Abstract
Respiratory complex I (NADH:ubiquinone oxidoreductase) is a large mitochondrial inner membrane enzyme consisting of 45 subunits and 8 iron-sulfur (Fe/S) clusters. While complex I dysfunction is the most common reason for mitochondrial diseases, the assembly of complex I and its Fe/S cofactors remains elusive. Here, we identify the human mitochondrial P-loop NTPase, designated huInd1, that is critically required for the assembly of complex I. huInd1 can bind an Fe/S cluster via a conserved CXXC motif in a labile fashion. Knockdown of huInd1 in HeLa cells by RNA interference technology led to strong decreases in complex I protein and activity levels, remodeling of respiratory supercomplexes, and alteration of mitochondrial morphology. In addition, huInd1 depletion resulted in massive decreases in several subunits (NDUFS1, NDUFV1, NDUFS3, and NDUFA13) of the peripheral arm of complex I, with the concomitant appearance of a 450-kDa subcomplex representing part of the membrane arm. By a novel radiolabeling technique, the amount of iron associated with complex I was also shown to reflect the dependence of this enzyme on huInd1 for assembly. Together, these data identify huInd1 as a new assembly factor for human respiratory complex I with a possible role in the delivery of one or more Fe/S clusters to complex I subunits.
In the mitochondria of mammalian cells, energy is harnessed from NADH through a series of electron transfer steps, involving NADH:ubiquinone oxidoreductase (complex I) (for a review, see Brandt [7]). This largest component of the respiratory chain comprises 45 protein subunits, 7 of which are encoded by the human mitochondrial genome. Additionally, complex I contains stably bound cofactors, including flavin mononucleotide and eight iron-sulfur clusters (ISC). Assembly of this highly composite enzyme is not well understood so far but presumably involves several ancillary proteins (30, 71). Deficiencies in complex I activity are the most common cause of diseases of mitochondrial etiology (17, 28, 56). As approximately 50% of patients with complex I deficiencies have no mutations in the individual subunits of complex I, it seems likely that a number of assembly factors are relevant for disease (27). Indeed, two assembly proteins, NDUFAF2/B17.2L (38) and NDUFAF1/CIA30 (14, 27, 69), have already been characterized, and mutations in the corresponding genes have been identified in patients with complex I disease. More recently described complex I assembly factors with (potential) relevance for disease are C6ORF66 (49, 50), C8ORF38 (43), Ecsit (70), C20ORF7 (63), and apoptosis-inducing factor (4, 67).
Besides fulfilling roles in the molecular chaperoning of complex I protein subunits, assembly factors are also expected to be necessary for the insertion of prosthetic groups into either monomeric complex I subunits, complex I subcomplexes, or the fully assembled apoenzyme. Fe/S clusters in particular are known to require specialized and dedicated molecular machinery for their synthesis and delivery to apoproteins (for a review, see Lill and Mühlenhoff [33]). The biogenesis components and the molecular mechanisms of Fe/S protein assembly were originally elucidated for bacteria and Saccharomyces cerevisiae. In yeast mitochondria, 15 components of the mitochondrial ISC assembly machinery are known to catalyze Fe/S protein maturation. In brief, sulfide is generated via the desulfuration of cysteine by the cysteine desulfurase complex Nfs1-Isd11 and then combined with ferrous iron on the scaffold protein Isu1. The Isu1-bound, labile Fe/S cluster is transferred to apoproteins, aided by dedicated chaperone proteins and a monothiol glutaredoxin. Generation of extramitochondrial Fe/S proteins requires an as yet unidentified product of the ISC biogenesis pathway. This compound is exported from mitochondria by the ISC export machinery, including the ABC transporter Atm1 (ABCB7 in humans). Construction of cytosolic and nuclear Fe/S proteins is then accomplished by components of the cytosolic iron-sulfur protein assembly (CIA) machinery. Analogous to Isu1 of the mitochondrial ISC pathway, the two cytosolic P-loop NTPase proteins, Nbp35 and Cfd1, serve as scaffolds for the assembly of Fe/S clusters (24, 37, 48, 61). The transfer of the assembled Fe/S clusters to apoproteins involves the essential Fe/S protein Nar1 and the WD40 repeat protein Cia1 (2, 3). Another recently discovered member of the CIA machinery is the essential Fe/S protein Dre2 (79). All the ISC and CIA proteins are highly conserved among eukaryotes, from yeast to humans, suggesting similar mechanisms of Fe/S protein assembly in lower and higher eukaryotes. Findings from studies of several mammalian ISC, ISC export, and CIA components have supported this idea (5, 10, 44, 58, 60, 61, 65). Small amounts of several ISC assembly machinery components have been detected in the cytosol and nuclei of mammalian cells (47). The extramitochondrial form of human Isu1 was shown to assist Fe/S protein regeneration and/or repair.
Interestingly, an additional mitochondrial protein with sequence similarity to cytosolic Nbp35/Cfd1 has been identified in eukaryotes, and the form of the protein in the obligate respiratory yeast Yarrowia lipolytica has been characterized previously (8). This protein, termed Ind1, for iron-sulfur protein required for NADH dehydrogenase, contains the conserved nucleotide binding domain characteristic of this subclass of P-loop NTPases (known as the Mrp/NBP35 subclass [31]). In addition, Ind1 harbors a C-terminal domain including the conserved CXXC motif that is believed to provide the ligands for a transiently bound Fe/S cluster (37). We recently demonstrated that the IND1 gene is present almost exclusively in the genomes of species that retain functional complex I, including humans (8). Deletion of the IND1 gene in Y. lipolytica results in a specific decrease in the levels and therefore the activity of complex I. In the present study, we used RNA interference (RNAi)-mediated depletion of human Ind1 (huInd1) to demonstrate the requirement of this mitochondrial Fe/S protein for complex I assembly and hence activity. This demonstration shows the functional conservation of Ind1 among eukaryotes from yeast to humans. In addition to the previous findings with Y. lipolytica, we observed ultrastructural changes in huInd1-depleted mitochondria, including the loss of crista membranes, massive remodeling of the respiratory supercomplexes, and increased lactate production, all likely resulting from the respiratory chain defect in huInd1-depleted cells. A novel subcomplex consisting of parts of the membrane arm of complex I accumulates in the absence of huInd1. Importantly, the levels of several subunit proteins, NDUFS1, NDUFV1, NDUFS3, and NDUFA13, in the complex I peripheral arm hosting the eight Fe/S clusters were dramatically decreased in huInd1-deficient cells, indicating that this part of complex I was not assembled. As the first two proteins of this list directly coordinate Fe/S clusters and huInd1 binds a labile Fe/S cluster, the functional role of huInd1 may be in Fe/S cluster incorporation into complex I subunits, reminiscent of the function of Cfd1 and Nbp35.
MATERIALS AND METHODS
Reagents and cell lines.
Antibodies against recombinant huInd1 were raised in rabbits (22). Antibodies against purified bovine complex I, complex III dimer (complex III2), and ATP synthase α and β subunits; the chemical 2-decyl-4-quinazolinyl amine (DQA); and bovine heart mitochondria were generous gifts of H. Schägger (Frankfurt, Germany). The anti-complex I antibody was raised in rabbits by Schägger's group using purified bovine complex I as described by Okun et al. (40). Abnova antibodies against NDUFS1 (MaxPab polyclonal) and NDUFV1 (monoclonal) were purchased from Biozol (Eching, Germany); monoclonal anti-NDUFA9, anti-NDUFB6, anti-NDUFA13, and anti-NDUFS3 antibodies were purchased from MitoSciences (Eugene, OR). H. Puccio kindly supplied us with antiserum against glutamine phosphoribosylpyrophosphate amidotransferase (GPAT). Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G antibodies were from Bio-Rad. Small interfering RNAs (siRNAs) with predesigned sequences were purchased from Ambion. Phusion DNA polymerase (Finnzymes, Espoo, Finland) was exclusively used for PCR amplification. INSTA-blot, a polyvinylidene difluoride (PVDF) membrane containing blotted proteins from various human tissues, was purchased from Merck Biosciences. All cell culture reagents were purchased from PAA Laboratories (Cölbe, Germany). All other reagents were obtained from Sigma, unless otherwise noted.
HeLa cells were maintained in Dulbecco's modified Eagle medium containing 4.5 g/liter glucose and supplemented with 7.5% fetal calf serum, 1 mM glutamine, and 1% penicillin-streptomycin.
Plasmids.
