Abstract
During normal development as well as in diseased states such as cancer, extracellular “niches” often provide cues to proximal cells and activate intracellular pathways. Activation of such signaling pathways in turn instructs cellular proliferation and differentiation. In the Caenorhabditis elegans gonad, GLP-1/Notch signaling instructs germ line stem cells to self-renew through mitotic cell division. As germ cells progressively move out of the niche, they differentiate by entering meiosis and eventually form gametes. In this model system, we uncovered an unexpected role for the dynein motor complex in promoting normal differentiation of proliferating germ cells. We demonstrate that dynein light chain 1 (DLC-1) and its partner, dynein heavy chain 1, inhibit the proliferative cell fate, in part through regulation of METT-10, a conserved putative methyltransferase. We show that DLC-1 physically interacts with METT-10 and promotes both its overall levels and nuclear accumulation. Our results add a new dimension to the processes controlled by the dynein motor complex, demonstrating that dynein can act as an antiproliferative factor.
Caenorhabditis elegans germ cells proceed through a well-orchestrated developmental program on their way to producing specialized gametes (23). The C. elegans gonad functions as an assembly line organized with a distal-to-proximal polarity with respect to the uterus (Fig. 1A). Proliferating germ cells, including the germ line stem cells, reside in the distal region of the gonad that forms the stem cell niche. These cells self-renew through mitosis or, at a certain frequency, differentiate by entering meiosis and give rise to gametes after an extended meiotic prophase (Fig. 1B). Proliferation of the distal germ cells results in a net flux of germ cells toward the proximal end, and it is thought that differentiation is a consequence of progressive displacement away from the stem cell niche (9, 33, 36).
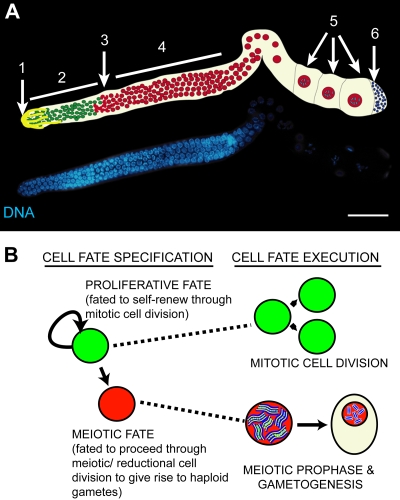
FIG. 1.
(A) Fluorescence micrograph of a dissected, DAPI (4′,6-diamidino-2-phenylindole)-stained adult C. elegans hermaphrodite germ line, with schematic. In the distal region, proliferating germ cells (2) reside in close contact with the somatic distal tip cell (1). At the transition zone (3), germ cells enter meiosis and proceed through meiotic prophase (4) to give rise to sperm (6) and oocytes (5). (B) Scheme depicting the decision that C. elegans germ line stem cells make between maintaining a proliferative fate (green) and differentiating (red) to produce gametes.
The primary signal instructing germ cells to proliferate is activation of the GLP-1/Notch signaling pathway in germ cells through interaction with the somatic distal tip cell that expresses the Notch ligand LAG-2, which provides the stem cell niche (19, 20, 27). Mutations that disrupt GLP-1/Notch receptor function result in premature differentiation and meiotic entry of germ line stem cells (1), while gain-of-function mutations that hyperactivate GLP-1/Notch cause germ line tumors characterized by germ cell overproliferation (5, 43).
Multiple factors act to restrain GLP-1-dependent germ cell proliferation (19, 27). The C. elegans mett-10 gene was initially characterized as an inhibitor of germ cell proliferative fate but was also shown to play a role in promoting progression through mitotic cell division and meiotic development (13). mett-10 encodes a putative methyltransferase that is conserved in higher eukaryotes and a loss-of-function mutation that disrupts the binding pocket for S-adenosylmethionine and compromises the balance between proliferative and meiotic fates (13, 35; http://www.rcsb.org/pdb/explore.do?structureId=2H00). METT-10 accumulates in nuclei as germ cells enter meiosis, consistent with its role in inhibiting proliferative fate and promoting meiotic progression (13). However, it is unclear how METT-10 nuclear accumulation is controlled or if it is essential for METT-10 activity.
We find that inhibition of proliferative fate is also regulated by dynein light chain 1 (DLC-1) and its partner, dynein heavy chain 1 (DHC-1); DLC-1 upregulates mett-10 mRNA levels and promotes the nuclear accumulation of the METT-10 protein. Cytoplasmic dynein is a large, microtubule-associated motor complex that traffics organelles, proteins, and RNAs toward microtubule minus ends (7, 25, 58). Dynein is required for many cellular processes, including execution of mitotic cell division, a process necessary for cell proliferation (17, 25, 51). Thus, the unexpected role of dynein in the inhibition of germ cell proliferative fate demonstrates that proliferative fate is genetically separable from the act of mitotic cell division itself and addresses how cellular machinery can implement developmental programs.
MATERIALS AND METHODS
General worm culture and genetics.
Standard procedures for culture and genetic manipulation of C. elegans strains were followed (6). Strain constructions were verified by single-worm PCR and sequencing of alleles. To this end, we identified a molecular lesion in dhc-1(js319) (29) as an AG→AA mutation in the splice acceptor for exon 13.
Alleles used in this study are listed by chromosome as follows. For chromosome I, rrf-1(pk1417), dhc-1(js319), and dhc-1(or195) were used; for chromosome III, mett-10(oz36) (antimorph), mett-10(g38) (hypomorph), mett-10(tm2697) (hypomorph), mett-10(oj32) (hypomorph), mett-10(ok2204) (null), glp-1(oz264gf), and glp-1(bn18ts) were used.
Integrated transgenes used in this study are listed as follows: ozIs7[pmett-10:mett-10::gfp; unc-119(+)], ozIs9[pmett-10:mett-10(5A)::gfp; unc-119(+)] (low protein and mRNA expression, as measured by immunoprecipitation [IP] and quantitative reverse transcription-PCR [qRT-PCR]) (data not shown), ozIs11[pmett-10:mett-10(5A)::gfp; unc-119(+)] (high protein and mRNA expression, as measured by IP and qRT-PCR) (data not shown), ozIs15[pmett-10:mett-10(oj32, 5A)::gfp; unc-119(+)] (line 1), ozIs16[pmett-10:mett-10(oj32, 5A)::gfp; unc-119(+)] (line 2), ozIs20[pmett-10:mett-10(-NLS, 5A)::gfp; unc-119(+)] (line 1), and ozIs21[pmett-10:mett-10(-NLS, 5A)::gfp; unc-119(+)] (line 2).
Extrachromosomal arrays used in this study are listed as follows: ozEx66 [pmett-10:mett-10::gfp; unc-119(+)], ozEx69 [pmett-10:mett-10(-NLS)::gfp; unc-119(+)], ozEx67 [pmett-10:mett-10(5A)::gfp; unc-119(+)], ozEx72 [pmett-10:mett-10(-NLS, 5A)::gfp; unc-119(+)], and ozEx73 [pmett-10:mett-10(oz36)::gfp; unc-119(+)].
Immunohistochemistry.
Germ line dissection, fixation, and staining were carried out as previously described (31). The antibodies used were as follows: monoclonal mouse anti-green fluorescent protein (anti-GFP) (mAb3E6) from Invitrogen (catalog no. A-11120) at a dilution of 1/100 and rabbit anti-DLC-1 at a dilution of 1/10 (see below), donkey anti-mouse antibody-Alexa 594, goat anti-rabbit antibody-Alexa 488, and goat anti-rabbit antibody-Alexa 594 at a dilution of 1/400, obtained from Molecular Probes (Invitrogen, Carlsbad, CA). Labeling of DNA synthesis with the nucleotide analog 5-ethynyl-2′-deoxyuridine (EdU) (49) was done by feeding worms with bacteria containing EdU (13). EdU detection and costaining with antibodies were carried out as previously described (13).
Generation of anti-DLC-1 antibody and quantification of DLC-1 levels.
