Abstract
Programmed DNA double-strand breaks (DSBs) in meiosis are formed by Spo11 (Rec12 in fission yeast), a topoisomerase II-like protein, which becomes covalently attached to DNA 5′ ends. For DSB repair through homologous recombination, the protein must be removed from these DNA ends. We show here that Rec12 is endonucleolytically removed from DSB ends attached to a short oligonucleotide (Rec12-oligonucleotide complex), as is Spo11 in budding yeast. Fission yeast, however, has only one size class of Rec12-oligonucleotide complexes, whereas budding yeast has two size classes, suggesting different endonucleolytic regulatory mechanisms. Rec12-oligonucleotide generation strictly requires Ctp1 (Sae2 nuclease homolog), the Rad32 (Mre11) nuclease domain, and Rad50 of the MRN complex. Surprisingly, Nbs1 is not strictly required, indicating separable roles for the MRN subunits. On the basis of these and other data, we propose that Rad32 nuclease has the catalytic site for Rec12-oligonucleotide generation and is activated by Ctp1, which plays an additional role in meiotic recombination.
The repair of DNA double-strand breaks (DSBs) is essential for living cells. Faithful repair requires processing of the DSB for creation of a singe-stranded DNA end that can invade an intact homologous DNA template for repair. In some cases, a protein is bound to the DSB end and must be removed for repair to proceed. One notable example occurs during meiosis, when programmed DSBs are made by Spo11 (or its homolog), which becomes covalently linked to the 5′ DSB ends (17). Removal of the protein is essential for repair of the DSBs and subsequent formation of crossovers, which are important for the proper segregation of homologs at the first meiotic division as well as for the generation of genetic diversity. Removal of topoisomerases from DNA ends is also required for faithful repair when the topoisomerase reaction is aborted midway, as when cells are treated with topoisomerase inhibitors. Here, we address the mechanism of protein removal.
Meiotic recombination in the fission yeast Schizosaccharomyces pombe is initiated by the formation of programmed DSBs by Rec12, its Spo11 homolog (4). To date, DSBs have been demonstrated by direct analysis of DNA only for S. pombe and the budding yeast Saccharomyces cerevisiae. This mechanism of recombination initiation (by DSB introduction) is thought to occur in most eukaryotes, since Spo11 homologs are widely conserved evolutionarily (17). After their formation, DSBs are repaired via homologous recombination. At least in S. cerevisiae, the DNA ends are subjected to 5′-to-3′ exonucleolytic resection to create 3′ OH single-stranded DNA (ssDNA) overhangs (29). The resected DNA ends are then coated with an ssDNA-binding protein and invade homologous duplex DNA by the action of Rad51, a RecA homolog, and its accessory proteins.
Spo11 and its homologs, including Rec12, have sequence similarity to a type II topoisomerase from archaea, TopoVI (17). Consistent with this similarity, Spo11 and Rec12 introduce breaks in the DNA by a mechanism similar to that of topoisomerases: a dimer of the protein breaks both strands of the DNA molecule, and the conserved tyrosine of each monomer is covalently bound to the 5′ end of each DNA strand. The S. pombe Rec12 protein, including its highly conserved tyrosine residue thought to be at the active site, is essential for meiotic recombination and DSB formation (7).
After the introduction of DSBs, Rec12, like Spo11, remains covalently attached to the DNA end (15). In order to process these meiotic DSBs into a substrate capable of strand invasion, Rec12 has to be removed from the DNA end. A candidate for this removal is the MRN (Rad32-Rad50-Nbs1) nuclease complex, which is required for meiotic recombination. The requirement for the MRN complex in Spo11-mediated DSB formation in S. cerevisiae (3) complicates determination of its role in DSB processing. The MRN complex is not, however, required for DSB formation in S. pombe (34), an outcome that enables us to determine the role of this complex in Rec12 removal from meiotic DSBs. A recent study implicated two putative nucleases, Ctp1 and Rad32 (Mre11 homolog), along with its partner protein Rad50, in the removal of Rec12 from the DNA end (14), but the mechanism of Rec12 removal was not determined, nor were other proteins tested for possible roles. In budding yeast, Spo11 is removed by an endonucleolytic process, producing oligonucleotides of two distinct size classes attached to Spo11 (23). We asked whether the same mechanism applies in S. pombe Rec12 removal. Here, we use an assay for the Rec12-oligonucleotide complex to determine the mechanism of this crucial step of meiotic recombination and discuss a model for the initial steps of this process in S. pombe.
MATERIALS AND METHODS
Induction and analysis of meiosis.
The strains used are listed in Table 1. Strains were grown, induced for meiosis, and analyzed for cellular DNA content and meiotic DNA breakage as described by Young et al. (35). Meiotic recombination and viable-spore yields were conducted as described by Young et al. (34) and Ellermeier et al. (9).
TABLE 1.
S. pombe strains
| Strain | Genotypea |
|---|---|
| GP13 | h−ade6-52 |
| GP1293 | h+ade6-M26 arg1-14 |
| GP4117 | h−pat1-114 ade6-3049 |
| GP5374 | h−ade6-52 rad50S |
| GP6010 | h+rec12-201::His6-FLAG2pat1-114 ade6-3049 |
| GP6013 | h+rec12-201::His6-FLAG2pat1-114 rad50S ade6-3049 |
| GP6232 | h−/h−ade6-3049/ade6-3049 pat1-114/pat1-114 rec12-201::His6-FLAG2/rec12-201::His6-FLAG2+/his4-239 lys4-95/+ |
| GP6653 | h−rec12-201::His6-FLAG2pat1-114 ade6-3049 exo1-1::ura4+ |
| GP6654 | h+rec12-201::His6-FLAG2pat1-114 ade6-3049 ctp1::kanMX6 |
| GP6677 | h+rec12-201::His6-FLAG2pat1-114 ade6-3049 rad32-D65N |
| GP6730 | h+rec12-201::His6-FLAG2pat1-114 ade6-3049 rad50::kanMX6 |
| GP6733 | h+rec12-201::His6-FLAG2pat1-114 ade6-3049 rec10-175::kanMX6 |
| GP6848 | h−ade6-52 nbs1::kanMX6 |
| GP6849 | h−ade6-52 rad50::kanMX6 |
| GP6853 | h+ade6-M26 arg1-14 nbs1::kanMX6 |
| GP6892 | h+rec12-201::His6-FLAG2pat1-114 ade6-3049 nbs1::kanMX6 |
| GP6927 | h+ade6-M26 arg1-14 ura4-D18 rad32::ura4+ |
| GP6928 | h−ade6-52 ura4-D18 rad32::ura4+ |
| GP6930 | h+ade6-M26 arg1-14 ctp1::kanMX6 |
| GP6932 | h−ade6-52 ctp1::kanMX6 |
| GP6934 | h+ade6-M26 arg1-14 rad50S |
| GP6950 | h+ade6-M26 arg1-14 rad50::kanMX6 |
Isolation of Rec12-oligonucleotide complexes.