All relevant oligonucleotide sequences are listed in Table 1. A cDNA clone of the huInd1 gene (NUBPL; RefSeq accession no. NM_025152) in pCMV-Sport6 was obtained from Geneservice. The protein encoded by this vector did not contain the first 7 amino acid residues. We therefore employed PCR amplification (with oligonucleotide set 1) to add the appropriate nucleotides, along with BglII and SalI restriction sites for subcloning of the huInd1 gene into pEGFP-N3 (Clontech) to generate a mammalian expression vector (pEGFP-IND1) encoding full-length huInd1 fused to enhanced green fluorescent protein (EGFP) at the C terminus. For the expression of N-terminally His-tagged recombinant huInd1 in Escherichia coli, we amplified a portion of the huInd1 gene from the commercially obtained vector by PCR using oligonucleotide set 2. The construct lacks the first 51 codons (i.e., the putative mitochondrial targeting sequence) and contains BamHI and PstI restriction sites for cloning into pETDuet-1 (Novagen). Constructs for vector-based RNAi were generated by using oligonucleotides 3 to 5, based on the three short hairpin RNA sequences acquired from Ambion, and by cloning these oligonucleotides into pSUPERIOR.puro (OligoEngine) using BglII and HindIII restriction sites. The first and third RNA targets (oligonucleotides 3 and 5) elicited strong knockdown of huInd1 (data not shown). The sequence of oligonucleotide 3 was used for all subsequent experiments (and all of the results presented in this paper). A rescue construct that renders transcripts resistant to RNAi was generated by a single PCR amplification protocol using overlapping, mismatched primers (oligonucleotide set 6) to linearly amplify the entire pEGFP-IND1 vector with silent mutations in the RNAi-targeted regions, yielding vector smIND1 (80). The latter construct was further mutated using oligonucleotide set 7 to yield an RNAi-resistant transcript (vector smIND1CC) that would generate a version of huInd1 with both of the conserved cysteine residues, at positions 244 and 247, changed to alanine residues. For microscopic subcellular localization experiments, we cotransfected cells with a vector that encodes a stable version of Dicosoma sp. red fluorescent protein preceded by the presequence of subunit VIII of cytochrome c oxidase (pDsRed2-Mito; Clontech).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Purpose(s) or description |
|---|---|---|
| Set 1 | To add region encoding first 7 amino acid residues, remove stop codon, and add BglII and SalI restriction sites for cloning into pEGFP-N3 | |
| Forward | TATAGATCTACCATGGGGATTTGGCAGCGTCTGCTGCTTTTTGGTGGGGTGTCG | |
| Reverse | TATGTCGACTTCTGAAGGTGATGGCAATCTTCTTAC | |
| Set 2 | To remove putative mitochondrial localization sequence and add BamHI and PstI restriction sites for cloning into pETDuet-1 | |
| Forward | ATGGTAGGATCCGTCCCGAGGACTTCCAAAGC | |
| Reverse | ATGGTACTGCAGTCATTCTGAAGGTGATGGC | |
| 3 | 5′-GCAGAAACCGATAGAAGGT-3′ | huInd1 siRNA 1 |
| 4 | 5′-GGCCTTATGGTAATGTCGG-3′ | huInd1 siRNA 2 |
| 5 | 5′-GGTTCACAAAACTTTGATG-3′ | huInd1 siRNA 3 |
| Set 6 | To generate smIND1 by site-directed mutagenesis | |
| Forward | CAAAaCAaAAgCCaATtGAgGGaGTTAAACAAGTTATAGTTGTGGCT | |
| Reverse | TAACtCCcTCaATtGGcTTtTGtTTTGGCAGTCCTCGGGACATGATT | |
| Set 7 | To generate smIND1CC by site-directed mutagenesis | |
| Forward | CCAGgcgCCAAAAgcTAAACACAAAACTCATATTTTTGG | |
| Reverse | TGTTTAgcTTTTGGcgcCTGGAAAACACTCATGTTTTGG |
Bases corresponding to mutations are in lowercase.
Purification and chemical reconstitution of recombinant huInd1.
huInd1 was produced in E. coli according to a procedure described previously (37) and purified anaerobically by nickel-nitrilotriacetic acid affinity chromatography. The protein was immediately desalted on a PD-10 column and subsequently chemically reconstituted (37). UV-visible spectra were recorded using a V-550 spectrophotometer (Jasco Instruments).
Transfection.
After human HeLa cells were harvested by trypsinization and washed in electroporation buffer (21 mM HEPES, pH 7.15, 137 mM NaCl, 5 mM KCl, 0.7 mM sodium phosphate, 6 mM glucose), 4 × 106 cells (for 75-cm2 culture flasks) or 8 × 106 cells (for 175-cm2 flasks) were resuspended in 550 μl electroporation buffer supplemented with 25 μg RNAi vector and 5 μg rescue vector. Transfection was performed using an EASYJect+ electroporator (Equibio) set to 250 V/1,500 μF and a pulse of 25 to 30 ms. After electroporation, cells were immediately transferred into growth medium supplemented with 25 to 30% HeLa cell-conditioned medium. The cells were retransfected every third day and assayed 3 days after the third transfection (i.e., on day 9) unless otherwise indicated. Since the rescue vector encodes a huInd1-EGFP fusion protein, we were able to monitor the transfection efficiency by flow cytometry with a FACScan instrument (Becton Dickinson) and found it to be consistently around 90% (60).
Microscopy.
After transfection, cells were seeded onto coverslips in 24-well plates and incubated under standard culture conditions. After 3 days, the samples were washed three times in phosphate-buffered saline (PBS), incubated for 20 min in 4% paraformaldehyde in PBS, washed again in PBS, and mounted onto microscope slides. Microscopy was performed with a Leica TCS SP2 AOBS confocal microscope. Samples prepared for and visualized by transmission electron microscopy were processed and examined as described previously (15).
BN-PAGE, hrCNE, and analysis of native PAGE-resolved complexes.
Sample preparation and blue native polyacrylamide gel electrophoresis (BN-PAGE) were performed as described by Wittig et al. (73). Briefly, washed HeLa cell pellets were resuspended in 10 volumes of BN mitobuffer (83.3 mM sucrose, 6.7 mM sodium phosphate, pH 7.0), homogenized in a motorized Dounce homogenizer (40 passes at 2,000 rpm), and aliquoted into microcentrifuge tubes (∼20 mg [wet weight] of cells/tube). Crude mitochondrial fractions were pelleted by centrifugation of the homogenates for 10 min at 13,000 × g at 4°C and flash frozen in liquid nitrogen until further use. Mitochondrion-enriched sediment was suspended in 35 μl BN solubilization buffer (50 mM NaCl, 50 mM imidazole, 2 mM 6-aminocaproic acid, 1 mM EDTA, pH 7.0) and then solubilized by the addition of 20% digitonin (dissolved in water at 95°C) to a final concentration of 3.3% with immediate mixing by pipetting. The samples were clarified by centrifugation (15 min at 100,000 × g and 4°C), and the protein content was determined by the method of Bradford. Equal amounts (150 to 300 μg) of protein were supplemented with 10 μl BN loading buffer (5% Coomassie blue G250 in 0.5 M 6-aminocaproic acid) and up to 5% glycerol, and samples were resolved on a nondenaturing, Tricine-acrylamide gradient (4 to 13% polyacrylamide) gel as described previously (52). The sizes of BN-PAGE-resolved proteins were estimated by resolving digitonin- or dodecyl-β-d-maltoside-solubilized bovine heart mitochondria as molecular mass markers as described previously. High-resolution clear native PAGE (hrCNE) was performed identically to BN-PAGE except that hrCNE cathode buffer (50 mM tricine, 7.5 mM imidazole, 0.02% dodecyl-β-d-maltoside, 0.05% deoxycholate [pH ∼7.0; no pH adjustment]) was used instead of the BN cathode buffers and a solution of 0.01% Ponceau S with 5% glycerol was used as the sample buffer (75).
Complex I activity was determined in-gel as described previously (75). Briefly, BN-PAGE gels were immersed in a solution of 5 mM Tris-HCl, pH 7.4, containing 2.5 mg/ml nitrotetrazolium blue chloride (Roth GmbH) and 150 μM NADH and incubated until visible bands appeared (10 to 30 min). The gels were then fixed in 50% methanol-10% acetic acid (which also removes residual blue dye from the gel).
Immunostaining of native gels was accomplished by soaking the gels in three volumes of 2× blotting buffer (300 mM Tris, 100 mM acetic acid) supplemented with 1% sodium dodecyl sulfate (SDS) for 20 min, with the gels being turned over every 5 min. The gels were next sandwiched between two glass plates and incubated for 1 h at room temperature (75) and then blotted onto PVDF membranes with a semidry blotting apparatus using 1× blotting buffer for 18 to 24 h at 0.5 mA/cm2 (maximum of 15 V). The PVDF membranes were blocked using PBS supplemented with 0.5% Tween 20 and washed or probed using PBS plus 0.1% Tween 20.
For further resolution, native gel strips were used for two-dimensional (2D) Tricine-SDS-PAGE (52, 73). Briefly, lanes were excised from native gels and incubated for 30 min in 1% SDS. The gel strips were laid horizontally over 10% polyacrylamide-SDS-Tricine gels, according to the formulation and running conditions described previously, with a cathode (top) buffer consisting of 0.1 M Tris, 0.1 M Tricine, and 0.1% SDS (the pH was not adjusted) (53) and an anode buffer of 0.2 M Tris-HCl, pH 8.9. 2D gels analyzed by immunoblotting were handled identically to native gels except that the 1-h incubation between glass plates was omitted.
For 55Fe-labeling experiments, 1 μM 55Fe-labeled transferrin was prepared as described previously (35) and added to the incubation medium after the third transfection and the cells were grown for another 3 days. Cells were harvested and subjected to BN-PAGE as detailed above. The gels were dried on a gel dryer and subjected to phosphorimaging using a Fuji FLA-3000 instrument (Fujifilm).
Measurement of complex I activity by spectrophotometry.
Solubilized, mitochondrion-enriched samples were generated using the same procedure used for native electrophoresis. Thirty to 50 μg of protein was added to a cuvette containing CI reaction buffer (20 mM sodium MOPS [morpholinepropanesulfonic acid], pH 7.4, 50 mM NaCl, 2.5 mg/ml bovine serum albumin, 2 mM KCN, 60 μM decylubiquinone, and 100 μM deamino-NADH), and the mixture was gently stirred with a plastic stirring spatula. An identical sample additionally containing a 27 μM concentration of the specific complex I inhibitor DQA (25, 39) was prepared in parallel. Complex I activity was assessed by determining the rate differences in absorption changes at 340 nm (i.e., the rate of oxidation of deamino-NADH) between inhibited and noninhibited samples.
Assessment of lactate production.
Culture media from transfected cells were diluted 20-fold in water, and 11-μl aliquots of the dilutions were loaded onto a 96-well UV-visible microtiter plate. To each sample, 238 μl of lactate assay buffer (112 mM glycylglycine, pH 10.0, 225 mM glutamate, 3.2 mg/ml NAD+, 40 U/ml glutamate-pyruvate transaminase) was added, and the plate was preanalyzed by using an Infinite 200 plate reader (Tecan) measuring at 340 nm. Once a stable reading was achieved, 3 μl of a 5,500 U/ml lactate dehydrogenase (LDH) solution was added to each well and the absorption at 340 nm (i.e., the reduction of NAD+) was monitored until a plateau was reached. The amount of lactate present in the sample was evaluated by comparing the difference in absorption immediately before the addition of LDH and after the plateau in absorption was reached to a standard curve generated with standard calcium lactate solutions. This value was then normalized with respect to the total number of cells in the culture flask from which the medium was extracted.
Common biochemical and molecular biological methods.
