Abstract
The prevalence of the currently known Acinetobacter species and related trends of antimicrobial resistance in a Dutch university hospital were studied. Between 1999 and 2006, Acinetobacter isolates from clinical samples were collected prospectively. Isolates were analyzed by amplified fragment length polymorphism fingerprinting. For species identification, a profile similarity cutoff level of 50% was used, and for strain identification, a cutoff level of 90% was used. Susceptibility for antimicrobial agents was tested by disk diffusion by following the CLSI guideline.
The incidences of Acinetobacter isolates ranged from 1.7 to 3.7 per 10,000 patients per year, without a trend of increase, during the study years. Twenty different species were distinguished. Acinetobacter baumannii (27%) and Acinetobacter genomic species (gen. sp.) 3 (26%) were the most prevalent. Other species seen relatively frequently were Acinetobacter lwoffii (11%), Acinetobacter ursingii (4%), Acinetobacter johnsonii (4%), and Acinetobacter junii (3%). One large cluster of A. baumannii, involving 31 patients, and 16 smaller clusters of various species, involving in total 39 patients, with at most 5 patients in 1 cluster, occurred. Overall, 37% of the A. baumannii isolates were fully susceptible to the tested antibiotics. There was a borderline significant (P = 0.059) trend of decreasing susceptibility. A. baumannii was the Acinetobacter species causing the largest burden of multiple-antibiotic resistance and transmissions in the hospital.
More than 30 named and unnamed species of Acinetobacter have been described (14), some of which are of clinical importance, including A. baumannii, Acinetobacter gen. sp. 3, and Acinetobacter gen. sp. 13TU, while other species, like A. junii, A. johnsonii, A. ursingii, and Acinetobacter schindleri, can also incidentally be associated with infections (8). Much attention has been paid to outbreaks caused by acinetobacters (28), which in most cases are caused by A. baumannii (15, 23). Notably, in diagnostic microbiology, isolates identified as A. baumannii may also include the closely related species Acinetobacter gen. sp. 3 or Acinetobacter gen. sp. 13TU. Bacteria belonging to other Acinetobacter species are frequently not further identified as or designated Acinetobacter species, as this would require genotypic methods that are usually not available in clinical diagnostic microbiology. These difficulties in identification explain why, overall, not much is known about the occurrences of the different Acinetobacter species in the hospital.
The aim of the present study was to determine the prevalences of the currently known Acinetobacter species and related trends of antimicrobial resistance in our hospital through the years. To this aim, we identified all available Acinetobacter isolates obtained from our hospital in the period between 1999 and 2006 to the species level by amplified fragment length polymorphism (AFLP) analysis, a well-validated method for Acinetobacter species identification (7, 8). Furthermore, we tested the susceptibilities of the isolates to antibiotics and used AFLP analysis to assess strain relatedness as an indication of transmission of strains in the hospital.
MATERIALS AND METHODS
Setting.
The study was performed in the Leiden University Medical Centre, The Netherlands, between 1999 and 2006. In 2006, the hospital had 18,908 admissions, with a mean duration of stay of 7.5 days and a total of 141,128 patient-days. In addition, for the purpose of day care, 11,957 patients were admitted.
Isolates.
Acinetobacter strains were isolated from clinical specimens in the diagnostic microbiology laboratory of the hospital. Identification of the organisms as belonging to the genus Acinetobacter or, presumptively, as Acinetobacter species was done with the Vitek 2 system (bioMerieux, Boxtel, The Netherlands). For each patient, each colony morphotype possibly representing a unique strain was analyzed by AFLP typing (see below), and depending on the outcome, each unique strain per patient was included. Isolates were collected prospectively and stored at minus 80°C in glycerol broth.
Isolates were classified as hospital related when the specimen from which the organism was cultured was taken during the stay in the hospital or within 14 days after discharge of the patient from the hospital.
AFLP analysis for species and strain identification.
Selective amplification of genomic restriction fragments by AFLP was performed as described previously (12). Briefly, purified DNA was digested using EcoRI and MseI, and amplification was done with a Cy5-labeled EcoRI+A primer and an MseI+C primer (A and C are selective bases). The ALF express II DNA analysis system (Amersham Biosciences, Roosendaal, The Netherlands) was used for fragment separation. The fingerprints of fragments of 50 to 500 bp were investigated by cluster analysis using the Bionumerics 4.5 software package (Applied Maths, Sint-Martens-Latem, Belgium), with the Pearson product moment coefficient (r) as a similarity measure and the unweighted-pair group method using arithmetic averages for grouping. For species identification, isolates were compared to a library of strains of all described (genomic) species, using a similarity cutoff level of 50%, above which strains were considered to belong to the same species (7, 12). Isolates were considered to belong to the same clone or strain if they grouped together at a level greater than or equal to 80% or 90%, respectively (7).