To generate the glutathione S-transferase (GST)-DLC-1 fusion protein, full-length DLC-1 was cloned into the pGEX-5x-3 vector (catalog no. 27-4586-01; GE Healthcare Life Sciences). Inductions were carried out in Escherichia coli strain BL21(DE3) (Sigma, MO). From an overnight starter culture grown at 30°C, cultures were diluted and grown at 37°C until they reached an optical density at 600 nm of 0.5. Cells were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. Cells were harvested, and GST-DLC-1 was then extracted essentially as described previously (50) and injected into rabbits obtained from Pocono Rabbit Farm & Laboratory (Canadensis, PA). Sera were affinity purified against a His6-DLC-1 fusion protein. To make His6-DLC-1, DLC-1 was amplified from pGEX(DLC-1) and cloned into pTRCHIS (catalog no. K4410-01 and K4410-40; Invitrogen). pTRCHIS(DLC-1) was transformed into BL21(DE3) cells, and cultures were induced similarly to GST-DLC-1. His6-DLC-1 was extracted under native conditions using Ni-nitrilotriacetic acid agarose (catalog no. 30210; Qiagen). His6-DLC-1 was subsequently coupled to cyanogen bromide-activated Sepharose (catalog no. C9142; Sigma, MO), and antibody purification was carried out essentially as previously described (24). Specificity was determined both by Western blotting (where antibody recognizes a single 10-kDa band) (data not shown) and by antibody staining of germ lines and intestines from animals treated with and without dlc-1 RNA interference (RNAi).
For quantification of DLC-1 levels (Fig. 2J), raw images of DLC-1 staining were imported into ImageJ, germ lines were traced, and the average pixel intensity was calculated. All pixel intensities were then normalized to represent the percent knockdown by dividing the pixel intensity for the sample by the average DLC-1 pixel intensity for glp-1(oz264) germ lines fed control RNAi and multiplying by 100.
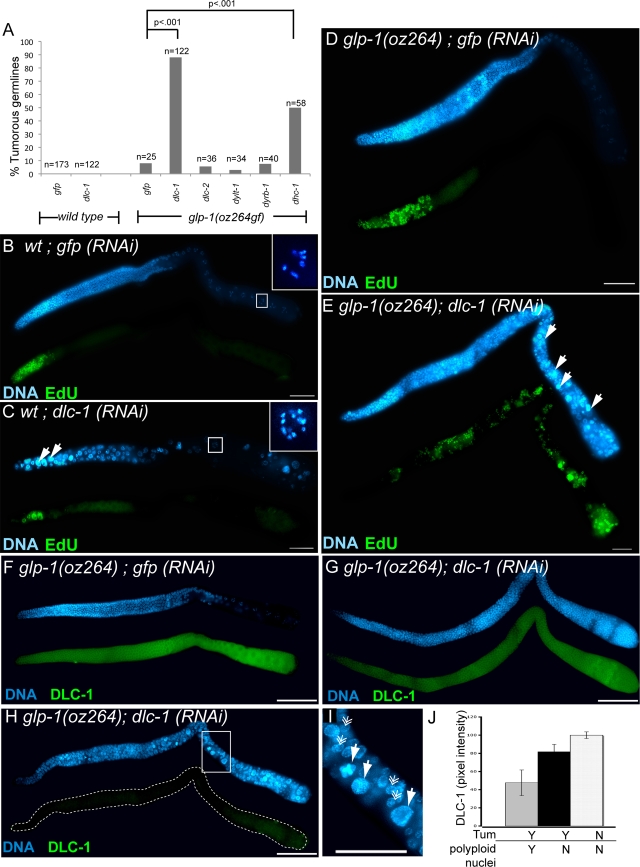
FIG. 2.
dlc-1 functions to inhibit proliferative fate in the C. elegans germ line. (A) RNAi against dlc-1 and dhc-1 enhances tumor formation in the glp-1(oz264gf) background at 20°C, while RNAi against other light chains does not. dlc-2, paralog of dlc-1; dylt-1, Tctex-type dynein light chain; dyrb-1, Roadblock-type light chain. Two-tailed P values were determined using Fisher's exact test. (B, C) In wild-type germ lines (B), as well as in dlc-1 RNAi of control animals (C), a 3-h pulse of the nucleotide analog EdU, which incorporates during DNA synthesis, labels (green) only the very distal end (left) of the germ line, although dlc-1 RNAi causes abnormal nuclei containing large, dense DNA bodies (white arrows) and unpaired chromosomes in diakinesis to occur. Insets show chromosomal morphology of diakinetic oocytes, with six sets of bivalents in oocytes from control germ lines (B) but 12 univalents for dlc-1 RNAi (C). (D, E) dlc-1 RNAi causes tumor formation and EdU incorporation throughout the germ line in glp-1(oz264gf) (E), while the glp-1(oz264gf) mutant on its own has only a marginal increase in EdU incorporation (D). (F to H) Staining against DLC-1 (green) in glp-1(oz264gf) animals treated with double-stranded RNA targeting either gfp as a control (F) or dlc-1 (G, H). (G) A tumorous germ line with partial DLC-1 knockdown and unaffected nuclear morphology; (H) a tumorous germ line with stronger DLC-1 knockdown and abnormal nuclear morphology. (I) This inset from panel H highlights the large size of abnormal nuclei. Solid arrows indicate large, DNA-dense (abnormal) nuclei, and feather arrows indicate nuclei of relatively normal size. (J) Quantification of DLC-1 levels by pixel intensity for glp-1(oz264gf); gfp(RNAi) germ lines (white bar), glp-1(oz264gf); dlc-1(RNAi) tumorous germ lines with normal nuclear morphology (black bar), and glp-1(oz264gf); dlc-1(RNAi) tumorous germ lines with abnormal nuclear morphology (gray bar). Error bars indicate the standard errors of the means for the results. All scale bars = 20 μm. Tum, tumorous; wt, wild type.
Image capture and processing.
Fluorescence micrographs were taken on a Zeiss compound microscope using Axioplan 2.0 imaging software and a Hamamatsu camera. Each dissected and stained gonad was captured as a montage, with overlapping cell boundaries. The montages were then assembled in Adobe Photoshop CS3 and processed identically. The boundary of each montage was processed using a feather tool of 20 pixels for each image. Confocal images were captured on a PerkinElmer UltraView confocal microscope using a z-step of 0.3 μm.
RNAi of METT-10 yeast two-hybrid interactors.
RNAi clones were either purchased from Open Biosystems or generated by PCR amplification and cloning of 1-kb exonic sequences into the pPD128.36 double T7 promoter vector, sequenced to verify identity, and transformed into the E. coli host HT115(DE3). RNAi feeding in rrf-1(pk1417), rrf-1(pk1417) glp-1(oz264gf), and rrf-1(pk1417) mett-10(oz36) backgrounds at the permissive temperature (20°C) was performed as previously described (31). F1 L4 hermaphrodites were transferred onto fresh RNAi plates and scored quantitatively for germ line tumors 48 h later by differential interference contrast (DIC) microscopy. In the course of the screening, it came to our attention that mett-10 mutants are relatively resistant to germ line RNAi.
Construction of transgenes and generation of transgenics.
The initial mett-10::gfp construct includes 1.6 kb upstream and 1.9 kb downstream of the METT-10 coding region, with GFP (intervening sequence):FLAG inserted immediately before the translational stop cloned into the pMM016 vector containing the transformation marker unc-119 (13). Mutations of the mett-10 coding region, including nuclear localization signal (NLS) deletion, DNASQ→AAAAA, methyltransferase mutation (G110R) mimicking the mett-10(oj32) mutation (13), and deletion of coding sequences downstream to the mett-10(oz36) premature stop codon, were done with sewing PCR (22). To create low-copy-number-integrated transgenic lines, the microparticle bombardment method was used (45). Generation of extrachromosomal arrays was done by injection of pMM016(mett-10) plasmids at a concentration of 40 ng/μl into unc-119(ed3) worms.
GST pulldowns.
Full-length METT-10 (C. elegans coding region ZK1128.2a), METT-10 (N terminal) (amino acids [aa] 1 to 271), or METT-10 (C terminal) (aa 272 to 479) was amplified from N2 Bristol cDNA and cloned into pTRCHIS (catalog no. K4410-01, K4410-40; Invitrogen), sequenced, and transformed into BL21(DE3) (Sigma, MO) cells for expression. Proteins were expressed under the same conditions as GST-DLC-1 (see above). For GST pulldowns, bacterial extracts were resuspended in phosphate-buffered saline plus protease inhibitors and sonicated. Cleared cell extracts were added directly to 5 μg of GST-DLC-1 already bound to glutathione-Sepharose in 20 mM Tris at pH 7.5, 500 mM NaCl, 0.5% NP-40, and 1× protease inhibitors. For binding to GST-METT-10, significantly less protein was used (100 to 200 ng), as METT-10 does not express well in vitro. Binding reactions were carried out overnight at 4°C and washed four times with 10 mM Tris at pH 7.5, 150 mM NaCl, and 0.1% NP-40. Western blot analysis using an anti-His6 antibody (catalog no. 27-4710-01; Amersham Biosciences) was performed with beads boiled in sodium dodecyl sulfate sample buffer with β-mercaptoethanol. Point mutations in METT-10 and DLC-1 were generated using sewing PCR or site-directed mutagenesis.
