Abstract
The TOM complex is the general mitochondrial entry site for newly synthesized proteins. Precursors of β-barrel proteins initially follow this common pathway and are then relayed to the SAM/TOB complex, which mediates their integration into the outer membrane. Three proteins, Sam50 (Tob55), Sam35 (Tob38/Tom38), and Sam37 (Mas37), have been identified as the core constituents of the latter complex. Sam37 is essential for growth at elevated temperatures, but the function of the protein is currently unresolved. To identify interacting partners of Sam37 and thus shed light on its function, we screened for multicopy suppressors of sam37Δ. We identified the small subunit of the TOM complex, Tom6, as such a suppressor and found a tight genetic interaction between the two proteins. Overexpression of SAM37 suppresses the growth phenotype of tom6Δ, and cells lacking both genes are not viable. The ability of large amounts of Tom6 to suppress the sam37Δ phenotype can be linked to the capacity of Tom6 to stabilize Tom40, an essential β-barrel protein which is the central component of the TOM complex. Our results suggest that Sam37 is required for growth at higher temperatures, since it enhances the biogenesis of Tom40, and this requirement can be overruled by improved stability of newly synthesized Tom40 molecules.
The vast majority of mitochondrial proteins are nuclearly encoded and are synthesized in the cytosol. The precursor proteins that contain targeting and sorting signals are then imported into the organelle and sorted into the correct intramitochondrial compartment. The translocase of the mitochondrial outer membrane (TOM complex) serves as the mitochondrial entry gate for the vast majority of protein precursors. The TOM complex mediates the translocation of precursor proteins across the mitochondrial outer membrane. Upon exit from the TOM import pore, the precursor proteins are distributed to different import routes according to their final destination and their sorting signals (5, 34).
Two forms of the TOM complex were defined: the holocomplex and the core complex. The TOM core complex comprises Tom40, Tom22 and the three small Tom proteins, Tom5, Tom6, and Tom7 (2, 8). The holo-TOM complex additionally includes the primary import receptors, Tom20 and Tom70 (24, 32). Tom40, an essential protein which forms a membrane-embedded β-barrel structure, is the central component of the general import pore of the TOM complex (1, 16). Tom22 has a dual function: it can serve as a receptor protein, and it plays a critical role in the integrity of the TOM complex (21, 27, 46). The small Tom components Tom5, Tom6, and Tom7 are between 50 and 70 amino acids in length and are assumed to regulate the organization of the complex. None of these three small proteins is essential when deleted individually. Tom5 was reported to have a function in recognizing precursor proteins and may have in addition a general role in TOM complex stability (10, 42). Tom6 and Tom7 are suggested to act as antagonists in the regulation of TOM complex organization. Tom6 supports stability, probably by serving as an adaptor for interaction between Tom22 and Tom40 (3, 9). Tom7 in turn may be a negative regulator by promoting the dissociation of the subunits in the complex (17).
β-Barrel proteins are a group of mitochondrial outer membrane proteins which are characterized by their distinct topology. These proteins form a cylindrically shaped structure composed of β-strands (52). Notably, β-barrels are found only in the outer membranes of gram-negative bacteria and eukaryotic organelles of endosymbiotic origin, namely, chloroplasts and mitochondria (12, 38). Precursors of β-barrel proteins are recognized first by the receptors of the TOM complex before their transfer through the import channel to the intermembrane space (23, 39). Next, they are relayed to a specialized heterooligomeric protein complex in the outer membrane, termed either TOB complex, for topogenesis of outer membrane β-barrel proteins (35), or SAM complex, for sorting and assembly machinery (22, 50). The SAM/TOB complex mediates the membrane insertion of the β-barrel substrates.
In yeast, three proteins, Sam50 (Tob55/Omp85), Sam35 (Tob38/Tom38), and Sam37 (Mas37/Tom37), have been identified as core constituents of this complex. The central element of the SAM complex, Sam50, is a β-barrel protein and has homologous proteins in all known eukaryotes (11, 13). It is proposed to play a key role in facilitating the membrane integration of β-barrel precursors (13, 22, 35). The function of the remaining SAM subunits has not been resolved. For Sam35, it was suggested that it may stabilize the structure of Sam50 and contribute to precursor binding in cooperation with Sam50 (6, 15, 19, 30, 47). Very recently, a direct interaction of Sam35 with a sorting signal of β-barrel proteins was demonstrated (25). Unlike the case with the previous two components, which are essential for viability, sam37Δ strains exhibit only a slight growth defect at 30°C, whereas growth is strongly impaired at elevated temperatures (14, 50). Similar to Sam35, Sam37 behaves like a peripheral membrane protein. It is both extractable from the membrane under alkaline conditions and degradable by treatment of intact mitochondria with external protease (40, 50). Besides a general role in promoting SAM complex stability, recent reports proposed a potential function of Sam37 in the release of β-barrel precursors from the SAM complex (6, 15).
Currently the molecular mechanism of the function of the SAM complex is only poorly understood, and the mode of interaction between the TOM and SAM complexes is unclear. In particular, little is known of the distinct function of Sam37. To gain insight into the contribution of this protein to the overall function of the SAM complex and to the biogenesis of β-barrel proteins, we screened for multicopy suppressors of the sam37Δ phenotype. Surprisingly, we identified the small subunit of the TOM complex, Tom6, as such a suppressor and observed further tight genetic interactions between TOM6 and SAM37. Collectively, our results shed new light on the mode of action of Sam37 and demonstrate functional interactions between the TOM and SAM complexes.
MATERIALS AND METHODS
Yeast strains and growth.
Standard genetic techniques were used for growth and manipulation of Saccharomyces cerevisiae cells. Unless stated otherwise, the strains used were isogenic to YPH499, YPH500, and YPH501 (43). The sam37Δ strain on the YPH499 background was described previously (15), and that on the BY4741 background was purchased from Research Genetics. Transformation of yeast was carried out in most cases according to the lithium acetate method. Yeast cells were grown under aerobic conditions on YPD (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto peptone, 2% glucose), YPG (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto peptone, 3% glycerol), or synthetic medium (SD). For growth tests on plates, cells were grown to log phase and diluted to an optical density at 600 nm of 1.0. Cells were then diluted in fivefold increments, and 5 μl of each was spotted onto the appropriate solid medium.
Yeast genes were deleted by a PCR-based approach using the HIS3 marker amplified from the pFA6a-HIS3MX6 plasmid or the kanamycin resistance cassette amplified from the pFA6a-kanMX4 plasmid. The complete coding sequences including the start and stop codons were deleted by homologous recombination. The TOM6 and TOM7 genes were deleted in strain YPH500 using the kanMX4 cassette. For construction of double mutants, deletion strains carrying the kanMX4 marker were mated with sam37Δ cells of the opposite mating type carrying the HIS3MX6 cassette. Heterozygous diploids were selected on medium lacking histidine and containing G418, sporulated, and subjected to tetrad dissection. Genotypes were determined by selection with the markers and confirmed by PCR. To confirm the synthetic lethality phenotype, the heterozygous diploids were transformed prior to sporulation with a plasmid containing the URA3 marker gene and carrying either TOM6 or SAM37. Double mutant spores harboring a plasmid were streaked on YPD or medium supplemented with 5-fluoroorotic acid (5-FOA). PCR-mediated gene manipulation was used to fuse sequences encoding a hemagglutinin (HA) tag at the end of the chromosomal copy of TOM6 (26).
Selection of high-copy-number suppressors.
Sam37Δ cells were transformed by electroporation with a high-copy-number library constructed by F. Lacroute into the URA3 2μm vector pFL44L (44). [Ura+] clones were selected and replica plated onto glycerol-containing medium at 37°C. [Gly+] clones were streaked again on glycerol-containing medium, and total DNA was extracted from those clones that grew at 37°C. The extracted DNA was used to transform Escherichia coli cells. Plasmids were recovered from the E. coli cells, and the DNA inserts were analyzed by sequencing.
For overproduction of proteins in yeast cells, the corresponding DNA was introduced into the multicopy plasmid, pRS426xTPIp-URA3 (a kind gift of K. Dietmeier).
Biochemical procedures.