For protein extraction, cells containing Rec12-FLAG were opened by vigorous shaking with glass beads in ice-cold 10% trichloroacetic acid (1 ml/gm of wet cell paste) as described by Neale et al. (23). The precipitated proteins were solubilized in sodium dodecyl sulfate (SDS) extraction buffer (1 ml/gm of wet cell paste) (23). Soluble protein (0.5 to 2.0 mg) was diluted twofold with 2× immunoprecipitation (IP) buffer (23) and incubated overnight at 4°C with 5 μg of monoclonal anti-FLAG antibody (clone M2; Sigma-Aldrich) bound to 25 μl of magnetic protein G-agarose beads (Dynabeads; Invitrogen) prewashed with bovine serum albumin (5 mg/ml) in phosphate-buffered saline. After being collected and washed with 1× IP buffer, the immune complexes were washed with terminal deoxynucleotidyl transferase (TdT) buffer (New England Biolabs) and incubated for 1 h at 37°C in 12.5 μl 1× TdT buffer containing 0.5 mM CoCl2, 5 units of TdT (New England Biolabs), and 8 μCi of [α-32P]dCTP (3,000 Ci/mmol). Immune complexes were washed twice with 1× IP buffer and eluted by boiling them in 2× Laemmli buffer (23). Eluted proteins were fractionated by SDS-polyacrylamide gel electrophoresis using 10% precast gels in bis-Tris buffer (Invitrogen). Protein-oligonucleotide complexes were transferred to an Immobilon-P membrane (Millipore). To detect the protein-bound oligonucleotide, the membrane was exposed to X-ray film and to a Typhoon storage phosphor imaging system (GE Healthcare). For Western blot analysis, the membrane was probed with monoclonal anti-FLAG antibody conjugated to horseradish peroxidase (clone M2; 1:3,000 dilution; Sigma) and detected using an enhanced chemiluminescence detection kit (Supersignal; West Pico-Pierce).
Determination of oligonucleotide size.
To determine the length of the Rec12-associated oligonucleotides, radiolabeled complexes prepared as described above from 4.5 h after meiotic induction were treated with proteinase K (0.1 mg/ml; Roche) for 20 h at 50°C. An equal volume of 2× sequencing buffer (80% formamide, 10 mM EDTA, 0.1% xylene cyanol) was added to the proteinase K reaction mixture, which was boiled for 2 min and chilled on ice. Samples were fractionated on a “sequencing” gel (20% polyacrylamide [19:1 acrylamide-bisacrylamide], 7 M urea, 1× TBE, 18 cm long) at 700 V for 1.5 h. Gels were exposed to Biomax supersensitive film (Kodak) for 72 h as recommended by the manufacturer.
RESULTS
Rec12 is endonucleolytically removed from DSB ends.
To determine whether Rec12 is removed from DSBs in S. pombe by using a mechanism similar to that used for S. cerevisiae, we induced meiosis in a synchronized culture of cells expressing the functional Rec12-FLAG protein (5). At various times during meiosis, cells were broken open in the presence of 10% trichloroacetic acid to prevent degradation of intracellular intermediates and to precipitate proteins (23). After the proteins were dissolved in SDS, Rec12-FLAG was precipitated with anti-FLAG antibodies. To determine whether Rec12 had DNA attached to it, the immunoprecipitates were incubated with [α-32P]dCTP and TdT, which labels 3′ OH DNA ends. The components of the reaction mixture were resolved by SDS-polyacrylamide gel electrophoresis and transferred to a membrane, which was analyzed by Western blot analysis and autoradiography.
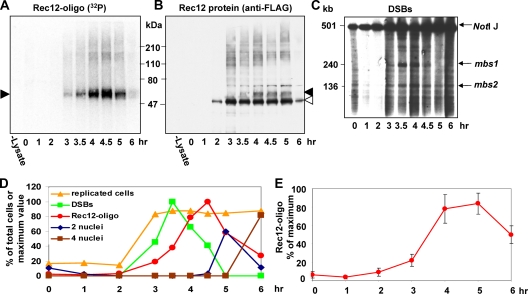
As expected, total Rec12-FLAG protein appeared 2 h after meiotic induction, and its level increased during meiosis; the protein's mobility indicated a mass of ∼47 kDa (Fig. 1). Three hours after induction of meiosis, a 32P-labeled radioactive species appeared and persisted until at least 5 or 6 h after induction (Fig. 1A). The major Rec12-FLAG species migrated faster than the 32P-labeled radioactive signal, which presumably represents Rec12 bound to a short oligonucleotide. A fainter band detected by the antibody migrated with the radioactive signal, which we designate the Rec12-oligonucleotide complex (Fig. 1B). Only about 10% of Rec12 protein was bound to the DNA. This result suggests that Rec12 is involved in steps of meiosis other than DSB formation, such as chromosome segregation (8, 27).
FIG. 1.