IP and Western analysis.
To generate enough worms for IP-Western analysis of METT-10:GFP (flag), we resuspended worms from 10 60-mm overgrown plates of each transgenic line in 5% lactose and 10× OP50 or 1× bacteria expressing double-stranded RNA and plated these worms onto 10 100-mm plates, which were shifted to 25°C to maximize transgene expression. After 2 days, worms were washed off in phosphate-buffered saline, and extracts and IPs were carried out essentially as described previously (18). Immunoprecipitated proteins were analyzed using Western blotting with both mouse monoclonal anti-FLAG M2 (catalog no. F1804; Sigma) and rabbit anti-GFP (catalog no. A11122; Invitrogen) antibodies. For loading controls, we used mouse anti-α-tubulin (catalog no. T9026; Sigma) and anti-CGH-1 (a gift from David Greenstein). CGH-1 stands for conserved germ line helicase and is a protein that is abundantly expressed throughout the germ line, localizing to both the cytoplasm and P granules (39).
mRNA quantification.
To measure mett-10:gfp transgene expression relative to endogenous, each mett-10::gfp transgene was crossed into the mett-10(ok2204) background, which contains a large deletion of the mett-10 5′ region. Total RNA was isolated using Trizol (Invitrogen), treated with DNase I (Epicentre), chloroform extracted, ethanol precipitated, and dissolved in water. cDNA was synthesized using random hexamer primers and SuperScript VILO reverse transcriptase (Invitrogen). Transcripts were quantified using Sybr green real-time PCR (Clontech) and gene-specific primers (see below) calibrated with genomic DNA. mRNA levels were calculated, adjusting for amplification efficiency (44) and normalizing to internal cgh-1 or act-3 transcripts and external genomic DNA standards. Standard deviations were calculated using all PCR replicates from all biological replicates.
The following primers were used for quantitative RT-PCR, with the forward primer sequence listed first, followed by the reverse primer sequence: mett-10, TGATATTGGCACCGGAACATCGTG and CATCTCCGTCAGTGGCAATGAACT; dlc-1, TGGCATTGCATCGTCGGAAGAAAC and AGACTTGAATAGCAGGATGGCGAC; act-3, TCTTGACTTGGCTGGACGTGATCT and TGATGTCACGGACGATTTCACGCT; and cgh-1, ATCAACTTCCTCCCAAAGGAGCG and TCATACGGCTTGTGCATGTGCTTC.
RESULTS
DLC-1 inhibits germ cell proliferative fate.
In a screen for proteins that inhibit germ cell proliferative fate, we discovered that partial RNAi depletion of dlc-1 enhanced germ line tumor formation in a genetic background sensitized by excessive glp-1 activity [glp-1(oz264gf)] (Fig. 2A) (26). DLC-1 is an LC8-type dynein light chain and a component of the dynein motor complex that participates in multiple processes, including mitotic cell division (16, 25, 37, 42, 58).
Although DLC-1 depletion causes germ line tumors in the glp-1(gf)-sensitized genetic background (Fig. 2B, D, and E), it disrupts mitotic and meiotic processes in wild-type worms. In addition to apparent polyploid nuclei, which are larger and contain more DNA, many oocytes contained unpaired chromosomes (compare Fig. 2B and C). Our observation of polyploidy is consistent with previously described roles for dynein in mitotic cell division across taxa (16, 25, 37, 42, 58), while the observation of unpaired diakinetic chromosomes extends those of previous studies that have demonstrated a role for dynein in mediating homologous chromosome pairing in fission yeast (12, 60, 61). At first glance, these findings appear paradoxical because the germ line tumors in the glp-1 gain-of-function background require additional mitotic divisions. However, when we assessed the degree of DLC-1 knockdown using antibody staining (Fig. 2F to H), we found that the role of DLC-1 in inhibiting proliferative fate is uncovered in the glp-1 mutant background because only a slight decrease in DLC-1 is needed to enhance the tumorous phenotype of the glp-1(oz264gf) mutant without causing mitotic defects. In contrast, more-severe knockdown affects both cell fate and cell division, leading to polyploidy (Fig. 2F to J).
While dynein light chains function within the context of the dynein motor complex, other functions independent of the motor complex have been described (3, 21, 28, 46). There are multiple light chains, but among the multiple dynein light chains we tested, including the closest dlc-1 paralog, dlc-2, the Roadblock-type light chain dyrb-1, and the Tctex-type light chain dylt-1, only RNAi of dlc-1 enhanced glp-1(oz264gf) overproliferation (Fig. 2A). It is possible that we did not achieve an optimal level of knockdown to reveal an overproliferation phenotype for the other light chains. Strikingly, RNAi-mediated depletion of DHC-1, the essential motor component of dynein motor complex activity (17), also increased tumor formation, suggesting that it plays a role in proliferative fate specification (Fig. 2A). We conclude, therefore, that DLC-1 affects the proliferative fate decision in the context of the motor complex, although it may be unique among the light chains in doing so.
The dynein motor complex promotes normal localization and levels of METT-10.
One reason we suspected that DLC-1 might play a role in the germ cell proliferative fate decision was that a genome-wide yeast two-hybrid screening of conserved worm proteins (32) suggested that DLC-1 may be a functional partner of another protein that inhibits proliferative fate, the METT-10 putative methyltransferase (13). Loss of METT-10 also enhances germ cell proliferation and formation of germ line tumors in the glp-1(oz264gf)-sensitized genetic background (13). METT-10 accumulates in nuclei as cells enter meiosis, in line with its functions in inhibiting proliferative fate and promoting meiotic development (Fig. 3A). Our finding that DLC-1 depletion enhances the glp-1(oz264gf) tumorous phenotype, coupled with the established role of dynein in trafficking proteins to the nucleus (16, 25, 37, 42), suggested that DLC-1 may inhibit germ cell proliferative fate through regulation of METT-10 nuclear accumulation. Using RNAi, we found that DLC-1 knockdown decreases accumulation of a METT-10::GFP fusion protein in nuclei throughout the germ line and also in somatic cells (Fig. 3A and B; also data not shown).
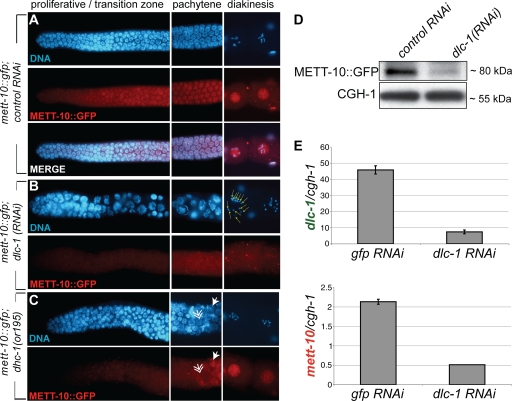
FIG. 3.
DLC-1 promotes METT-10 nuclear accumulation. (A) METT-10::GFP expression in a wild-type germ line. (B) dlc-1 RNAi causes a decrease in METT-10::GFP accumulation, abnormal nuclear morphology (left), and unpaired chromosomes in diakinesis (right, yellow arrows). (C) METT-10::GFP nuclear accumulation is delayed in the dhc-1(or195) mutant background, with nuclear accumulation observed only in late pachytene. Solid and feather arrows indicate nuclei positive and negative for METT-10::GFP, respectively. (D) IP-Western blotting for METT-10::GFP levels in whole animals subject to both control (empty vector) RNAi and dlc-1(RNAi). CGH-1 is a germ line ubiquitous protein. (E) qRT-PCR for dlc-1 (top) and mett-10 (bottom) normalized to cgh-1 mRNA levels. RNAi was carried out in the rrf-1(pk1417) background, which retains RNAi only in the germ line.