Susceptibility testing.
Resistance to 12 antimicrobial agents or combinations of agents which are effective against susceptible A. baumannii strains was determined by disk diffusion by following the CLSI guidelines. The semiautomated Biomic image analysis system, release 2005 (Giles Scientific, Inc., New York, NY), was used to read and interpret zone sizes. The control strains were Escherichia coli ATCC 35218, E. coli 25922, Enterococcus faecalis ATCC 29212, and Acinetobacter strains LUH 4624 and LUH 4576, received from A. Nemec (National Institute Public Health, Prague, Czech Republic). Bacterial isolates were suspended in 2 ml saline to a turbidity corresponding to a 0.5 McFarland standard. The agents (Oxoid) (values in parentheses represent content levels [μg/disc], susceptibility breakpoints [mm], and resistance breakpoints [mm], in that order) were gentamicin (10, ≥15, and ≤12), netilmicin (30, ≥15, and ≤12), tobramycin (10, ≥15, and ≤12), amikacin (30, ≥17, and ≤14), ampicillin and sulbactam (10 and 10, respectively; ≥15; and ≤11), piperacillin (100, ≥21, and ≤17), ceftazidime (30, ≥18, and ≤14), meropenem (10, ≥16, and ≤13), imipenem (10, ≥16, and ≤13), ofloxacin (5, ≥16, and ≤12), sulfamethoxazole and trimethoprim (23.75 and 1.25, respectively; ≥16; and ≤10), and tetracycline (30, ≥19, and ≤14). The plates were incubated 24 h at 37°C and then read by using the Biomic system. The category of susceptibility was determined from the zones of inhibition as susceptible, intermediate, or resistant. Trends in susceptibility over the years were analyzed by the chi-square test, with linear-by-linear association, using the statistical package SPSS.
RESULTS
Patients.
During the years 1999 to 2006, the numbers of patients with hospital-related Acinetobacter spp. varied between 28 and 58 (Table 1). The incidences ranged from 1.7 to 3.7 per 10,000 patient-days and were highest in 2001, 2003, and 2005. In 2001, an Acinetobacter baumannii cluster that started in November 2000 and lasted until October 2001 dominated. The cluster involved 31 patients (23). When the patients belonging to the cluster were omitted, the incidence was found to be 2.3 per 10,000 patient-days for both years. Apart from this big cluster, the years 2003 and 2005 were distinguished by relatively high numbers of secondary cases, defined as clusters of two or more patients with the same genotype at the same period at the same ward. Four small clusters of transmission, involving totals of nine and eight patients in 2003 and 2005, respectively, were observed. After correction for the secondary cases of these clusters, the incidences in 2003 and 2005 were 3.0 and 3.3 per 10,000 patient-days, respectively.
TABLE 1.
Overview of hospital-related Acinetobacter isolates in 331 patients
| Parameter | Value for indicated yr |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | |
| No. of patients | 28 | 49 | 58 | 36 | 54 | 46 | 57 | 37 |
| No. of patients/10,000 patient-daysa | 1.7 | 2.9 | 3.7 | 2.3 | 3.5 | 2.9 | 3.7 | 2.4 |
| No. of secondary casesb | 2 | 11 | 21 | 1 | 8 | 2 | 7 | 1 |
| No. of patients with isolate(s) typeda | 22 | 47 | 51 | 24 | 52 | 43 | 56 | 36 |
| No. of single strains | 25 | 48 | 56 | 25 | 59 | 45 | 61 | 40 |
| A. baumannii | 5 | 23 | 29 | 5 | 20 | 11 | 23 | 13 |
| A. junii | 3 | 4 | 2 | 2 | 1 | |||
| A. ursingii | 1 | 4 | 1 | 1 | 2 | 2 | 2 | 2 |
| A. lwoffii | 1 | 1 | 8 | 2 | 10 | 3 | 9 | 4 |
| Acinetobacter gen. sp. 13TU | 1 | 1 | 2 | 3 | 1 | 1 | ||
| Acinetobacter gen. sp. “close to 13TU” | 1 | |||||||
| Acinetobacter gen. sp. 15TU | 1 | 2 | ||||||
| Acinetobacter gen. sp. 3 | 4 | 12 | 8 | 10 | 15 | 16 | 14 | 14 |
| A. radioresistens | 1 | 2 | 1 | |||||
| A. calcoaceticus | 1 | 1 | 2 | 1 | 1 | |||
| Acinetobacter gen. sp. 10 | 2 | 1 | 1 | |||||
| Acinetobacter gen. sp. 11 | 1 | 1 | 1 | 1 | ||||
| A. johnsonii | 2 | 1 | 3 | 1 | 3 | 3 | ||
| A. haemolyticus | 1 | 2 | ||||||
| A. gyllenbergii | 1 | 1 | ||||||
| Acinetobacter gen. sp. 14BJ | 1 | 1 | 1 | |||||
| Acinetobacter gen. sp. “between 1 and 3” | 1 | |||||||
| A. beijerinckii | 2 | 3 | ||||||
| A. parvus | 1 | |||||||
| Acinetobacter gen. sp. 16 | 1 | |||||||
| Unidentified | 2 | 4 | 2 | 2 | 2 | |||
Secondary cases were included.