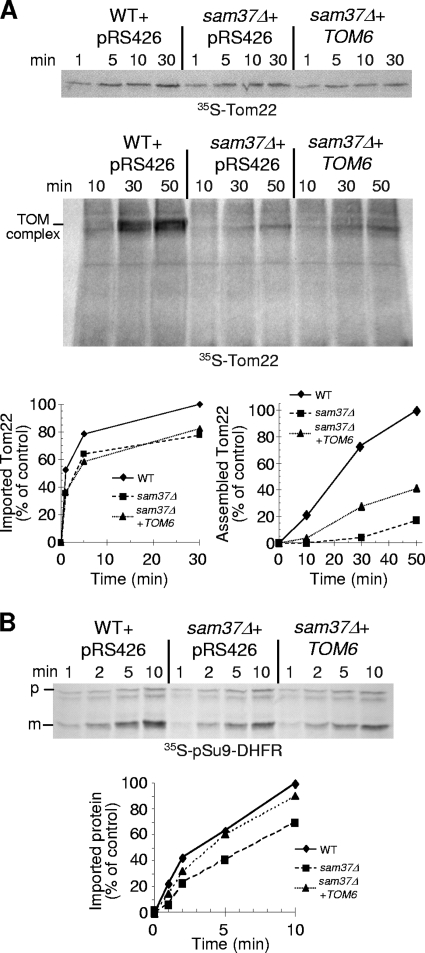
Mitochondria were isolated from yeast cells by differential centrifugation as described previously (7). For isolation of mitochondria from temperature-sensitive mutants and their parental strains, cells were grown at 25°C unless otherwise stated. Import experiments with radiolabeled precursor proteins and isolated mitochondria were performed in an import buffer containing 250 mM sucrose, 3 mg/ml bovine serum albumin, 80 mM KCl, 5 mM MgCl2, 10 mM morpholinepropanesulfonic acid (MOPS)-KOH, 2 mM NADH, and 2 mM ATP, pH 7.2. Organelles isolated from sam37Δ strains or from the corresponding parental strain were incubated at 37°C for 15 min and then equilibrated at 25°C for 2 min before the import reaction was initiated. Radiolabeled precursor proteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]methionine after in vitro transcription by SP6 polymerase from pGEM4 vectors containing the gene of interest.
Fluorescence microscopy.
For visualization of mitochondria, cells were transformed with a yeast expression vector harboring the mitochondrial presequence of subunit 9 of the Fo ATPase of Neurospora crassa fused to green fluorescent protein (GFP) (49). Microscopy images were acquired with an Axioskop20 fluorescence microscope equipped with an Axiocam MRm camera using the 43 Cy3 filter set and the AxioVision software program (Zeiss).
Blue native polyacrylamide gel electrophoresis (BN-PAGE).
Mitochondria were lysed in 30 μl digitonin buffer (1% digitonin, 20 mM Tris-HCl, 0.1 mM EDTA, 50 mM NaCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, pH 7.4). After incubation for 20 min at 4°C and a clarifying spin (36,670 × g, 15 min, 2°C), 5 μl sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM bis-Tris, 500 mM 6-aminocaproic acid, pH 7.0) was added, and the mixture was analyzed by electrophoresis in a 4 to 16% gradient blue native gel (41). Gels were blotted to polyvinylidene difluoride membranes, and proteins were further analyzed by autoradiography and/or immunodecoration.
RESULTS
TOM6 is a high-copy-number suppressor of sam37Δ.
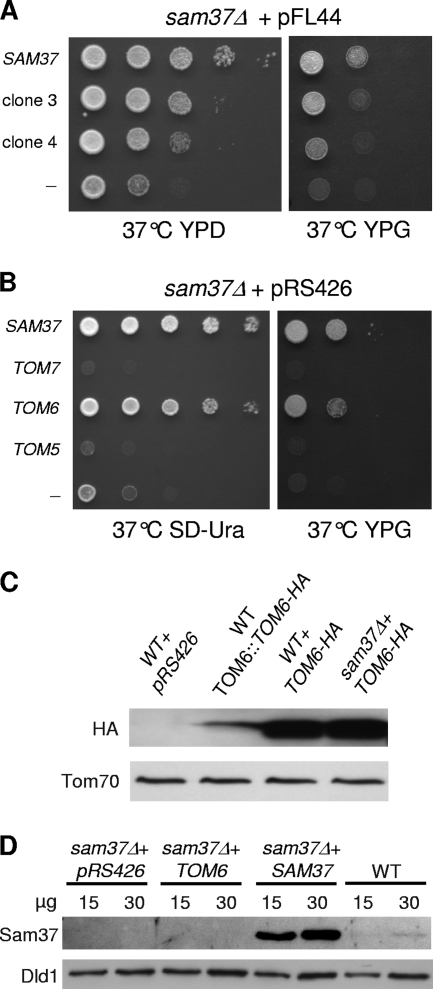
Yeast cells deleted for SAM37 are not viable at elevated temperatures on a nonfermentable carbon source. To search for proteins interacting with Sam37, we looked for high-copy-number suppressors of sam37Δ. Overproduction of the other subunits of the SAM complex (Sam50 and Sam35) was already shown to partially suppress the temperature-sensitive phenotype of sam37Δ cells only on a fermentable carbon source or at a normal temperature (6, 28). However, a strong suppressor of the thermosensitivity on a nonfermentable carbon source, where fully functional mitochondria are required, has not been found so far. We searched for high-copy-number suppressors by transforming sam37Δ cells with a yeast genomic library cloned on a high copy number, 2μm-based yeast expression vector. Transformants were first selected at 24°C on selective medium and then replica plated on glycerol containing rich medium and tested for their capacity to grow at 37°C. We screened in total 45,000 clones and identified among them 9 clones that exhibited normal growth at 37°C. Plasmids that complement the phenotype were isolated, and PCR amplification demonstrated that seven of the nine clones harbored the SAM37 gene. This observation suggests that we had good coverage of the yeast genome. Of note, sam37Δ cells overexpressing SAM37 displayed growth behavior similar to that of wild-type cells (data not shown). We verified that the remaining two plasmids lacking SAM37 could support normal growth at 37°C on both fermentable and nonfermentable carbon sources (Fig. 1A).
FIG. 1.
TOM6 is a high-copy-number suppressor of sam37Δ. (A) Cells lacking Sam37 were transformed with a plasmid containing SAM37, plasmids isolated from suppressor colonies 3 and 4, or an empty plasmid (−). The cells were tested by drop dilution assay for their ability to grow at 37°C on the indicated medium. (B) Sam37Δ cells harboring plasmids expressing the indicated genes or an empty plasmid (−) were tested by drop dilution assay for their ability to grow at 37°C on the indicated medium. (C) Expression of TOM6-HA from a multicopy plasmid results in elevated amounts of the protein. Crude mitochondria were isolated from wild-type (WT) cells transformed with an empty plasmid, cells where the chromosomal copy of TOM6 was replaced by TOM6-HA, and from wild-type and sam37Δ cells transformed with a multicopy plasmid containing TOM6-HA. All cells were grown at 30°C. Equal amounts were analyzed by sodium dodecyl sulfate (SDS)-PAGE and immunodecoration with an antibody against either the HA tag or the outer membrane protein Tom70 as a control. (D) The multicopy SAM37 plasmid results in overproduction of Sam37. Sam37Δ cells were transformed with either an empty vector (pRS426) or a plasmid carrying TOM6 or SAM37. Mitochondria were isolated from wild-type cells and the transformed cells, and the indicated amounts of the organelles were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies. Dld1 is anchored to the inner membrane and serves here as a control protein.
These two plasmids were analyzed by sequencing, and both were found to harbor a genomic DNA fragment which covers the IRC23, TOM6, and DPB5 genes (open reading frames YOR044w, YOR045w, and YOR046c, respectively). since the first and last genes encode proteins which do not have a clear link to mitochondrial function, we concentrated on TOM6, which encodes one of the small subunits of the outer membrane translocase. We subcloned the gene into a high-copy-number plasmid and observed that the resulting plasmid was indeed able to complement the growth defects of sam37Δ cells at 37°C (Fig. 1B). We also obtained similar complementation when SAM37 was deleted from the wild-type strain BY4741 (data not shown). Such a complementation was not observed when the control plasmid (without an insert) was employed. Thus, TOM6 is a high-copy-number suppressor of sam37Δ. A direct verification of the overproduction of Tom6 is not possible due to lack of an appropriate antibody. However, several lines of evidence strongly suggest that Tom6 was indeed overproduced under these conditions. We replaced the chromosomal copy of TOM6 by a C-terminally HA-tagged variant and in parallel introduced this variant into the overexpression vector. An immunodecoration with an antibody against the HA tag demonstrated that the levels of expression of the plasmid-encoded protein were severalfold higher than those of chromosomally encoded TOM6-HA (Fig. 1C). Similarly, reverse transcription-PCR analysis suggested that the amounts of TOM6-HA mRNA are substantially larger in the case of the plasmid-encoded protein (data not shown). Moreover, when we used the same plasmid to overexpress other genes, either previously (36, 48) or in the current study for SAM37, TOM20, TOM22, TOM70, and TOM40, we were always able to confirm expression levels that were severalfold higher than those in wild-type cells (Fig. 1D and 2A; see also Fig. 8A).
FIG. 2.
