. Amino acid residues 166 to 183 of S. oralis GAPDH exhibited the strongest binding activity toward rFimA; confocal laser scanning microscopy revealed that the synthetic peptide corresponding to amino acid residues 166 to 183 of S. oralis GAPDH (pep166-183, DNFGVVEGLMTTIHAYTG) inhibits S. oralis-P. gingivalis biofilm formation in a dose-dependent manner. Moreover, pep166-183 inhibited interbacterial biofilm formation by several oral streptococci and P. gingivalis strains with different types of FimA. These results indicate that the binding domain of S. oralis GAPDH for P. gingivalis fimbriae exists within the region encompassing amino acid residues 166 to 183 of GAPDH and that pep166-183 may be a potent inhibitor of P. gingivalis colonization in the oral cavity.
Coadhesion and coaggregation of Porphyromonas gingivalis, which is a predominant periodontopathic bacterium, and other oral bacteria are considered to be important with respect to colonization in the oral cavity (21, 33). P. gingivalis possesses several components as adhesins on the cell surface, such as FimA fimbriae and Mfa1 fimbriae (2, 6, 12, 14, 17, 23, 25), vesicles (10, 15, 19, 31), hemagglutinin (24), and Arg- and Lys-specific cysteine proteinases (1). Interactions between the following P. gingivalis cell surface components and oral gram-positive bacteria have been documented: (i) FimA fimbriae and Actinomyces viscosus (12), (ii) FimA and Mfa1 fimbriae and Streptococcus gordonii (6, 23), (iii) Arg- and Lys-specific cysteine proteinases and A. viscosus (1), and (iv) vesicles and Actinomyces naeslundii (10), A. viscosus (15, 31), and Streptococcus mutans (19). Additionally, P. gingivalis FimA fimbriae have been shown to interact with epithelial cells (17), cultured human fibroblasts (14), and salivary proteins (2), which indicates that P. gingivalis FimA fimbriae play an important role as a main adhesive component in bacterial colonization. Previously, we reported that P. gingivalis FimA fimbriae bind to Streptococcus oralis, an early colonizer in dental plaque (3). Moreover, we demonstrated that oral streptococcal cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) binds to P. gingivalis FimA fimbriae; furthermore, this interaction exhibits high affinity and high specificity (25, 26).
GAPDH is a classical glycolytic protein which is responsible for the phosphorylation of glyceraldehyde-3-phosphate, leading to the generation of 1,3-bisphosphoglycerate; however, recently, a number of diverse functions including roles in membrane fusion, microtubule binding, phosphotransferase activity, nuclear RNA export, DNA replication and DNA repair, apoptosis, and viral pathogenesis have been reported (34). An adherent function in several microorganisms has been demonstrated. For example, Mycoplasma suis GAPDH is involved in adhesion to erythrocytes (16); surface GAPDH proteins from group A streptococci bind a number of human proteins, including plasmin(nogen) (8, 35) and lysozyme, myosin, actin, and fibronectin (32), and adhere to human pharyngeal cells (18); Staphylococcus aureus and Staphylococcus epidermidis surface-localized GAPDHs bind transferrin (28); secreted GAPDH from Escherichia coli binds human plasminogen and fibrinogen (9); Candida albicans GAPDH binds fibronectin and laminin (13); and Lactobacillus plantarum GAPDH adheres to human colonic mucin (20). Moreover, we have shown previously that the cell surface GAPDHs of several oral streptococci including S. oralis, S. gordonii, Streptococcus sanguinis, and Streptococcus parasanguinis bind P. gingivalis recombinant fimbrillin (rFimA), a monomeric structural subunit of fimbriae (27).
GAPDH encoded by the gapdh gene, which is one of the most common housekeeping genes, is well conserved in eubacteria and eukaryotes (11). Our previous study demonstrated that S. oralis ATCC 9811 GAPDH possesses a high degree of homology to several other bacterial GAPDHs; its deduced amino acid sequence shares approximately 90% identity with those of Streptococcus pneumoniae TIGR4, S. gordonii FSS2, Streptococcus pyogenes M1, and Streptococcus equisimilis H46A GAPDHs and approximately 70% identity with those of S. epidermidis ATCC 12228, S. aureus N315, and Fusobacterium nucleatum ATCC 25586 GAPDHs (25). Therefore, we hypothesized that a common site of oral streptococcal GAPDHs may contribute to P. gingivalis colonization through the interaction with FimA fimbriae.
The present investigation identified the P. gingivalis FimA fimbria binding domain of S. oralis ATCC 9811 GAPDH. In addition, we examined the inhibitory effect of the synthetic peptide corresponding to the P. gingivalis FimA fimbria binding site of S. oralis GAPDH on interbacterial biofilm formation by various streptococci and P. gingivalis strains with different types of FimA by confocal laser scanning microscopy (CLSM).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis strains ATCC 33277, OMZ 314, and W 50, S. oralis ATCC 9811 and ATCC 10557, S. gordonii G9B, S. sanguinis ATCC 10556, and S. parasanguinis ATCC 15909 were maintained as frozen stocks in our laboratory. P. gingivalis 6/26 and HNA 99 were provided by Atsuo Amano (Osaka University). P. gingivalis strains were cultured in prereduced Trypticase soy broth (Becton, Dickinson and Company [BD], Sparks, MD) containing 1 mg of yeast extract (BD)/ml, 5 μg of hemin (Sigma-Aldrich Japan K. K., Tokyo, Japan)/ml, and 1 μg of menadione (Sigma-Aldrich)/ml for 24 h in anaerobic system 1025 (Forma, Marietta, OH) in an atmosphere of 80% N2-10% CO2-10% H2 at 35°C. The bacterial cells were harvested by centrifugation at 5,000 × g for 6 min at 4°C in a high-speed refrigerated centrifuge (SRX-201; Tomy Seiko Co., Ltd., Tokyo, Japan), washed, and suspended in sterile 10 mM phosphate buffer containing 0.15 M NaCl (phosphate-buffered saline [PBS], pH 7.4). Oral streptococci were cultured at 37°C for 16 h in Todd-Hewitt broth (BD), harvested by centrifugation at 6,000 × g for 15 min at 4°C, washed, and suspended in chemically defined medium modified with sucrose (0.8%) as the carbon source (mCDM) (22). E. coli M15(pREP4) (Qiagen GmbH, Hilden, Germany) was cultured in Luria-Bertani broth (BD), and when necessary, 50 μg of ampicillin (Wako Pure Chemical Industries, Ltd., Osaka, Japan)/ml was included.