Previously described methods were used for harvesting and fractionating HeLa cells with digitonin (5, 12) and measurements of the catalytic activities of aconitase (12), succinate dehydrogenase (SDH) using the dichlorophenolindophenol (DCIP) method (23, 60), cytochrome c oxidase (66), and citrate synthase (CS) and LDH (5, 26, 59). Cross contamination of mitochondrial and cytosolic fractions was evaluated by comparing the CS and LDH activities of the two fractions. On average, less than 9% of the specific LDH activity was present in the mitochondrial preparations and less than 0.4% of the specific CS activity was present in the cytosolic fractions. All data shown are representative of at least three separate experiments. Where included, error bars represent standard deviations.
RESULTS
HuInd1 is a mitochondrial protein capable of binding an Fe/S cluster.
The amino acid sequence of huInd1 (listed in the human genome database as “nucleotide binding protein-like” [RefSeq accession no. NM_025152]) aligns perfectly to those of its counterparts in other eukaryotic species (8; also data not shown). The Ind1 proteins contain a highly conserved nucleotide binding domain and a putative Fe/S binding signature. The Tetrahymena thermophila (57), Y. lipolytica (8), Arabidopsis (9), and murine (43) Ind1 homologues have already been shown to localize to mitochondria. In keeping with such localization, the sequence of huInd1 is predicted to have a mitochondrial presequence at its N terminus. To verify that huInd1 is in fact a mitochondrial protein, HeLa cells were transfected with an expression vector encoding huInd1 fused to EGFP at the C terminus. Confocal laser microscopy analysis of the cells, cotransfected with a vector encoding mitochondrion-targeted Dicosoma sp. red protein, revealed mitochondrial localization of huInd1 (Fig. 1A). As an independent localization method, we performed subcellular fractionation of HeLa cells by differential centrifugation followed by immunoblotting with antibodies raised against recombinant huInd1. A band with an apparent size of 31 kDa was present in the pellet fraction (containing mitochondria) (Fig. 1B) and in the total cell lysate but was undetectable in the supernatant fraction (the cytosol). This size is consistent with processing of the mitochondrial presequence from the full-length protein (34 kDa). A band smaller than that for mature huInd1 was detected in the cytosolic fraction by the huInd1 antibody (Fig. 1B, NS). This protein was not affected by our RNAi treatment, explained below (data not shown), and therefore is not related to a cytosolic variant of huInd1. To analyze the tissue distribution of huInd1, we probed an immunoblot containing various human tissue samples with anti-huInd1 antibodies. The analysis revealed highest expression of the protein in liver and kidney tissues and significant levels in small intestine and brain tissues (data not shown).
FIG. 1.
huInd1 is a mitochondrial Fe/S protein. (A) HeLa cells cotransfected with constructs encoding a huInd1-EGFP fusion protein and a mitochondrion-targeted Dicosoma sp. red protein (dsRed-mito) show virtually complete colocalization of the two fluorescent labels. (B) HeLa cells were permeabilized by digitonin treatment. The total cell lysate (L) was fractionated into crude mitochondria (M) and the cytosolic fraction (C) by centrifugation at 15,000 × g. Immunostaining of the samples with antibodies raised against huInd1 revealed a specific band in the mitochondrial fraction that represents endogenous huInd1, while a cross-reacting (nonspecific [NS]) protein band with greater electrophoretic mobility was present in the cytosol and cell lysate. Immunostaining of the same membranes for alpha-tubulin (localized in the cytosol) and mitochondrial aconitase (mAco; localized in the mitochondria) was used to verify the purity of the fractions. (C) UV-visible spectrum of chemically reconstituted, recombinant huInd1. The protein was produced in E. coli as a truncated version which does not include the predicted mitochondrial presequence but contains an N-terminal His6 tag. The purified protein was chemically reconstituted under anaerobic conditions with sulfide and ferrous iron. The resulting UV-visible spectrum of 26 μM protein contained a dithionite-sensitive (2 mM) absorption shoulder at approximately 400 nm, which is indicative of a [4Fe-4S] cluster.
For biochemical investigation, we generated recombinant huInd1 in E. coli. Purification yielded a colorless protein that apparently did not contain any significant proportion of coordinated metal ions. We therefore performed chemical reconstitution with iron and sulfide under anaerobic conditions, a procedure which is known to elicit spontaneous Fe/S cluster formation on potential cluster-harboring proteins (37). Spectrophotometric analysis of the reconstituted huInd1 demonstrated a UV-visible absorption spectrum characteristic of a [4Fe-4S] cluster (3, 41), with an absorption shoulder around 400 nm that was decreased by 30% following dithionite reduction (Fig. 1C). Brown coloring originates from the [4Fe-4S]2+ cluster charge transfer transitions. When a chemical reductant, such as dithionite, reduces the [4Fe-4S]2+ cluster to a [4Fe-4S]1+ cluster, the iron atoms become reduced and exhibit severely diminished spectral intensity around 400 nm (41). Reconstituted huInd1 was silent in electron paramagnetic resonance analysis (data not shown), an observation which was also made with ApbC, a bacterial member of the Nbp35/Mrp family (6). The technical constraints of using cultured human cells precluded us from performing immunoprecipitations of radioiron-bound huInd1, as are commonly used in the yeast model to show Fe/S cluster incorporation in vivo. Nonetheless, considering both our data and the conservation of the Fe/S binding motif in huInd1 relative to that in Yarrowia, which we have already shown to bind an Fe/S cluster (8), we deduce that huInd1 is a mitochondrial protein which can assemble a [4Fe-4S] cluster.
HuInd1 depletion interferes with mitochondrial morphology and cellular respiration.
To analyze the phenotypical consequences of huInd1 deficiency and the cellular function of huInd1, a vector-based RNAi strategy was used to deplete HeLa cells of endogenous huInd1. As shown in Fig. 2A, the level of huInd1, as detected by immunostaining, was strongly decreased in mitochondria isolated from cells repeatedly transfected with the short hairpin RNA vector termed siIND1 (61). After three transfections, corresponding to 9 days of depletion, huInd1 was barely detectable, while the protein pattern of the whole-cell extract did not change (data not shown). We examined huInd1-deficient cells by electron microscopy, since several studies concerned with altered mitochondrial respiratory function (20, 38, 46) and one concerned with decreased activity of the mitochondrial ISC assembly machinery (5) report alterations in mitochondrial morphology. RNAi depletion of huInd1 resulted in abnormally large mitochondria with enlarged matrix spaces and few crista membranes, which appeared to be tightly associated in a parallel arrangement (Fig. 2B, middle panels). To assess the specificity of the siRNA approach, RNAi-treated HeLa cells were cotransfected with the vector smIND1, which contains silent mutations that afford its transcript escape from RNAi. Synthesis of huInd1 (as a GFP fusion protein) from this vector fully restored normal mitochondrial morphology (Fig. 2B, bottom panels). These results document the specificity of our RNAi method (see also Fig. 3) and suggest that huInd1 function, directly or indirectly, is important for the maintenance of normal mitochondrial ultrastructure.
FIG. 2.
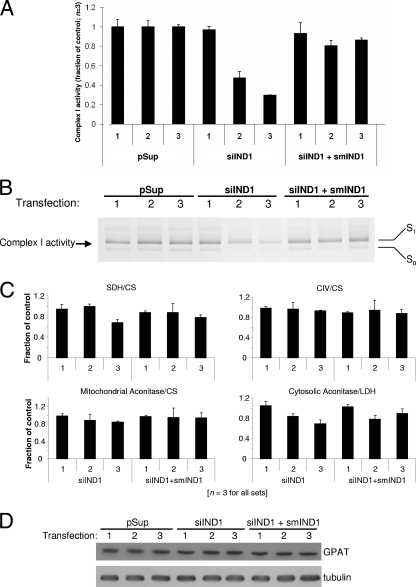
huInd1 depletion results in abnormal mitochondrial morphology and lactate production. (A) HeLa cells were transfected with an empty vector (pSup) or the RNAi vector siIND1. After growth for 3 days, cells were harvested and a fraction was retransfected. This procedure was repeated twice. Three days after each transfection with siIND1, samples (designated 1 to 3) were collected and fractionated using digitonin and the mitochondrial fractions were analyzed by SDS-PAGE and immunostaining for huInd1. Loading of equal amounts was verified by probing the membrane with an antibody that recognizes the α and β subunits of ATP synthase F1 (F1 α/β). (B) HeLa cells were transfected as described in the legend to panel A except that an additional sample in which cells were cotransfected with vectors siIND1 and smIND1 was generated. The latter vector contains silent mutations which render its mRNA resistant to RNAi depletion and thus allows RNAi-resistant synthesis of huInd1. Three days after the third transfection, cells were fixed directly in culture dishes and examined by electron microscopy (5). Two representative images are shown for each condition. Scale bars, 1 μm. (C) Lactate levels in culture medium conditioned for 3 days by HeLa cells transfected three times with either the empty vector (pSup), the RNAi vector (siIND1), or the RNAi and rescue vectors (siIND1+smIND1). Values were adjusted with respect to the total cell number and then normalized according to the lactate level in medium from cells transfected with the empty vector (pSup).
FIG. 3.
RNAi-mediated depletion of huInd1 specifically affects complex I. HeLa cells were transfected up to three times (as indicated by numbers 1, 2, and 3) with the indicated vectors as described in the legend to Fig. 2 and further analyzed. (A) Digitonin-solubilized crude mitochondrial fractions were assayed spectrophotometrically for complex I activity (i.e., DQA-sensitive deamino-NADH oxidation). Samples treated with RNAi (siIND1) show a three- to fourfold decrease in complex I activity after the third transfection. This decreased activity was recovered upon coexpression of an RNAi-resistant version of huInd1 (from smIND1). Error bars for the control samples (transfected with the empty vector pSup) reflect the standard deviations in a representative set of transfections. For the RNAi-treated samples, data from each transfection set were normalized to the control values within that set and the normalized values from three sets of transfections were then averaged. Error bars for these values thus represent the standard deviations across three transfection sets. (B) In-gel complex I enzymatic activity of HeLa mitochondrial preparations after resolution of complex I by BN-PAGE. S1 and S0 are the supercomplexes containing complex I-complex III2-complex IV and complex I-complex III2, respectively (74). (C) The indicated enzyme activities were assayed spectrophotometrically using digitonin-solubilized mitochondrial fractions prepared identically to those used for BN-PAGE. The activities of mitochondrial enzymes were normalized to that of CS, while those of cytosolic fractions were normalized to LDH activity. The data are shown as fractions of the control values (i.e., those for cells transfected with the empty vector in each set of transfections). SDH, complex II; CIV, cytochrome c oxidase. (D) Total cellular lysate samples described above were analyzed by immunostaining of the cytosolic Fe/S protein GPAT and of tubulin after SDS-PAGE.