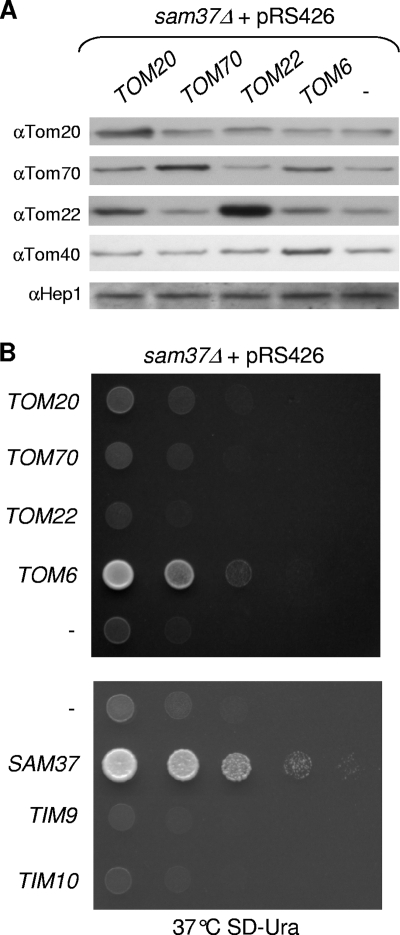
Receptor subunits of the TOM complex and the small Tim chaperones cannot rescue the sam37Δ growth phenotype. (A) sam37Δ cells transformed with either an empty pRS426 vector (−) or a plasmid encoding the indicated Tom subunit were used. Cells were grown at 30°C and ruptured by vortexing in the presence of glass beads. Crude mitochondria were obtained by differential centrifugation and analyzed by sodium dodecyl sulfate-PAGE. Proteins were blotted onto a membrane, which was then immunodecorated with antibodies (indicated by “α”) against the indicated Tom subunit or with antibodies against the matrix protein Hep1. (B) sam37Δ cells harboring plasmids expressing the indicated genes or an empty vector (−) were tested by drop dilution assay for their ability to grow at 37°C on the indicated medium.
FIG. 8.
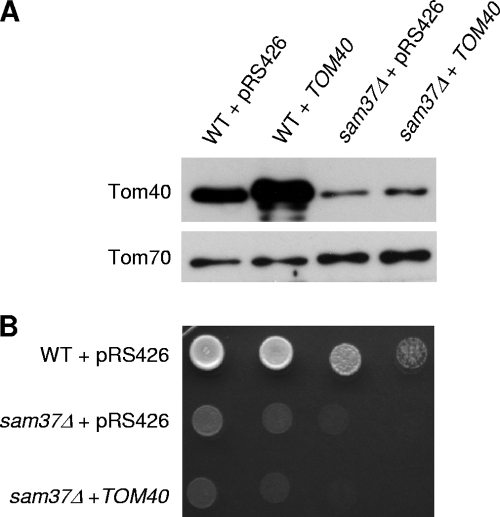
An overexpression plasmid encoding Tom40 cannot suppress the growth phenotype of sam37Δ cells. (A) Crude mitochondria were isolated from both wild-type (WT) and sam37Δ cells transformed with either an empty vector (pRS426) or a plasmid carrying TOM40. Cells were grown at 30°C. Equal amounts of mitochondria were analyzed by sodium dodecyl sulfate-PAGE and immunodecoration with antibodies against either Tom40 or Tom70. (B) Wild-type and sam37Δ cells harboring an empty pRS426 and sam37Δ cells transformed with a plasmid carrying TOM40 were tested by drop dilution assay for their ability to grow at 37°C on synthetic medium lacking Ura.
Next, we asked whether the suppression capacity of TOM6 is shared by the other small subunits of the TOM complex, although we did not find them to be suppressors in our screen. To that end, a high-copy-number plasmid encoding either Tom5 or Tom7 was transformed into sam37Δ cells, and the growth of the transformed cells was monitored. None of these overproduced proteins could rescue the growth phenotype of sam37Δ cells (Fig. 1B), suggesting that the capacity of Tom6 is unique among the small subunits of the TOM complex. Similarly, we observed that overproduction of the receptor subunits of the TOM complex (Tom20, Tom22, or Tom70) could not suppress the sam37Δ phenotype (Fig. 2B). The small Tim proteins were found to interact with precursors of β-barrel proteins upon their transfer through the intermembrane space (15, 18, 51). Therefore, we investigated whether TIM9 or TIM10 can function as a multicopy suppressor of the sam37Δ phenotype. The overproduction of either small Tim protein did not support normal growth of sam37Δ cells (Fig. 2B). Collectively, it appears that Tom6 has the unique capacity to suppress the sam37Δ phenotype while other TOM components or the small Tims do not share this ability.
Of note, Tom6 was originally identified as a high-copy-number suppressor of a TOM40 temperature-sensitive allele (20). Further studies revealed that this protein can stabilize the interaction of Tom40 with the receptor proteins, whereas Tom7 was found to have an opposite activity and to promote dissociation of the TOM complex (3, 9, 17). These earlier observations are in agreement with our current results, where overexpression of TOM7 in sam37Δ cells resulted in a growth phenotype which is even more severe than that of sam37Δ cells transformed with an empty vector (Fig. 1B).
Overexpression of SAM37 can rescue a tom6Δ growth defect.
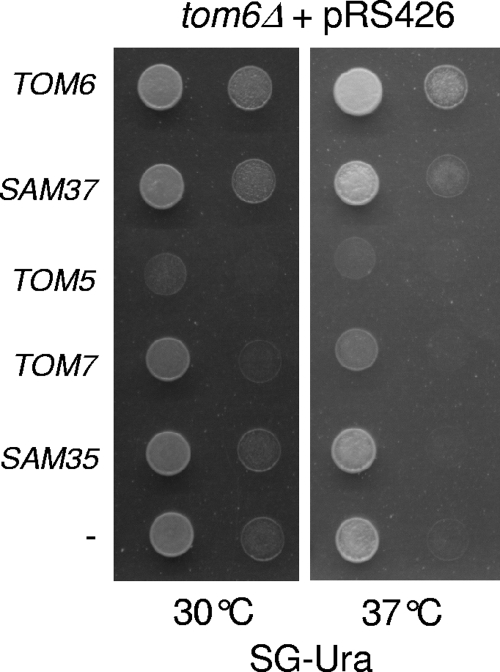
To better understand the functional link between the two proteins, we tested the reciprocal relationships, namely, whether SAM37 can function as a high-copy-number suppressor of tom6Δ. The deletion of TOM6 alone was originally reported to result in no obvious growth defect (20). Later, Alconada et al. observed a slightly reduced growth of tom6Δ cells on a nonfermentable carbon source at 37°C (3). We reproduced this observation and showed that this mild growth phenotype is rescued upon overexpression of SAM37 (Fig. 3). An overexpression plasmid encoding Sam35, the second peripheral subunit of the SAM complex, or an empty plasmid did not have such an effect. Similarly, large amounts of the other small subunits of the TOM complex, Tom5 and Tom7, could not rescue and rather worsened the growth phenotype of tom6Δ cells (Fig. 3). Thus, the genetic interaction between SAM37 and TOM6 is specific and observed in both directions.
FIG. 3.
Overexpression of SAM37 can rescue a tom6Δ growth defect. Cells devoid of TOM6 were transformed with a plasmid carrying the indicated gene or with an empty plasmid (−). The transformed cells were tested by drop dilution assay for their ability to grow at the indicated temperatures on glycerol-containing synthetic medium (SG-Ura).
Tom6 is essential for viability of sam37Δ cells.
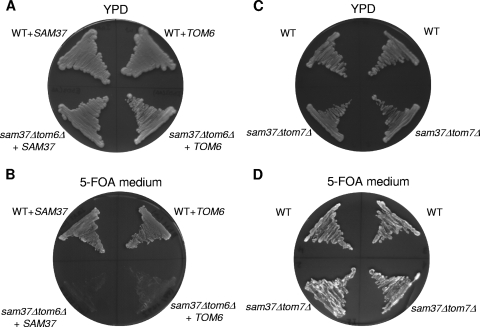
To construct a double deletion mutant, we mated a tom6Δ strain with a sam37Δ strain, induced sporulation, and performed tetrad dissection. Upon dissecting 30 tetrads, we could not recover any colony where the two genes were disrupted, suggesting a synthetic lethality phenotype. The synthetic lethality could be confirmed by the observation that a double deletion strain containing either TOM6 or SAM37 carried on a plasmid is viable (Fig. 4A). However, when the URA3-containing plasmid was eliminated by plating the cells on a medium containing 5-FOA, the cells lost their viability (Fig. 4B). Hence, we conclude that the double mutation tom6Δ sam37Δ is lethal. As a further control, we tested whether disruption of SAM37 in combination with deletion of TOM7 would also lead to synthetic lethality. In contrast to the situation with TOM6, the sam37Δ tom7Δ double deletion strain was viable (Fig. 4C and D). Thus, the synthetic lethality phenotype of SAM37 and TOM6 is not shared by all TOM components and provides additional evidence for the unique functional interaction between the two proteins.
FIG. 4.
Double deletion of SAM37 and TOM6 results in synthetic lethality. Wild-type (WT) and sam37Δ tom6Δ cells containing plasmid-borne SAM37 or TOM6 were plated at 30°C on glucose-containing rich medium (YPD) (A) or 5-FOA-containing medium (5-FOA medium) (B). Wild-type (WT) and sam37Δ tom7Δ cells were plated in a similar manner (C and D). The pRS426 vector used in this experiment contains a URA3 gene, which upon its expression converts the nontoxic 5-FOA compound to toxic 5-fluorouracil. Thus, a loss of this plasmid is favored under these conditions.