Preparation of S. oralis rGAPDH.
S. oralis ATCC 9811 genomic DNA was purified with an AquaPure genomic DNA isolation kit according to the instructions of the manufacturer (Bio-Rad Laboratories, Hercules, CA) as a template for amplifying the GAPDH gene by PCR. The primer sequences (with restriction sequences underlined) were as follows: forward primer, 5′-CGCCGCGGATCCAAAGTAGTTAAAGTTGGTATTAACGGT-3′, and reverse primer, 5′-GGCGCCGAATTCGTCGACATTATTTAGCGATTTTTGCG-3′. The forward primer incorporated the BamHI and lysine (AAA) sites, whereas the reverse primer incorporated the SalI site and a translational stop site. PCR using an iCycler thermal cycler (Bio-Rad) was performed with reaction mixtures containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 1.0 μM primer, 10 ng of template DNA, and 0.025 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) in a final volume of 100 μl. The following cycling program was run: preheating at 94°C for 9 min and 33 cycles at 94°C for 45 s, 50°C for 1 min, and 72°C for 40 s. The PCR fragment was checked by DNA sequencing and cloned into plasmid pQE-30, and E. coli M15(pREP4) was transformed with the resulting plasmid according to the instructions of the pQE-30 manufacturer (Qiagen). His-tagged recombinant GAPDH (rGAPDH) from S. oralis ATCC 9811 was purified with a HisTrap HP kit (GE Healthcare UK Ltd., Buckinghamshire, United Kingdom). Purified S. oralis rGAPDH was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 5 to 15% Ready Gel J gel (Bio-Rad). The gel was stained with 0.1% Coomassie brilliant blue (CBB) in 40% methanol-10% acetic acid and destained by treatment with 40% methanol-1% acetic acid. A low-molecular-mass calibration kit (consisting of phosphorylase b, 97 kDa; albumin, 66 kDa; ovalbumin, 45 kDa; carbonic anhydrase, 30 kDa; trypsin inhibitor, 20.1 kDa; and α-lactalbumin, 14.4 kDa [Amersham Pharmacia Biotech, Buckinghamshire, England]) was used to estimate molecular masses. For the Western blot assay, rGAPDH subjected to SDS-PAGE was transferred onto a nitrocellulose membrane, after which the membrane was blocked with Block Ace (casein solution prepared from homogenized milk; Snow Brand Co., Ltd., Sapporo, Japan) for 1 h at room temperature. After three washes with PBS containing 0.05% Tween 20, the membrane was incubated with 1:1,000 penta-His horseradish peroxidase (HRP) conjugate antibodies (Qiagen) for 2 h at room temperature. His-tagged rGAPDH was visualized with an HRP conjugate substrate kit (Bio-Rad). Prestained SDS-PAGE low-range standards (phosphorylase b, 112 kDa; bovine serum albumin, 81 kDa; ovalbumin, 49.9 kDa; carbonic anhydrase, 36.3 kDa; soybean trypsin inhibitor, 29.9 kDa; and lysozyme, 21.3 kDa [Bio-Rad]) were used for molecular mass calibration for the Western blot assay.
Preparation of S. oralis rGAPDH fragments.
Purified S. oralis ATCC 9811 rGAPDH (100 μg) was dissolved in 100 μl of 100 mM Tris-HCl (pH 8.5) containing 6 M urea (Wako) and 0.5% (wt/vol) EDTA (Dojindo Laboratories, Kumamoto, Japan), and reduction was carried out using 1 μmol of dithiothreitol at 37°C for 3 h in a nitrogen atmosphere. Alkylation was conducted by the addition of 2 μmol of iodoacetic acid (Wako) in 1 μl of 0.25 M NaOH at 25°C for 30 min in the dark. Following adjustment to pH 8.5 with Tris base (Sigma-Aldrich), rGAPDH was diluted into a 4 M urea solution; subsequently, a 400-μl solution of lysyl endopeptidase (5 μg; Wako) dissolved in 0.05% polyoxyethylene(10) octylphenyl ether (Triton X-100; Wako) was added. The mixture was incubated at 37°C for 12 h in the dark, after which 200 μl of 10% trifluoroacetic acid (TFA) was introduced to terminate the digestion. The separation of rGAPDH fragments was effected by employing reverse-phase high-pressure liquid chromatography (HPLC) involving an L-6200/6000 liquid chromatograph (Hitachi Ltd., Tokyo, Japan) equipped with a Symmetry300 C18 column (4.6 by 250 mm with a 5-μm particle size; Waters, Milford, MA); the column was equilibrated with 0.1% TFA in HPLC-grade water. The elution of rGAPDH fragments was performed at room temperature with a linear gradient of 0 to 60% acetonitrile in 0.1% TFA (1%/min) at a flow rate of 1 ml/min. The fractionated fragments were collected manually by monitoring the absorbance at 210 nm with an L-4000 detector (Hitachi). The peak exhibiting the strongest binding activity toward P. gingivalis rFimA was resubjected to chromatography on the same HPLC system utilizing an Xterra phenyl column (4.6 by 250 mm with a 5-μm particle size; Waters) equilibrated with 0.1% TFA and 10% acetonitrile in HPLC-grade water. The elution of fragments was conducted with a linear gradient of 10 to 60% acetonitrile in 0.1% TFA (1%/min) at a flow rate of 1 ml/min at room temperature. The fractionated peak displaying the strongest binding activity toward rFimA was lyophilized and digested with three proteinases.