Secondary cases are defined as patients with the same genotype at the same period at the same ward.
The ratio of men to women was 1.6, varying from 1.1 to 2.7. The mean ratio of men to women admitted to the hospital during the study years was 0.96 (range, 0.91 to 1), indicating that Acinetobacter is seen more often in men than in women. The median ages of the patients per year ranged from 47 to 52 years. The youngest patients were newborns a few days old in the neonatology intensive care unit. The oldest patient was 91 years old. Forty-six percent of the patients were admitted to one of the intensive care units of the hospital. Next in frequency were patients from the departments of urology and hematology.
Acinetobacter isolates.
Of the 365 patients with documented Acinetobacter cultures, isolates from 331 patients were available for further investigation. If multiple isolates per patient were available, the first isolate of a series of samples from each positive body site was taken for study. If patients had recurrent episodes of positive cultures, the first sample of each period was studied. For each patient, isolates were compared by AFLP analysis for strain relatedness (similarity, ≥90%) and, next, replicate isolates were excluded. All together, a total of 359 single strains were found in the 331 patients (including the secondary cases, i.e., patients with the same AFLP type in the same period and ward).
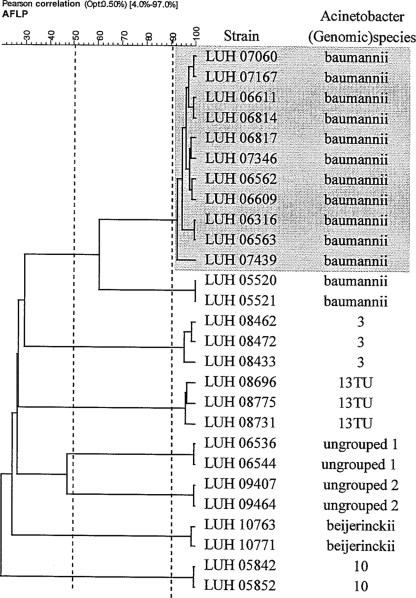
At the species cutoff level of 50% (Fig. 1), as determined by AFLP analysis, 20 different species were distinguished (Table 1), whereas 12 isolates could not be identified. The most prevalent species were A. baumannii (27%, with the large cluster not included) and Acinetobacter gen. sp. 3 (26%) (Table 1). Other species seen relatively frequently were A. lwoffii (11%), A. ursingii (4%), A. johnsonii (4%), and A. junii (3%).
FIG. 1.
Dendrogram of cluster analysis of AFLP profiles of sets of strains possibly involved in transmission. The horizontal axis represents the similarity level at which strains are grouped. This level is determined by Pearson's product moment correlation coefficient. Grouping was obtained by the unweighted-pair group method using average linkages. Similarity levels of 50% and 90% (dotted lines) are considered to represent the species and strain cutoff levels, respectively. “Ungrouped 1” and “ungrouped 2” represent strains that could not be identified as the described (genomic) species. Strains marked in gray represent a prolonged cluster in the hospital. Opt, optimization (shift allowed between any two patterns for the best possible matching).
Thirty-two percent of the 359 single strains were isolated from airway secretions, 16% from wounds and wound drains, 15% from blood or intravascular lines, and 9% from urine. A. baumannii and Acinetobacter gen. sp. 3 were most frequently isolated from sputum samples (48 and 32%, respectively) and wound samples (19 and 18%, respectively). A. lwoffii was mainly isolated from blood samples or intravascular lines (42%).