Significant depletion of mitochondrial crista membranes is frequently observed in cells with defective oxidative phosphorylation. Cells and patients with compromised oxidative phosphorylation rely more heavily on glycolysis for energy production and thus accumulate pyruvate, which in turn is converted into lactate by LDH. Hence, patients with mitochondrial diseases are often afflicted by lactic acidosis. Since the growth medium used in this study contained phenol red, we consistently noticed increased acidity in the medium of cells depleted of huInd1 compared to that in the medium of mock-transfected controls. In line with this observation, huInd1-deficient HeLa cells indeed produced approximately twofold more lactate than control cells (Fig. 2C). This increased lactate content is consistent with the idea that huInd1 deficiency impaired oxidative phosphorylation.
The major role of huInd1 is in complex I assembly.
To directly examine the effects of huInd1 depletion on the respiratory complexes, we first measured complex I activity both by a spectrophotometric enzyme activity assay (Fig. 3A) and by an in-gel assay of complex I activity after resolution by BN-PAGE (Fig. 3B) of digitonin-solubilized crude mitochondrial fractions. The latter analysis revealed two major bands, presumably consisting of supercomplexes comprising complex I-complex III2-complex IV and complex I-complex III2, known as S1 and S0, respectively (74). By both methods, the activity of complex I in the RNAi-treated cells was found to be decreased three to fourfold compared to that in vector-only controls. It is important that the spectrophotometric assay employed here is highly specific for complex I, revealing only DQA-sensitive electron transfer using deamino-NADH as a substrate (25, 39). Concurrent transfection of HeLa cells with the vector encoding the RNAi sequence and vector smIND1 almost completely restored complex I activities to control values, demonstrating the specificity of the RNAi treatment in both assay systems.
We next determined the activities of some cellular Fe/S proteins (respiratory complex II [SDH] and mitochondrial and cytosolic aconitases) to analyze the specificity of huInd1 depletion for complex I activity. In contrast to the potent effects of huInd1 depletion on complex I, no significant consequences of decreased huInd1 levels on the activities of respiratory complex IV (a heme- and copper-containing enzyme) or the mitochondrial Fe/S enzyme aconitase were observed (Fig. 3C). Slight decreases (1.4-fold relative to control values) in SDH and cytosolic aconitase activities were observed in huInd1-depleted cells after three, but not after only two, transfections with the siRNA vector. Effects on both activities were reversed by the synthesis of huInd1 from the RNAi-resistant gene. To rule out general effects of huInd1 depletion on cytosolic Fe/S protein biogenesis, we also measured the levels of the Fe/S protein GPAT, which is rapidly degraded in the absence of a bound Fe/S cluster, an established method for estimating the integrity of extramitochondrial Fe/S cluster generation (34, 61, 81). The levels of GPAT were unchanged upon the depletion of huInd1, documenting normal cytosolic Fe/S protein biogenesis in huInd1-depleted cells (Fig. 3D). Collectively, these results suggest a high degree of specificity of huInd1 depletion for complex I enzyme activity.
The level of fully assembled complex I is decreased upon huInd1 depletion.
We next analyzed whether the decrease in complex I activity elicited by huInd1 knockdown can be attributed to compromised assembly of the complex. To this end, we examined the levels of fully assembled complex I by immunoblotting and by separation of the samples from BN-PAGE by second-dimension SDS-PAGE. For sample preparation, we used digitonin, a relatively gentle detergent, yielding conditions under which respiratory supercomplexes remain intact (55). Immunodetection of complex I in blots from BN-PAGE gels was achieved by using a polyclonal antiserum raised against purified complex I (54). The analysis revealed a major immunoreactive band (Fig. 4A) corresponding to the position of the major band appearing in the complex I in-gel activity assay. This band corresponds to a supercomplex of complexes I, III2, and IV (S1) (Fig. 3B) (13, 55). Depletion of huInd1 resulted in a pronounced decrease of this band, which could be restored by cotransfection with vector smIND1 for RNAi-resistant production of huInd1. Consistent with these data, 2D electrophoresis (BN-PAGE followed by SDS-PAGE) and silver staining revealed a column of protein spots corresponding to a supercomplex comprising complexes I, III2, and IV that strongly decreased upon huInd1 knockdown (Fig. 4B, black arrows). Immunostaining of the 2D gels with the anti-complex I antibody reemphasized the severe decrease in assembled complex I (Fig. 4C).
FIG. 4.
Depletion of huInd1 interferes with the assembly of complex I and leads to respiratory chain remodeling. (A) HeLa cells were transfected up to three times (1 to 3) with the indicated vectors as described in the legend to Fig. 2. Mitochondrial preparations were solubilized with digitonin, resolved by BN-PAGE, and immunostained with a polyclonal anti-complex I antibody (α-complex I). S1 is the supercomplex containing complex I-complex III2-complex IV (74). (B) BN-PAGE-resolved mitochondrial samples from triply transfected cells described in the legend to panel A were run in a second dimension (SDS-PAGE) and were subsequently silver stained. Whereas levels of complex I-containing supercomplex protein (S1; black arrows) decreased in RNAi-treated samples, the levels of complex III dimer (III2; white arrowheads) increased. V, respiratory complex V. (C) Samples were treated as described in the legend to panel B. Instead of being silver stained, the gels were blotted onto PVDF membranes and probed with the polyclonal antibody against complex I. The lanes corresponding to complex I subunits are indicated by black arrows, and the positions of complex I Fe/S proteins NDUFS1 and NDUFV1 are indicated. The cross-reacting spots corresponding to the abundant ATP synthase F1 α and β subunits (white arrowheads) show the equal degrees of exposure of the two membranes. (D) 55Fe autoradiography analysis of BN-PAGE-resolved mitochondrial samples from HeLa cells transfected three times as described in the legend to panel A. In this experiment, growth after the third transfection was in the presence of 1 μM 55Fe-transferrin. After BN-PAGE, the radioactivity was evaluated by phosphorimaging. The positions of 55Fe binding proteins or complexes are indicated. (E) Immunostained BN-PAGE-resolved mitochondrial preparations. Samples were probed with an antibody raised against complex III2 (top) or an antibody that recognizes the α and β subunits of ATP synthase F1 (bottom). The positions of respiratory complexes or supercomplexes are indicated.
To directly examine the incorporation of iron into complex I and other respiratory complexes, we developed a novel in vivo method employing the radioisotope 55Fe. After the third transfection with the vectors as described above, HeLa cells were grown in the presence of 1 μM 55Fe-transferrin for 3 days. Cell extracts were then subjected to BN-PAGE followed by autoradiography. This labeling procedure allowed us to directly visualize the incorporation of 55Fe into the supercomplex I-III2-IV (Fig. 4D). The migration of this supercomplex in the gel was identical to that of the major reactive species revealed by the in-gel activity assay (Fig. 3B). The other respiratory complexes/supercomplexes were identified by comparing their positions in the gel to those of a bovine heart mitochondrion standard preparation, which was run in a parallel lane. Bands corresponding to the positions of complex II, the complex III dimer, and the complex III2-complex IV supercomplex (all of which contain Fe/S clusters and/or heme) were apparent. Upon RNAi treatment, the 55Fe signal of the complex I-containing supercomplex was barely detectable (Fig. 4D), whereas no diminution of other complex III-containing assemblages was observed. A slight decrease in the 55Fe labeling of complex II in huInd1-depleted cells was observed (Fig. 4D), closely resembling the diminished enzyme activity of complex II observed as described above (Fig. 3C). Not surprisingly, the iron storage protein ferritin, the major site of iron delivery in tumor cell lines, was the most prominent band in these autoradiograms. The identity of the ferritin protein complex was verified by gel supershift analysis using polyclonal antiferritin antibodies (data not shown). Shorter exposure times for the gel shown in Fig. 4D revealed no changes in the ferritin signal. Upon the synthesis of huInd1 from the RNAi-resistant vector smIND1, the incorporation of 55Fe into the various iron-containing complexes was at wild-type levels. Collectively, these findings show a strong defect in complex I activity as the result of defective assembly under conditions of huInd1 depletion. As a consequence, complex I can no longer be integrated into the supercomplex with respiratory complexes III and IV.
HuInd1 depletion results in remodeling of respiratory supercomplexes and the appearance of a subcomplex.
The 2D gel analyses described above indicated characteristic changes in bands other than that for complex I, even though the majority of the bands were unaffected (Fig. 4B and C). For instance, the intensities of the protein spots within the complex III column of silver-stained 2D gels exhibited behavior reciprocal to that of complex I subunits upon RNAi treatment (Fig. 4B, white arrowheads), suggesting that the fraction of complex III that normally associates with complex I remains present despite the disappearance of its supercomplex partner. This notion is further supported by immunoblot analyses of BN-PAGE gels probed with an antibody against complex III2 (Fig. 4E). Three major bands were observed in the blots: the topmost is in the migratory position corresponding to the above-described stable complex I-complex III2-complex IV supercomplex species (13, 55). The middle band does not contain any complex I activity and migrates more slowly than the band corresponding to the complex V monomer (∼700 kDa), suggesting that this band corresponds to the supercomplex comprising the complex III homodimer (∼490 kDa) and complex IV (∼200 kDa) (55). The lowest band matches the expected size of the complex III homodimer. As the amount of protein in the complex I-complex III2-complex IV band decreased, a reciprocal increase in both of the lower bands was observed in cells deficient in huInd1 (Fig. 4E). In other words, the fraction of complex III associated with complex I-complex IV decreased while that present in the complex III2 and complex III2-complex IV supercomplexes increased. These data indicate that the diminution of complex I assembly leads to altered interaction behavior of its binding partner, complex III, even though quantitation of the bands revealed that the overall content in complex III did not appear to change.