Expression level of TOM6 does not affect the SAM complex.
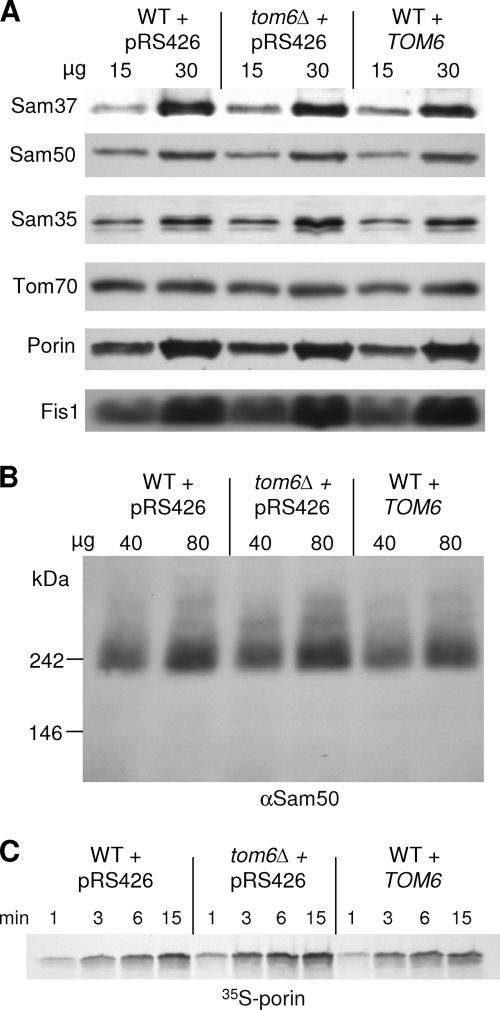
The genetic interaction of TOM6 with SAM37 might point to a physical interaction between the two gene products. However, we could not observe such an interaction between the two proteins (data not shown), and such physical interaction has not been reported so far. Sam37 is known to be required for optimal biogenesis of Tom40, and in its absence smaller amounts of Tom40 are detected in mitochondria (50). Tom6 was suggested to interact directly with Tom40 and to stabilize the assembly of the latter (3, 9, 20). Hence, Tom6 can exert its effect directly on the SAM complex and/or overexpression of TOM6 in sam37Δ cells can stabilize Tom40 molecules. To test the first alternative, we monitored the steady-state levels of SAM components and the stability of the SAM complex in cells overexpressing TOM6 or deleted for the protein. In both cases, levels of Sam37, Sam50, and Sam35 and those of control proteins were similar to those in wild-type cells (Fig. 5A). Next, we analyzed the SAM complex by BN-PAGE and found that changes in the level of Tom6 affect neither the amounts nor the migration behavior of the SAM complex (Fig. 5B). Finally, we monitored the in vitro import of the β-barrel precursor porin into mitochondria isolated from the above strains and did not observe any difference compared to import into mitochondria isolated from wild-type cells (Fig. 5C). Taken together, these results suggest that Tom6 affects neither the stability nor the activity of the SAM complex.
FIG. 5.
Expression levels of TOM6 do not affect the SAM complex. (A) Mitochondria were isolated from wild-type (WT) or tom6Δ cells transformed with an empty vector (pRS426) or from wild-type cells transformed with a plasmid carrying TOM6. The given amounts of mitochondria were analyzed by sodium dodecyl sulfate (SDS)-PAGE and immunodecoration with antibodies against the indicated mitochondrial outer membrane proteins. (B) The indicated amounts of mitochondria were analyzed by BN-PAGE and immunodecoration with antibodies against Sam50. (C) Mitochondria isolated as described above were incubated with radiolabeled precursor of porin at 25°C for various time periods. Samples were then treated with proteinase K, and mitochondria were reisolated. Imported porin was analyzed by SDS-PAGE and autoradiography.
Overexpression of TOM6 stabilizes Tom40.
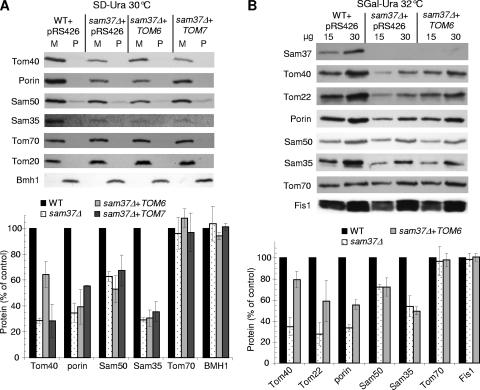
To characterize the effect of overexpression of TOM6 on the steady-state level of Tom40, we grew the cells at 30°C, isolated crude mitochondria, and analyzed their protein content. The results shown in Fig. 6A demonstrate that overexpression of TOM6 in sam37Δ cells caused an increase in the steady-state levels of Tom40, whereas porin, another β-barrel protein, was not affected. Incidentally, overexpression of TOM7 in cells lacking Sam37 resulted in a further reduction in the levels of Tom40, in agreement with the retarded growth rate of this strain and with a destabilizing effect of Tom7 (Fig. 1B and 6A). On the other hand, steady-state levels of porin were increased when TOM7 was overexpressed in sam37Δ cells. This observation is compatible with previous reports suggesting that Tom7 is important for optimal import of porin (17). As reported before, the deletion of SAM37 resulted in a significant reduction of the levels of Sam35 (6, 47) independently of the overexpression of either TOM6 or TOM7 (Fig. 6A and B).
FIG. 6.
Overexpression of TOM6 stabilizes Tom40. (A) Wild-type (WT) cells transformed with an empty vector (pRS426) and sam37Δ cells transformed with either an empty vector (pRS426) or a plasmid carrying TOM6 or TOM7 were used. Cells were grown at 30°C and ruptured by vortexing in the presence of glass beads. Crude mitochondria were obtained by differential centrifugation (M) and were analyzed together with the postmitochondrial supernatant fraction (P) by sodium dodecyl sulfate (SDS)-PAGE. Proteins were blotted onto a membrane, which was then immunodecorated with antibodies against the indicated mitochondrial outer membrane proteins or with antibodies against the cytosolic marker protein Bmh1. The results, for each protein, of at least three experiments were quantified, and the average values are presented. Error bars represent standard deviations. For each protein, the amount in wild-type mitochondria was set at 100%. (B) Mitochondria were isolated from cells grown at 32°C. The indicated amounts of mitochondria were analyzed by SDS-PAGE and immunodecoration with antibodies against the indicated mitochondrial outer membrane proteins. The intensities of the bands corresponding to the various proteins were quantified. For each amount of mitochondria, the intensity of the bands in the altered mitochondria was calculated as a percentage of the signal obtained from the wild-type organelle. For each protein, the average value of the two different amounts of organelles from at least three experiments is presented. Error bars represent standard deviations.
To better observe the effect of TOM6 overexpression, we isolated mitochondria from cells grown at 32°C, since at this temperature sam37Δ cells exhibit a more severe growth phenotype than at 30°C. We monitored the steady-state amounts of various proteins and observed that proteins like Fis1, Tom70, and Tom20, anchored to the outer membrane via a single helical segment, were not affected by the deletion of Sam37 (Fig. 6B and data not shown). In contrast, the levels of β-barrel proteins are affected to various extents upon deletion of SAM37 (50). Tom40 amounts were highly reduced in sam37Δ cells. Porin was significantly affected, whereas levels of Sam50 were only slightly decreased (Fig. 6B). Overexpression of TOM6 resulted in a significant increase in the levels of Tom40 but importantly had a more limited or no effect on the amounts of the other β-barrel proteins porin and Sam50, respectively (Fig. 6B). Like all β-barrel precursors, precursor molecules of porin are initially translocated across the outer membrane by the TOM import pore, which is composed mainly by Tom40 molecules. Thus, the increased levels of porin upon overexpression of TOM6 in sam37Δ cells are probably the consequence of enhanced levels of Tom40 in these cells. On the basis of these results and those of in vitro import experiments (see below), we propose Tom40 to be the primary target explaining the suppression of the growth phenotype. Of note, as reported before, the absence of Sam37 results in reduced amount of Tom22, which contains a helical transmembrane domain (45). Importantly, these low levels were considerably recovered upon overexpression of TOM6 in sam37Δ cells (Fig. 6B).