Inspection of AFLP profiles for possible strain relatedness (≥90%) showed a number of clusters possibly indicative of transmission, a subset of which is shown in Fig. 1. Apart from the large cluster of A. baumannii in 31 patients observed in 2000 and 2001, 16 small clusters of isolates involving 39 patients were observed. Nine of the clusters involved A. baumannii; two clusters A. ursingii and Acinetobacter gen. sp. 3; and one cluster Acinetobacter gen. sp. 13TU, Acinetobacter gen. sp. 10, and Acinetobacter beyerinckii. Twelve clusters involved two patients, two clusters three patients, one cluster four patients, and one cluster five patients. Two clusters (the big cluster observed in 2000 and 2001 and a smaller cluster observed in 2003) were recognized while occurring. The other clusters were identified through the investigations of the present study.
Antimicrobial susceptibility.
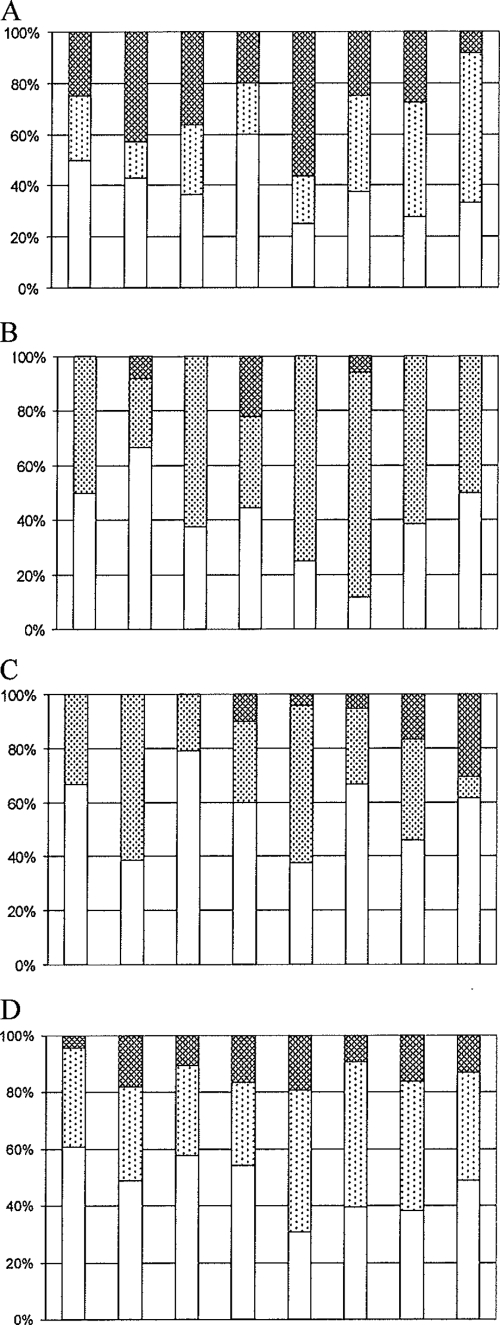
The percentages of fully susceptible A. baumannii strains varied between 25 and 60% (overall 35%), with the lower percentages observed during the more-recent years (Fig. 2A). Only one isolate was completely resistant. Piperacillin, tetracycline, cotrimoxazole, ofloxacin, gentamicin, and ceftazidime resistances were most frequently observed (Table 2). Two A. baumannii isolates were carbapenem resistant.
FIG. 2.
Percentages of Acinetobacter isolates susceptible to all (□), resistant to <3 (░⃞), and resistant to ≥3 (▩) of the 12 tested antibiotics. Isolates with a disk zone in the intermediary range were counted as resistant. Of the isolates occurring in clusters, only one isolate was counted. (A) A. baumannii; (B) Acinetobacter gen. sp. 3; (C) Acinetobacter species other than A. baumannii and Acinetobacter gen. sp. 3; (D) all Acinetobacter species.
TABLE 2.
Antimicrobial susceptibility of A. baumanniia
| Antibiotic(s) | No. of isolates (%) |
||
|---|---|---|---|
| Sensitive | Intermediate | Resistant | |
| Ceftazidime | 63 (71.6) | 11 (12.5) | 14 (15.9) |
| Piperacillin | 45 (51.1) | 16 (18.2) | 27 (30.7) |
| Meropenem | 86 (97.7) | 0 | 2 (2.3) |
| Imipenem | 87 (98.9) | 0 | 1 (1.1) |
| Amoxicillin-sulbactam | 81 (92.0) | 0 | 7 (8.0) |
| Gentamicin | 72 (81.8) | 2 (2.3) | 14 (15.9) |
| Netilmicin | 78 (88.6) | 4 (4.6) | 6 (6.8) |
| Tobramycin | 79 (89.8) | 3 (3.4) | 6 (6.8) |
| Amikacin | 79 (89.8) | 0 | 9 (10.2) |
| Ofloxacin | 70 (79.6) | 1 (1.1) | 17 (19.3) |
| Tetracyclin | 43 (48.9) | 22 (25.0) | 23 (26.1) |
| Cotrimoxazole | 68 (77.3) | 2 (2.3) | 18 (20.4) |
Secondary cases with identical antimicrobial sensitivities were excluded (n = 88).
