Abstract
Campylobacter is a major cause of human gastroenteritis worldwide. Risk of Campylobacter infection in humans has been associated with many sources, including dogs. This study aimed to investigate whether C. jejuni carried by dogs could potentially be a zoonotic risk for humans and if there were common sources of C. jejuni infection for both humans and dogs. Multilocus sequence typing (MLST) together with macrorestriction analysis of genomic DNA using SmaI and pulsed-field gel electrophoresis (PFGE) were both used to analyze 33 C. jejuni isolates obtained from various dog populations, including those visiting veterinary practices and from different types of kennels. MLST data suggested that there was a large amount of genetic diversity between dog isolates and that the majority of sequence types found in isolates from these dogs were the same as those found in isolates from humans. The main exception was ST-2772, which was isolated from four samples and could not be assigned to a clonal complex. The most commonly identified clonal complex was ST-45 (11 isolates), followed by ST-21 (4 isolates), ST-508 (4 isolates), and ST-403 (3 isolates). The profiles obtained by macrorestriction PFGE were largely in concordance with the MLST results, with a similar amount of genetic diversity found. The diversity of sequence types found within dogs suggests they are exposed to various sources of C. jejuni infection. The similarity of these sequence types to C. jejuni isolates from humans suggests there may be common sources of infection for both dogs and humans. Although only a small number of household dogs may carry C. jejuni, infected dogs should still be considered a potential zoonotic risk to humans, particularly if the dogs originate from kennelled or hunt kennel dog populations, where the prevalence may be higher.
Campylobacter species are among the most commonly reported bacterial cause of human gastroenteritis (2, 5, 10, 44). The majority of such infections are caused by C. jejuni and C. coli. There is some evidence of an elevated risk of Campylobacter infection in humans associated with dog or pet ownership (1, 17, 38).
Multilocus sequence typing (MLST) is a tool used to investigate relationships between Campylobacter spp., particularly C. jejuni. It is useful for investigating possible reservoir hosts and host associations and also for studying the molecular epidemiology of the disease (11, 28). The C. jejuni clonal complexes ST-21 and ST-45 and others, such as ST-61, are often isolated from human cases of campylobacteriosis (12, 31). The complexes ST-21 and ST-45 are commonly found in chickens, cattle, water, and wildlife, and ST-61 has been particularly associated with cattle (7, 18, 22, 36).
Some other C. jejuni sequence types isolated from humans have not yet been associated with cattle and poultry, and the sources of these need further investigation (31). In contrast, some sequence types are strongly associated with certain animal hosts (6) but are not usually isolated from humans (12, 13, 15, 31).
Whether or not dogs are a possible source of C. jejuni infection for humans is not fully understood. Macrorestriction pulsed-field gel electrophoresis (PFGE) has been used in studies to compare C. jejuni isolates found in both dogs and humans (29), and in some cases the dog and the owner did share an identical strain (8). There is currently little MLST data available for Campylobacter spp. isolated from dogs (16; http://pubmlst.org/campylobacter [accessed 5 February 2009]), although ST-45 infection in humans has also been significantly associated with contact with pet cats and dogs (22).
The aims of this study were to examine 33 C. jejuni isolates obtained from dogs from various populations using MLST in order to determine whether strains of C. jejuni carried by dogs appear to be different from those found in humans and other species and to determine possible sources of canine infection. PFGE with SmaI digestion was also performed with the isolates in order to investigate the epidemiology of the disease within the different dog populations.
MATERIALS AND METHODS
Thirty-three isolates of C. jejuni were isolated in the United Kingdom between 2005 and 2008; the origins of the isolates and locations are shown in Table 1. Essentially, two were obtained from household pets (43) (Table 1), three were from a cross-sectional study of dogs visiting veterinary practices (30), and 11 were from longitudinal and cross-sectional studies of boarding and rescue kennels. A further 17 isolates were obtained from three hunt kennels. Samples were cultured using several different isolation methods as previously described (30). Briefly, the methods were as follows: (i) campylobacter selective agar (Lab M) with the addition of cefoperazone and amphotericin (Lab M) and (ii) filtration through a 0.7-μm nitrocellulose membrane onto campylobacter-selective agar as described previously, with the addition of teicoplanin (Oxoid Ltd.) supplement, were used to isolate Campylobacter spp. in all studies except with hunt kennel 3, where only the second method with prior filtration was used. Additionally, campylobacter enrichment broth (Lab M), along with 10% lysed horse blood (Southern Group Labs Ltd.), was used for studies A and B (Table 1) (30, 43). All plates were incubated for 96 h at 37°C under microaerophilic conditions with an atmosphere of N2 (74%), O2 (11%), H2 (3%), and CO2 (12%) in a variable-atmosphere incubator (Don Whitely Scientific Ltd.). Although up to four colonies and a sweep were taken from each plate, only one colony was chosen per sample for further molecular typing by MLST and PFGE.