We further analyzed the effects of decreased huInd1 by hrCNE. This technique, first described by Wittig et al. (75), is identical to BN-PAGE except that it employs a cathode buffer that contains the detergents deoxycholate and dodecyl-β-d-maltoside for solubilizing membrane protein complexes yet lacks Coomassie brilliant blue. hrCNE separation, followed by immunostaining, revealed two major bands (Fig. 5A, S and NS). The depletion of huInd1 by RNAi treatment decreased one of the bands (S), an effect fully reversed by the synthesis of RNAi-resistant huInd1. The lower band (NS) does not correspond in position to a reactive band in a complex I in-gel activity assay, suggesting that this band is either an inactive subcomplex or a cross-reacting protein artifact. Interestingly, we also observed an increase in a pair of more quickly migrating bands in RNAi-treated cells (Fig. 5A, subcomplex). These protein complexes had higher mobility levels than the 490-kDa bovine complex III dimer. To further verify that these bands are related to complex I and may represent a subcomplex, we subjected lanes from hrCNE gels to SDS-PAGE in a second dimension, followed by immunoblotting. In the position corresponding to the putative subcomplex, the Fe/S cluster binding subunits NDUFS1 (75 kDa) and NDUFV1 (51 kDa; see also below) were not detectable while the intensity of a smaller complex I subunit of about 15 kDa increased (Fig. 5B, white arrowheads). In addition, probing blots with a specific antibody against NDUFA9 showed a decrease in this protein's levels in the position of the supercomplex (Fig. 5B, middle panels; see also below). On the other hand, using antibodies against the membrane subunit NDUFB6 revealed a shift in the presence of this protein from the fully assembled complex to the subcomplex (Fig. 5B, bottom panels). We conclude from these data that in the absence of huInd1, only a subcomplex of complex I can be assembled. This subcomplex has an approximate size of 450 kDa and does not contain the peripheral arm subunits NDUFS1, NDUFV1, NDUFA13, or NDUFA9 but does contain the membrane arm subunit NDUFB6.
FIG. 5.
Appearance of a subcomplex of complex I upon depletion of huInd1. (A) HeLa cells were transfected as described in the legend to Fig. 2A. Mitochondrial preparations were resolved by hrCNE and immunostained with the polyclonal antibody against complex I (α-complex I). Two major immunoreactive bands are apparent near the top of the gel (arrows); the higher of the two (specific [S]) significantly decreased after RNAi treatment. The lower band (nonspecific [NS]) does not correspond to a positive band in an in-gel complex I activity assay (data not shown). The amount of a subcomplex of complex I migrating at approximately 450 kDa (bracket) is largely increased upon the depletion of huInd1. (B, top panels) Immunostaining of complex I proteins in samples resolved first by hrCNE, as described in the legend to panel A, and then by SDS-PAGE. The white arrowheads denote a complex I subunit (approximately 15 kDa) which is present in the 450-kDa subcomplex and increases following huInd1 depletion. The subcomplex does not contain the NDUFS1 and NDUFV1 Fe/S proteins. (Middle and bottom panels) Immunostaining of a 2D gel blot (prepared as described for the top panels) with antibodies against NDUFA9 and NDUFB6 (α-NDUFA9 and α-NDUFB6). The level of NDUFA9 in fully assembled complex I decreases upon huInd1 depletion, but NDUFA9 does not appear in a subcomplex under these conditions. In contrast, NDUFB6 is enriched in a subcomplex when huInd1 is depleted.
Defective assembly of the peripheral arm of complex I containing the Fe/S subunits NDUFS1 and NDUFV1 upon huInd1 depletion.
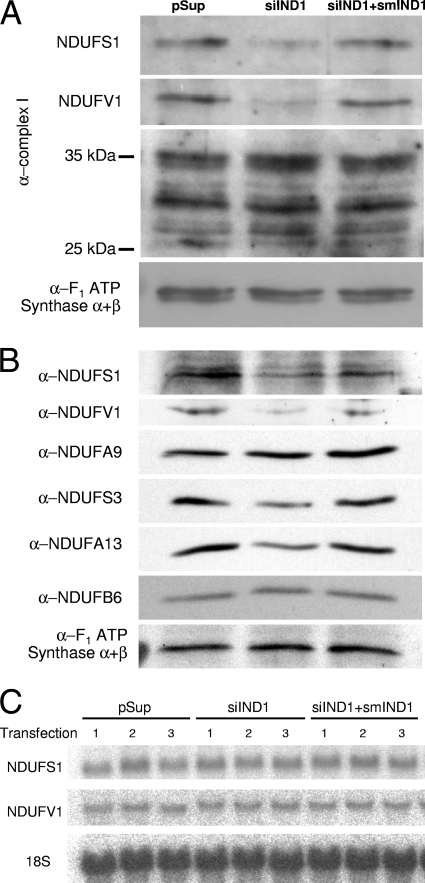
To more closely examine the effects of huInd1 depletion on individual complex I subunits, we examined the protein levels and compared them with the levels of the respective mRNAs. Mitochondrial fractions from mock-treated, huInd1-depleted, or complemented cells were subjected to SDS-PAGE and immunostaining using an antiserum raised against purified complex I. This antiserum detects, in addition to numerous other complex I components, the Fe/S cluster binding subunits NDUFS1 and NDUFV1 (54) (Fig. 5B). Upon huInd1 knockdown, we noted a specific decrease in the levels of NDUFS1 (75 kDa) and NDUFV1 (50 kDa) (Fig. 6A). On the contrary, several other subunits detected by the serum were unchanged (Fig. 6A and data not shown). These results were confirmed by probing additional, similarly prepared blots with commercial antibodies against NDUFS1 or NDUFV1 (Fig. 6B). Northern blotting established that neither NDUFS1 nor NDUFV1 gene expression levels were affected, excluding a transcriptional cause for the observed decrease in NDUFS1 and NDUFV1 protein levels (Fig. 6C). These data suggest a posttranscriptional role of huInd1 in the synthesis, assembly, and/or stabilization of these two Fe/S cluster complex I subunits.
FIG. 6.
huInd1 depletion results in severely decreased levels of constituents of the peripheral arm of complex I. (A) HeLa cells, triply transfected with vectors as described in the legend to Fig. 2A, were fractionated using digitonin, and the mitochondrial fractions were resolved by SDS-PAGE. Immunostaining was with the polyclonal antibody against complex I (α-complex I) or an antibody that recognizes the α and β subunits of ATP synthase F1 (α-F1 ATP synthase α+β). (B) Blots prepared as described in the legend to panel A were probed with antibodies against NDUFS1 (α-NDUFS1), NDUFV1, NDUFA9, NDUFS3, NDUFA13 (GRIM-19), or NDUFB6 or an antibody that recognizes the α and β subunits of ATP synthase F1. (C) The decreases in NDUFS1 and NDUFV1 upon huInd1 depletion are posttranscriptional. mRNAs were extracted from HeLa cells transfected as described in the legend to panel A and were analyzed by Northern blotting using probes generated from cDNA encoding NDUFS1 or NDUFV1 or an oligonucleotide complementary to 18S rRNA.
To further analyze the consequences of huInd1 depletion for complex I proteins and to test the specificity of the diminution of the Fe/S protein subunits NDUFS1 and NDUFV1, we probed immunoblots with antibodies against NDUFS3 and NDUFA13 (GRIM-19) of the peripheral arm and NDUFA9 and NDUFB6 of the membrane arm. The levels of the peripheral arm constituents NDUFS3 and NDUFA13 (71) also decreased upon huInd1 depletion (Fig. 6B). Neither NDUFS3 nor NDUFA13 contains Fe/S clusters, making it likely that the formation of the entire peripheral arm is severely hindered in the absence of huInd1. It seems feasible that, as a consequence of the functional defect of huInd1, the Fe/S cluster-containing subunits of the peripheral arm are not assembled, leading to their instability and proteolytic degradation. This in turn may lead to an assembly defect in the peripheral arm and cause the decay of unassembled constituents of this part of complex I.
No changes in the levels of the membrane constituents NDUFA9 and NDUFB6 were observed (Fig. 6B). While the result for NDUFB6 is consistent with our 2D gel immunoblot analysis, we noted an interesting difference regarding the lack of NDUFA9 in the 2D gel immunoblots (Fig. 5B). Apparently, NDUFA9 and NDUFB6 behave differently upon the depletion of huInd1. NDUFB6 is integrated into the 450-kDa subcomplex detectable by hrCNE, whereas NDUFA9 is detectable only in intact isolated mitochondria (Fig. 5B [results obtained using mitochondrial membranes] and 7B [results obtained using intact mitochondria]). Thus, the latter protein does not appear to be integrated into a high-molecular-mass complex detectable by hrCNE. We conclude from these findings that the lack of huInd1 results in an assembly defect in the peripheral arm of complex I but that a subcomplex containing the membrane protein NDUFB6 is still formed.
The twin-cysteine-residue motif is required for the function of huInd1.
Ind1 proteins contain conserved cysteine residues (at positions 244 and 247 in huInd1) that are also present in Cfd1/Nbp35 P-loop NTPases (37). To analyze the possibility that these two residues are ligands for Fe/S cluster binding, we mutated them to alanine residues in the RNAi-resistant construct (smIND1) and examined whether cotransfection with this DNA and the RNAi construct could rescue the observed complex I defect phenotype. Whereas smIND1 was able to restore both complex I activity (Fig. 7A and B) and levels of fully assembled complex I (Fig. 7C and D), the rescue construct encoding huInd1 with mutated cysteine residues (smIND1CC) was unable to do so. Using the C-terminal EGFP tag fused to the vector-encoded huInd1 proteins, we could demonstrate that both the levels and the subcellular localization patterns of the proteins were similar (Fig. 7E to G). This finding suggests that the two conserved cysteine residues are essential for the function of huInd1 in the assembly of complex I. Notably, while both the expression and localization of the huInd1 protein encoded by smIND1CC were comparable to those of the protein encoded by smIND1 (Fig. 7E and F), the stability of the double cysteine mutant protein was significantly decreased after cell lysis. Immunoblotting of cell extracts revealed decreased levels of the mutated protein. These levels depended on the speed of preparation of the cell extracts (Fig. 7G and data not shown).
FIG. 7.