To confirm that DLC-1 functions with the motor complex to promote METT-10 nuclear accumulation, we tested whether reductions in DHC-1 activity also resulted in a decrease in nuclear METT-10. We found that in a genetic background with decreased DHC-1 function [dhc-1(or195)] (51), METT-10::GFP nuclear accumulation is delayed, with a strong nuclear signal only in late pachytene rather than near the transition zone as occurs in wild-type germ lines, in more than 50% of germ lines (Fig. 3C). This finding indicates that the dynein motor complex promotes METT-10 nuclear accumulation.
In addition to diminished nuclear localization, overall staining intensity for METT-10::GFP is reduced upon DLC-1 depletion, suggesting that DLC-1 also promotes or maintains the total METT-10 protein level (Fig. 3B). Indeed, IP of METT-10::GFP from animals treated with control RNAi or RNAi against dlc-1 revealed that total METT-10::GFP levels are reduced by partial DLC-1 depletion (Fig. 3D). This reduction does not reflect a decrease in germ line size, because the levels of a ubiquitous germ line protein, CGH-1 (39), were unchanged (Fig. 3D).
DLC-1 regulates mett-10 RNA levels.
We considered two potential mechanisms by which DLC-1 could regulate total METT-10 protein accumulation. LC8-type dynein light chains regulate transcription (46, 47) and also interact directly with various proteins to regulate their subcellular localization and/or dimerization status, which could alternatively affect protein stability (2, 10, 38, 52). To address whether DLC-1 regulates METT-10 at the mRNA level, we tested the effect of DLC-1 depletion on levels of METT-10 mRNA using qRT-PCR and found that knockdown of DLC-1 results in four- to fivefold-higher reductions in mett-10 mRNA relative to the germ line ubiquitous gene cgh-1 (Fig. 3E). Thus, DLC-1 promotes overall METT-10 mRNA levels potentially through a transcriptional mechanism.
DLC-1 binds METT-10 in vitro.
We tested if DLC-1 interacts directly with METT-10 using GST pulldown affinity chromatography and found that His6-tagged METT-10 binds GST-DLC-1 in vitro (Fig. 4A). We used a series of amino acid substitution mutations in DLC-1 to see if its interaction with METT-10 requires the same amino acid domain shown to mediate LC8-type dynein light chain interaction with potential “cargo” proteins and dynein intermediate chains (10, 38). Indeed, disruption of amino acid residues F62, T67, and F73 greatly reduced binding to METT-10, while mutation of H68 did not affect binding (Fig. 3A), showing that the interaction between DLC-1 and METT-10 is similar to previously characterized interactions (10, 38).
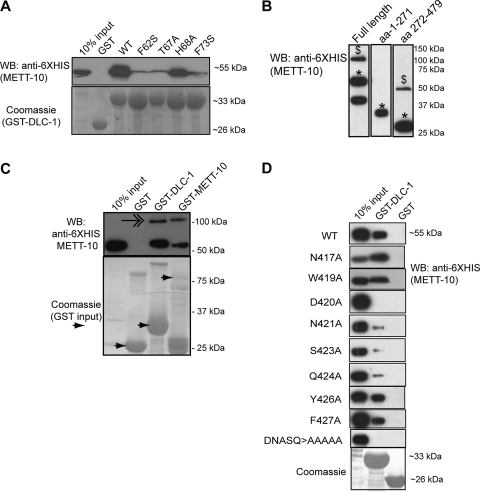
FIG. 4.
METT-10 binds DLC-1 in vitro. (A) GST-DLC-1 point mutants (Coomassie) were assayed for binding to full-length His6-METT-10 proteins. WT, wild type. (B) METT-10 can form multimers. Western blot (WB) analysis of bacterially expressed His6-METT-10 fusion proteins reveals bands that run at the size of monomers (*) and dimers ($). The lowest band in lane 1 likely represents a METT-10 degradation product. (C) A dimeric band (feathered arrow) is observed when full-length His6-METT-10 is pulled down with GST-DLC-1 and full-length GST-METT-10. (D) Fine mapping of the DLC-1 binding domain of METT-10 using GST-pulldown analysis analogous to that shown in panel C. The identity of individually mutated METT-10 residues are labeled on the left.
In the course of analyzing the physical interaction between METT-10 and DLC-1, we discovered that METT-10 strongly interacts with itself. We noticed a sodium dodecyl sulfate-stable supershifted band in the gel electrophoresis of some METT-10 proteins equivalent in size to the expected size of a dimer (Fig. 4B), in line with previous reports that some methyltransferases require multimerization for function (34, 56). The stable self-interaction domain of METT-10 maps to the C-terminal region (aa 272 to 479), and a supershift was never seen with a METT-10 N-terminal fragment (aa 1 to 271) alone (Fig. 4B). To confirm that METT-10 binds to itself, we carried out GST pulldown analysis between HIS6-tagged and GST-tagged METT-10. We observed the supershifted band for HIS6-tagged METT-10 pulled down by GST-tagged METT-10, suggesting that METT-10 may exist in an oligomeric form (Fig. 4C). Interestingly, the putative METT-10 dimer is also pulled down by DLC-1, indicating that the C-terminal interface by which METT-10 interacts with itself is independent of the interface used to bind DLC-1 (Fig. 4C).
DLC-1 and the METT-10 NLS act redundantly to ensure METT-10 nuclear accumulation.
The finding that DLC-1 interacts with METT-10 raised the possibility that DLC-1 participates directly in METT-10 nuclear accumulation. To address whether DLC-1 promotes METT-10 localization through direct binding, we needed to specifically test the contribution of DLC-1 interaction to METT-10 nuclear accumulation in vivo. To this end, we mapped the residues that interact with DLC-1 in METT-10. Previous work showed that LC8-type light chains interact with a short peptide sequence in their target proteins, usually conforming to either a (S/K)XTQT or GIQVD sequence motif (2, 4), but METT-10 does not have either of these motifs. We thus tested a series of METT-10 C-terminal truncations (data not shown), followed by individual amino acid replacements in an eight-amino-acid region, for their ability to interact with DLC-1 in GST pulldown experiments. Mutation of METT-10 amino acid residues D420, N421, S423, and Q424 to alanine severely compromised binding to DLC-1, thus defining the METT-10 motif that interacts with DLC-1 as “DNASQ” (Fig. 4D).
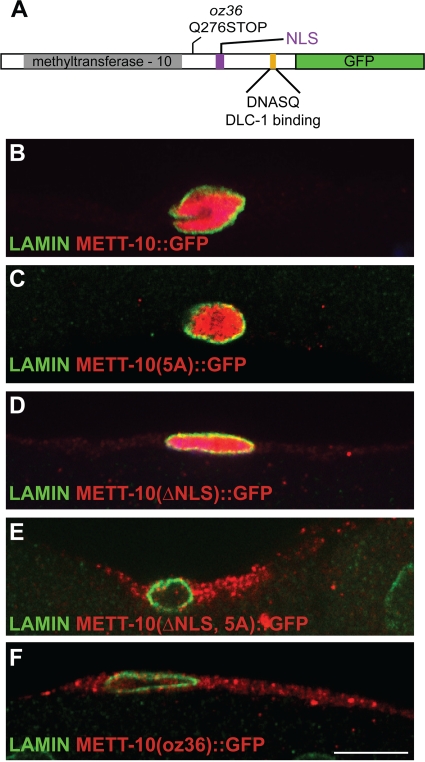
To determine the effect of DLC-1 binding on METT-10 localization in vivo, we constructed a transgenic version of METT-10::GFP in which the “DNASQ” motif was mutated to “AAAAA” (5A mutant) (Fig. 5A) and compared the localization of METT-10::GFP constructs within gonadal sheath cells, which are large cells that express METT-10 at higher levels and thus are easy to visualize. Wild-type METT-10::GFP is concentrated in the nucleus (Fig. 5B), and disruption of the DNASQ motif that interacts with DLC-1 reduces, but does not eliminate, nuclear METT-10::GFP (Fig. 5C).
FIG. 5.
Dual mechanism ensures METT-10 nuclear accumulation. (A) Domain architecture of METT-10 marked with protein sequence change caused by mett-10(oz36) and location of the DLC-1 binding motif. Gray shading highlights the conserved methyltransferase-10 domain, and purple indicates the NLC sequence. (B to F) Confocal images (0.3 μm) of wild-type and mutant METT-10::GFP expression in gonadal sheath cells stained with antibodies against GFP and lamin to visualize nuclear membrane.