TABLE 1.
Origins and multilocus sequence types of all C. jejuni isolates from dogsa
| Isolate | Origin | Study | % Prevalence (CI) | Location | Yr | ST | CC | Allele no. |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | ||||||||
| 1 | Household dog | B | 0.5 (0.0-3) | Northwest | 2005 | 403 | 403 | 10 | 27 | 16 | 19 | 10 | 5 | 7 |
| 2 | Household dog | C | NA | Northwest | 2007 | 1326 | 45 | 104 | 7 | 10 | 4 | 1 | 7 | 1 |
| 3 | Boarding dog | E | 9.1 (1.9-26) | Northwest | 2007 | 508 | 508 | 1 | 6 | 60 | 24 | 12 | 28 | 1 |
| 4 | Boarding dog | E | 9.1 (1.9-26) | Northwest | 2007 | 508 | 508 | 1 | 6 | 60 | 24 | 12 | 28 | 1 |
| 5 | Vet-visiting dog | A | 1.2 (0.3-3) | Southwest | 2006 | 273 | 206 | 2 | 21 | 5 | 37 | 60 | 1 | 5 |
| 6# | Vet-visiting dog | A | 1.2 (0.3-3) | Southwest | 2006 | 132 | 508 | 1 | 6 | 22 | 24 | 12 | 28 | 1 |
| 7 | Vet-visiting dog | A | 1.20 (.3-3) | Glasgow | 2006 | 312 | 658 | 14 | 45 | 2 | 4 | 19 | 3 | 6 |
| 8 | Rescue dog | G | 1.9 (0.2-8.6) | Birmingham | 2007 | 45 | 45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 |
| 9 | Rescue dog | D | 20 (8-36) | Northwest | 2007 | 257 | 257 | 9 | 2 | 4 | 62 | 4 | 5 | 6 |
| 10 | Rescue dog | D | 20 (8-36) | Northwest | 2007 | 137 | 45 | 4 | 7 | 10 | 4 | 42 | 7 | 1 |
| 11 | Rescue dog | D | 20 (8-36) | Northwest | 2007 | 45 | 45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 |
| 12 | Rescue dog | D | 20 (8-36) | Northwest | 2007 | 45 | 45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 |
| 13 | Rescue dog | D | 20 (8-36) | Northwest | 2007 | 3613 | 45 | 4 | 296 | 10 | 4 | 1 | 7 | 1 |
| 14 | Rescue dog | F | 3.4 (0.4-15) | Cambridge | 2007 | 1044 | 658 | 2 | 10 | 2 | 4 | 19 | 3 | 6 |
| 15† | Rescue dog* | D | 20 (8-36) | North West | 2007 | 267 | 283 | 4 | 7 | 40 | 4 | 42 | 51 | 1 |
| 16 | Rescue dog* | D | 20 (8-36) | North West | 2007 | 45 | 45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 |
| 17 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 2772 | - | 10 | 4 | 43 | 19 | 6 | 18 | 7 |
| 18 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 104 | 21 | 2 | 1 | 1 | 3 | 7 | 1 | 5 |
| 19 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 45 | 45 | 4 | 7 | 10 | 4 | 1 | 7 | 1 |
| 20 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 403 | 403 | 10 | 27 | 16 | 19 | 10 | 5 | 7 |
| 21 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 104 | 21 | 2 | 1 | 1 | 3 | 7 | 1 | 5 |
| 22† | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 334 | 45 | 4 | 7 | 40 | 4 | 42 | 7 | 1 |
| 23 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 2772 | - | 10 | 4 | 43 | 19 | 6 | 18 | 7 |
| 24 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 403 | 403 | 10 | 27 | 16 | 19 | 10 | 5 | 7 |
| 25 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 2772 | - | 10 | 4 | 43 | 19 | 6 | 18 | 7 |
| 26 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 2772 | - | 10 | 4 | 43 | 19 | 6 | 18 | 7 |
| 27† | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 334 | 45 | 4 | 7 | 40 | 4 | 42 | 7 | 1 |
| 28† | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 334 | 45 | 4 | 7 | 40 | 4 | 42 | 7 | 1 |
| 29 | Hunt dog | H | 26.5 (16-40) | North Wales | 2008 | 206 | 206 | 2 | 21 | 5 | 37 | 2 | 1 | 5 |