A portion of the fragment (500 pmol) was dissolved in 100 μl of 10 mM Tris-HCl (pH 7.5) and then digested with 0.2 μg of endoproteinase Asp-N (Roche Diagnostics GmbH, Mannheim, Germany) at 37°C for 4 h. The fragment (1 nmol) was digested with trypsin (Promega, Madison, WI) in a 100-μl volume containing 50 mM ammonium bicarbonate (pH 7.8) and 0.4 μg of trypsin at 37°C for 4 h. The resulting tryptic fragment corresponding to amino acid residues 163 to 193 of S. oralis GAPDH (600 pmol) was further digested with 0.2 μg of endoproteinase Glu-C (Roche Diagnostics) in 100 μl of 50 mM ammonium bicarbonate (pH 7.8) at 25°C for 4 h. Three products of digestion with the aforementioned proteinases were separated by HPLC with a Symmetry300 C18 column as described above, and the binding activity of each fragment was measured.
Binding of S. oralis rGAPDH fragments to P. gingivalis rFimA.
P. gingivalis rFimA (type I fimbriae) was prepared in accordance with the methods reported previously (29). Interactions between P. gingivalis rFimA and the cleaved fragments of S. oralis rGAPDH were analyzed with a BIAcore 2000 apparatus (GE Healthcare). The carboxymethylated dextran matrix on the CM5 sensor chip (GE Healthcare) was activated with N-hydroxysuccinimide and N-ethyl-N-[(3-dimethylamino)propyl]carbodiimide hydrochloride (1:1) at a flow rate of 5 μl/min at 37°C. P. gingivalis rFimA (20 μg/ml) in 10 mM sodium acetate buffer (pH 4.5) was immobilized at 2,500 resonance units (RU) on the matrix according to the instructions in the manual from the sensor chip manufacturer (GE Healthcare). Excess active sites on the matrix were blocked with 1 M ethanolamine-HCl, and the matrix was washed with 10 mM NaOH. All materials were dissolved in 10 mM HEPES buffer containing 3 mM EDTA and 0.005% surfactant P20 (GE Healthcare), which also served as a running buffer in the experiments. Each cleaved fragment of S. oralis rGAPDH was injected across the active CM5 surface (which had rFimA) and an empty control CM5 surface at a flow rate of 20 μl/min at 37°C. The binding of each fragment was monitored, and results are expressed as RU in a sensorgram. A result of 1,000 RU corresponded to a change in the surface concentration of 1 ng/mm2 on the sensory chip. At the end of each run, the surface was regenerated by successive injections with 10 mM NaOH. Specific profiles of GAPDH fragment binding to the immobilized rFimA were obtained following subtraction of the control surface signal from the response signal. Analysis of these kinetic parameters was conducted with BIAevaluation 3.1, a software package (GE Healthcare), according to the operator's manual.
Amino acid analysis of S. oralis rGAPDH fragments.
Amino acid sequencing, amino acid analysis, and mass spectrometry analysis of these fragments were performed with a Procise-cLC sequencer (Applied Biosystems), an L-8500A amino acid analyzer (Hitachi), and a Proteomics 4700 mass spectrometer (Applied Biosystems). The instruments were operated according to the manufacturers’ protocols. All fragments were unambiguously identified as the peptides listed in Table 1.
TABLE 1.
Binding activities of S. oxalis ATCC 9811 rGAPDH fragments with P. gingivalis rFimAa
| Proteinase(s) used for digestion or type of fragment |
S. oralis ATCC 9811 GAPDH fragment |
Ka (M−1) | |
|---|---|---|---|
| Amino acid residues | Amino acid sequence | ||
| Lysyl endopeptidase | 163-216 | ALQDNFGVVEGLMTTIHAYTGDQMILDGPHRGGDLRRARAGAANIVPNSTGAAK | 3.54 × 107 |
| Endoproteinase Asp-N | 166-183 | ---DNFGVVEGLMTTIHAYTG--------------------------------- | 4.51 × 107 |
| 184-188 | ---------------------DQMIL---------------------------- | 0 | |
| 189-195 | --------------------------DGPHRGG--------------------- | 0 | |
| 196-216 | ---------------------------------DLRRARAGAANIVPNSTGAAK | 0 | |
| Trypsin | 163-193 | ALQDNFGVVEGLMTTIHAYTGDQMILDGPHR----------------------- | 3.67 × 107 |
| 202-216 | ---------------------------------------AGAANIVPNSTGAAK | 0 | |
| Trypsin and endoproteinase Glu-C | 163-172 | ALQDNFGVVE-------------------------------------------- | 0 |
| 173-193 | ----------GLMTTIHAYTGDQMILDGPHR----------------------- | 0 | |
| Synthetic peptide | 166-183 | ---DNFGVVEGLMTTIHAYTG--------------------------------- | 3.84 × 108 |
| 163-179 | ALQDNFGVVEGLMTTIH------------------------------------- | 3.54 × 106 | |
The binding activities (Ka values) of S. oxalis ATCC 9811 rGAPDH and native GAPDH were determined to be 1.19×107 and 4.34×107 M−1, respectively. Dashes in amino acid sequences represent missing amino acids.
Synthesis of peptides.
Peptides corresponding to amino acid residues 166 to 183 and 163 to 179 of S. oralis ATCC 9811 GAPDH (pep166-183 [DNFGVVEGLMTTIHAYTG] and pep163-179 [ALQDNFGVVEGLMTTIH], respectively) and a control peptide containing Ala substitutions for hydrophobic amino acids of pep166-183 (DNAGAAEGAATTAHAYTG) were obtained from Invitrogen Corp. (Carlsbad, CA). The purity of each synthetic peptide was more than 96% as determined by reverse-phase HPLC.
Biofilm formation by P. gingivalis with streptococci.
Analysis of biofilm formation by P. gingivalis with streptococci was conducted as described previously (22). Briefly, samples of 1.5 × 109 CFU/ml of streptococci were stained with 15 μg of hexidium iodide (HI; Molecular Probes, Carlsbad, CA) for 15 min and washed with mCDM three times. Streptococci (5 × 107 CFU) were inoculated into mCDM in individual chambers coated with human whole saliva and cultured anaerobically in a CultureWell chambered coverglass system (Grace Bio-Labs, Bend, OR) at 37°C for 16 h. P. gingivalis (1.5 × 109 CFU/ml) was stained with 16 μg of fluorescein isothiocyanate (FITC; Molecular Probes) for 30 min and washed with PBS three times; subsequently, samples of 5 × 106 CFU of P. gingivalis were added to the wells exhibiting streptococcal biofilm formation. The mixtures were incubated anaerobically at 37°C for 24 h in the dark on a rotator (Mini-Shaker 3D; BIOSAN, Riga, Latvia). In experiments investigating the inhibition of biofilm formation, P. gingivalis was preincubated with pep166-183 or the control peptide at room temperature for 30 min.