For Acinetobacter gen. sp. 3, the percentages of full susceptibility ranged from 20 to 50% (overall, 37%). Through the years, the proportion of resistant isolates seemed to increase, with the highest levels of resistance observed in 2003 and 2004. This trend did not persist in 2005 and 2006 (Fig. 2B). Piperacillin resistance was most frequently observed (Table 3). Forty percent of the isolates showed intermediate susceptibility to piperacillin.
TABLE 3.
Antimicrobial susceptibilities of Acinetobacter gen. sp. 3a
| Antibiotic(s) | No. of isolates (%) |
||
|---|---|---|---|
| Sensitive | Intermediate | Resistant | |
| Ceftazidime | 83 (93.3) | 5 (5.6) | 1 (1.1) |
| Piperacillin | 41 (46.1) | 36 (40.4) | 12 (13.5) |
| Meropenem | 89 (100) | 0 | 0 |
| Imipenem | 89 (100) | 0 | 0 |
| Amoxicillin-sulbactam | 89 (100) | 0 | 0 |
| Gentamicin | 85 (95.5) | 0 | 4 (4.5) |
| Netilmicin | 85 (95.5) | 2 (2.2) | 2 (2.2) |
| Tobramycin | 88 (98.9) | 0 | 1 (1.1) |
| Amikacin | 87 (97.8) | 2 (2.2) | 0 |
| Ofloxacin | 88 (98.9) | 0 | 1 (1.1) |
| Tetracyclin | 76 (85.4) | 9 (10.1) | 4 (4.5) |
| Cotrimoxazole | 89 (100) | 0 | 0 |
Secondary cases with identical antimicrobial sensitivities were excluded (n = 89).
Of the remainder of Acinetobacter species, 40 to 80% (overall 58%) were susceptible. Through the years, the proportions of susceptible and resistant isolates varied without a trend of increasing resistance, although more multiple resistant strains were seen in the last 2 years of the study (Fig. 2C). Of the 12 multiple-resistant strains, 6 were identified as A. ursingii. Forty percent of the A. ursingii isolates were multiple resistant, compared to 15% of the A. Johnsonii isolates and none of the A. junii isolates.
Overall, 82 isolates (23%) were multiresistant, defined as resistant to three or more antibiotics. Sixty-five of these 82 multiresistant isolates were A. baumannii. For all Acinetobacter isolates together, there was a borderline significant trend (P = 0.059) of decreasing susceptibility between 1999 and 2006. For A. baumannii (P = 0.302), Acinetobacter gen. sp. 3 (P = 0.225), and the other Acinetobacter species (P = 0.46), analyzed separately, no trend was demonstrated.
Comparison of isolates with European clones I to III.
Previous studies have shown that three major groups of highly similar strains which are considered to represent three lineages of descent (European clones I to III) occur among epidemic strains of A. baumannii in Europe (6, 13, 24). These studies have shown that strains of these clones can be separated by AFLP profiling, with ∼80% being considered the cutoff level, above which strains are considered to belong to the same clone. In the current study, we compared the AFLP profiles of the A. baumannii strains to those of sets of strains of the three clones available in the Leiden University Medical Center culture collection. Thus, five isolates from 2003 were identified as belonging to European clone I (data not shown). These isolates showed a high level of similarity to each other (>95%) and were from the same time-space frame, indicating epidemic spread. Five other isolates were identified as belonging to European clone II; two of these, obtained in 1999, were considered to represent a common strain that had spread between two patients (data not shown). Altogether, strains of the European clones were relatively rare among the A. baumannii strains of our study.
DISCUSSION
The present study shows that through the years, the incidences of patients with an Acinetobacter isolate from a clinical specimen varied in our hospital under conditions of endemicity between 1.7 and 3.7 per 10.000 patient-days with secondary cases included and between 1.7 and 3.3 with secondary cases excluded. Between 1999 and 2006, the incidences showed no trend of increase.
Acinetobacter was found half as frequently in men as in women. A small study of Acinetobacter anitratus blood culture isolates reported a male/female ratio of 2.7:1 (16). Our study substantiates the overrepresentation of men, which is not explained by a higher admission rate. The opposite is true; slightly more women than men are admitted to our hospital.
A variety of species was found. Only a quarter of the isolates were A. baumannii. Acinetobacter gen. sp. 3 was seen as often as A. baumannii.