In addition to the DLC-1 binding motif, METT-10 also contains a potential nuclear localization sequence (ARKRAKA), raising the possibility that this may act redundantly with the DLC-1 binding site to enable METT-10 nuclear accumulation. Supporting this idea, deletion of the putative METT-10 NLS alone does not block nuclear entry (Fig. 5D), but simultaneous disruption of both the NLS and the DLC-1 binding site makes METT-10 predominantly cytoplasmic (Fig. 5E).
Cytoplasmic METT-10 rescues most mett-10 mutant phenotypes.
Identification of at least two pathways promoting METT-10 nuclear accumulation enabled us to address if METT-10 nuclear accumulation is essential in vivo and whether the METT-10:DLC-1 physical interaction is vital to METT-10 function. To this end, we compared the abilities of the integrated transgenes expressing various mutant forms of METT-10 to rescue mett-10 mutant phenotypes. These included METT-10 with a mutant DLC-1 binding site [mett-10(5A)::gfp], a mutant DLC-1 binding motif and deletion of the NLS [mett-10(ΔNLS, 5A)::gfp], and METT-10 with a mutant DLC-1 binding site and a mutation in the catalytic core that should reduce the methyltransferase activity [mett-10(G110R, 5A)::gfp] (see Materials and Methods). We compared expression of mRNA and protein by the various transgenes using qRT-PCR and Western blots (data not shown; see Materials and Methods). The mRNA and protein levels of wild-type and mutant transgenes are similarly correlated, suggesting that METT-10(5A)::GFP is not especially unstable and agreeing with the finding described above that DLC-1 promotes METT-10 levels primarily by regulating mett-10 mRNA levels.
Each mett-10 transgene was tested for its ability to rescue several mett-10 mutant phenotypes, including abnormalities in somatic and germ cell development observed in the mett-10 null background and defective proliferative fate specification caused by loss-of-function mett-10 mutations (13). Loss of mett-10 function causes protrusion of the vulva, irregular gonadal shape, and defects in germ cell development, leading to a sterile phenotype when animals are raised at 25°C (13). While a methyltransferase-defective transgene failed to rescue any defects, mett-10(5A)::gfp and mett-10(ΔNLS, 5A)::gfp transgenes fully rescued the protruding vulva phenotype and partially rescued abnormalities in gonad shape and germ cell development (Table 1). This partial rescue (81% to 86% reduction in phenotype penetrance as opposed to 100% reduction in that for the wild-type transgene) suggests that the degree of METT-10 nuclear accumulation, promoted by both DLC-1 interaction and the NLS, facilitates METT-10 functions but is not essential. Moreover, even in the absence of both the DLC-1 interaction site and the NLS, some residual METT-10 may still enter the nucleus [some nuclear puncta can be seen for mett-10(ΔNLS, 5A)::gfp] (Fig. 5E), enabling partial rescue. The fact that the methyltransferase-defective transgene does not rescue any of these phenotypes demonstrates that enzymatic activity is required for METT-10 function, and it is possible that even if the critical METT-10 targets are nuclear proteins, METT-10 may also modify them in the cytoplasm before they enter the nucleus.
TABLE 1.
Transgenic rescue of mett-10 phenotypese
| Transgenec | % Tumorous germ lines (no.)d |
Transgenic rescue of mett-10(ok2204) at 25°Cb |
||||
|---|---|---|---|---|---|---|
| mett-10 (ok2204) | glp-1(oz264) mett-10(g38) | Animals scored |
% Abnormal gonad shape (no.)a | |||
| % Pvl | % Ste | No. | ||||
| No transgene | 0 (96) | 89 (47) | 98 | 99 | 82 | 30 (44) |
| met-10(wt)::gfp | 0 (136) | 0 (56) | 5 | 0** | 38 | 0 (35) |
| mett-10(5A)::gfp (LOW) | 0 (86) | 0 (54) | 65 | 19** | 43 | ND |
| mett-10(5A)::gfp (HIGH) | 0 (36) | ND | 1 | 0** | 137 | 3 (32)** |
| mett-10 (enzymatic-defective, 5A)::gfp (line 1) | 0 (36) | 91 (84) | 92 | 98 | 46 | 30 (43) |
| mett-10 (enzymatic-defective, 5A)::gfp (line 2) | 0 (48) | 94 (83) | 97 | 99 | 97 | ND |
| mett-10(ΔNLS, 5A)::gfp (line 1) | 0 (37) | 4.3 (94) | 0 | 19** | 42 | 10 (31)** |
| mett-10(ΔNLS, 5A)::gfp (line 2) | ND | ND | 0 | 14** | 36 | 12 (25) |
Gonads with one or more ectopic distal protrusions.
**, two-tailed P value and Fisher's exact test value of <0.05 were significantly different from no transgene. Pvl, protruding vulva; Ste, sterile. Scored by DIC.
LOW and HIGH refer to low and high mRNA and protein expression levels, respectively, as measured by IP and qRT-PCR (data not shown).
Tumorous includes proliferation throughout the germ line, although some germ cells in various meiotic stages may be observed. Scored by DIC.
ND, not determined.
Loss of mett-10 function compromises the balance between germ cell proliferation and differentiation, which similar to this balance in dlc-1 and dhc-1, is revealed in a glp-1 hyperactivation mutant background (13). Thus, to evaluate the role of METT-10 nuclear accumulation in proliferative fate specification, we tested for rescue of the tumorous phenotype of the glp-1(oz264gf) mett-10(g38) double mutants. Expression of METT-10(5A)::GFP and METT-10(ΔNLS, 5A)::GFP rescued the tumorous phenotype of glp-1(oz264) mett-10(g38) animals, but a methyltransferase-defective version of METT-10 did not rescue the mutant phenotype (Table 1). Thus, METT-10 inhibition of proliferative fate, similar to other METT-10 developmental functions, requires enzymatic activity but not the full extent of nuclear accumulation that is mediated by its NLS and DLC-1.
Dynein also regulates proliferative fate independently of METT-10.
Based on the above-mentioned observations, one possible explanation for the enhancement of glp-1(oz264gf) tumor formation by DLC-1 depletion is the reduced METT-10 accumulation caused by decreased mett-10 mRNA, combined with decreased transport of METT-10 to the nucleus. However, genetic interactions among mett-10, dlc-1, and dhc-1 indicate that dynein also plays roles in proliferative fate specification independent of its effects on METT-10 protein accumulation.
The role of METT-10 in proliferative fate specification was initially revealed by the antimorphic mett-10(oz36) truncation mutant, which exhibits germ line tumors even in the presence of wild-type glp-1 (13). mett-10(oz36) mutants exhibit, with respect to germ line proliferation, a phenotype that is more severe than those exhibited by mutants lacking all mett-10 activity (13). The dose dependence of the proliferative effect of the METT-10(oz36) truncated protein showed that it is a “poisonous” protein that disrupts the function of at least one other unknown factor that normally inhibits proliferative fate (13). One prediction of this model is that depletion of this poisoned factor should enhance mett-10(oz36) tumor formation. Interestingly, we found that RNAi knockdown of DLC-1 enhances tumor formation in the mett-10(oz36) genetic background (Table 2). Moreover, two different weak loss-of-function heavy chain alleles, dhc-1(or195) (51) and dhc-1(js319) (29, 51), also specifically enhanced the mett-10(oz36) tumorous phenotype (Table 2), consistent with a genetic model in which METT-10(oz36) poisons DLC-1 activity. However, METT-10(oz36) lacks both the DLC-1 binding site and the canonical NLS and thus is predominantly cytoplasmic (Fig. 5A and F), indicating that DLC-1 cannot be the poisoned factor. Thus, the ability of DLC-1 knockdown and dhc-1 reduction-of-function mutants to further enhance tumor formation indicates that DLC-1 also regulates proliferative fate independently of its effects on METT-10, potentially by promoting the activity of a factor normally poisoned by the METT-10(oz36) mutant protein.
TABLE 2.