Biofilm analysis by CLSM.
Analysis of biofilm formation was accomplished with a CLSM (LSM510 version 3.2; Carl Zeiss Co., Ltd., Oberkochen, Germany). After the wells were washed with PBS, monospecies or two-species biofilms formed on the bottoms of the wells were observed at a magnification of less than ×40 by using an argon laser (488-nm wavelength) to visualize HI-stained streptococci and an HeNe laser (543-nm wavelength) for visualization of FITC-stained P. gingivalis. Six fields from each CLSM image were selected randomly; biovolumes of streptococci and P. gingivalis were calculated using Imaris software version 5.0.1 (Bitplane AG, Zurich, Switzerland). The level of inhibition of biofilm formation by peptides was calculated as follows: percent inhibition = (1 − A/B) × 100 (A, volume ratio of P. gingivalis to streptococci when the test peptide was added; B, volume ratio of P. gingivalis to streptococci when the control peptide was added). The statistical difference was analyzed by one-way analysis of variance followed by Dunnett's test; moreover, dose dependency was analyzed by utilizing the Jonckheere-Terpstra test with 2006 Excel statistics software (SSRI, Tokyo, Japan).
RESULTS
Identification of P. gingivalis fimbria binding domain in S. oralis GAPDH.
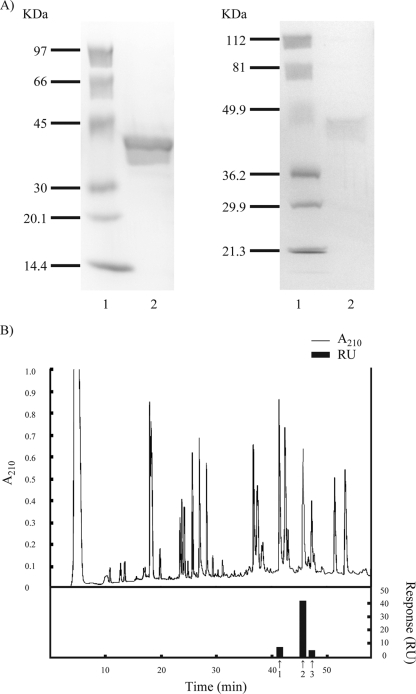
S. oralis rGAPDH was prepared based on the amino acid sequence of S. oralis ATCC 9811 GAPDH. A lysine site was incorporated in the forward primer in order to facilitate the removal of the His tag with lysyl endopeptidase. Purified rGAPDH was subjected to SDS-PAGE; a band displaying a molecular mass of approximately 40 kDa was detected (Fig. 1A, left, lane 2). The purity of rGAPDH was greater than 97% according to SDS-PAGE gel staining with CBB. A Western blot assay demonstrated reaction of the band with anti-penta-His HRP conjugate antibodies, which indicated that the band was His-tagged rGAPDH (Fig. 1A, right, lane 2). Moreover, the identity of the band as S. oralis GAPDH was confirmed by N-terminal amino acid sequencing and mass spectrometry. The binding of S. oralis rGAPDH and native GAPDH to P. gingivalis rFimA was confirmed by the BIAcore instrument; the equilibrium association constants (Ka values) were 1.19 × 107 and 4.34 × 107 M−1, respectively.
FIG. 1.
(A) Results of SDS-PAGE analysis and Western blot assay of purified S. oralis ATCC 9811 rGAPDH. The sample was subjected to SDS-PAGE (5 to 15% gel) and electrotransferred onto a nitrocellulose membrane. After being blocked with Block Ace, the membrane was incubated with penta-His conjugate. Bound antibodies were visualized by employing an HRP conjugate substrate kit. (Left) SDS-PAGE gel with CBB staining; (right) Western blot. Lanes: 1, molecular mass standard proteins; 2, S. oralis ATCC 9811 rGAPDH. (B) HPLC profile and binding activity of lysyl endopeptidase-digested S. oralis rGAPDH with P. gingivalis rFimA. S. oralis rGAPDH was digested with lysyl endopeptidase and subjected to HPLC involving a Symmetry300 C18 column. The elution of rGAPDH fragments was effected with a linear gradient of 0 to 60% acetonitrile at a flow rate of 1 ml/min. Fractionated fragments were collected manually by monitoring the absorbance at 210 nm. The binding activity of each peak toward P. gingivalis rFimA was measured by a BIAcore apparatus. 1, absorbance peak eluted at 41.5 min; 2, absorbance peak eluted at 46 min; 3, absorbance peak eluted at 47.5 min.