Three-quarters of the isolates belonged to species known to be associated with infections in humans. Next to A. baumannii and Acinetobacter gen. sp. 3, these are A. junii (2), A. ursingii (5, 12), Acinetobacter gen. sp. 13TU (11, 25), and A. johnsonii (18). A. lwoffii, a well-known colonizer of the human skin, made up 11% of the isolates. It was frequently found on tips of intravascular catheters and in blood cultures. The question is whether this must be considered contamination of the cultures or represents infection.
Only a few studies have investigated the distribution of acinetobacters in clinical specimens at the species level, and considerable differences in outcome have been reported. For example, our data differ from the observations made by Struelens et al. (20) in a Belgian hospital. Under circumstances of endemicity, about half of their Acinetobacter isolates were identified as A. baumannii, twice as many as we found. Acinetobacter gen. sp. 3 was found in 5% of their isolates, compared to 24% in our survey. This inconsistency may be explained either by the use of biweekly surveillance cultures in the Belgian study or by the use of different methods for species identification. We used AFLP analysis, a well-validated method for species identification (8), while Struelens et al. used the phenotypic system developed by Bouvet and Grimont (4). It is of note that the identification of Acinetobacter gen. sp. 3 with this system is not perfect (65%) (9). Seifert et al. (17), also using the system of Bouvet and Grimont, identified 73% of the Acinetobacter isolates from clinical samples as A. baumannii. Acinetobacter gen. sp. 3 was found in 9%, A. johnsonii in 5%, and A. lwoffii in 4% of the isolates.
Traub and Bauer (22) identified 45% of 2,359 clinical isolates of Acinetobacter as gen. sp. 3, 15% as gen. sp. 13, and 14% as A. baumannii by phenotypic identification. The origin of their isolates is not described, and therefore, it is not clear whether their collection is representative of the situation of endemicity or concerns a selection of strains.
Tjernberg and Ursing (21), who used DNA-DNA hybridization, a robust method for species identification, also found a predominance of Acinetobacter gen. sp. 3 among consecutive clinical Acinetobacter isolates in a Swedish hospital. In a recent study from Ireland, Acinetobacter gen. sp. 3 was the most frequent species found, representing 39% of 114 isolates over a 30-month period (3). A. baumannii was second, with 22%. For A. baumannii, our findings are in line with this Irish study; however, for Acinetobacter gen. sp. 3, the difference is considerable. Whether these differences are due to geographic, methodological, or epidemiological differences between the studies is hard to say.
Antimicrobial resistance is one of the major problems associated with Acinetobacter. In our survey, resistance was evidently present, although not at the scale often reported (27). Multiple resistance, defined as resistance to three or more antibiotics, occurred predominantly in A. baumannii. Carbapenem resistance was seen in only two of the A. baumannii isolates. A likely explanation for the relatively low level of resistant Acinetobacter isolates in the hospital is the restricted use of antibiotics, which is in line with the Dutch practice regarding antimicrobial therapy, the strict isolation of patients with multiresistant isolates, and the immediate beginning of infection control measures when several patients are infected with the same strain.
During the 8 years of our survey, one large cluster of A. baumannii involving 31 patients occurred. Fifteen smaller clusters were seen, most of them caused by A. baumannii. A variety of other species were involved: Acinetobacter gen. sp. 3, Acinetobacter gen. sp. 10, Acinetobacter gen. sp. 13TU, and the novel species A. beyerinckii (14). Although A. baumannii and Acinetobacter gen. sp. 3 clinical isolates were found in almost equal numbers, the former species was involved in nine clusters, among which the largest clusters occurred, and Acinetobacter gen. sp. 3 was observed in only two small clusters, involving a total of five patients. Outbreaks caused by A. baumannii are well known and get a lot of attention in the literature (15, 23). Outbreaks caused by other species have been reported much less often (1, 10, 25, 26), which may, in part, be due to underreporting. Indeed, identification of Acinetobacter isolates to the species level, as we did in the present study, is not usual in routine microbiological diagnostics. The Vitek 2 system is unreliable for species identification of Acinetobacter (3). The question is whether speciation should be done routinely. Easy methods for species identification do not exist, so the costs of species identification should be outweighed by the benefit of it for the management of patients and the timely institution of control measures. For daily practice, the relevance of knowing the species is limited. Treatment is primarily determined by the susceptibility pattern of the isolate and not by the species to which it belongs. This may be different for infection control measures. In The Netherlands, it is common practice to put patients infected by multiresistant bacteria in strict isolation, for which an isolation room is needed in the case of A. baumannii. Patients infected by species with a lower risk of transmission can be treated with less radical precautions, like contact isolation (see WIP [www.wip.nl/], a guideline for highly resistant microorganisms). This is in the interest of the patients because nursing in isolation rooms is accompanied by inferior care (19). The results of the present study show that the majority of the multidrug-resistant Acinetobacter isolates are A. baumannii. Therefore, identification at the species level will have a minor impact on isolation of patients infected with multiresistant Acinetobacter. Species identification is indispensable for analysis of outbreaks. The types of measures differ, dependent on whether multiple species are found or different strains of one species or a single strain of one species is involved (23).