The conserved cysteine residues of the CXXC motif are essential for huInd1 function. (A to D) HeLa cells were transfected with vectors as described in the legend to Fig. 2A. Where indicated, an additional, smIND1-based construct encoding a form of huInd1 with both conserved cysteine residues (C244 and C247) mutated to alanine (smIND1CC) was used. Mitochondrial fractions from the cells were then resolved on BN-PAGE gels and assessed for complex I activity (A) or assayed spectrophotometrically for complex I activity (B). The BN-PAGE-separated samples were immunostained for complex I protein (C) or run in a second dimension (SDS-PAGE) and silver stained (D). The complex I-containing supercomplex lanes are labeled S. ATP synthase F1Fo (V) served as an internal control. The samples in panels A and C were run in the same respective gels, but irrelevant lanes were removed. α-complex I, anti-complex I antibody. (E and F) Analyses of the fluorescence of wild-type and mutated huInd1-EGFP fusion proteins in HeLa cells indicate that both proteins are present in situ. (E) Confocal micrographs of HeLa cells transfected thrice with either the vector encoding the rescue mutant (smIND1), which contains silent mutations that render the mRNA resistant to RNAi treatment, or a version of this rescue construct with C244 and C247 mutated to alanine residues (smIND1CC). (F) Flow cytometry analysis of HeLa cells transfected as described the legend to Fig. 2A. In the panels on the left, cells were analyzed already after the first transfection. (G) Western blot analysis of cells transfected three times with the empty vector (pSup), the huInd1 RNAi construct (siIND1), the RNAi construct together with the silent-mutation rescue construct (siIND1+smIND1), or the RNAi construct together with the C244A/C247A mutant rescue construct (siIND1+smIND1CC). The blot was probed with anti-huInd1 antiserum (α-huInd1). Because the rescue constructs also encode C-terminal EGFP, the rescue proteins (huInd1-EGFP) are detected at a position of slower mobility in the gel. Note that the endogenous level of huInd1 is only slightly lower than the level of huInd1 from the rescue mutant (smIND1).
DISCUSSION
The present study establishes the mitochondrial P-loop NTPase huInd1 as a crucial component for the assembly of human respiratory complex I. Depletion of huInd1 levels by RNAi technology strongly decreased the enzyme activity of this largest complex of the respiratory chain, while other respiratory complexes and several mitochondrial (aconitase) and cytosolic (GPAT) Fe/S proteins were not affected. Small decreases in SDH and cytosolic aconitase were observed. However, in contrast to the 50 and 70% decreases in complex I activity after two and three transfections, respectively, with the huInd1 RNAi construct, those Fe/S proteins were hardly changed and decreased by only 30%, respectively. While we cannot completely rule out a role of huInd1 in SDH and/or cytosolic aconitase maturation, it is more likely that these moderate effects are secondary (late) adaptations of the cells to the chronic complex I deficiency brought on by huInd1 depletion. The apparent specificity for complex I is consistent with the fact that most, but not all, species lacking this respiratory complex do not have a viable copy of the gene for Ind1 in their genomes (8). We therefore propose that the function of this mitochondrial protein in complex I assembly is conserved in complex I-containing lower and higher eukaryotes.
huInd1 is a member of the Mrp/NBP35 subfamily of P-loop NTPase proteins, which includes Nbp35 and Cfd1 (31). While the latter proteins are crucial components of the CIA machinery required for cytosolic and nuclear Fe/S protein maturation (37, 61), we show here that the function of huInd1 is more specific in being dedicated to the assembly of mitochondrial complex I. What might be its particular function in the assembly process? Several lines of evidence support a function similar to that of Cfd1 and Nbp35. These proteins form a heterotetrameric complex which serves a scaffold function in assembling an Fe/S cluster before its transfer to target Fe/S proteins in the cytosol (37). Hence, our hypothesis is that huInd1 provides Fe/S clusters to complex I subunits. In this respect, we can envision huInd1 as a specialized biogenesis factor which donates Fe/S clusters to specific apoproteins at the final stage of Fe/S protein maturation. Another interesting group of proteins playing a similar specialized role is yeast Isa1, Isa2, and Iba57 and the bacterial counterparts, all of which are dedicated to the maturation of aconitase-type and radical S-adenosylmethionine Fe/S proteins (18, 32, 64). It is likely that future studies will reveal several more proteins apart from the core Fe/S cluster biogenesis components that perform specialized functions, e.g., in specifically delivering Fe/S clusters to apoproteins both inside and outside mitochondria.
Similar to Cfd1 and Nbp35, Ind1 is able to bind a [4Fe-4S] cluster in vitro (this work) and in vivo (8). Presumably, this Fe/S cluster is coordinated by the two conserved cysteine residues of the CXXC motif in the C-terminal domain of Ind1. As shown here, these residues are essential for huInd1 function in vivo. Likewise, these two of the four conserved cysteine residues in the C-terminal domains of Cfd1 and Nbp35 are indispensable for Fe/S cluster binding to these proteins and for yeast cell viability (48; D. J. A. Netz, unpublished data). This similarity suggests that the modes of Fe/S cluster binding to the three proteins may be comparable. Since the CXXC motif can provide only two (of the three to four required) ligands for Fe/S cluster coordination, we speculate that huInd1 forms a homodimer. Consistent with this possibility, the related CXXC motif-containing P-loop NTPase from Salmonella enterica, ApbC, has been shown previously to be a homodimer and to bind a [4Fe-4S] cluster, possibly in a bridged coordination (6).
All of the proposed Fe/S subunits of complex I, NDUFS1, NDUFV1, NDUFV2, NDUFS7, and NDUFS8, reside in the peripheral arm and are encoded by the nuclear genome. The levels of several subunits of the complex I peripheral arm, including the large Fe/S cluster-coordinating subunits NDUFS1 (75 kDa) and NDUFV1 (51 kDa), are greatly decreased in huInd1-depleted cells. This finding provides strong support for the direct involvement of huInd1 in the assembly of this part of complex I, possibly by playing a direct role in Fe/S cluster assembly. The following scenario integrates our observations made upon the RNAi knockdown of huInd1 in HeLa cells. Upon the depletion of huInd1, one or more of the Fe/S clusters fail to become inserted into the Fe/S cluster-coordinating subunits, including NDUFS1 and NDUFV1, which are related in amino acid sequence to the bacterial Fe/S subunits Nqo3 and Nqo1 (51, 77, 78). In turn, the maturation defect of complex I Fe/S proteins may lead to the observed failure to assemble parts or all of the peripheral arm, thus leading to the complex I assembly and activity defects. It is well established that proteins with non-covalently linked metallocofactors are not able to properly fold and/or assemble in the absence of their cofactors and, hence, are sensitive to intracellular proteolysis (76). For instance, plant ferredoxin has been shown to absolutely require Fe/S cluster incorporation for adoption of the protein's appropriate 3D structure (42). Furthermore, the levels of the cytosolic Fe/S proteins IRP1 and GPAT, both of which contain cubane Fe/S clusters, are decreased when Fe/S cluster biogenesis is compromised (34, 61). Defective folding, as the result of a missing Fe/S cofactor, may also explain why huInd1 with mutations in the CXXC cysteine residues (encoded by vector smIND1CC) was less stable than the wild-type huInd1 protein, despite its adequate level of expression (Fig. 7G).
While the peripheral arm of complex I was not assembled upon huInd1 depletion, a subcomplex of approximately 450 kDa was detected by hrCNE. Our results suggest that this subcomplex does not contain NDUFS1, NDUFV1, and NDUFA9 subunits but does include the integral membrane protein NDUFB6 (Fig. 5B) and therefore may be related to a membrane-integrated subcomplex. Although somewhat controversial, several reports suggest that, in humans, the membrane arm to which some of the peripheral arm subunits are anchored forms a subcomplex of 400 to 500 kDa from which, together with the remainder of the peripheral arm subunits, the fully assembled complex I is constructed (reviewed by Vogel et al. [71]). Despite the similarity in molecular masses and the absence of NDUFA9 (68), several experimental observations allow us to distinguish this complex (termed the 400/500-kDa complex) from our 450-kDa subcomplex seen upon huInd1 depletion. First, the 400/500-kDa complex was detected by BN gel electrophoresis, while the 450-kDa complex was not visible on BN gels and could be visualized only by hrCNE. Further, the depletion of huInd1 resulted in a strong decrease in the subunit NDUFS3, which reportedly is part of the 400/500-kDa complex (1, 71). Thus, we suggest that the 450-kDa subcomplex constitutes a novel entity comprising predominantly a membrane part of complex I and is distinct from the previously described 400/500-kDa complex. Even though it is unclear at present whether the 450-kDa subcomplex may be a physiological complex I assembly intermediate, we note that the subcomplex is also seen at low levels under conditions of normal huInd1 function (Fig. 5). Hence, the 450-kDa subcomplex may be a bona fide assembly intermediate rather than an artificial assemblage forming as a result of the interference of Fe/S cluster maturation in complex I subunits that we produced in our experimental system.
We report here abnormal mitochondrial ultrastructure in cells depleted of huInd1. While we cannot completely rule out the possibility that huInd1 has some direct role in the maintenance of mitochondrial morphology, our results suggest a simpler explanation in that the altered respiratory competence of huInd1-depleted mitochondria is causative of the morphological changes. These findings are consistent with those of previous studies demonstrating altered mitochondrial morphology not only in cells depleted of mitochondrial DNA (19, 62) but more specifically also in patients with complex I deficiency (16, 36). Furthermore, it is well established that mitochondria in cells that utilize primarily glycolysis for energy production tend to assume the so-called orthodox conformation, in which narrow cristae form few intercristae junctions in a large matrix, as opposed to a condensed state, characterized by large cristae that make numerous junctions in each crista compartment (11). Since huInd1-depleted HeLa cells are deficient in complex I and generate more lactate than controls, it appears that the unusual ultrastructure is best explained as a morphological adaptation to severely decreased respiratory metabolism. Notably, the ultrastructural alterations described here differ characteristically from those observed upon depletion of the mitochondrial cysteine desulfurase Nfs1 (5). In the latter case, the general defect in cellular Fe/S protein maturation is accompanied by the appearance of concentric, onion-shaped mitochondria which exhibit some similarity to structures found in Drosophila cells expressing the “fuzzy onion” mutant of the mitochondrial fusion GTPase Fzo1 (21).