To identify the P. gingivalis fimbria binding domain in S. oralis GAPDH, rGAPDH was digested with lysyl endopeptidase and subjected to reverse-phase HPLC employing a Symmetry300 C18 column. The binding activity of each peak toward P. gingivalis rFimA was measured by the BIAcore apparatus. As shown in Fig. 1B, the strongest binding activity toward P. gingivalis rFimA was observed with the absorbance peak eluted at 46 min. The peak was resubjected to HPLC utilizing an Xterra phenyl column and was separated into 10 fractions (data not shown). The binding activity of each fraction was also measured by the BIAcore instrument. The fraction exhibiting the strongest binding activity toward rFimA was subjected to amino acid sequencing, amino acid analysis, and mass spectrometric analysis. The amino acid sequence of this fragment was ALQDNFGVVEGLMTTIHAYTGDQMILDGPHRGGDLRRARAGAANIVPNSTGAAK, which corresponds to amino acid residues 163 to 216 of S. oralis ATCC 9811 GAPDH. The Ka of the fragment according to BIAcore analysis was 3.54 × 107 M−1 (Table 1). The fragment was further digested with endopeptidase Asp-N; four fragments were isolated by HPLC. A fragment corresponding to amino acid residues 166 to 183 (DNFGVVEGLMTTIHAYTG) exhibited strong binding to rFimA (Ka = 4.51 × 107 M−1), whereas three other fragments (DQMIL, DGPHRGG, and DLRRARAGAANIVPNSTGAAK) demonstrated no binding activity toward rFimA as detected by the BIAcore system (Table 1). The fragment corresponding to amino acid residues 163 to 216 of S. oralis ATCC 9811 GAPDH was also digested with trypsin; subsequently, two fragments were isolated by HPLC. The fragment corresponding to amino acid residues 163 to 193 (ALQDNFGVVEGLMTTIHAYTGDQMILDGPHR) exhibited strong binding activity toward rFimA (Ka = 3.67 × 107 M−1); in contrast, the fragment corresponding to amino acid residues 202 to 216 (AGAANIVPNSTGAAK) displayed no binding activity toward rFimA. The fragment corresponding to amino acid residues 163 to 193 (ALQDNFGVVEGLMTTIHAYTGDQMILDGPHR) was further digested with endopeptidase Glu-C. Two fragments (ALQDNFGVVE and GLMTTIHAYTGDQMILDGPHR) were isolated by HPLC; however, neither peptide demonstrated binding activity toward rFimA (Table 1). These results suggest that a binding domain for P. gingivalis FimA fimbriae may exist within amino acid residues 166 to 183 of S. oralis ATCC 9811 GAPDH.
The peptide corresponding to amino acid residues 166 to 183 of S. oralis ATCC 9811 GAPDH (pep166-183; DNFGVVEGLMTTIHAYTG) was synthesized; subsequently, the binding specificity of the peptide for P. gingivalis rFimA was characterized. BIAcore characterization revealed that the resonance response reflecting the P. gingivalis rFimA-pep166-183 interaction occurred in an analyte concentration-dependent manner. The Ka value for the interaction was 3.84 × 108 M−1, demonstrating high-affinity binding (Table 1).
Inhibitory effect of pep166-183 on biofilm formation by P. gingivalis ATCC 33277 with S. oralis ATCC 9811.
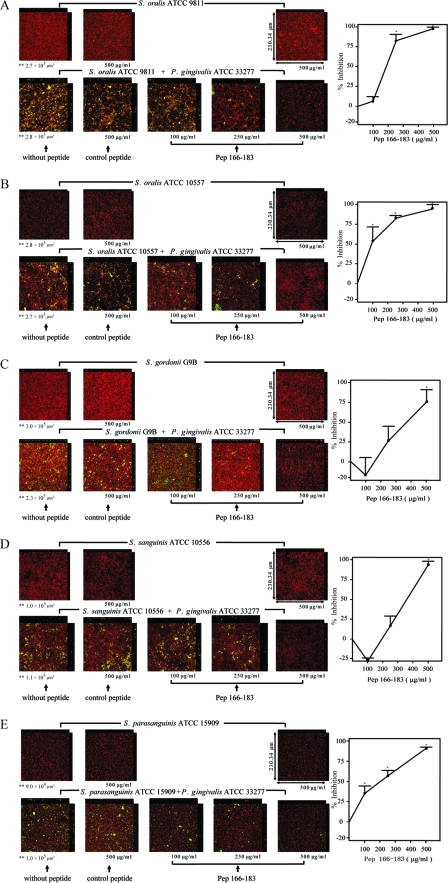
The inhibitory effect of pep166-183 on biofilm formation by P. gingivalis ATCC 33277 and S. oralis ATCC 9811 was examined. As shown in Fig. 2A, pep166-183 inhibited biofilm formation by P. gingivalis ATCC 33277 and S. oralis ATCC 9811 in a dose-dependent manner (P value for trend, <0.001). P. gingivalis ATCC 33277-S. oralis ATCC 9811 biofilm formation was significantly inhibited by pep166-183 at 250 and 500 μg/ml (P < 0.001); the percentages of inhibition were approximately 83 and 98%, respectively. A control peptide in which hydrophobic amino acid residues were replaced with Ala (DNAGAAEGAATTAHAYTG) was synthesized. The control peptide at 500 μg/ml exerted no effect on either P. gingivalis ATCC 33277-S. oralis ATCC 9811 biofilm formation or S. oralis ATCC 9811 monospecies biofilm formation (Fig. 2A). Pep166-183 at 500 μg/ml exerted no effect on S. oralis ATCC 9811 monospecies biofilm formation.
FIG. 2.
Inhibitory effects of pep166-183 on biofilm formation by P. gingivalis ATCC 33277 with various streptococci as determined by CLSM. HI-stained streptococci (5 × 107 CFU/well; red) were inoculated into individual chambers coated with human whole saliva and cultured anaerobically at 37°C for 16 h. FITC-stained P. gingivalis ATCC 33277 (5 × 106 CFU/well; green) was added to the wells in which streptococcal biofilm was observed. The mixtures were incubated anaerobically at 37°C for 24 h and analyzed by CLSM. In experiments evaluating the inhibition of biofilm formation, P. gingivalis ATCC 33277 was preincubated aerobically with pep166-183 or the control peptide at room temperature for 30 min. Magnification, ×40. The following streptococci were tested: S. oralis ATCC 9811 (A), S. oralis ATCC 10557 (B), S. gordonii G9B (C), S. sanguinis ATCC 10556 (D), and S. parasanguinis ATCC 15909 (E). Representative photographs are shown. In each panel, the upper section displays the monospecies streptococcal biofilm, and the lower section displays the biofilm formed by various streptococci and P. gingivalis ATCC 33277. Data are the means and standard errors of results for six fields in CLSM images. *, P < 0.001 for comparison to results obtained for the control peptide; **, volume of streptococci.
Inhibitory effect of pep166-183 on biofilm formation by P. gingivalis ATCC 33277 with various streptococci.