Not many A. baumannii isolates that belonged to one of the three major European clones were found (6, 24), but most of those found were associated with cross-infection, which emphasizes the epidemiological potential of these clones. Four of the eight secondary cases observed in 2003 were due to clone I. Clone II was observed five times, one of which involved a secondary case.
In conclusion, we found that under endemic conditions, a wide range of Acinetobacter species was isolated from clinical samples in our hospital. The incidence of Acinetobacter remained stable over the study years, while there was a borderline significant trend toward decreased susceptibility to antimicrobial drugs. Small clusters of transmission occurred regularly. A. baumannii was seen as often as Acinetobacter gen. sp. 3. Half of the isolates belonged to these two species. A. baumannii causes the largest burden of multiple antimicrobial resistance and transmissions in the hospital.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Bernards, A. T., A. J. de Beaufort, L. Dijkshoorn, and C. P. van Boven. 1997. Outbreak of septicaemia in neonates caused by Acinetobacter junii investigated by amplified ribosomal DNA restriction analysis (ARDRA) and four typing methods. J. Hosp. Infect. 35:129-140. [DOI] [PubMed] [Google Scholar]
- 2.Bernards, A. T., H. I. Harinck, L. Dijkshoorn, T. van der Reijden, and P. J. van den Broek. 2004. Persistent Acinetobacter baumannii? Look inside your medical equipment. Infect. Control Hosp. Epidemiol. 25:1002-1004. [DOI] [PubMed] [Google Scholar]
- 3.Boo, T. W., F. Walsh, and B. Crowley. 2009. Molecular characterization of carbapenem-resistant Acinetobacter species in an Irish university hospital: predominance of Acinetobacter genomic species 3. J. Med. Microbiol. 58:209-216. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228-240. [Google Scholar]
- 5.de la Tabla Ducasse, V. O., C. M. Gonzalez, J. A. Saez-Nieto, and F. Gutierrez. 2008. First case of post-endoscopic retrograde cholangiopancreatography bacteraemia caused by Acinetobacter ursingii in a patient with choledocholithiasis and cholangitis. J. Med. Microbiol. 57:1170-1171. [DOI] [PubMed] [Google Scholar]
- 6.Dijkshoorn, L., H. Aucken, P. Gerner-Smidt, P. Janssen, M. E. Kaufmann, J. Garaizar, J. Ursing, and T. L. Pitt. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dijkshoorn, L., and A. Nemec. 2008. The diversity of the genus Acinetobacter, p. 1-34. In U. Gerischer (ed.), Acinetobacter molecular microbiology. Caister Academic Press, Norfolk, United Kingdom.
- 8.Dijkshoorn, L., A. Nemec, and H. Seifert. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939-951. [DOI] [PubMed] [Google Scholar]
- 9.Gerner-Smidt, P., I. Tjernberg, and J. Ursing. 1991. Reliability of phenotypic tests for identification of Acinetobacter species. J. Clin. Microbiol. 29:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horrevorts, A., K. Bergman, L. Kollee, I. Breuker, I. Tjernberg, and L. Dijkshoorn. 1995. Clinical and epidemiological investigations of Acinetobacter genomospecies 3 in a neonatal intensive care unit. J. Clin. Microbiol. 33:1567-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald, A., S. G. Amyes, and R. Paton. 1999. The persistence and clonal spread of a single strain of Acinetobacter 13TU in a large Scottish teaching hospital. J. Chemother. 11:338-344. [DOI] [PubMed] [Google Scholar]
- 12.Nemec, A., T. De Baere, I. Tjernberg, M. Vaneechoutte, T. van der Reijden, and L. Dijkshoorn. 2001. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 51:1891-1899. [DOI] [PubMed] [Google Scholar]
- 13.Nemec, A., L. Dijkshoorn, and T. van der Reijden. 2004. Long-term predominance of two pan-European clones among multi-resistant Acinetobacter baumannii strains in the Czech Republic. J. Med. Microbiol. 53:147-153. [DOI] [PubMed] [Google Scholar]