Dynein disruption enhances tumor formation in the background of the mett-10(oz36) cytoplasmic mutant
| Genotype | % Tumorousc | No. of tumors |
|---|---|---|
| dlc-1a | ||
| rrf-1(pk1417) mett-10(oz36) gfp(RNAi) | 3 | 117 |
| rrf-1(pk1417) dlc-1(RNAi) | 0 | 122 |
| rrf-1(pk1417) mett-10(oz36) dlc-1(RNAi) | 62 | 130 |
| Dynein heavy chainb | ||
| dhc-1(or195) | 0 | 37 |
| mett-10(oz36) | 43 | 65 |
| glp-1(oz264) | 1 | 99 |
| dhc-1(or195) glp-1(oz264) | 9 | 22 |
| dhc-1(or195) mett-10(tm2697) | 0 | 35 |
| dhc-1(or195) mett-10(g38) | 0 | 18 |
| dhc-1(or195) mett-10(oj32) | 0 | 26 |
| dhc-1(or195) mett-10(ok2204) | 0 | 65 |
| dhc-1(or195) mett-10(oz36) | 100** | 46 |
| dhc-1(or195) glp-1(bn18) mett-10(oz36) | 0 | 38 |
| dhc-1(js319) | 0 | 26 |
| dhc-1(js319) glp-1(oz264) | 6 | 17 |
| dhc-1(js319) mett-10(tm2697) | 0 | 59 |
| dhc-1(js319) mett-10(g38) | 0 | 40 |
| dhc-1(js319) mett-10(oj32) | 0 | 30 |
| dhc-1(js319) mett-10(ok2204) | 0 | 24 |
| dhc-1(js319) mett-10(oz36) | 76** | 42 |
| dhc-1(js319) glp-1(bn18) mett-10(oz36) | 0 | 52 |
For all genotypes, animals were m− z−, i.e., came from homozygous mutant mothers. rrf-1(pk1417) enables germ line restricted RNAi. All RNAi and scoring was carried out at 20°C.
For all genotypes, all animals were m+ z−, i.e., came from mothers carrying a wild-type copy of mett-10, and were scored at 2 days past L4 at 20°C by dissection and DAPI (4′,6′-diamidino-2-phenylindole) staining.
**, significance as calculated by Fisher's exact test, using mett-10(oz36) alone for expected values.
Dynein and METT-10 may function together in cell division and meiosis.
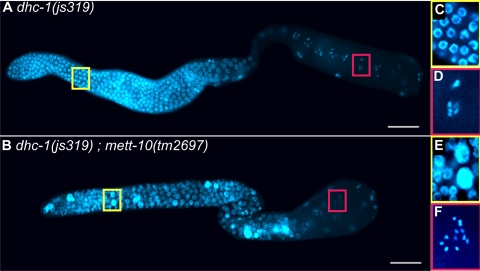
The dhc-1(js319) dynein heavy chain mutation did not cause tumors when combined with loss-of-function mett-10 alleles, but the double mutant animals had a fully penetrant sterility not seen in either mutant alone. On closer examination, dhc-1(js319) mett-10 germ lines exhibited the following two primary defects reminiscent of the phenotypes caused by RNAi knockdown of DLC-1: polyploidy in the distal region and unpaired chromosomes in diakinesis (Fig. 6 and Table 3). We also saw these phenotypes at low levels of penetrance in dhc-1(js319) animals, but they are distinct from the cell cycle progression defects observed in mett-10 single mutants (13). Thus, loss of mett-10 function enhances a hypomorphic dhc-1 allele, causing phenotypes typical of dynein loss-of-function mutations. This indicates that METT-10 also facilitates the function of the dynein motor complex in cell division and meiosis. Complete loss of mett-10 function alone does not cause these phenotypes, indicating that mett-10 function in cell division and meiotic events is not essential unless dynein function is partially compromised.
FIG. 6.
mett-10 mutants enhance dynein loss-of-function phenotypes of the dhc-1(js319) allele. (A, C, and D) Most dhc-1(js319) mutant germ lines are relatively normal at both 20 and 25°C, although they exhibit polyploidy (large DNA-dense nuclei) and univalents (unpaired chromosomes in diakinesis) at very low penetrance. (B, E, and F) Double mutations between dhc-1(js319) and any mett-10 allele lead to significant enhancement of both these phenotypes, and all double mutant animals are sterile at 20°C. This set of phenotypes closely phenocopies loss of dlc-1 function in the germ line.
TABLE 3.
mett-10 loss-of-function alleles enhance mitotic and meiotic defects of a dhc-1 allele
| Genotype | % Germ lines with: |
Total no. of germ lines | |
|---|---|---|---|
| Polyploid nucleia | Univalents in diakinesis | ||
| dhc-1(js319) | 4 | 4 | 26 |
| dhc-1(js319) glp-1(oz264) | 5 | 0 | 19 |
| dhc-1(js319) mett-10(tm2697) | 100 | 42 | 59 |
| dhc-1(js319) mett-10(g38) | 100 | 30 | 40 |
| dhc-1(js319) mett-10(ok2204) | 92 | 8 | 24 |
| dhc-1(js319) mett-10(oj32) | 97 | 3 | 30 |
| dhc-1(js319) mett-10(oz36) | 100 | NAb | 42 |
| dhc-1(js319) glp-1(bn18) mett-10(oz36) | 100 | 12 | 52 |
Polyploid nuclei are very large and stain intensely with the DNA dye DAPI. These are distinct from the enlarged, weakly DAPI-staining nuclei indicative of cell cycle arrest observed in mett-10 single mutants (13). All animals were m+ z−, i.e., came from mothers carrying a wild-type copy of mett-10, and were scored at 2 days past L4.
NA, not applicable; as germ lines are tumorous.
Reduction in glp-1 activity suppresses germ line overproliferation but not the polyploidy and unpaired chromosome phenotypes of dhc-1 mett-10(oz36) double mutants (Tables 2 and 3). Thus, the roles of mett-10 and dynein in cell division and meiotic events can be separated from their roles in regulation of the proliferative fate decision. We conclude, therefore, that regulation of proliferative fate, perhaps through negative regulation of glp-1 signaling, is only one cellular function in which dynein and METT-10 cooperate.
DISCUSSION
Dynein inhibits specification of proliferative fate in the C. elegans germ line.
The genetic and molecular studies presented here demonstrate that the dynein motor protein complex inhibits the decision of germ cells to proliferate in the C. elegans germ line, despite being necessary for the execution of mitotic cell division in those cells that do proliferate. The evidence further indicates that dynein regulates this cell fate decision in part by promoting the expression and function of the METT-10 putative methyltransferase that inhibits proliferative fate and in part through a METT-10-independent pathway.
DLC-1 regulates METT-10 RNA levels and protein subcellular localization.
DLC-1 knockdown reduces both METT-10 mRNA and protein levels. The mammalian LC8-type dynein light chain, DLC-1, localizes to chromatin and acts in a transcriptional transactivation complex with the estrogen receptor (46), raising the possibility that DLC-1 could directly regulate mett-10 transcription. It is also possible that DLC-1 normally promotes mett-10 mRNA stability or enables the nuclear localization of other proteins that ensure adequate mett-10 transcription. Regulation of mett-10 mRNA levels is likely to be the primary mode of METT-10 regulation by DLC-1 and may significantly contribute to the overproliferation phenotype observed when dlc-1 is depleted in the glp-1(oz264) background.
The direct physical interaction between DLC-1 and METT-10 reported here appears to play a redundant, nonessential role in METT-10 function. We showed that NLS-mediated and dynein-mediated pathways act redundantly to ensure normal METT-10 nuclear accumulation. The role of dynein and the microtubule network, and specifically LC8-type light chains, in facilitating the nuclear import of proteins is well documented (11, 16, 37, 48, 54, 57). However, in cases where it has been examined, dynein mediates nuclear accumulation by enhancing the efficiency of NLS-dependent nuclear import (37, 48). In these cases, unlike the example of METT-10, nuclear accumulation still requires an intact NLS. Thus, our work reveals a qualitatively different role for dynein, and LC8/DLC-1 interaction, in mediating nuclear accumulation. We cannot rule out the possibility, however, that DLC-1 also specifically promotes the function of a secondary, noncanonical NLS in METT-10.
When we specifically disrupted NLS- and DLC-1-mediated METT-10 nuclear accumulation in vivo, we found that it plays a nonessential role in most mett-10 developmental functions. Because our double mutants still retain some residual nuclear METT-10, a small nuclear pool may be sufficient to carry out its function as a methyltransferase. It is also possible that METT-10 association with dynein is transient and promotes modification of target molecules on their way to the nucleus, with METT-10 nuclear accumulation as a by-product. In this scenario, cytoplasmic METT-10 would be able to modify targets in the cytoplasm before their transport to the nucleus.