The development of a combined 55Fe-radiolabeling and immunoprecipitation assay for yeast has allowed the in vivo investigation of Fe/S protein biogenesis (29). So far, no comparable assay system to detect mammalian Fe/S proteins in a living cell by radiolabeling methods has been reliably introduced. This lack is due mainly to the fact that the added radioactive iron is subject to isotope dilution by the cellular iron stores, thus resulting in low signal-to-noise ratios. Here, we were able to evaluate the iron incorporation into several respiratory chain complexes by performing BN-PAGE analyses of mitochondrial preparations from HeLa cells treated with 55Fe-labeled transferrin, the physiological iron source for mammalian cells. We observed a strong decrease in 55Fe-labeled complex I, whereas 55Fe binding to the other complexes was unaffected. To the best of our knowledge, this is the first study to estimate, by autoradiography, the presence of iron in these Fe/S proteins. Previously, electrophoresis techniques have revealed only such abundant proteins as transferrin, hemoglobin, and ferritin (45, 72). We anticipate this novel technique to be useful for future studies, not only for the in vivo analysis of Fe/S cluster incorporation into respiratory chain complexes but also into other cellular Fe/S proteins.
In conclusion, we have identified huInd1 as an Fe/S protein required for the proper assembly of complex I in humans. Our findings are consistent with the idea that huInd1 plays a critical role in Fe/S cluster incorporation into the Fe/S-containing subunits of complex I. The phenotype induced in HeLa cells by the depletion of huInd1 resembles that of patients with complex I disease: decreased complex I catalytic activity, irregular mitochondrial morphology, increased lactate production, and the formation of a 450-kDa subcomplex. As approximately half of complex I patients have no mutations in complex I subunits or known assembly factors, it is likely that patients harboring mutations in huInd1 will be identified.
Acknowledgments
We gratefully acknowledge the advice on BN-PAGE and hrCNE from H. Schägger and C. Bach (Frankfurt) and gifts of antibodies, bovine heart mitochondria, and DQA. We thank Sabine Molik for assistance with antibody generation.
A.D.S. was supported by postdoctoral fellowships from the Alexander-von-Humboldt Foundation and FRSQ, Canada. R.L. acknowledges generous support from the Deutsche Forschungsgemeinschaft (SFB 593 and TR1, Gottfried-Wilhelm Leibniz Program, and GRK 1216), the German-Israeli Foundation (GIF), and Fonds der chemischen Industrie. U.B. acknowledges generous support from the Deutsche Forschungsgemeinschaft, Cluster of Excellence, Macromolecular Complexes (EXC 115). J.B. is funded by a university research fellowship from the Royal Society.
Footnotes
Published ahead of print on 14 September 2009.
REFERENCES
- 1.Antonicka, H., I. Ogilvie, T. Taivassalo, R. P. Anitori, R. G. Haller, J. Vissing, N. G. Kennaway, and E. A. Shoubridge. 2003. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278:43081-43088. [DOI] [PubMed] [Google Scholar]
- 2.Balk, J., D. J. Aguilar Netz, K. Tepper, A. J. Pierik, and R. Lill. 2005. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 25:10833-10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balk, J., A. J. Pierik, D. J. Netz, U. Mühlenhoff, and R. Lill. 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23:2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benit, P., S. Goncalves, E. P. Dassa, J. J. Briere, and P. Rustin. 2008. The variability of the harlequin mouse phenotype resembles that of human mitochondrial-complex I-deficiency syndromes. PLoS ONE 3:e3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biederbick, A., O. Stehling, R. Rösser, B. Niggemeyer, Y. Nakai, H. P. Elsässer, and R. Lill. 2006. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 26:5675-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, J. M., A. J. Pierik, D. J. Netz, R. Lill, and D. M. Downs. 2008. Bacterial ApbC can bind and effectively transfer iron-sulfur clusters. Biochemistry 47:8195-8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandt, U. 2006. Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75:69-92. [DOI] [PubMed] [Google Scholar]
- 8.Bych, K., S. Kerscher, D. J. Netz, A. J. Pierik, K. Zwicker, M. A. Huynen, R. Lill, U. Brandt, and J. Balk. 2008. The iron-sulphur protein Ind1 is required for effective complex I assembly. EMBO J. 27:1736-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bych, K., D. J. Netz, G. Vigani, E. Bill, R. Lill, A. J. Pierik, and J. Balk. 2008. The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J. Biol. Chem. 283:35797-35804. [DOI] [PubMed] [Google Scholar]
- 10.Cavadini, P., G. Biasiotto, M. Poli, S. Levi, R. Verardi, I. Zanella, M. Derosas, R. Ingrassia, M. Corrado, and P. Arosio. 2007. RNA silencing of the mitochondrial ABCB7 transporter in HeLa cells causes an iron-deficient phenotype with mitochondrial iron overload. Blood 109:3552-3559. [DOI] [PubMed] [Google Scholar]
- 11.Detmer, S. A., and D. C. Chan. 2007. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8:870-879. [DOI] [PubMed] [Google Scholar]
- 12.Drapier, J. C., and J. B. Hibbs, Jr. 1996. Aconitases: a class of metalloproteins highly sensitive to nitric oxide synthesis. Methods Enzymol. 269:26-36. [DOI] [PubMed] [Google Scholar]
- 13.Dudkina, N. V., H. Eubel, W. Keegstra, E. J. Boekema, and H. P. Braun. 2005. Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc. Natl. Acad. Sci. USA 102:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunning, C. J., M. McKenzie, C. Sugiana, M. Lazarou, J. Silke, A. Connelly, J. M. Fletcher, D. M. Kirby, D. R. Thorburn, and M. T. Ryan. 2007. Human CIA30 is involved in the early assembly of mitochondrial complex I and mutations in its gene cause disease. EMBO J. 26:3227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsässer, H. P., U. Lehr, B. Agricola, and H. F. Kern. 1993. Structural analysis of a new highly metastatic cell line PaTu 8902 from a primary human pancreatic adenocarcinoma. Virchows Arch. B 64:201-207. [DOI] [PubMed] [Google Scholar]
- 16.Folgero, T., K. Bertheussen, S. Lindal, T. Torbergsen, and P. Oian. 1993. Mitochondrial disease and reduced sperm motility. Hum. Reprod. 8:1863-1868. [DOI] [PubMed] [Google Scholar]
- 17.Fukui, H., and C. T. Moraes. 2008. The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? Trends Neurosci. 31:251-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelling, C., I. W. Dawes, N. Richhardt, R. Lill, and U. Mühlenhoff. 2008. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol. Cell. Biol. 28:1851-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilkerson, R. W., D. H. Margineantu, R. A. Capaldi, and J. M. Selker. 2000. Mitochondrial DNA depletion causes morphological changes in the mitochondrial reticulum of cultured human cells. FEBS Lett. 474:1-4. [DOI] [PubMed] [Google Scholar]
- 20.Hackenbrock, C. R. 1966. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J. Cell Biol. 30:269-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hales, K. G., and M. T. Fuller. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90:121-129. [DOI] [PubMed] [Google Scholar]
- 22.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Hatefi, Y., and D. L. Stiggall. 1978. Preparation and properties of succinate: ubiquinone oxidoreductase (complex II). Methods Enzymol. 53:21-27. [DOI] [PubMed] [Google Scholar]
- 24.Hausmann, A., D. J. Aguilar Netz, J. Balk, A. J. Pierik, U. Mühlenhoff, and R. Lill. 2005. The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. USA 102:3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollingworth, R. M., K. I. Ahammadsahib, G. Gadelhak, and J. L. McLaughlin. 1994. New inhibitors of complex I of the mitochondrial electron transport chain with activity as pesticides. Biochem. Soc. Trans. 22:230-233. [DOI] [PubMed] [Google Scholar]
- 26.Howell, B. F., S. McCune, and R. Schaffer. 1979. Lactate-to-pyruvate or pyruvate-to-lactate assay for lactate dehydrogenase: a re-examination. Clin. Chem. 25:269-272. [PubMed] [Google Scholar]
- 27.Janssen, R., J. Smeitink, R. Smeets, and L. van den Heuvel. 2002. CIA30 complex I assembly factor: a candidate for human complex I deficiency? Hum. Genet. 110:264-270. [DOI] [PubMed] [Google Scholar]
- 28.Kirby, D. M., M. Crawford, M. A. Cleary, H. H. Dahl, X. Dennett, and D. R. Thorburn. 1999. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology 52:1255-1264. [DOI] [PubMed] [Google Scholar]
- 29.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazarou, M., D. R. Thorburn, M. T. Ryan, and M. McKenzie. 2009. Assembly of mitochondrial complex I and defects in disease. Biochim. Biophys. Acta 1793:78-88. [DOI] [PubMed] [Google Scholar]
- 31.Leipe, D. D., Y. I. Wolf, E. V. Koonin, and L. Aravind. 2002. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317:41-72. [DOI] [PubMed] [Google Scholar]
- 32.Lill, R. 2009. Function and biogenesis of iron-sulphur proteins. Nature 460:831-838. [DOI] [PubMed] [Google Scholar]
- 33.Lill, R., and U. Mühlenhoff. 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77:669-700. [DOI] [PubMed] [Google Scholar]
- 34.Martelli, A., M. Wattenhofer-Donze, S. Schmucker, S. Bouvet, L. Reutenauer, and H. Puccio. 2007. Frataxin is essential for extramitochondrial Fe-S cluster proteins in mammalian tissues. Hum. Mol. Genet. 16:2651-2658. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Medellin, J., and H. M. Schulman. 1972. The kinetics of iron and transferrin incorporation into rabbit erythroid cells and the nature of stromal-bound iron. Biochim. Biophys. Acta 264:272-274. [DOI] [PubMed] [Google Scholar]