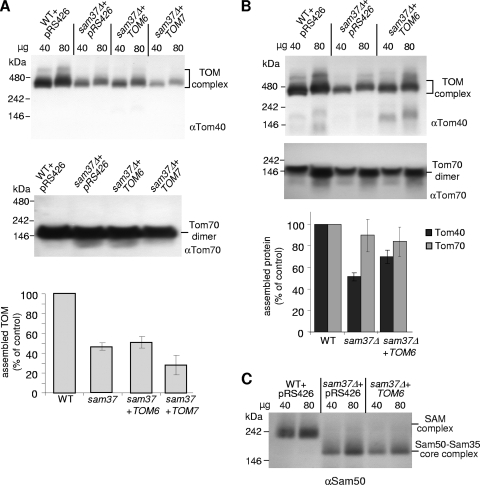
Next, we monitored whether the larger amounts of Tom6 in sam37Δ cells can also improve the assembly of Tom40 molecules into TOM complexes. To that end, we analyzed the TOM complex in the various strains by using a native gel system (BN-PAGE). As reported before, the TOM complex is diminished in sam37Δ cells (50). Overexpression of TOM6 partially restored the steady-state levels of the TOM complex in cells grown at 30°C or at 32°C (Fig. 7A and B, respectively). The level of assembled TOM complex in cells grown at 32°C was improved from 51% of the wild-type level in sam37Δ cells to 69% in sam37Δ cells overexpressing TOM6. In agreement with the growth phenotype, higher levels of Tom7 destabilized the TOM complex even further (Fig. 7A). In contrast to the effect on the TOM complex, overexpression of either TOM6 or TOM7 did not influence the stability of the SAM core complex, which is formed by Sam35 and Sam50 in the absence of Sam37 (Fig. 7C and data not shown). These results are in line with our observations that Tom6 does not affect the SAM complex (Fig. 5). Taken together, the elevated levels of Tom6 have a stabilizing effect on the TOM complex but do not affect the structure and function of the SAM complex.
FIG. 7.
Overexpression of TOM6 stabilizes the TOM complex (A). Mitochondria were isolated from cells grown at 30°C and solubilized in 1% digitonin before their analysis by BN-PAGE and immunodecoration with antibodies (indicated by “α”) against either Tom40 or Tom70 (as a control). The migration behavior of molecular mass markers and bands corresponding to assembled TOM complex are indicated. Of note, employing two different amounts of mitochondria resulted in two different ratios of detergent/protein. This difference is reflected in a minor change in the migration behavior of the complexes. The bands corresponding to the assembled TOM complex in samples containing 80 μg mitochondria were quantified. The amount in wild-type (WT) organelle was set to 100%, and the average values of at least three experiments are presented. Error bars represent standard deviations. (B) Mitochondria were isolated from cells grown at 32°C and analyzed by BN-PAGE as in part A. The bands corresponding to the assembled TOM complex or dimeric Tom70 (as a control) in samples containing 80 μg mitochondria were quantified. The amount in wild-type organelle was set to 100%, and the average values for at least three experiments are presented. Error bars represent standard deviations. (C) Mitochondria at the indicated amounts were solubilized in 0.5% Triton X-100 and analyzed by BN-PAGE and immunodecoration with antibodies against Sam50. The migration behavior of molecular mass markers and bands corresponding to either SAM or Sam50-Sam35 core complexes are indicated.
Since overexpressing TOM6 appears to rescue the sam37Δ phenotype by stabilizing Tom40, we asked whether overexpressing TOM40 itself can have a similar outcome. As shown in Fig. 8A, transforming a plasmid that can lead to overexpression of Tom40 in wild-type cells did not enhance the levels of the protein in sam37Δ cells, probably due to degradation of unassembled precursor molecules. This observation suggests that the defect in the biogenesis of Tom40 in sam37Δ cells cannot be repaired by synthesizing more molecules of the protein. As expected from the protein levels, transforming pRS426-TOM40 into sam37Δ cells did not suppress the growth phenotype of these cells (Fig. 8B). These findings are also in accordance with the fact that we did not find TOM40 or any other gene besides TOM6 in our initial genetic screen.
Overexpression of TOM6 suppresses various phenotypes of sam37Δ cells.
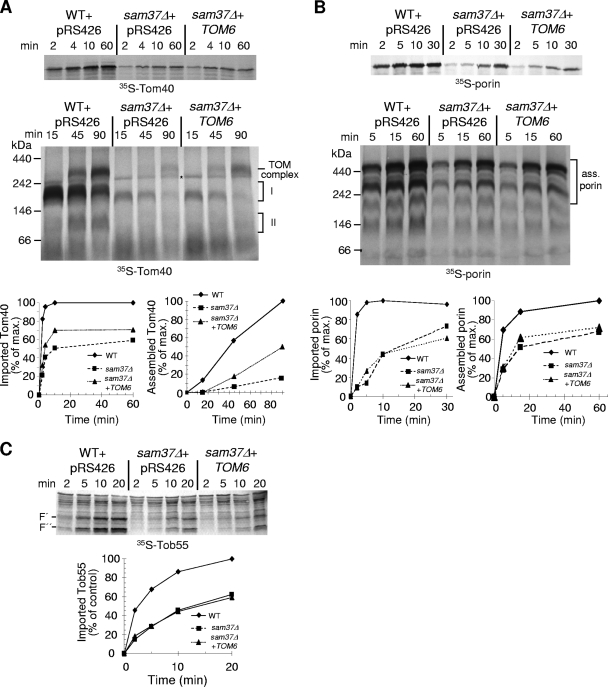
The deletion of SAM37 results in various phenotypes in addition to the reduced steady-state amounts of Tom40. Among them are a reduction in mitochondrial protein import efficiency (6, 14), impaired biogenesis of TOM proteins with an α-helical transmembrane segment (45), and altered mitochondrial morphology (29). Currently it is not resolved to what extent these phenotypes are secondary to the defects in function or assembly of the TOM complex observed in sam37Δ cells. To address this question, we asked whether the overexpression of TOM6 in sam37Δ cells can suppress these phenotypes. First, we monitored the membrane integration of β-barrel precursors. Several distinct assembly intermediates can be followed over time when radiolabeled Tom40 precursors are imported into isolated mitochondria and subsequently analyzed by BN-PAGE. Following translocation through the TOM pore, an assembly intermediate associated with the SAM complex of approximately 250 kDa, termed intermediate I, is formed. After release from the SAM complex, a species referred to as intermediate II, of approximately 100 kDa, is observed. This second intermediate consists of a Tom40 precursor with small Tom subunits (31). Finally, the recruitment of additional Tom subunits is thought to result in the formation of the TOM core complex, with a molecular mass of 400 to 450 kDa. Of note, in the absence of Sam37, there are only minor amounts of Tom40 precursor molecules associated with the SAM complex (Fig. 9A, intermediate I). This observation can be explained by two possible scenarios: first, the SAM complex without Sam37 has a lower capacity to bind Tom40 precursors; alternatively, Sam37 promotes the release of precursor proteins from the SAM complex, and as the downstream steps are impaired in mitochondria lacking Sam37, the turnover of precursor molecules at intermediate I is enhanced. Naturally, the two possibilities are not mutually exclusive, since Sam37 might have a dual role. Similar observations were made with mitochondria lacking Mdm12 or Mmm1, where in vitro import of Tom40 resulted in a reduction in the levels of all assembly intermediates (28). Since both proteins are suggested to act in the biogenesis of Tom40 downstream of the SAM complex, these previous observations demonstrate the feasibility of the second possibility.
FIG. 9.
Overexpression of TOM6 suppresses the Tom40 import phenotype of sam37Δ cells. (A) Mitochondria were isolated from wild-type (WT) and sam37Δ cells harboring an empty vector and from sam37Δ cells overexpressing TOM6. Mitochondria were incubated with radiolabeled precursor of Tom40 at 25°C for various time periods. Upper panel: samples were treated with proteinase K (PK), and mitochondria were reisolated. Imported Tom40 was analyzed by sodium dodecyl sulfate (SDS)-PAGE and autoradiography. Lower panel: mitochondria were solubilized with 1% digitonin, and samples were analyzed by BN-PAGE and autoradiography. Assembly intermediates I and II and the assembled TOM complex are indicated. An unspecific band is marked with an asterisk. The bands corresponding to PK-protected Tom40 (import) and assembled Tom40 (assembly) were quantified, and an experiment representative of three independent experiments is presented. The amount of protein imported into or assembled within wild-type mitochondria after the longest incubation period was set to 100%. (B) Mitochondria as for part A were incubated with radiolabeled precursor of porin at 25°C for various time periods. Further treatment and analysis were as described in the legend to part A. The bands corresponding to assembled porin are indicated. The bands corresponding to the PK-protected and the assembled porin were quantified. (C) Mitochondria as for part A were incubated with radiolabeled precursor of Sam50 at 25°C for various time periods. Samples were treated with PK, and mitochondria were reisolated. Imported Sam50 was analyzed by SDS-PAGE and autoradiography. Two characteristic proteolytic fragments with apparent molecular masses of 30 and 25 kDa are indicated with F′ and F″, respectively (see reference 15). The bands corresponding to F′ were quantified, and the intensity of this fragment formed upon import into mitochondria isolated from wild-type cells for the longest time period was set to 100%. An experiment representative of three independent experiments is presented.