While DLC-1 may inhibit proliferative fate in part through promotion of normal METT-10 activity, it is clear that DLC-1 (and dynein) inhibit proliferative fate by other means as well, since disruptions in dynein function enhance germ line tumor formation of the mett-10(oz36) cytoplasmic mutant. Interestingly, disruptions of dynein function do not cause germ line tumors in other mett-10 mutant backgrounds, including the null. Since METT-10(oz36) is proposed to “poison” the function of another gene product through an unproductive physical association (13), it is possible that dynein promotes the normal function of the gene product(s) poisoned by METT-10(oz36), thus sensitizing mett-10(oz36) to perturbations in dynein function. Moreover, it is possible that METT-10, dynein, and this “poisoned factor” function to negatively regulate the proproliferative GLP-1/Notch signaling functions, as the mett-10(oz36) dhc-1(or195) tumors are exquisitely sensitive to the levels of glp-1 activity.
Dynein and METT-10 function together in multiple contexts.
While METT-10 and the dynein motor complex function together to inhibit germ cell proliferative fate, we also uncovered a role for METT-10 in other dynein-dependent processes. Specifically, decreases in METT-10 activity enhance the mitotic and meiotic defects of a weak dynein heavy chain loss-of-function allele, leading to highly penetrant sterility, polyploidy, and unpaired chromosomes in diakinesis. Interestingly, O'Connell and colleagues identified a phenotype in mett-10(oj32) loss-of-function mutants similar to those seen in dynein loss-of-function mutants (41). Specifically, mett-10(oj32) embryos have a visible defect in the flattening of the posterior centrosome during the first asymmetric division (41). The function of centrosome flattening is unclear but may be caused by the dynein-dependent force that pulls the posterior centrosome toward the posterior pole, thus ensuring asymmetric division (40, 53). Importantly, defects in centrosome flattening are also observed for perturbations in dynein and dynactin function, as well as genetic disruption of dynein-interacting proteins (8, 40, 51, 53, 55).
The genetic interactions between mett-10, dlc-1, and dhc-1 in proliferative fate specification, mitotic cell division, and meiosis suggest that these different cellular functions may have a common underlying mechanism. However, the experiments presented here also delineate a role for DLC-1 in promoting METT-10 activity during proliferative fate specification and a role for METT-10 in promoting DLC-1 activity during cell division and meiotic pairing and/or recombination. The most parsimonious interpretation of these findings is that METT-10 promotes DLC-1 activity, which in turn promotes METT-10 function in a positive feedback loop.
How could METT-10 affect DLC-1 function? DLC-1 is an LC8-type dynein light chain, which exists as a dimer when associated with the dynein motor complex (58). Two models have been proposed for the role of the LC8 dynein light chain with respect to the dynein motor complex. One model suggests that LC8 dimers act as dimerization hubs, binding to regions of intrinsic disorder in a diverse set of proteins, leading to increased structural organization and dimerization (2, 59). This model proposes that LC8 promotes normal dynein function by promoting dimerization of the dynein intermediate chain (2, 28). Indeed, since METT-10 can simultaneously bind to itself and to DLC-1, it is possible that DLC-1 promotes METT-10 dimerization in vivo. A second model proposes that DLC-1/LC8 functions as a cargo adaptor molecule, enabling the association of the dynein motor complex with a wide spectrum of proteins (25, 30, 38). The unifying theme of both models is that DLC-1 carries out multiple cellular functions by binding to and regulating the function of a diverse set of target proteins. It is possible that METT-10 methylates targets of the dynein motor complex and that this methylation event promotes their association with DLC-1 or the motor complex. Alternatively, DLC-1 and METT-10 could coordinately regulate target proteins by coupling multiple modes of regulation, such as posttranslational modification and subcellular localization. In both cases, association with dynein would enhance METT-10 function but may not be essential.
Proliferative and antiproliferative roles of dynein.
The dynein motor complex participates in a diversity of processes, including nuclear movements, spindle assembly and orientation, mitotic checkpoint inactivation, organelle movement, and endocytosis (20, 25). Here we show that dynein inhibits the specification of germ cell proliferative fate, despite also being necessary for mitotic cell division in proliferating cells. These “antiproliferative” (cell fate) and “proproliferative” (cell fate execution/cell division) functions of dynein are not unique to the maintenance of the balance between proliferation and differentiation in the C. elegans germ line. The essential roles of dynein in the execution of cell division are well known (25), as is its role in promoting the activity of tumor suppressors, including p53, by facilitating their nuclear transport (14, 16, 48, 57). Moreover, in keeping with its many cellular functions, the dynein motor complex influences multiple cell fate decisions by a diversity of mechanisms. These include the trafficking of cell fate determinants, such as egalitarian and its associated oskar mRNA (38), spindle positioning during asymmetric division (8, 40), ensuring the normal timing of mitotic cell division (15), and now, promoting RNA levels and protein nuclear accumulation of a factor influencing cell fate. Thus, dynein acts as a “handyman,” performing many of the jobs necessary to execute a developmental program.
Acknowledgments
We thank the Caenorhabditis Genetics Center, the Japanese National BioResource Project, the C. elegans gene knockout consortium, and Mike Nonet for providing strains. We thank Ziva Misulovin for assistance with the qRT-PCR described in the legend to Fig. 4E and Siqun Xu for technical assistance in generating mett-10::gfp arrays. We extend our sincerest thanks to Dale Dorsett, Swathi Arur, Yair Dorsett, Justin Fay, Kevin O'Connell, Jim Skeath, Paul Fox, Doug Chalker, Mike Nonet, Jim Havranek, John Cooper, and four anonymous reviewers for experimental suggestions, provisions of reagents, or comments on the manuscript.
This work was supported by grant GM63310 awarded to T.S.
Footnotes
Published ahead of print on 14 September 2009.