Rec12 is endonucleolytically removed from DNA at the time expected for a meiotic recombination intermediate. (A and B) Equal amounts of protein from meiotic cell extracts prepared at the indicated time after meiotic induction of strain GP6232 (rec12-201::His6-FLAG2) were immunoprecipitated and labeled with TdT and [α-32P]dCTP. The membrane was exposed to a Phosphorimager to detect 32P (A) and then probed with an anti-FLAG antibody conjugated to horseradish peroxidase to detect Rec12-FLAG (B). Filled arrowhead, Rec12 covalently bound to a short oligonucleotide (Rec12-oligo); open arrowhead, unbound Rec12 protein. The species with mobilities lower than those indicated by arrowheads may be Rec12 bound to DNA broken at nearly random sites (Fig. 2). (C) DNA was digested with NotI, subjected to pulsed-field gel electrophoresis, and Southern blot-hybridized with a probe from the left end of NotI fragment J, as described by Young et al. (35). Arrows indicate chromosome I NotI fragment J (top) and its two prominent DNA fragments broken at meiotic break sites mbs1 and mbs2. (D) Kinetics of the appearance of the Rec12-oligonucleotide complex (red circles) during meiosis. Orange triangles, percentage of cells with G2 DNA content, as measured by flow cytometry (data not shown); green squares, DSBs at meiotic break site mbs1 on NotI fragment J of chromosome I; blue diamonds, (nonnormalized) percentage of cells with two nuclei (meiosis I); brown squares, percentage of cells with four nuclei (meiosis II). (E) Reproducible kinetics of the appearance of the Rec12-oligonucleotide complex during meiosis. Data were normalized to the highest observed value for each experiment (at 4 or 5 h), set at 100%. Data points are the mean ± standard error of the mean (n = 4).
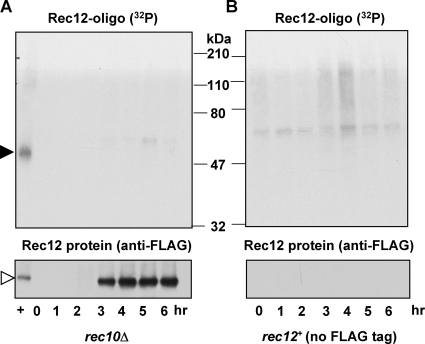
To determine if the appearance of the Rec12-oligonucleotide complex is dependent on DSB formation, we tested a mutant lacking Rec10, a linear element protein essential for DSB formation (10). Although Rec12 protein was readily detected by anti-FLAG antibody, no radioactive species was detectable (Fig. 2A). Thus, as expected, Rec12-oligonucleotide formation (“clipping”) depends on DSB formation.
FIG. 2.
Rec12-mediated DSB formation is required for formation of the Rec12-oligonucleotide complex. Meiotic inductions of a rec10Δ mutant strain (GP6733) (A) and a control strain (GP4117) with wild-type Rec12 (no FLAG tag) (B) are shown. Samples were treated as described for Fig. 1. Filled arrowhead, 32P-labeled Rec12-oligonucleotide complex; open arrowhead, Rec12-free protein. The faint, low-mobility 32P-labeled species in panel B may be Rec12-DNA complexes broken at nearly random sites and bound nonspecifically to the antibody beads (Fig. 1). “+” indicates rec10+ rec12-FLAG cell extract (strain GP6010 at 4 h).
The Rec12-oligonucleotide complex is kinetically competent to be an intermediate of meiotic recombination.
To be a kinetically competent intermediate of meiotic recombination, the Rec12-oligonucleotide complex should appear after the formation of DSBs and before the first meiotic division. To test this expectation, we studied the kinetics of Rec12-oligonucleotide formation in relation to premeiotic DNA replication, DSB formation, and the two meiotic divisions (Fig. 1D). DNA replication, monitored by flow cytometry (data not shown), occurred between 2 and 3 h after meiotic induction. DSBs at the meiotic hot spot mbs1, detected by Southern blot hybridization, were first visible 3 h after meiotic induction, were maximal at 3.5 h, and largely disappeared by 5 h (Fig. 1C). As assayed by microscopy of stained nuclei, meioses I and II were nearly completed by ∼5 h and ∼6 h, respectively (Fig. 1D). These events occurred at approximately the times reported previously (6). The maximal abundance of the Rec12-oligonucleotide complex occurred about an hour after maximal DSB abundance and about a half hour before meiosis I (Fig. 1C, D, and E). The appearance of this species between DSB formation and meiosis I makes it kinetically competent to be a recombination intermediate. From this outcome and the genetic requirements for its production (see also below), we conclude that Rec12-oligonucleotide formation is an essential step in meiotic recombination.
A single size distribution of the Rec12-oligonucleotide complex suggests a nonconserved regulation of endonucleolytic cleavage in meiosis.
In S. cerevisiae meiosis, Spo11 attached to oligonucleotides appears as two distinct nucleoprotein complexes with different electrophoretic mobilities (23). After release from Spo11 by proteolysis, the oligonucleotides migrate during gel electrophoresis also as two distinct size classes, approximately 12 to 26 nucleotides (nt) and 28 to 34 nt long. In contrast, a similar analysis of S. pombe Rec12 revealed only one size class of Rec12-oligonucleotide complexes (Fig. 1A).
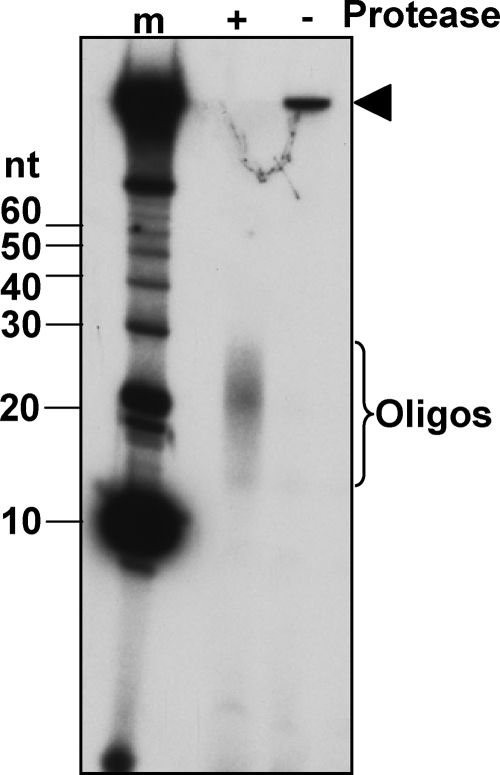
To test further that there is indeed only one size class of complexes and to determine the lengths of the attached oligonucleotides, we treated the 32P-labeled Rec12-oligonucleotide complexes with proteinase K to degrade the covalently attached Rec12 protein and resolved the 32P-labeled oligonucleotides on a denaturing (“sequencing”) gel. After proteinase K treatment, we detected only one size class of oligonucleotides (Fig. 3). The mode of the distribution was ∼22 nt, and the range was ∼13 to 29 nt. These results show that indeed only one size class of Rec12-oligonucleotide complex is formed in S. pombe. We discuss the significance of this result later.