Next, the inhibitory effect of pep166-183 on biofilm formation by P. gingivalis ATCC 33277 with various streptococci was examined by the same method applied in the inhibitory experiment involving P. gingivalis ATCC 33277 and S. oralis ATCC 9811. Pep166-183 inhibited biofilm formation by P. gingivalis ATCC 33277 with S. oralis ATCC 10557, S. gordonii G9B, S. sanguinis ATCC 10556, or S. parasanguinis ATCC 15909 in a dose-dependent manner (P value for trend, <0.001) (Fig. 2B to E). Pep166-183 significantly inhibited P. gingivalis ATCC 33277-S. oralis ATCC 10557 biofilm formation at 100, 250, and 500 μg/ml (P < 0.001); percentages of inhibition were approximately 69, 83, and 94%, respectively (Fig. 2B). Pep166-183 also inhibited P. gingivalis ATCC 33277-S. gordonii G9B biofilm formation; however, significant inhibition was observed solely at 500 μg/ml (P < 0.001), and the percent inhibition was approximately 75%. Biofilm formation by P. gingivalis ATCC 33277-S. sanguinis ATCC 10556 (Fig. 2D) and P. gingivalis ATCC 33277-S. parasanguinis ATCC 15909 (Fig. 2E) was significantly inhibited by pep166-183 at 500 μg/ml (P < 0.001); the percent inhibition was greater than 90% in both cases. Pep166-183 significantly inhibited P. gingivalis ATCC 33277-S. parasanguinis ATCC 15909 biofilm formation, even at a concentration of 100 μg/ml (P < 0.001) (Fig. 2E). Neither the tested streptococcus-P gingivalis ATCC 33277 biofilm formation nor monospecies streptococcal biofilm formation was affected by the control peptide at 500 μg/ml (Fig. 2B to E). Pep166-183 at 500 μg/ml exerted no inhibitory effect on monospecies biofilm formation by these streptococci (Fig. 2B to E).
Inhibitory effect of pep166-183 on biofilm formation by S. oralis ATCC 9811 and P. gingivalis strains with different types of fimbriae.
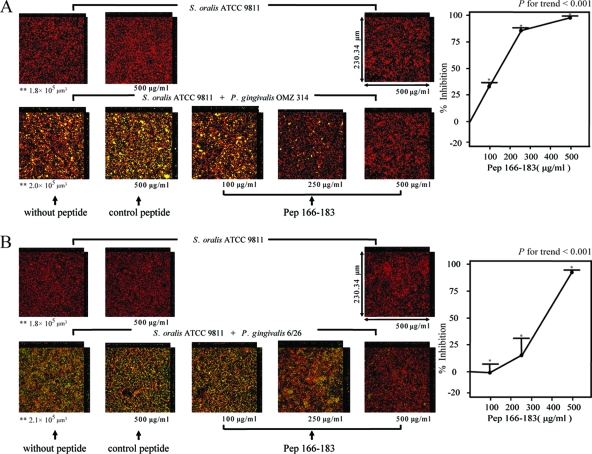
The inhibitory effect of pep166-183 on biofilm formation by S. oralis ATCC 9811 and P. gingivalis in the presence of different types of fimbriae was examined. P. gingivalis ATCC 33277 (type I fimbriae), OMZ 314 (type II), 6/26 (type III), W 50 (type IV), and HNA 99 (type V) were selected as representative strains possessing different types of fimbriae. Pep166-183 inhibited biofilm formation by P. gingivalis OMZ 314 and S. oralis ATCC 9811 in a dose-dependent manner (P value for trend, <0.001) (Fig. 3A). The volume of biofilm formation by P. gingivalis 6/26 with S. oralis ATCC 9811 was less than that by P. gingivalis ATCC 33277 with S. oralis ATCC 9811; however, dose-dependent inhibition by pep166-183 was observed (P value for trend, <0.001) (Fig. 3B). Although P. gingivalis W 50 and HNA 99 exhibited biofilm formation with S. oralis ATCC 9811, the volume of biofilm formation was much less than that by P. gingivalis ATCC 33277 with S. oralis ATCC 9811; therefore, the inhibitory effects of pep166-183 on P. gingivalis W 50-S. oralis ATCC 9811 and P. gingivalis HNA 99-S. oralis ATCC 9811 biofilm formation were not determined (data not shown).
FIG. 3.
Inhibitory effects of pep166-183 on biofilm formation by S. oralis ATCC 9811 and P. gingivalis strains with different types of fimbriae as determined by CLSM. HI-stained S. oralis ATCC 9811 (5 × 107 CFU/well; red) was inoculated into individual chambers coated with human whole saliva and cultured anaerobically at 37°C for 16 h. FITC-stained P. gingivalis OMZ 314 cells (A) and 6/26 cells (B) (5 × 106 CFU/well; green) were added to the wells with S. oralis ATCC 9811 biofilm. The mixtures were incubated anaerobically at 37°C for 24 h and analyzed by CLSM. In experiments evaluating the inhibition of biofilm formation, P. gingivalis was preincubated with pep166-183 or control peptide aerobically at room temperature for 30 min. In each panel, the upper section displays the S. oralis ATCC 9811 monospecies biofilm and the lower section exhibits the biofilm formed by S. oralis ATCC 9811 and P. gingivalis. Magnification, ×40. Representative photographs are shown. Data are the means and standard errors of results for six fields in CLSM images. *, P < 0.001 for comparison to results obtained for the control peptide; **, volume of streptococci.