Importantly, overexpressing TOM6 in the absence of Sam37 did not have any effect on the binding of Tom40 precursor to the SAM complex (Fig. 9A, intermediate I). In contrast, mitochondria isolated from sam37Δ cells overexpressing TOM6 displayed an improvement in both the import and assembly of Tom40 precursor, although both processes were still rather reduced compared to results with wild-type organelles (Fig. 9A). Thus, it appears that under the suboptimal conditions of the in vitro system, overproduction of Tom6 cannot fully complement the impaired capacity of sam37Δ organelles to import Tom40 precursors. Of note, these results are similar to observations made for sam37Δ cells overexpressing SAM35. Although such overexpression can complement the sam37Δ growth phenotype under certain growth conditions, mitochondria isolated from these cells are still impaired in the in vitro import of porin (6). Importantly, import and assembly of other β-barrel proteins, such as porin and Sam50, were not affected by larger amounts of Tom6 (Fig. 9B and C). We conclude that elevated amounts of Tom6 improve the import and assembly of newly synthesized Tom40 precursor molecules in sam37Δ cells.
It was recently reported that the absence of Sam37 resulted in impaired import and assembly of Tom22 (45). We tested whether overexpression of TOM6 can counteract this impairment and observed that membrane insertion was not improved but assembly of Tom22 precursors into the TOM complex was increased (Fig. 10A). These findings are in line with a proposed role of Tom6 in promoting the interaction between Tom40 and Tom22 rather than mediating membrane insertion of Tom22 (8, 9). Mitochondria lacking Sam37 are also hampered in their ability to import newly synthesized matrix-destined precursor proteins (Fig. 10B) (6). Mitochondria isolated from sam37Δ cells overexpressing TOM6 displayed an improvement in the import of this type of precursor protein, such as pSu9-DHFR (Fig. 10B) and F1β (data not shown). Similarly to other effects, we suggest that this improvement is due to enhanced stability of the TOM complex, which plays a crucial role in the import of these precursor proteins.
FIG. 10.
Overproduction of Tom6 suppresses Tom22 assembly and improves import phenotypes of sam37Δ cells. (A) Radiolabeled precursor of Tom22 was incubated with various mitochondria as described in the legend to Fig. 9A. Either samples were extracted with carbonate and pellets were analyzed by sodium dodecyl sulfate (SDS)-PAGE and autoradiography, or they were solubilized with 1% digitonin and analyzed by BN-PAGE and autoradiography. Bands corresponding to assembled TOM complex are indicated. The bands corresponding to Tom22 in the carbonate pellet (import) and assembled Tom22 (assembly, assayed by BN-PAGE) were quantified. The amount of protein imported into or assembled within mitochondria isolated from wild-type (WT) cells after the longest incubation period was set to 100%. An average for three experiments is presented. (B) Radiolabeled precursor of pSu9-DHFR was incubated with various mitochondria as described in the legend to Fig. 9A. Samples were treated with proteinase K, mitochondria were reisolated, and imported proteins were analyzed by SDS-PAGE and autoradiography. The precursor and mature forms are indicated by p and m, respectively. The bands corresponding to the mature form were quantified, and the amount of protein imported into mitochondria isolated from wild-type cells for the longest time period was set to 100%.
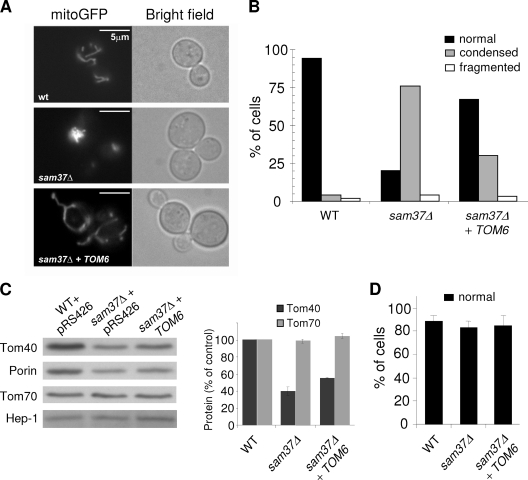
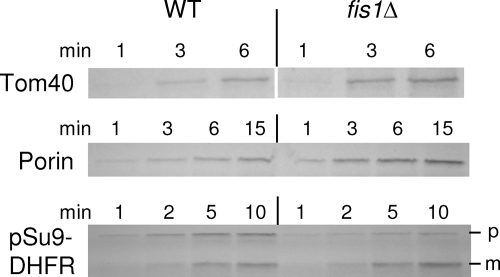
Finally, we examined whether the altered mitochondrial morphology observed in the absence of Sam37 can be restored by overexpressing TOM6. The vast majority of sam37Δ cells contained condensed mitochondria instead of the regular tubular shape (Fig. 11A and B ) (29). This situation was altered dramatically in sam37Δ cells overexpressing TOM6. About 70% of these cells harbored wild-type-like tubular organelles (Fig. 11A and B). Next, we wanted to verify that the morphology defect is secondary to the impaired biogenesis in sam37Δ cells. To that end, we analyzed sam37Δ cells grown at 25°C, where a clear growth phenotype is not observed. These cells already have reduced levels of the β-barrel proteins, Tom40, and porin (Fig. 11C) (6), and the in vitro assembly of Tom40 in mitochondria isolated from these cells is hampered (data not shown). In contrast, the mitochondrial morphology of these cells is normal (Fig. 11D). These results suggest that the biogenesis defect occurs before morphology changes. Moreover, to demonstrate that morphology changes per se are unlikely to cause the observed changes in Tom40 assembly, we monitored the import of Tom40, porin, and pSu9-DHFR into mitochondria lacking the morphology protein Fis1. Although mitochondria from fis1Δ cells have a very severe morphology phenotype (33), their capacity to import these precursor proteins is not altered in comparison with that of wild-type mitochondria (Fig. 12). Taken together, these results demonstrate that stabilizing the TOM complex can complement the morphology defect in these cells and support the notion that the altered mitochondrial morphology in cells deleted for SAM37 is secondary to impaired function of the TOM complex.
FIG. 11.
Large amounts of Tom6 suppress the morphology phenotype of sam37Δ cells. (A) Wild-type cells (wt), sam37Δ cells, and sam37Δ cells overexpressing TOM6 were transformed with a vector expressing mitochondrially localized GFP. Cells were grown at 32°C on liquid glucose-containing medium and were observed by fluorescence microscopy. Mitochondrial morphology (mitoGFP) and bright-field images are presented (bar, 5 μm). (B) Quantification of the phenotypes of at least 100 cells from each strain. Cells were grown at 30°C and classified into those containing normal tubular (normal), condensed, or fragmented mitochondria. WT, wild type. (C) Cells grown at 25°C harbor reduced levels of Tom40 and porin. Crude mitochondria were isolated from both wild-type (WT) and sam37Δ cells transformed with an empty vector (pRS426) and from sam37Δ cells overexpressing TOM6. Equal amounts of mitochondria were analyzed by sodium dodecyl sulfate-PAGE and immunodecoration with antibodies against the indicated mitochondrial proteins. The bands corresponding to Tom40 and Tom70 from three experiments were quantified. The amount in wild-type mitochondria was set to 100%. Error bars represent standard deviations. (D) Cells as in panel A were grown at 25°C. For each strain, more than 100 cells were analyzed by fluorescence microscopy for their mitochondrial morphology. The percentage of cells containing normal tube-like mitochondria is presented. WT, wild type.
FIG. 12.
In vitro import of precursor proteins is not reduced in mitochondria isolated from fis1Δ cells. Mitochondria were isolated from wild-type (WT) and fis1Δ cells. Organelles were incubated with the radiolabeled precursors of Tom40, porin, or pSu9-DHFR at 25°C for various time periods. Samples were treated with proteinase K, and mitochondria were reisolated. Imported proteins were analyzed by sodium dodecyl sulfate-PAGE and autoradiography. The precursor and mature forms of pSu9-DHFR are indicated as p and m, respectively.
Large amounts of Sam37 improve the in vitro assembly of Tom40 in tom6Δ cells.
The TOM complex is destabilized in the absence of Tom6, and a subcomplex of about 100 kDa is observed (8). Since overexpression of SAM37 partially suppressed the growth phenotype of tom6Δ cells on synthetic medium at 37°C (Fig. 3), we tested whether such overexpression can improve the stability of the TOM complex in these cells. Unfortunately, when we grew the cells under the conditions where complementation of the growth phenotype was observed (synthetic medium, 37°C), growth was so slow that we were not able to obtain sufficient cells for isolation of mitochondria, a process required for the BN-PAGE analysis. Therefore, we had to grow the cells using conditions under which the growth phenotype of tom6Δ cells is hardly observed (synthetic medium, 30°C) and overexpression of SAM37 does not result in an obvious change in the growth rate (Fig. 3). We could not observe any change in the dissociated TOM complex upon overexpression of SAM37 in these cells (Fig. 13A). Similar results were also obtained upon incubation of the isolated mitochondria at 37°C prior to the BN-PAGE analysis or upon analysis of mitochondria isolated from cells grown first at 25°C before being shifted for 10 h to grow at 37°C (data not shown). Thus, under these experimental conditions, the partial suppression of the growth phenotype at 37°C is not reflected in a change in the migration behavior of bands corresponding to the endogenous TOM complex.