FIG. 3.
Single size distribution of the Rec12-oligonucleotide complex. The Rec12-oligonucleotide complex was prepared 4.5 h after meiotic induction of strain GP6010 as in Fig. 1. The protein-DNA complexes were treated (+) or not treated (−) with proteinase K. The reaction mixture was analyzed on a “sequencing” gel alongside a set of 5′ 32P-labeled oligonucleotide markers (Invitrogen) (lane m) and autoradiographed. Filled arrowhead, Rec12 covalently bound to a short oligonucleotide (Rec12-oligonucleotide complex).
The nuclease activity of the MRN complex is required for Rec12 clipping.
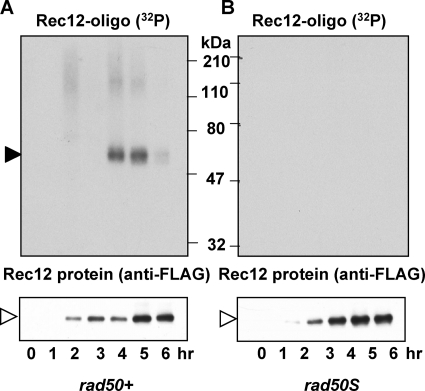
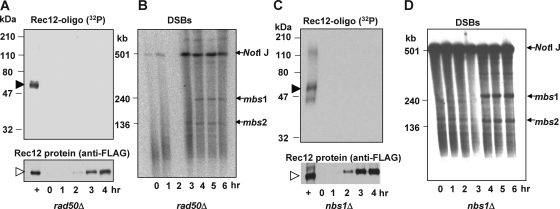
To determine the enzymes required for formation of the Rec12-oligonucleotide complex, we tested a series of mutants deficient in nucleases that were previously implicated in the initial steps of homologous recombination (7). The MRN complex, composed of Rad32, Rad50, and Nbs1, is a strong candidate for this reaction. In the rad50S missense mutant with an altered MRN complex, Rec12 forms DSBs but remains covalently bound to the DNA (15). As expected, we could not detect the Rec12-oligonucleotide complex in this mutant at any time after induction, although Rec12 protein was induced to the level seen in the wild type (Fig. 4A and B). We could, however, detect the radioactive signal in a wild-type strain analyzed in parallel (Fig. 4A).
FIG. 4.
The rad50S mutation blocks Rec12-oligonucleotide formation. Meiotic induction of a rad50+ control strain GP6010 (A) and rad50S mutant strain GP6013 (B) is shown. Samples were treated as described for Fig. 1. Filled arrowhead, 32P-labeled Rec12-oligonucleotide complex; open arrowhead, Rec12-free protein.
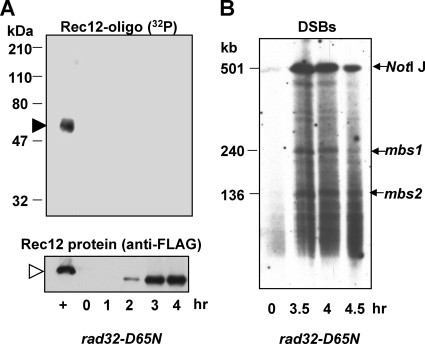
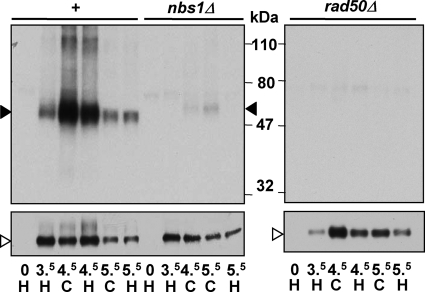
Next, we tested whether the putative nuclease active site of the MRN complex, which resides in Rad32 (33), is required for Rec12-oligonucleotide generation. The rad32-D65N mutant is equivalent to the S. cerevisiae mre11-D56N mutant, which lacks nuclease activity but forms an intact MRN (MRX) complex (19, 20); the S. pombe rad32-D65N mutant also forms the MRN complex (25). In this putative nuclease-deficient mutant, the Rec12-oligonucleotide complex was not detectable (Fig. 5A), although Rec12 was expressed and DSBs were formed and persisted to at least 6 h after induction (Fig. 5B and unpublished data). Rad32 nuclease activity per se is not sufficient for Rec12-oligonucleotide generation, since rad50Δ and nbs1Δ strains were also deficient in Rec12-oligonucleotide formation, although Rec12 was expressed and DSBs were formed in these mutants (Fig. 6A to D). In summary, the MRN subunits Rad50 and Nbs1 and the putative nuclease active site in Rad32 are required for clipping off Rec12. Below, we show that Nbs1 is not, however, strictly required for clipping.
FIG. 5.
The nuclease activity of the MRN complex is necessary for Rec12-oligonucleotide formation. (A) Meiotic induction of nuclease-deficient mutant rad32-D65N mutant strain GP6677. Samples were treated as described for Fig. 1. “+” indicates rad32+ cell extract of strain GP6010 at 4 h. Filled arrowhead, 32P-labeled Rec12-oligonucleotide complex; open arrowhead, Rec12-free protein. (B) DSB analysis of DNA from the meiotic induction shown in panel A. Arrows, chromosome I NotI fragment J (top) and its two prominent DNA fragments broken at meiotic break sites mbs1 and mbs2.
FIG. 6.