DISCUSSION
This study firstly identified the P. gingivalis FimA fimbria binding domain in S. oralis ATCC 9811 GAPDH. The domain corresponding to amino acid residues 166 to 183 (DNFGVVEGLMTTIHAYTG) exhibited the strongest binding activity toward rFimA (Ka = 4.51 × 107 M−1), indicating that the P. gingivalis FimA fimbria binding domain exists in this region. Consequently, pep166-183 was synthesized, and the binding of the peptide to rFimA was confirmed by BIAcore analysis (Ka = 3.84 × 108 M−1). A peptide corresponding to amino acid residues 163 to 179 (ALQDNFGVVEGLMTTIH) of S. oralis ATCC 9811 GAPDH was also synthesized; subsequently, binding activity toward rFimA was assessed. This peptide also bound to rFimA; however, the Ka was lower (3.54 × 106 M−1) than that of pep166-183 (3.84 × 108 M−1); this result indicated that the C-terminal amino acid residues of pep166-183 (AYTG) might be important for binding to rFimA. Nagy et al. (30) showed that the 43 amino acid residues of the N terminus of human GAPDH constitute the RNA binding domain; furthermore, Jin et al. (18) reported that group A streptococcal surface dehydrogenase possesses two binding sites for human pharyngeal cells, a C-terminal strong binding site and an N-terminal weak binding site. However, no report regarding GAPDH binding site involvement in the interbacterial interaction appears in the literature. The identification of the domain of S. oralis GAPDH which interacts with P. gingivalis FimA fimbriae and the finding that the synthetic peptide corresponding to the binding domain inhibits biofilm formation by various streptococci and P. gingivalis strains with different types of fimbriae are significant points of the present investigation.
Employing combinatorial libraries involving substitutions for several active-site amino acid residues in a region of the S. gordonii SspB polypeptide (residues 1167 to 1193) designated BAR, Daep et al. (7) reported adhesion to P. gingivalis minor fimbrial antigen Mfa1 and possible contributions of electrostatic and hydrophobic interactions to Mfa1-BAR binding. In the present study, the control peptide in which the hydrophobic amino acid residues were replaced with Ala exerted little inhibitory effect on P. gingivalis biofilm formation with streptococci; this finding indicates that the hydrophobic amino acid residues of pep166-183 are important with respect to biofilm formation. On the basis of data from Predict Protein (http://www.predictprotein.org/), pep166-183 is characterized by a β-sheet, whereas the control peptide is characterized by an α-sheet. Changes in the electric charge and conformation of the peptide as consequences of the replacement of hydrophobic amino acid residues with Ala may be the reason why the control peptide exerted no inhibitory effect on biofilm formation.
CLSM observation revealed pep166-183 inhibition of biofilm formation by P. gingivalis ATCC 33277 and S. oralis ATCC 9811 in a dose-dependent manner. Previously, Maeda et al. (27) reported a correlation between the streptococcal cell surface GAPDH activity and coaggregation activity with P. gingivalis; additionally, S. oralis ATCC 10557, S. gordonii G9B, S. sanguinis ATCC 10556, and S. parasanguinis ATCC 15909 exhibited high levels of cell surface GAPDH activity and coaggregation activity with P. gingivalis. The GAPDH amino acid sequences from these streptococci displayed identities of greater than 97% to that from S. oralis ATCC 9811; moreover, the amino acid sequences corresponding to residues 166 to 183 of S. oralis ATCC 9811 GAPDH were completely identical in these streptococci. Therefore, we hypothesized that pep166-183 might inhibit the interaction of these streptococci with P. gingivalis and repress interbacterial biofilm formation. As expected, pep166-183 inhibited biofilm formation by P. gingivalis ATCC 33277 with other tested streptococci (Fig. 2B to E). Given that the degree of homology among GAPDHs from oral streptococci was very high, the possibility that GAPDHs from other oral streptococci such as S. mutans and Streptococcus criceti might interact with P. gingivalis FimA fimbriae existed; however, cell surface GAPDH activities and coaggregation activities of S. mutans and S. criceti with P. gingivalis were very low (27). The amount of GAPDH expressed on the surfaces of streptococcal cells may be related to the degree of attachment to P. gingivalis.
P. gingivalis FimA fimbriae are classified into six types (I to V and Ib) based on the diversity of fimA genes encoding FimA (5). Amano et al. (4) reported that a majority of periodontitis patients carry type II fimA organisms. Based on the result that pep166-183 inhibited biofilm formation by S. oralis ATCC 9811 and P. gingivalis OMZ 314 (type II), as well as P. gingivalis ATCC 33277, pep166-183 may be beneficial as a potent inhibitor of P. gingivalis colonization with respect to the prevention of periodontitis.
In conclusion, this study demonstrated that the P. gingivalis FimA fimbria binding domain in S. oralis ATCC 9811 GAPDH exists within amino acid residues 166 to 183; furthermore, the present findings indicated that the hydrophobic amino acid residues play an important role in the binding interaction. The peptide corresponding to the binding domain for P. gingivalis FimA fimbriae inhibited biofilm formation by various streptococci and P. gingivalis strains with different types of FimA fimbriae in a dose-dependent manner. These results indicated that pep166-183 may play an important role in P. gingivalis biofilm formation; moreover, this peptide may be applicable as an inhibitor of P. gingivalis colonization.
Acknowledgments
This work was supported by grants-in-aid for scientific research (B; 17390564 and 20390534) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank Mitsuko Nakatani and Yoko Takada for the expert technical assistance and Kenji Kangawa of National Cardiovascular Center Research Institute for helpful advice.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 8 September 2009.