REFERENCES
- 1.Austin, J., and J. Kimble. 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51:589-599. [DOI] [PubMed] [Google Scholar]
- 2.Barbar, E. 2008. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 47:503-508. [DOI] [PubMed] [Google Scholar]
- 3.Benashski, S. E., A. Harrison, R. S. Patel-King, and S. M. King. 1997. Dimerization of the highly conserved light chain shared by dynein and myosin V. J. Biol. Chem. 272:20929-20935. [DOI] [PubMed] [Google Scholar]
- 4.Benison, G., P. A. Karplus, and E. Barbar. 2007. Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site. J. Mol. Biol. 371:457-468. [DOI] [PubMed] [Google Scholar]
- 5.Berry, L. W., B. Westlund, and T. Schedl. 1997. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 124:925-936. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carson, J. H., H. Cui, and E. Barbarese. 2001. The balance of power in RNA trafficking. Curr. Opin. Neurobiol. 11:558-563. [DOI] [PubMed] [Google Scholar]
- 8.Couwenbergs, C., J. C. Labbe, M. Goulding, T. Marty, B. Bowerman, and M. Gotta. 2007. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J. Cell Biol. 179:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crittenden, S. L., K. A. Leonhard, D. T. Byrd, and J. Kimble. 2006. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17:3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day, C. L., H. Puthalakath, G. Skea, A. Strasser, I. Barsukov, L. Lian, D. Huang, and M. G. Hinds. 2004. Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands. Biochem. J. 377:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desfarges, S., B. Salin, C. Calmels, M. L. Andreola, V. Parissi, and M. Fournier. 2009. HIV-1 integrase trafficking in S. cerevisiae: a useful model to dissect the microtubule network involvement of viral protein nuclear import. Yeast 26:39-54. [DOI] [PubMed] [Google Scholar]
- 12.Ding, D. Q., A. Yamamoto, T. Haraguchi, and Y. Hiraoka. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 6:329-341. [DOI] [PubMed] [Google Scholar]
- 13.Dorsett, M., B. Westlund, and T. Schedl. 2009. METT-10, a putative methyltransferase, inhibits germ cell proliferative fate in Caenorhabditis elegans. Genetics 183:233-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabbro, M., and B. R. Henderson. 2003. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 282:59-69. [DOI] [PubMed] [Google Scholar]
- 15.Fan, S. S., and D. F. Ready. 1997. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development 124:1497-1507. [DOI] [PubMed] [Google Scholar]
- 16.Giannakakou, P., D. L. Sackett, Y. Ward, K. R. Webster, M. V. Blagosklonny, and T. Fojo. 2000. p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2:709-717. [DOI] [PubMed] [Google Scholar]
- 17.Gonczy, P., S. Pichler, M. Kirkham, and A. A. Hyman. 1999. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147:135-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guang, S., A. F. Bochner, D. M. Pavelec, K. B. Burkhart, S. Harding, J. Lachowiec, and S. Kennedy. 2008. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321:537-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, D., and T. Schedl. 2006. The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line. Curr. Top. Dev. Biol. 76:185-215. [DOI] [PubMed] [Google Scholar]
- 20.Henderson, S. T., D. Gao, E. J. Lambie, and J. Kimble. 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120:2913-2924. [DOI] [PubMed] [Google Scholar]
- 21.Herzig, R. P., U. Andersson, and R. C. Scarpulla. 2000. Dynein light chain interacts with NRF-1 and EWG, structurally and functionally related transcription factors from humans and drosophila. J. Cell Sci. 113(Pt. 23):4263-4273. [DOI] [PubMed] [Google Scholar]
- 22.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 23.Hubbard, E. J., and D. Greenstein. 2005, posting date. Introduction to the germ line, part 1-4. In The C. elegans Research Community (ed.), WormBook. http://www.wormbook.org.43. [DOI] [PMC free article] [PubMed]
- 24.Jones, A. R., R. Francis, and T. Schedl. 1996. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180:165-183. [DOI] [PubMed] [Google Scholar]
- 25.Karki, S., and E. L. Holzbaur. 1999. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 11:45-53. [DOI] [PubMed] [Google Scholar]
- 26.Kerins, J. 2006. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Ph.D. dissertation. Washington University, Saint Louis, MO. [DOI] [PMC free article] [PubMed]
- 27.Kimble, J., and S. L. Crittenden. 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23:405-433. [DOI] [PubMed] [Google Scholar]
- 28.King, S. M. 2008. Dynein-independent functions of DYNLL1/LC8: redox state sensing and transcriptional control. Sci. Signal 1:pe51. [DOI] [PubMed] [Google Scholar]
- 29.Koushika, S. P., A. M. Schaefer, R. Vincent, J. H. Willis, B. Bowerman, and M. L. Nonet. 2004. Mutations in Caenorhabditis elegans cytoplasmic dynein components reveal specificity of neuronal retrograde cargo. J. Neurosci. 24:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K. H., S. Lee, B. Kim, S. Chang, S. W. Kim, J. S. Paick, and K. Rhee. 2006. Dazl can bind to dynein motor complex and may play a role in transport of specific mRNAs. EMBO J. 25:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, M. H., M. Ohmachi, S. Arur, S. Nayak, R. Francis, D. Church, E. Lambie, and T. Schedl. 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177:2039-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, S., C. M. Armstrong, N. Bertin, H. Ge, S. Milstein, M. Boxem, P. O. Vidalain, J. D. Han, A. Chesneau, T. Hao, D. S. Goldberg, N. Li, M. Martinez, J. F. Rual, P. Lamesch, L. Xu, M. Tewari, S. L. Wong, L. V. Zhang, G. F. Berriz, L. Jacotot, P. Vaglio, J. Reboul, T. Hirozane-Kishikawa, Q. Li, H. W. Gabel, A. Elewa, B. Baumgartner, D. J. Rose, H. Yu, S. Bosak, R. Sequerra, A. Fraser, S. E. Mango, W. M. Saxton, S. Strome, S. Van Den Heuvel, F. Piano, J. Vandenhaute, C. Sardet, M. Gerstein, L. Doucette-Stamm, K. C. Gunsalus, J. W. Harper, M. E. Cusick, F. P. Roth, D. E. Hill, and M. Vidal. 2004. A map of the interactome network of the metazoan C. elegans. Science 303:540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, H. 1997. The tao of stem cells in the germline. Annu. Rev. Genet. 31:455-491. [DOI] [PubMed] [Google Scholar]
- 34.Mallam, A. L., and S. E. Jackson. 2007. The dimerization of an alpha/beta-knotted protein is essential for structure and function. Structure 15:111-122. [DOI] [PubMed] [Google Scholar]
- 35.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 36.Morrison, S. J., and J. Kimble. 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441:1068-1074. [DOI] [PubMed] [Google Scholar]
- 37.Moseley, G. W., D. M. Roth, M. A. DeJesus, D. L. Leyton, R. P. Filmer, C. W. Pouton, and D. A. Jans. 2007. Dynein light chain association sequences can facilitate nuclear protein import. Mol. Biol. Cell 18:3204-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navarro, C., H. Puthalakath, J. M. Adams, A. Strasser, and R. Lehmann. 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 6:427-435. [DOI] [PubMed] [Google Scholar]
- 39.Navarro, R. E., E. Y. Shim, Y. Kohara, A. Singson, and T. K. Blackwell. 2001. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development 128:3221-3232. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen-Ngoc, T., K. Afshar, and P. Gonczy. 2007. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9:1294-1302. [DOI] [PubMed] [Google Scholar]
- 41.O'Connell, K. F., C. M. Leys, and J. G. White. 1998. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics 149:1303-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Rourke, S. M., M. D. Dorfman, J. C. Carter, and B. Bowerman. 2007. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 3:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pepper, A. S., D. J. Killian, and E. J. Hubbard. 2003. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 163:115-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157:1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rayala, S. K., P. den Hollander, S. Balasenthil, Z. Yang, R. R. Broaddus, and R. Kumar. 2005. Functional regulation of oestrogen receptor pathway by the dynein light chain 1. EMBO Rep. 6:538-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rayala, S. K., P. den Hollander, B. Manavathi, A. H. Talukder, C. Song, S. Peng, A. Barnekow, J. Kremerskothen, and R. Kumar. 2006. Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. J. Biol. Chem. 281:19092-19099. [DOI] [PubMed] [Google Scholar]
- 48.Roth, D. M., G. W. Moseley, D. Glover, C. W. Pouton, and D. A. Jans. 2007. A microtubule-facilitated nuclear import pathway for cancer regulatory proteins. Traffic 8:673-686. [DOI] [PubMed] [Google Scholar]
- 49.Salic, A., and T. J. Mitchison. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105:2415-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, p.15.36-15.39, 3rd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 51.Schmidt, D. J., D. J. Rose, W. M. Saxton, and S. Strome. 2005. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol. Biol. Cell 16:1200-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnorrer, F., K. Bohmann, and C. Nusslein-Volhard. 2000. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2:185-190. [DOI] [PubMed] [Google Scholar]
- 53.Severson, A. F., and B. Bowerman. 2003. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J. Cell Biol. 161:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shrum, C. K., D. Defrancisco, and M. K. Meffert. 2009. Stimulated nuclear translocation of NF-kappaB and shuttling differentially depend on dynein and the dynactin complex. Proc. Natl. Acad. Sci. USA 106:2647-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srinivasan, D. G., R. M. Fisk, H. Xu, and S. van den Heuvel. 2003. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 17:1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomikawa, C., A. Ochi, and H. Hori. 2008. The C-terminal region of thermophilic tRNA (m7G46) methyltransferase (TrmB) stabilizes the dimer structure and enhances fidelity of methylation. Proteins 71:1400-1408. [DOI] [PubMed] [Google Scholar]
- 57.Trostel, S. Y., D. L. Sackett, and T. Fojo. 2006. Oligomerization of p53 precedes its association with dynein and nuclear accumulation. Cell Cycle 5:2253-2259. [DOI] [PubMed] [Google Scholar]
- 58.Vale, R. D. 2003. The molecular motor toolbox for intracellular transport. Cell 112:467-480. [DOI] [PubMed] [Google Scholar]
- 59.Wang, L., M. Hare, T. S. Hays, and E. Barbar. 2004. Dynein light chain LC8 promotes assembly of the coiled-coil domain of swallow protein. Biochemistry 43:4611-4620. [DOI] [PubMed] [Google Scholar]
- 60.Wells, J. L., D. W. Pryce, and R. J. McFarlane. 2006. Homologous chromosome pairing in Schizosaccharomyces pombe. Yeast 23:977-989. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto, A., R. R. West, J. R. McIntosh, and Y. Hiraoka. 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145:1233-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]