The intact MRN complex is required for Rec12-oligonucleotide formation. Meiotic inductions of a rad50Δ mutant strain (GP6730) (A and B) and an nbs1Δ mutant strain (GP6892) (C and D) are shown. Samples were treated as described for Fig. 1. Filled arrowhead, 32P-labeled Rec12-oligonucleotide complex; open arrowhead, Rec12-free protein. “+” indicates rad50+ nbs1+ extracts of strain GP6010 at 4 h. Panels B and D show DSB analysis of DNA from the meiotic inductions shown in panels A and C, respectively. Arrows, chromosome I NotI fragment J (top) and its two prominent DNA fragments broken at meiotic break sites mbs1 and mbs2.
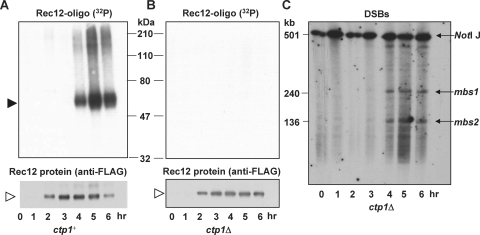
The putative nuclease Ctp1 is also essential for Rec12-oligonucleotide generation.
Ctp1, also called Nip1 (1, 22), is a conserved protein important for DSB repair through homologous recombination in S. pombe. Its homolog Sae2 in budding yeast has nuclease activity in vitro (21) and is required for Spo11-oligonucleotide formation in S. cerevisiae (23). It is not known, however, whether the nucleolytic activity of Sae2 is required for Spo11 clipping. CtIP, the human homolog of Ctp1, is essential for DSB resection in mitotic cells (26). To determine if Ctp1 is also essential for the formation of the Rec12-oligonucleotide complex, we tested a ctp1Δ mutant and found that the Rec12-oligonucleotide complex was not generated, although, as for the MRN mutants, Rec12 was expressed and DSBs were formed and persisted beyond their normal time of repair (Fig. 7). These results show that two putative nucleolytic proteins, Ctp1 and Rad32, are required for Rec12 clipping. We discuss the implications of this unexpected outcome later.
FIG. 7.
Ctp1 is a second putative nuclease necessary for Rec12-oligonucleotide formation. Meiotic induction of ctp1+ control strain GP6010 (A) and ctp1Δ mutant strain GP6654 (B) is shown. Samples were treated as described for Fig. 1. Filled arrowhead, 32P-labeled Rec12-oligonucleotide complex; open arrowhead, Rec12-free protein. (C) DSB analysis of DNA from the meiotic induction shown in panel B. Arrows, chromosome I NotI fragment J (top) and its two prominent DNA fragments broken at meiotic break sites mbs1 and mbs2.
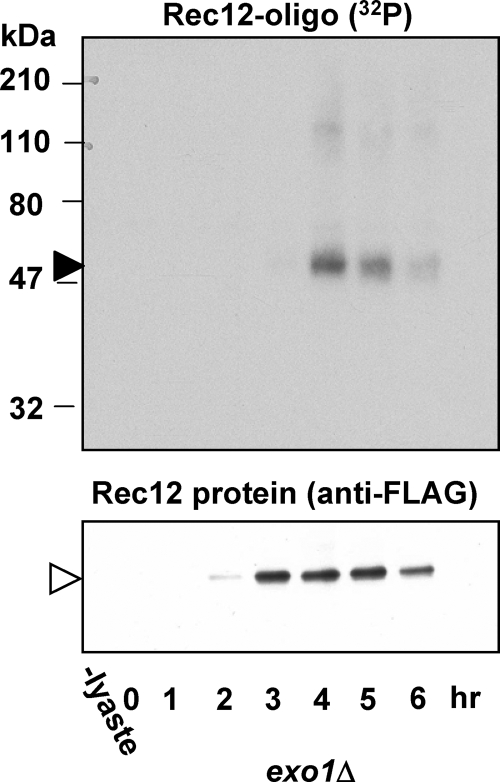
We tested another nuclease, exonuclease I (ExoI), which specifically digests double-stranded DNA in the 5′-to-3′ direction (30), as would be required for conversion of meiotic DSBs into invasive DNA ends with long 3′ single-stranded tails. We found that an exo1Δ mutant was fully proficient in forming the Rec12-oligonucleotide complex (Fig. 8). We discuss below a possible role for ExoI in meiotic recombination.
FIG. 8.
Exonuclease I is dispensable for Rec12-oligonucleotide formation. Meiotic induction of exo1Δ mutant strain GP6653 is shown. Samples were treated as described for Fig. 1. Filled arrowhead, 32P-labeled Rec12-oligonucleotide complex; open arrowhead, Rec12-free protein.
Nbs1 and Rad50 are differentially required for viable-spore formation and meiotic recombination.
Since MRN and Ctp1 are required for Rec12 clipping, we would expect each of the four polypeptides to be required for meiotic recombination and generation of viable spores. As expected, rad50Δ, rad32Δ, and ctp1Δ mutants produced <0.2% of the viable-spore yield of the wild type (10, 22, 34) (Table 2). Unexpectedly, the nbs1Δ mutant produced 26% of the wild-type viable-spore yield. These crosses were done at 25°C, the standard temperature for S. pombe meiotic crosses (13). Since we assayed DNA intermediates, including the Rec12-oligonucleotide complex, in synchronized meiotic cultures at 34°C, we repeated these crosses at 34°C. At this temperature, nbs1Δ yielded markedly fewer viable spores (1.2% of the wild-type level) but still more than eightfold more than rad50Δ, rad32Δ, or ctp1Δ at 25°C. Thus, the Rad50 and Rad32 subunits of the MRN complex and Ctp1 are more stringently required for producing viable spores than is Nbs1, although all four subunits appear indispensable for producing the Rec12-oligonucleotide complex, at least at 34°C (Fig. 5A, 6A and C, and 7B; see also “Nbs1 is not strictly required for Rec12-oligonucleotide generation,” below). Our assay for the Rec12-oligonucleotide complex may not be sufficiently sensitive, however, to detect the low level of the Rec12-oligonucleotide complex expected in the nbs1Δ mutant at 34°C (a small percentage of the wild-type level, based on the viable-spore yield).