REFERENCES
- 1.Abe, N., A. Baba, R. Takii, K. Nakayama, A. Kamaguchi, Y. Shibata, Y. Abiko, K. Okamoto, T. Kadowaki, and K. Yamamoto. 2004. Roles of Arg- and Lys-gingipains in coaggregation of Porphyromonas gingivalis: identification of its responsible molecules in translation products of rgpA, kgp, and hagA genes. Biol. Chem. 385:1041-1047. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., H. T. Sojar, J. Y. Lee, A. Sharma, M. J. Levine, and R. J. Genco. 1994. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect. Immun. 62:3372-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amano, A., T. Fujiwara, H. Nagata, M. Kuboniwa, A. Sharma, H. T. Sojar, R. J. Genco, and S. Shizukuishi. 1997. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J. Dent. Res. 76:852-857. [DOI] [PubMed] [Google Scholar]
- 4.Amano, A., M. Kuboniwa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 79:1664-1668. [DOI] [PubMed] [Google Scholar]
- 5.Amano, A., I. Nakagawa, N. Okahashi, and N. Hamada. 2004. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontal Res. 39:136-142. [DOI] [PubMed] [Google Scholar]
- 6.Chung, W. O., D. R. Demuth, and R. J. Lamont. 2000. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect. Immun. 68:6758-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daep, C. A., D. M. James, R. J. Lamont, and D. R. Demuth. 2006. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect. Immun. 74:5756-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Costa, S. S., and M. D. Boyle. 2000. Interaction of group A streptococci with human plasmin(ogen) under physiological conditions. Methods 21:165-177. [DOI] [PubMed] [Google Scholar]
- 9.Egea, L., L. Aquilera, R. Giménez, M. A. Sorolla, J. Aguilar, J. Badía, and L. Baldoma. 2007. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int. J. Biochem. Cell Biol. 39:1190-1203. [DOI] [PubMed] [Google Scholar]
- 10.Ellen, R. P., and D. A. Grove. 1989. Bacteroides gingivalis vesicles bind to and aggregate Actinomyces viscosus. Infect. Immun. 57:1618-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figge, R. M., M. Schubert, H. Brinkmann, and R. Cerif. 1999. Glyceraldehyde-3-phosphate dehydrogenase gene diversity in eubacteria and eukaryotes: evidence for intra- and inter-kingdom gene transfer. Mol. Biol. Evol. 16:429-440. [DOI] [PubMed] [Google Scholar]
- 12.Goulbourne, P. A., and R. P. Ellen. 1991. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J. Bacteriol. 173:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gozalbo, D., I. Gil-Navarro, I. Azorín, J. Renau-Piqueras, J. P. Martínez, and M. L. Gil. 1998. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 66:2052-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanazawa, S., K. Hirose, Y. Ohmori, S. Amano, and S. Kitano. 1988. Bacteroides gingivalis fimbriae stimulate production of thymocyte-activating factor by human gingival fibroblasts. Infect. Immun. 56:272-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiratsuka, K., Y. Abiko, M. Hayakawa, T. Ito, H. Sasahara, and H. Takiguchi. 1992. Role of Porphyromonas gingivalis 40-kDa outer membrane protein in the aggregation of P. gingivalis vesicles and Actinomyces viscosus. Arch. Oral Biol. 37:717-724. [DOI] [PubMed] [Google Scholar]
- 16.Hoelzle, L. E., K. Hoelzle, M. Helbling, H. Aupperle, H. A. Schoon, M. Ritzmann, K. Heinritzi, K. M. Felder, and M. M. Wittenbrink. 2007. MSG1, a surface-localised protein of Mycoplasma suis is involved in the adhesion to erythrocytes. Microbes Infect. 9:466-474. [DOI] [PubMed] [Google Scholar]
- 17.Isogai, H., E. Isogai, F. Yoshimura, T. Suzuki, W. Kagota, and K. Takano. 1988. Specific inhibition of adherence of an oral strain of Bacteroides gingivalis 381 to epithelial cells by monoclonal antibodies against the bacterial fimbriae. Arch. Oral Biol. 33:479-485. [DOI] [PubMed] [Google Scholar]
- 18.Jin, H., Y. P. Song, G. Boel, J. Kochar, and V. Pancholi. 2005. Group A streptococcal surface GAPDH, SDH, recognizes uPAR/CD87 as its receptor on the human pharyngeal cell and mediates bacterial adherence to host cells. J. Mol. Biol. 350:27-41. [DOI] [PubMed] [Google Scholar]
- 19.Kamaguchi, A., H. Baba, M. Hoshi, and K. Inomata. 1995. Effect of Porphyromonas gingivalis ATCC 33277 vesicle on adherence of Streptococcus mutans OMZ 70 to the experimental pellicle. Microbiol. Immunol. 39:521-524. [DOI] [PubMed] [Google Scholar]
- 20.Kinoshita, H., H. Uchida, Y. Kawai, T. Kawasaki, N. Wakahara, H. Matsuo, M. Watanabe, H. Kitazawa, S. Ohnuma, K. Miura, A. Horii, and T. Saito. 2008. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase(GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 104:1667-1674. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Richard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000. 42:47-79. [DOI] [PubMed] [Google Scholar]
- 22.Kuboniwa, M., G. D. Tribble, C. E. James, A. O. Kilic, L. Tao, M. C. Herzberg, S. Shizukuishi, and R. J. Lamont. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60:121-139. [DOI] [PubMed] [Google Scholar]
- 23.Lamont, R. J., C. A. Bevan, S. Gil, R. E. Persson, and B. Rosan. 1993. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol. Immunol. 8:272-276. [DOI] [PubMed] [Google Scholar]
- 24.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda, K., H. Nagata, Y. Yamamoto, M. Tanaka, J. Tanaka, N. Minamino, and S. Shizukuishi. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda, K., H. Nagata, M. Kuboniwa, K. Kataoka, N. Nishida, M. Tanaka, and S. Shizukuishi. 2004. Characterization of binding of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase to Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:5475-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda, K., H. Nagata, A. Nonaka, K. Kataoka, M. Tanaka, and S. Shizukuishi. 2004. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes Infect. 6:1163-1170. [DOI] [PubMed] [Google Scholar]
- 28.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata, H., A. Sharma, H. T. Sojar, A. Amano, M. J. Levine, and R. J. Genco. 1997. Role of the carboxyl-terminal region of Porphyromonas gingivalis fimbrillin in binding to salivary proteins. Infect. Immun. 65:422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagy, E., T. Henics, M. Eckert, A. Miseta, R. N. Lightowlers, and M. Kellermayer. 2000. Identification of the NAD+-binding fold of glyceraldehyde-3-phosphate dehydrogenase as a novel RNA-binding domain. Biochem. Biophys. Res. Commun. 275:253-260. [DOI] [PubMed] [Google Scholar]
- 31.Okuda, K. 1993. Attachment mechanisms and colonization, p. 139-157. In H. N. Shah, D. Mayland, and R. J. Genco (ed.), Biology of species Porphyromonas gingivalis. CRC Press Inc., Boca Raton, FL.
- 32.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 34.Sirover, M. A. 1999. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim. Biophys. Acta 1432:159-184. [DOI] [PubMed] [Google Scholar]
- 35.Winram, S. B., and R. Lottenberg. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 142:2311-2320. [DOI] [PubMed] [Google Scholar]