- 36.Moreadith, R. W., M. L. Batshaw, T. Ohnishi, D. Kerr, B. Knox, D. Jackson, R. Hruban, J. Olson, B. Reynafarje, and A. L. Lehninger. 1984. Deficiency of the iron-sulfur clusters of mitochondrial reduced nicotinamide-adenine dinucleotide-ubiquinone oxidoreductase (complex I) in an infant with congenital lactic acidosis. J. Clin. Investig. 74:685-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Netz, D. J., A. J. Pierik, M. Stümpfig, U. Mühlenhoff, and R. Lill. 2007. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3:278-286. [DOI] [PubMed] [Google Scholar]
- 38.Ogilvie, I., N. G. Kennaway, and E. A. Shoubridge. 2005. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Investig. 115:2784-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okun, J. G., P. Lummen, and U. Brandt. 1999. Three classes of inhibitors share a common binding domain in mitochondrial complex I (NADH:ubiquinone oxidoreductase). J. Biol. Chem. 274:2625-2630. [DOI] [PubMed] [Google Scholar]
- 40.Okun, J. G., V. Zickermann, K. Zwicker, H. Schagger, and U. Brandt. 2000. Binding of detergents and inhibitors to bovine complex I—a novel purification procedure for bovine complex I retaining full inhibitor sensitivity. Biochim. Biophys. Acta 1459:77-87. [DOI] [PubMed] [Google Scholar]
- 41.Orme-Johnson, W. H., and N. R. Orme-Johnson. 1978. Overview of iron-sulfur proteins. Methods Enzymol. 53:259-268. [DOI] [PubMed] [Google Scholar]
- 42.Pagani, S., G. Vecchio, S. Iametti, R. Bianchi, and F. Bonomi. 1986. On the role of the 2Fe-2S cluster in the formation of the structure of spinach ferredoxin. Biochim. Biophys. Acta 870:538-544. [Google Scholar]
- 43.Pagliarini, D. J., S. E. Calvo, B. Chang, S. A. Sheth, S. B. Vafai, S. E. Ong, G. A. Walford, C. Sugiana, A. Boneh, W. K. Chen, D. E. Hill, M. Vidal, J. G. Evans, D. R. Thorburn, S. A. Carr, and V. K. Mootha. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pondarré, C., B. B. Antiochos, D. R. Campagna, S. L. Clarke, E. L. Greer, K. M. Deck, A. McDonald, A. P. Han, A. Medlock, J. L. Kutok, S. A. Anderson, R. S. Eisenstein, and M. D. Fleming. 2006. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron-sulfur cluster biogenesis. Hum. Mol. Genet. 15:953-964. [DOI] [PubMed] [Google Scholar]
- 45.Richardson, D. R., P. Ponka, and D. Vyoral. 1996. Distribution of iron in reticulocytes after inhibition of heme synthesis with succinylacetone: examination of the intermediates involved in iron metabolism. Blood 87:3477-3488. [PubMed] [Google Scholar]
- 46.Rossignol, R., R. Gilkerson, R. Aggeler, K. Yamagata, S. J. Remington, and R. A. Capaldi. 2004. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64:985-993. [DOI] [PubMed] [Google Scholar]
- 47.Rouault, T. A., and W. H. Tong. 2005. Iron-sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat. Rev. Mol. Cell Biol. 6:345-351. [DOI] [PubMed] [Google Scholar]
- 48.Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine, and W. E. Walden. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saada, A., S. Edvardson, M. Rapoport, A. Shaag, K. Amry, C. Miller, H. Lorberboum-Galski, and O. Elpeleg. 2008. C6ORF66 is an assembly factor of mitochondrial complex I. Am. J. Hum. Genet. 82:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saada, A., R. O. Vogel, S. J. Hoefs, M. A. van den Brand, H. J. Wessels, P. H. Willems, H. Venselaar, A. Shaag, F. Barghuti, O. Reish, M. Shohat, M. A. Huynen, J. A. Smeitink, L. P. van den Heuvel, and L. G. Nijtmans. 2009. Mutations in NDUFAF3 (C3ORF60), encoding an NDUFAF4 (C6ORF66)-interacting complex I assembly protein, cause fatal neonatal mitochondrial disease. Am. J. Hum Genet. 84:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sazanov, L. A., and P. Hinchliffe. 2006. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311:1430-1436. [DOI] [PubMed] [Google Scholar]
- 52.Schägger, H. 2003. Blue native electrophoresis, p. 105-130. In C. Hunte, G. von Jagow, and H. Schägger (ed.), Membrane protein purification and crystallization. A practical guide, 2nd ed. Academic Press, New York, NY.
- 53.Schägger, H. 2006. Tricine-SDS-PAGE. Nat. Protoc. 1:16-22. [DOI] [PubMed] [Google Scholar]
- 54.Schägger, H., R. de Coo, M. F. Bauer, S. Hofmann, C. Godinot, and U. Brandt. 2004. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279:36349-36353. [DOI] [PubMed] [Google Scholar]
- 55.Schägger, H., and K. Pfeiffer. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smeitink, J., L. van den Heuvel, and S. DiMauro. 2001. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2:342-352. [DOI] [PubMed] [Google Scholar]
- 57.Smith, D. G., R. M. Gawryluk, D. F. Spencer, R. E. Pearlman, K. W. Siu, and M. W. Gray. 2007. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J. Mol. Biol. 374:837-863. [DOI] [PubMed] [Google Scholar]
- 58.Song, D., and F. S. Lee. 2008. A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 283:9231-9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srere, P. A., H. Brazil, and L. Gonen. 1963. The citrate condensing enzyme of pigeon breast muscle and moth flight muscle. Acta Chem. Scand. 17(Suppl.):S129-S134. [Google Scholar]
- 60.Stehling, O., H. P. Elsässer, B. Brückel, U. Mühlenhoff, and R. Lill. 2004. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum. Mol. Genet. 13:3007-3015. [DOI] [PubMed] [Google Scholar]
- 61.Stehling, O., D. J. Netz, B. Niggemeyer, R. Rösser, R. S. Eisenstein, H. Puccio, A. J. Pierik, and R. Lill. 2008. Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Mol. Cell. Biol. 28:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stevens, B. 1981. Mitochondrial structure, p. 471-488. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: life cycle and inheritance, 1st ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 63.Sugiana, C., D. J. Pagliarini, M. McKenzie, D. M. Kirby, R. Salemi, K. K. Abu-Amero, H. H. Dahl, W. M. Hutchison, K. A. Vascotto, S. M. Smith, R. F. Newbold, J. Christodoulou, S. Calvo, V. K. Mootha, M. T. Ryan, and D. R. Thorburn. 2008. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 83:468-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan, G., J. Lu, J. P. Bitoun, H. Huang, and H. Ding. 2009. IscA/SufA paralogues are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem. J. 420:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tong, W. H., and T. A. Rouault. 2006. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 3:199-210. [DOI] [PubMed] [Google Scholar]
- 66.Trounce, I. A., Y. L. Kim, A. S. Jun, and D. C. Wallace. 1996. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 264:484-509. [DOI] [PubMed] [Google Scholar]
- 67.Vahsen, N., C. Cande, J. J. Briere, P. Benit, N. Joza, N. Larochette, P. G. Mastroberardino, M. O. Pequignot, N. Casares, V. Lazar, O. Feraud, N. Debili, S. Wissing, S. Engelhardt, F. Madeo, M. Piacentini, J. M. Penninger, H. Schagger, P. Rustin, and G. Kroemer. 2004. AIF deficiency compromises oxidative phosphorylation. EMBO J. 23:4679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vogel, R. O., C. E. Dieteren, L. P. van den Heuvel, P. H. Willems, J. A. Smeitink, W. J. Koopman, and L. G. Nijtmans. 2007. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 282:7582-7590. [DOI] [PubMed] [Google Scholar]
- 69.Vogel, R. O., R. J. Janssen, C. Ugalde, M. Grovenstein, R. J. Huijbens, H. J. Visch, L. P. van den Heuvel, P. H. Willems, M. Zeviani, J. A. Smeitink, and L. G. Nijtmans. 2005. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 272:5317-5326. [DOI] [PubMed] [Google Scholar]
- 70.Vogel, R. O., R. J. Janssen, M. A. van den Brand, C. E. Dieteren, S. Verkaart, W. J. Koopman, P. H. Willems, W. Pluk, L. P. van den Heuvel, J. A. Smeitink, and L. G. Nijtmans. 2007. Cytosolic signaling protein Ecsit also localizes to mitochondria where it interacts with chaperone NDUFAF1 and functions in complex I assembly. Genes Dev. 21:615-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel, R. O., J. A. Smeitink, and L. G. Nijtmans. 2007. Human mitochondrial complex I assembly: a dynamic and versatile process. Biochim. Biophys. Acta 1767:1215-1227. [DOI] [PubMed] [Google Scholar]
- 72.Vyoral, D., and J. Petrak. 1998. Iron transport in K562 cells: a kinetic study using native gel electrophoresis and 59Fe autoradiography. Biochim. Biophys. Acta 1403:179-188. [DOI] [PubMed] [Google Scholar]
- 73.Wittig, I., H. P. Braun, and H. Schägger. 2006. Blue native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]
- 74.Wittig, I., R. Carrozzo, F. M. Santorelli, and H. Schägger. 2007. Functional assays in high-resolution clear native gels to quantify mitochondrial complexes in human biopsies and cell lines. Electrophoresis 28:3811-3820. [DOI] [PubMed] [Google Scholar]
- 75.Wittig, I., M. Karas, and H. Schägger. 2007. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6:1215-1225. [DOI] [PubMed] [Google Scholar]
- 76.Wittung-Stafshede, P. 2002. Role of cofactors in protein folding. Acc. Chem. Res. 35:201-208. [DOI] [PubMed] [Google Scholar]
- 77.Yano, T., V. D. Sled', T. Ohnishi, and T. Yagi. 1996. Expression and characterization of the flavoprotein subcomplex composed of 50-kDa (NQO1) and 25-kDa (NQO2) subunits of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. J. Biol. Chem. 271:5907-5913. [DOI] [PubMed] [Google Scholar]
- 78.Yano, T., T. Yagi, V. D. Sled', and T. Ohnishi. 1995. Expression and characterization of the 66-kilodalton (NQO3) iron-sulfur subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. J. Biol. Chem. 270:18264-18270. [DOI] [PubMed] [Google Scholar]
- 79.Zhang, Y., E. R. Lyver, E. Nakamaru-Ogiso, H. Yoon, B. Amutha, D. W. Lee, E. Bi, T. Ohnishi, F. Daldal, D. Pain, and A. Dancis. 2008. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28:5569-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng, L., U. Baumann, and J. L. Reymond. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou, G., S. S. Broyles, J. E. Dixon, and H. Zalkin. 1992. Avian glutamine phosphoribosylpyrophosphate amidotransferase propeptide processing and activity are dependent upon essential cysteine residues. J. Biol. Chem. 267:7936-7942. [PubMed] [Google Scholar]