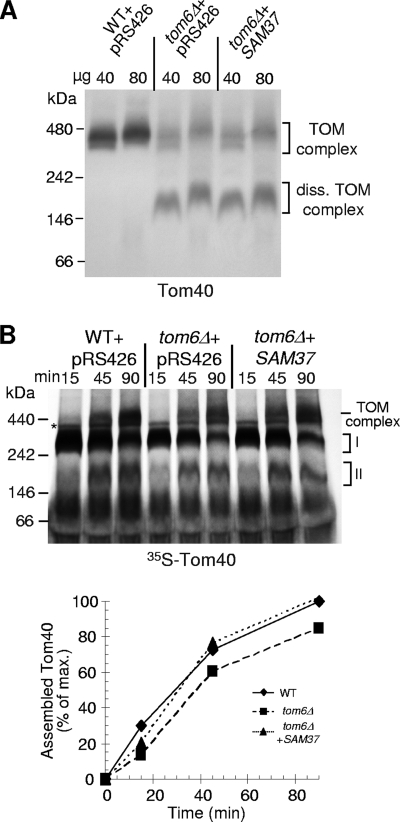
FIG. 13.
Overexpression of SAM37 in tom6Δ cells does not stabilize the endogenous TOM complex but improves the assembly of newly synthesized Tom40 precursor molecules. (A) Mitochondria were isolated from the indicated strains, solubilized in 1% digitonin, and analyzed by BN-PAGE and immunodecoration with antibodies against Tom40. The migration behavior of molecular mass markers and bands corresponding to assembled and dissociated (diss.) TOM complex are indicated. WT, wild type. (B) Mitochondria isolated as described above were incubated with radiolabeled precursor of Tom40 at 25°C for various time periods. Mitochondria were reisolated and solubilized with 1% digitonin, and samples were analyzed by BN-PAGE and autoradiography. Assembly intermediates I and II and the assembled TOM complex are indicated. An unspecific band is marked with an asterisk. The bands corresponding to assembled Tom40 for three independent experiments were quantified, and a representative experiment is presented. The amount of protein assembled within wild-type mitochondria after the longest incubation period was set to 100%.
On the other hand, the in vitro assembly of newly synthesized Tom40 molecules was enhanced when mitochondria isolated from tom6Δ cells harboring elevated amounts of Sam37 were used (Fig. 13B). Collectively, it appears that overexpression of SAM37 in tom6Δ cells does enhance the assembly of newly synthesized Tom40 molecules. However, since Sam37 is not a component of the TOM complex, its overexpression does not affect the stability of the TOM complex once the complex is already assembled.
DISCUSSION
The mechanism by which the SAM complex mediates the membrane integration of β-barrel substrates is only partially understood, and similarly unclear is the distinct function of Sam37, a peripheral subunit of the SAM complex. In the present study, we investigated the role of this component. Most of our knowledge about the assembly of β-barrel proteins results from studies of the biogenesis of Tom40, which often serves as a paradigm for the biogenesis of other β-barrel proteins. After its initial interaction with the SAM core complex, the precursor of Tom40 is released from that complex in order to interact with the partially assembled TOM complex. We propose that Sam37 has at least two functions. First, it stabilizes the SAM complex, and in its absence the steady-state levels of the remaining Sam50-Sam35 subcomplex are reduced (47). Second, Sam37 promotes the dissociation of substrate proteins from the SAM complex. In the case of the Tom40 precursor, Tom6 supports the association of the precursor protein with the TOM complex. Thus, the two proteins contribute to the relay of the Tom40 precursor from the SAM complex to the TOM complex. Such a scenario can easily explain why overexpression of one of the two proteins can compensate for the absence of the other: reduced release can be compensated by better association with the target complex, and impaired association can be overcome by better release. Double deletion is lethal, since the transfer process is hampered at both ends. The observations with mitochondria that lack Tom6 and harbor elevated amounts of Sam37 support the above proposal. Elevated amounts of Sam37 can enhance the biogenesis of newly synthesized Tom40 molecules. However, since Sam37 is not a component of the TOM complex, larger amounts of this protein cannot stabilize the TOM complex once it is already formed. Overexpression of TOM6 in sam37Δ cells could not completely restore the levels of assembled Tom40. However, we propose that as long as the amount of functional TOM complex is above a certain threshold level, even reduced levels of the complex (compared to wild-type levels) are sufficient to support nearly wild-type growth under the tested conditions.
Sam37 involvement in the release of Tom40 precursor from the SAM complex is supported by recent studies suggesting the involvement of Sam37 in discharging the SAM complex. One supportive argument for this notion is the observation that the biogenesis of Sam50 itself relies on Sam37 to a lesser extent than that of other β-barrel proteins (15). This difference may result from the necessity of most β-barrel proteins of leaving the SAM complex before their final integration into the outer membrane, whereas precursor molecules of Sam50 have already reached their destination upon association with preexisting SAM complexes. Chan and Lithgow reported that overexpression of SAM37 can rescue a temperature-sensitive allele of SAM35 without affecting the amount of β-barrel precursors bound to the remaining SAM core complex (6). Therefore, they suggested that Sam37 acts in β-barrel assembly downstream of Sam35. Our proposal of a discharging function for Sam37 is also in agreement with such a downstream function. It is also supported by reports suggesting that Sam37 is not required for the initial interaction of the β-barrel precursor with the Sam35-Sam50 core complex (6, 25, 47).
Tom6 is not the only stabilizing component of the TOM core complex, and Tom22 is another subunit with such a capacity (46). Accordingly, overexpression of TOM22 was reported to partially suppress defects of sam37Δ cells (27). However, in contrast to TOM6, overexpression of TOM22 does not suppress the growth defect of Sam37Δ under all growth conditions. This suggests that the elevated levels of Tom22 in Sam37Δ cells overexpressing TOM6 are not the primary reason for the suppression of the growth phenotype. The tight genetic interaction of SAM37 with the TOM complex is further reflected by the synthetic lethality phenotype of SAM37 deletion with the absence of either receptor subunit of the TOM complex, Tom20 or Tom70 (14). None of these two import receptors is essential as long as the other receptor is present. However, each single receptor becomes essential if the functionality of the TOM complex is already impaired by the absence of the other receptor (37). We propose a similar explanation for the synthetic lethality of Sam37Δ with deletion of the genes encoding the import receptors. The organization and function of the TOM complex in sam37Δ cells are considerably hampered. Hence, additional deletion of an import receptor reduces the functionality of the TOM complex even further, to a level that is no longer compatible with viability. Of note, although the two receptors share with TOM6 the synthetic lethality phenotype with Sam37Δ, they differ from TOM6 since they cannot fulfill the demanding task of compensating the sam37Δ growth defects.
We further observed that stabilizing the TOM complex in cells lacking Sam37 can complement their morphology defects. Based on our results, we propose that the morphology defect is not related to a direct function of Sam37 in mitochondrial morphology but rather emerged from the reduced function of the TOM complex in these cells. Supporting this proposal are previous observations where cells harboring temperature-sensitive alleles of TOM40 or SAM50 and cells depleted for Sam35 demonstrated similar morphology defects (4, 29). Moreover, the biogenesis of β-barrel proteins like Tom40 is not affected by deletion of genes encoding morphology components, such as UGO1, FZO1, FIS1, MDV1, and DNM1, although all these deletion strains have a severe morphology phenotype (28; this study). Thus, morphology changes per se are unlikely to cause either import defects or the observed changes in Tom40 assembly.
Taken together, this study provides new insights on the distinct function of Sam37 and demonstrates functional interactions between the TOM and the SAM complexes.
Acknowledgments
We thank K. Rehn for technical support, E. Kracker for help in performing some experiments, C. Kemper for support in the initial stages of this project, and F. Lacroute for the genomic library.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG RA1028/2-1).
Footnotes
Published ahead of print on 21 September 2009.