TABLE 2.
Differential requirements for Nbs1 versus Rad50, Rad32, and Ctp1 in viable-spore formation and meiotic recombination
| Mutation (temp)a | Viable-spore yield (% of wild-type level)b,c | ade6-arg1 intergenic-recombination level (cM)c,d | ade6 intragenic-recombination level (no. of Ade+ recombinants/106 viable spores)c |
|---|---|---|---|
| None | 100 | 74 ± 13 | 3,500 ± 490 |
| None (34°C) | 100 | 54 | 4,500 |
| nbs1Δ | 26 ± 5 | 10 ± 1.6 | 580 ± 40 |
| nbs1Δ (34°C) | 1.2 ± 0.3 | 10 ± 2.5 | −e |
| rad50S | 30 ± 3 | 64 ± 21 | 3,700 ± 280 |
| rad50Δ | 0.15 | 0.7 ± 0.1 | − |
| rad32Δ | 0.09 | 1.8 ± 0.5 | − |
| ctp1Δ | 0.002 ± <0.0001 | 1 | − |
Crosses were conducted at 25°C, except as noted.
In the wild type, 2.0 ± 0.5 viable spores per viable cell were obtained at 25°C and 0.15 at 34°C.
Mean ± standard error of the mean of results from 4 to 33 crosses except for the wild type at 34°C.
Spore colonies (470 to 2,348) from 4 to 13 crosses were tested for ade6-arg1 recombinants, except for the wild type at 34°C (1 cross, 140 colonies) and ctp1Δ (33 crosses, 204 colonies) (the intergenic-recombination data were pooled). Frequencies were converted to cM by using Haldane's equation.
The viable-spore yield was too low for an accurate determination.
To determine the requirements for the MRN complex and Ctp1 in meiotic recombination, we measured both crossing over (as ade6-arg1 intergenic recombination) and gene conversion (as ade6 intragenic recombination). While at 25°C crossing over was reduced by a factor of more than 70 in rad50Δ, rad32Δ, and ctp1Δ mutants, it was reduced by a factor of only ∼6 in nbs1Δ at both low and high temperatures. Gene conversion was also reduced by a factor of ∼6 in nbs1Δ at 25°C (Table 2). The low viable-spore yields in the rad50Δ, rad32Δ, and ctp1Δ mutants did not allow a reliable estimate of gene conversion at either temperature; the same was true for nbs1Δ at a high temperature. In summary, Rad50, Rad32, and Ctp1 are strongly required for formation of viable spores and meiotic recombinants, but surprisingly, Nbs1 is less strictly required, especially at a low temperature.
Nbs1 is not strictly required for Rec12-oligonucleotide generation.
To test the suggestion that Nbs1 is not strictly required for Rec12 clipping at a low temperature, we induced a culture for meiosis at 34°C and then reduced the temperature to 25°C at 3.5 or 4 h, when most DSBs have formed and Rec12-oligonucleotide generation has begun (Fig. 9). With this regimen, we detected a low level of the Rec12-oligonucleotide complex in the nbs1Δ mutant at 25°C but none in the rad50Δ mutant (Fig. 9). The level of the Rec12-oligonucleotide complex at 5.5 h was similar to the level of the viable-spore yield, about one-fourth of that for the wild type (Table 2). These results show that, as predicted, Nbs1 is not strictly required for Rec12 clipping, although Rad50 appears to be. We propose below an explanation for the differential requirements for the MRN subunits in meiotic recombination and DNA break repair.
FIG. 9.
Nbs1 is not strictly required for Rec12-oligonucleotide formation. Strains GP6010 (nbs1+), GP6892 (nbs1Δ), and GP6730 (rad50Δ) were induced as shown in Fig. 1 at 34°C (H) and cooled to 25°C (C) at 3.5 h. Samples were treated as described for Fig. 1. Filled arrowhead, 32P-labeled Rec12-oligonucleotide complex; open arrowhead, Rec12 free protein. A repeat of this experiment gave similar results (data not shown).
DISCUSSION
We report here that fission yeast Rec12 is clipped off the end of meiotic DSBs by the action of an endonuclease, producing a short oligonucleotide covalently linked to Rec12. This endonucleolytic cleavage requires the nuclease activity of the Rad32 (Mre11) component of the MRN complex and the Ctp1 putative nuclease (Table 3). The regulation of this key step in meiotic recombination appears to be distinctly different from that reported for budding yeast (23). We discuss below the implications of these and other results for the processing of meiotic DSBs and the formation of recombinants (Fig. 10) .
TABLE 3.
Summary of gene products required for Rec12-oligonucleotide formationa
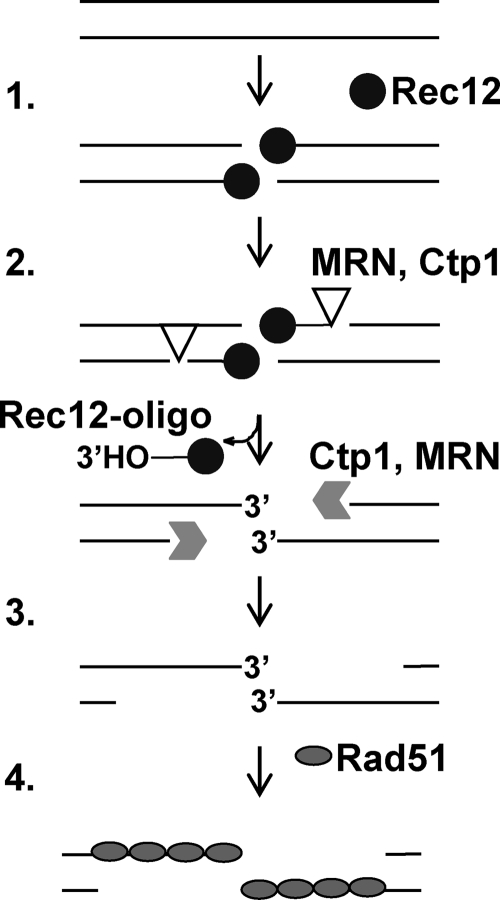
FIG. 10.