- 14.Nemec, A., M. Mulisek, M. Maixnerova, T. De Baere, T. van der Reijden, M. Vaneechoutte, and L. Dijkshoorn. 2009. Acinetobacter beijerinckii sp. nov. and Acinetobacter gyllenbergii sp. nov., haemoytic organisms isolated from humans. Int. J. Syst. Evol. Microbiol. 59:1376-1381. [DOI] [PubMed] [Google Scholar]
- 15.Peleg, A. Y., H. Seifert, and D. L. Paterson. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramphal, R., and R. M. Kluge. 1979. Acinetobacter calcoaceticus variety anitratus: an increasing nosocomial problem. Am. J. Med. Sci. 277:57-66. [DOI] [PubMed] [Google Scholar]
- 17.Seifert, H., R. Baginski, A. Schulze, and G. Pulverer. 1993. The distribution of Acinetobacter species in clinical culture materials. Zentralbl. Bakteriol. 279:544-552. [DOI] [PubMed] [Google Scholar]
- 18.Seifert, H., A. Strate, A. Schulze, and G. Pulverer. 1993. Vascular catheter-related bloodstream infection due to Acinetobacter johnsonii (formerly Acinetobacter calcoaceticus var. lwoffi): report of 13 cases. Clin. Infect. Dis. 17:632-636. [DOI] [PubMed] [Google Scholar]
- 19.Stelfox, H. T., D. W. Bates, and D. A. Redelmeier. 2003. Safety of patients isolated for infection control. JAMA 290:1899-1905. [DOI] [PubMed] [Google Scholar]
- 20.Struelens, M. J., E. Carlier, N. Maes, E. Serruys, W. G. Quint, and A. van Belkum. 1993. Nosocomial colonization and infection with multiresistant Acinetobacter baumannii: outbreak delineation using DNA macrorestriction analysis and PCR-fingerprinting. J. Hosp. Infect. 25:15-32. [DOI] [PubMed] [Google Scholar]
- 21.Tjernberg, I., and J. Ursing. 1989. Clinical strains of Acinetobacter classified by DNA-DNA hybridization. APMIS 97:595-605. [DOI] [PubMed] [Google Scholar]
- 22.Traub, W. H., and D. Bauer. 2000. Surveillance of nosocomial cross-infections due to three Acinetobacter genospecies (Acinetobacter baumannii, genospecies 3 and genospecies 13) during a 10-year observation period: serotyping, macrorestriction analysis of genomic DNA and antibiotic susceptibilities. Chemotherapy 46:282-292. [DOI] [PubMed] [Google Scholar]
- 23.van den Broek, P. J., J. Arends, A. T. Bernards, E. De Brauwer, E. M. Mascini, T. van der Reijden, L. Spanjaard, E. A. Thewessen, A. van der Zee, J. H. van Zeijl, and L. Dijkshoorn. 2006. Epidemiology of multiple Acinetobacter outbreaks in The Netherlands during the period 1999-2001. Clin. Microbiol. Infect. 12:837-843. [DOI] [PubMed] [Google Scholar]
- 24.van Dessel, H., L. Dijkshoorn, T. van der Reijden, N. Bakker, A. Paauw, P. van den Broek, J. Verhoef, and S. Brisse. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105-112. [DOI] [PubMed] [Google Scholar]
- 25.van Dessel, H., T. E. Kamp-Hopmans, A. C. Fluit, S. Brisse, A. M. de Smet, L. Dijkshoorn, A. Troelstra, J. Verhoef, and E. M. Mascini. 2002. Outbreak of a susceptible strain of Acinetobacter species 13 (sensu Tjernberg and Ursing) in an adult neurosurgical intensive care unit. J. Hosp. Infect. 51:89-95. [DOI] [PubMed] [Google Scholar]
- 26.Vaneechoutte, M., A. Elaichouni, K. Maquelin, G. Claeys, A. Van Liedekerke, H. Louagie, G. Verschraegen, and L. Dijkshoorn. 1995. Comparison of arbitrarily primed polymerase chain reaction and cell envelope protein electrophoresis for analysis of Acinetobacter baumannii and A. junii outbreaks. Res. Microbiol. 146:457-465. [DOI] [PubMed] [Google Scholar]
- 27.Van Looveren, M., and H. Goossens. 2004. Antimicrobial resistance of Acinetobacter spp. in Europe. Clin. Microbiol. Infect. 10:684-704. [DOI] [PubMed] [Google Scholar]
- 28.Villegas, M. V., and A. I. Hartstein. 2003. Acinetobacter outbreaks, 1977-2000. Infect. Control Hosp. Epidemiol. 24:284-295. [DOI] [PubMed] [Google Scholar]