REFERENCES
- 1.Ahting, U., M. Thieffry, H. Engelhardt, R. Hegerl, W. Neupert, and S. Nussberger. 2001. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J. Cell Biol. 153:1151-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahting, U., C. Thun, R. Hegerl, D. Typke, F. E. Nargang, W. Neupert, and S. Nussberger. 1999. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alconada, A., M. Kübrich, M. Moczko, A. Hönlinger, and N. Pfanner. 1995. The mitochondrial receptor complex: the small subunit Mom8b/Isp6 supports association of receptors with the general insertion pore and transfer of preproteins. Mol. Cell. Biol. 15:6196-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altmann, K., and B. Westermann. 2005. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell 16:5410-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolender, N., A. Sickmann, R. Wagner, C. Meisinger, and N. Pfanner. 2008. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, N. C., and T. Lithgow. 2008. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell 19:126-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daum, G., S. Gasser, and G. Schatz. 1982. Import of proteins into mitochondria: energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast mitochondria. J. Biol. Chem. 257:13075-13080. [PubMed] [Google Scholar]
- 8.Dekker, P. J. T., M. T. Ryan, J. Brix, H. Müller, A. Hönlinger, and N. Pfanner. 1998. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell. Biol. 18:6515-6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dembowski, M., K.-P. Künkele, F. E. Nargang, W. Neupert, and D. Rapaport. 2001. Assembly of Tom6 and Tom7 into the TOM core complex of Neurospora crassa. J. Biol. Chem. 276:17679-17685. [DOI] [PubMed] [Google Scholar]
- 10.Dietmeier, K., A. Hönlinger, U. Bömer, P. J. T. Dekker, C. Eckerskorn, F. Lottspeich, M. Kübrich, and N. Pfanner. 1997. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature 388:195-200. [DOI] [PubMed] [Google Scholar]
- 11.Eckart, K., L. Eichacker, K. Sohrt, E. Schleiff, L. Heins, and J. Soll. 2002. A Toc75-like protein import channel is abundant in chloroplasts. EMBO Rep. 3:557-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel, K., S. K. Buchanan, and T. Lithgow. 2001. The alpha and beta: protein translocation across mitochondrial and plastid outer membranes. Trends Biochem. Sci. 26:36-40. [DOI] [PubMed] [Google Scholar]
- 13.Gentle, I., K. Gabriel, P. Beech, R. Waller, and T. Lithgow. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164:19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gratzer, S., T. Lithgow, R. E. Bauer, E. Lamping, F. Paltauf, S. D. Kohlwein, V. Haucke, T. Junne, G. Schatz, and M. Horst. 1995. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 129:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habib, S. J., T. Waizenegger, M. Lech, W. Neupert, and D. Rapaport. 2005. Assembly of the TOB complex of mitochondria. J. Biol. Chem. 280:6434-6440. [DOI] [PubMed] [Google Scholar]
- 16.Hill, K., K. Model, M. T. Ryan, K. Dietmeier, F. Martin, R. Wagner, and N. Pfanner. 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395:516-521. [DOI] [PubMed] [Google Scholar]
- 17.Hönlinger, A., U. Bömer, A. Alconada, C. Eckerskorn, F. Lottspeich, K. Dietmeier, and N. Pfanner. 1996. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 15:2125-2137. [PMC free article] [PubMed] [Google Scholar]
- 18.Hoppins, S. C., and F. E. Nargang. 2004. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279:12396-12405. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa, D., H. Yamamoto, Y. Tamura, K. Moritoh, and T. Endo. 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J. Cell Biol. 166:621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassenbrock, C. K., W. Cao, and M. G. Douglas. 1993. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J. 12:3023-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiebler, M., P. Keil, H. Schneider, I. van der Klei, N. Pfanner, and W. Neupert. 1993. The mitochondrial receptor complex: a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell 74:483-492. [DOI] [PubMed] [Google Scholar]
- 22.Kozjak, V., N. Wiedemann, D. Milenkovic, C. Lohaus, H. E. Meyer, B. Guiard, C. Meisinger, and N. Pfanner. 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278:48520-48523. [DOI] [PubMed] [Google Scholar]
- 23.Krimmer, T., D. Rapaport, M. T. Ryan, C. Meisinger, C. K. Kassenbrock, E. Blachly-Dyson, M. Forte, M. G. Douglas, W. Neupert, F. E. Nargang, and N. Pfanner. 2001. Biogenesis of the major mitochondrial outer membrane protein porin involves a complex import pathway via receptors and the general import pore. J. Cell Biol. 152:289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Künkele, K.-P., S. Heins, M. Dembowski, F. E. Nargang, R. Benz, M. Thieffry, J. Walz, R. Lill, S. Nussberger, and W. Neupert. 1998. The preprotein translocation channel of the outer membrane of mitochondria. Cell 93:1009-1019. [DOI] [PubMed] [Google Scholar]
- 25.Kutik, S., D. Stojanovski, L. Becker, T. Becker, M. Meinecke, V. Kruger, C. Prinz, C. Meisinger, B. Guiard, R. Wagner, N. Pfanner, and N. Wiedemann. 2008. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132:1011-1024. [DOI] [PubMed] [Google Scholar]
- 26.Lafontaine, D., and D. Tollervey. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lithgow, T., T. Junne, K. Suda, S. Gratzer, and G. Schatz. 1994. The mitochondrial outer membrane protein Mas22p is essential for protein import and viability of yeast. Proc. Natl. Acad. Sci. USA 91:11973-11977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meisinger, C., S. Pfannschmidt, M. Rissler, D. Milenkovic, T. Becker, D. Stojanovski, M. J. Youngman, R. E. Jensen, A. Chacinska, B. Guiard, N. Pfanner, and N. Wiedemann. 2007. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 26:2229-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meisinger, C., M. Rissler, A. Chacinska, L. K. Szklarz, D. Milenkovic, V. Kozjak, B. Schonfisch, C. Lohaus, H. E. Meyer, M. P. Yaffe, B. Guiard, N. Wiedemann, and N. Pfanner. 2004. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell 7:61-71. [DOI] [PubMed] [Google Scholar]
- 30.Milenkovic, D., V. Kozjak, N. Wiedemann, C. Lohaus, H. E. Meyer, B. Guiard, N. Pfanner, and C. Meisinger. 2004. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 279:22781-22785. [DOI] [PubMed] [Google Scholar]
- 31.Model, K., C. Meisinger, T. Prinz, N. Wiedemann, K. N. Truscott, N. Pfanner, and M. T. Ryan. 2001. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8:361-370. [DOI] [PubMed] [Google Scholar]
- 32.Model, K., T. Prinz, T. Ruiz, M. Radermacher, T. Krimmer, W. Kuhlbrandt, N. Pfanner, and C. Meisinger. 2002. Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J. Mol. Biol. 316:657-666. [DOI] [PubMed] [Google Scholar]
- 33.Mozdy, A. D., J. M. McCaffery, and J. M. Shaw. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neupert, W., and J. M. Herrmann. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723-749. [DOI] [PubMed] [Google Scholar]
- 35.Paschen, S. A., T. Waizenegger, T. Stan, M. Preuss, M. Cyrklaff, K. Hell, D. Rapaport, and W. Neupert. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426:862-866. [DOI] [PubMed] [Google Scholar]
- 36.Popov-Celeketic, J., T. Waizenegger, and D. Rapaport. 2008. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J. Mol. Biol. 376:671-680. [DOI] [PubMed] [Google Scholar]
- 37.Ramage, L., T. Junne, K. Hahne, T. Lithgow, and G. Schatz. 1993. Functional cooperation of mitochondrial protein import receptors in yeast. EMBO J. 12:4115-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rapaport, D. 2003. How to find the right organelle—targeting signals in mitochondrial outer membrane proteins. EMBO Rep. 4:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapaport, D., and W. Neupert. 1999. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 146:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan, M. T., H. Muller, and N. Pfanner. 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274:20619-20627. [DOI] [PubMed] [Google Scholar]
- 41.Schägger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt, S., U. Ahting, L. Eichacker, B. Granvogl, N. E. Go, F. E. Nargang, W. Neupert, and S. Nussberger. 2005. Role of TOM5 in maintaining the structural stability of the TOM complex of mitochondria. J. Biol. Chem. 280:14499-14506. [DOI] [PubMed] [Google Scholar]
- 43.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stettler, S., N. Chiannilkulchai, S. Hermann-Le Denmat, D. Lalo, F. Lacroute, A. Sentenac, and P. Thuriaux. 1993. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 239:169-176. [DOI] [PubMed] [Google Scholar]
- 45.Stojanovski, D., B. Guiard, V. Kozjak-Pavlovic, N. Pfanner, and C. Meisinger. 2007. Alternative function for the mitochondrial SAM complex in biogenesis of alpha-helical TOM proteins. J. Cell Biol. 179:881-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Wilpe, S., M. T. Ryan, K. Hill, A. C. Maarse, C. Meisinger, J. Brix, P. J. Dekker, M. Moczko, R. Wagner, M. Meijer, B. Guiard, A. Hönlinger, and N. Pfanner. 1999. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401:485-489. [DOI] [PubMed] [Google Scholar]
- 47.Waizenegger, T., S. J. Habib, M. Lech, D. Mokranjac, S. A. Paschen, K. Hell, W. Neupert, and D. Rapaport. 2004. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 5:704-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waizenegger, T., T. Stan, W. Neupert, and D. Rapaport. 2003. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J. Biol. Chem. 278:42064-42071. [DOI] [PubMed] [Google Scholar]
- 49.Westermann, B., and W. Neupert. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16:1421-1427. [DOI] [PubMed] [Google Scholar]
- 50.Wiedemann, N., V. Kozjak, A. Chacinska, B. Schönfish, S. Rospert, M. T. Ryan, N. Pfanner, and C. Meisinger. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424:565-571. [DOI] [PubMed] [Google Scholar]
- 51.Wiedemann, N., K. N. Truscott, S. Pfannschmidt, B. Guiard, C. Meisinger, and N. Pfanner. 2004. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 279:18188-18194. [DOI] [PubMed] [Google Scholar]
- 52.Wimley, W. C. 2003. The versatile β-barrel membrane protein. Curr. Opin. Struct. Biol. 13:404-411. [DOI] [PubMed] [Google Scholar]