Model for the initial steps of meiotic DSB repair in S. pombe. (1) Rec12 and other proteins bind to the DNA after premeiotic replication and generate a DSB. Rec12 remains covalently bound to the DNA ends. (2) The Rad32 nuclease, as part of the MRN complex and activated by Ctp1, removes Rec12 bound to a short oligonucleotide. Failure to remove Rec12 from the DNA ends causes persistence of the breaks. (3) The DSB ends are resected by Ctp1, activated by MRN, to create 3′ OH ssDNA tails. (4) The ssDNA tails are coated by Rad51 and invade intact DNA (not shown) to repair the DSB and produce homologous recombinants.
Rec12 is removed from DSB ends via endonucleolytic cleavage.
Although the biochemical mechanism of meiotic DSB formation in S. pombe seems to be similar or identical to that in S. cerevisiae, many aspects of the regulation of DSB formation and repair are not conserved between these two distantly related yeasts (7). S. cerevisiae Spo11 is endonucleolytically clipped from the DNA end with a spectrum of short oligonucleotides attached to it; this spectrum is bimodal in length, with modes of ∼15 and ∼30 nt (23). We found that S. pombe Rec12 is also removed from DNA attached to a spectrum of oligonucleotides, suggesting that the basic endonucleolytic mechanism of removal is conserved. However, we found a unimodal distribution of oligonucleotide lengths, with a mode of ∼22 nt (Fig. 3). While the molecular basis of this difference is not currently clear, it may be that in S. cerevisiae some protein excludes endonucleolytic cleavage of DNA ∼20 to 25 nt from Spo11; this protein may not be present at meiotic DSBs in S. pombe. This suggestion is consistent with the idea that certain proteins, notably the MRN complex, are required for DSB formation in S. cerevisiae but not in S. pombe (7).
An additional dramatic difference in the regulation of DSB end processing in these yeasts is the time between DSB formation and removal of Spo11 or Rec12. The S. pombe Rec12-oligonucleotide complex was most abundant about an hour after maximal DSB abundance (Fig. 1D and E), indicating a distinct lag between DSB formation and clipping off of Rec12. In S. cerevisiae, Spo11-oligonucleotide formation occurs nearly immediately after DSB formation (23). This difference in timing accounts for the ability to detect Rec12 covalently bound to DNA in wild-type (i.e., rad50+) S. pombe (15) but apparently not in S. cerevisiae (18). These differences parallel the requirement for the MRN complex in DSB formation in S. cerevisiae but not in S. pombe (3, 34). Thus, in S. cerevisiae the MRN complex is in place to remove Spo11 immediately after the DSB is formed, whereas in S. pombe, DSBs are formed without MRN, which may take an hour to be recruited to DSBs or activated to remove Rec12.
Two putative nucleases are required for Rec12 clipping.
Two proteins with a putative nuclease activity, Ctp1 and Rad32, are required for the formation of the Rec12-oligonucleotide complex (Fig. 5A and 7B). Hartsuiker et al. (14) recently reported that both proteins are required for the disappearance of Rec12-DNA complexes detected by antibody to epitope-tagged Rec12, but the product of this reaction was not determined. In their study, as in ours (Fig. 5A), the rad32-D65N mutant, which lacks the putative nuclease active site (33) but forms the MRN complex (25), was as defective as a rad32Δ mutant. Ctp1 was also required in both studies. Ctp1 is a distant homolog of S. cerevisiae Sae2, which has nuclease activity in vitro (21), and of human CtIP, which in combination with a low level of Mre11-Rad50 (MR) has nuclease activity lacking with each single-protein preparation (26). Thus, we infer that two nucleases are required for a simple endonucleolytic cleavage.
To account for this conundrum, we propose that Rad32, as part of the MRN complex, is the endonuclease that clips off Rec12 and that Ctp1 is required for the activity of Rad32. This proposal is based in part on our recent report that meiotic recombination stimulated by a protein-free DSB is not blocked by the rad32-D65N mutation but is blocked by ctp1Δ (11). In this case, DSB resection but not protein removal is required for recombination. We propose that Ctp1 is the major resecting nuclease in S. pombe meiosis and that its binding near a DSB end enables Rad32 to clip off Rec12. We further propose that the MRN complex activates Ctp1, because Exo1, rather than Ctp1, is the major resecting nuclease in the absence of Rad50 (11). In S. pombe, Rec12 makes DSBs in the absence of MRN or Ctp1 (1, 34) (Fig. 5B, 6B and D, and 7C). We propose that after DSB formation, MRN and Ctp1 bind and positively regulate each other. This mechanism would coordinate Rec12 clipping and DNA resection for rapid and efficient DSB repair and recombination.
The role of Nbs1 differs from that of Rad50 and Rad32.
As expected from the failure of Rec12 to be removed from DSB ends in rad50Δ and rad32-D65N mutants (Fig. 5A and 6A), rad50Δ and rad32Δ mutants formed <0.2% as many viable spores as the wild type (Table 2) (10, 34). Unexpectedly, however, nbs1Δ mutants formed at least eight times more viable spores than rad50Δ or rad32Δ mutants; at a low temperature, the yield in nbs1Δ was even greater, about one-fourth that of the wild type and >150 times higher than that of rad50Δ or rad32Δ. At a low temperature, the nbs1Δ mutant produced readily detectable Rec12-oligonucleotide complexes, whereas the rad50Δ mutant did not; at a high temperature, neither mutant produced detectable Rec12-oligonucleotide complexes (Fig. 6A and C and 9). To our knowledge, this is the first report of distinct phenotypes of null (deletion) mutations in the MRN genes in S. pombe. In S. cerevisiae, xrs2Δ strongly reduces binding of Mre11 to DNA at a meiotic DSB hot spot, but rad50Δ only partially reduces this binding (2).
The curious temperature sensitivity of nbs1Δ suggests that some function can at least partially substitute for Nbs1 at a low temperature but not at a high temperature. One possibility, which we favor, is that MR can at least partially function without Nbs1 at a low temperature but that MR requires Nbs1 for stabilization or activation of MR at a high temperature. The MRN complexes of the two yeasts discussed here may differ in their requirements for an additional subunit for stabilization or activation of the MR complex at a low temperature. Consistent with our proposal, purified human Nbs1 stabilizes MR complexes (24). Additional experiments may support this suggestion and reveal other differences in the roles of the MRN subunits in DSB repair and recombination.
Acknowledgments
We are especially grateful to Scott Keeney and Matthew Neale for detailed protocols and advice on detecting Rec12-oligonucleotide complexes. We thank Paul Russell for ctp1Δ and nbs1Δ mutants; Randy Hyppa for unpublished data and technical help; and Sue Amundsen, Gareth Cromie, Randy Hyppa, and Naina Phadnis for comments on the manuscript.
This work was supported by research grant GM032194 from the National Institutes of Health to G.R.S.
Footnotes
Published ahead of print on 14 September 2009.
REFERENCES
- 1.Akamatsu, Y., Y. Murayama, T. Yamada, T. Nakazaki, Y. Tsutsui, K. Ohta, and H. Iwasaki. 2008. Molecular characterization of the role of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double-strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol. Cell. Biol. 28:3639-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borde, V., W. Lin, E. Novikov, J. H. Petrini, M. Lichten, and A. Nicolas. 2004. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell 13:389-401. [DOI] [PubMed] [Google Scholar]
- 3.Cao, L., E. Alani, and N. Kleckner. 1990. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61:1089-1101. [DOI] [PubMed] [Google Scholar]
- 4.Cervantes, M. D., J. A. Farah, and G. R. Smith. 2000. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5:883-888. [DOI] [PubMed] [Google Scholar]
- 5.Cromie, G. A., R. W. Hyppa, H. P. Cam, J. A. Farah, S. I. Grewal, and G. R. Smith. 2007. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genet. 3:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cromie, G. A., R. W. Hyppa, A. F. Taylor, K. Zakharyevich, N. Hunter, and G. R. Smith. 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127:1167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cromie, G. A., and G. R. Smith. 2008. Meiotic recombination in Schizosaccharomyces pombe: a paradigm for genetic and molecular analysis, p. 195-230. In R. Egel and D.-H. Lankenau (ed.), Genome dynamics and stability, vol. 3: recombination and meiosis. Springer-Verlag, Heidelberg, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, L., and G. R. Smith. 2003. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163:857-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellermeier, C., H. Schmidt, and G. R. Smith. 2004. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics 168:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellermeier, C., and G. R. Smith. 2005. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 102:10952-10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farah, J. A., G. A. Cromie, and G. R. Smith. 2009. Ctp1 and Exonuclease1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc. Natl. Acad. Sci. USA 106:9356-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farah, J. A., E. Hartsuiker, K. Mizuno, K. Ohta, and G. R. Smith. 2002. A 160-bp palindrome is a Rad50·Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 161:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe, p. 395-446. In R. King (ed.), Handbook of genetics, vol. 1. Plenum Press, New York, NY. [Google Scholar]
- 14.Hartsuiker, E., K. Mizuno, M. Molnar, J. Kohli, K. Ohta, and A. M. Carr. 2009. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell. Biol. 29:1671-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyppa, R. W., G. A. Cromie, and G. R. Smith. 2008. Indistinguishable landscapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genet. 4:e1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iino, Y., and M. Yamamoto. 1985. Negative control for the initiation of meiosis in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 82:2447-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 18.Keeney, S., and N. Kleckner. 1995. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc. Natl. Acad. Sci. USA 92:11274-11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogh, B. O., B. Llorente, A. Lam, and L. S. Symington. 2005. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 171:1561-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh, B. O., and L. S. Symington. 2004. Recombination proteins in yeast. Annu. Rev. Genet. 38:233-271. [DOI] [PubMed] [Google Scholar]
- 21.Lengsfeld, B. M., A. J. Rattray, V. Bhaskara, R. Ghirlando, and T. T. Paull. 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell 28:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Limbo, O., C. Chahwan, Y. Yamada, R. A. de Bruin, C. Wittenberg, and P. Russell. 2007. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 28:134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neale, M. J., J. Pan, and S. Keeney. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13:1276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porter-Goff, M. E., and N. Rhind. 2009. The role of MRN in the S-phase DNA damage checkpoint is independent of its Ctp1-dependent roles in double-strand break repair and checkpoint signaling. Mol. Biol. Cell 20:2096-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sartori, A. A., C. Lukas, J. Coates, M. Mistrik, S. Fu, J. Bartek, R. Baer, J. Lukas, and S. P. Jackson. 2007. Human CtIP promotes DNA end resection. Nature 450:509-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharif, W. D., G. G. Glick, M. K. Davidson, and W. P. Wahls. 2002. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steiner, W. W., and G. R. Smith. 2005. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics 169:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, H., D. Treco, and J. W. Szostak. 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64:1155-1161. [DOI] [PubMed] [Google Scholar]
- 30.Szankasi, P., and G. R. Smith. 1992. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J. Biol. Chem. 267:3014-3023. [PubMed] [Google Scholar]
- 31.Szankasi, P., and G. R. Smith. 1995. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science 267:1166-1169. [DOI] [PubMed] [Google Scholar]
- 32.Ueno, M., T. Nakazaki, Y. Akamatsu, K. Watanabe, K. Tomita, H. D. Lindsay, H. Shinagawa, and H. Iwasaki. 2003. Molecular characterization of the Schizosaccharomyces pombe nbs1+ gene involved in DNA repair and telomere maintenance. Mol. Cell. Biol. 23:6553-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, S., M. Tavassoli, and F. Z. Watts. 1998. Schizosaccharomyces pombe rad32 protein: a phosphoprotein with an essential phosphoesterase motif required for repair of DNA double strand breaks. Nucleic Acids Res. 26:5261-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young, J. A., R. W. Hyppa, and G. R. Smith. 2004. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young, J. A., R. W. Schreckhise, W. W. Steiner, and G. R. Smith. 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9:253-263. [DOI] [PubMed] [Google Scholar]