| 30 | Hunt dog | I | 10.3 (3-250 | Northwest | 2008 | 508 | 508 | 1 | 6 | 60 | 24 | 12 | 28 | 1 |
| 31 | Hunt dog | I | 10.3 (3-25) | Northwest | 2008 | 22 | 22 | 1 | 3 | 6 | 4 | 3 | 3 | 3 |
| 32 | Hunt dog | I | 10.3 (3-25) | Northwest | 2008 | 19 | 21 | 2 | 1 | 5 | 3 | 2 | 1 | 5 |
| 33 | Hunt dog | J | 5 (0.5-21) | Midlands | 2008 | 19 | 21 | 2 | 1 | 5 | 3 | 2 | 1 | 5 |
ST, sequence type; CC, clonal complex; CI, 95% confidence interval; hunt dog, hunt kennel dog; *, same dog (isolate 15 is from 2nd sample; isolate 16 is from 15th sample); #, rescue dog visiting a practice; †, isolate could not be digested using SmaI. A, national cross-sectional study of vet-visiting dogs (n = 249 dogs [30]); B, community-based household dogs (n = 183 dogs [43]); C, member of staff's dog (n = 1); D, longitudinal study in a rescue shelter (n = 30 dogs); E, longitudinal study in a boarding kennel (n = 22 dogs); F, cross-sectional study of a rescue shelter (n = 29 dogs); G, (stray block) boarding kennel (n = 52 dogs); H, hunt kennel 1 (n = 49 samples); I, hunt kennel 2 (n = 21 samples); J, hunt kennel 3 (n = 20 samples). All were cross-sectional studies except D and E, which were longitudinal studies. Prevalences were based on dogs except those for hunt kennels H to J, which were based on samples.
MLST.
Genomic DNA was prepared using a Promega kit according to the manufacturer's instructions. From this, internal fragments of seven housekeeping genes (encoding aspartase A [aspA], glutamine synthetase [glnA], citrate synthase [gltA], serine hydroxymethyl transferase [glyA], phosphoglucomutase [pgm], transketolase [tkt], and ATP synthase α subunit [uncA]) were sequenced as described by Dingle et al. (13), with some minor modifications. Primers described by Miller et al. (27) to amplify these loci from C. jejuni and C. coli isolates were substituted at loci for which no PCR products were observed in agarose gel electrophoresis. Nucleotide sequencing was carried out at least once on each DNA strand using the same primers as those employed to obtain the amplicon. Alleles, sequence types, and clonal complexes were assigned using the MLST database available at http://pubmlst.org/campylobacter.
Campylobacter PFGE.
Thirty-three C. jejuni isolates were examined by macrorestriction PFGE using a modified version of the method of Ribot et al. (32). Cells of C. jejuni were harvested from blood agar plates into 2-ml sterile phosphate-buffered saline (chemicals were supplied by Sigma unless otherwise stated). Twenty-five microliters of a 20-mg ml−1 proteinase K solution was added to the cell suspension. PFGE-grade agarose (Bio-Rad), 1% in 1× Tris-EDTA buffer (TE) (10 mM Tris-Hcl, 1 mM EDTA, pH 8.0), was added (400 μl) and transferred to duplicate plug molds; the plugs were allowed to set at 4°C. Plugs were transferred to sterile 5-ml bijou bottles containing 3 ml cell lysis buffer (50 mM Tris, 50 mM EDTA, 1% [wt/vol] N-lauryl sarcosine, pH 8.0) containing 25 μl 20-mg ml−1 proteinase K and were incubated with shaking at 54°C for 15 min. Plugs were washed four times at 54°C for 20 min: once with 3 ml sterile distilled water and three times with 3 ml 1× TE. They were then washed once in 500 μl 0.1× TE buffer for 20 min at 25°C. Blocks were equilibrated in 200 μl 1× restriction buffer for 20 min at 25°C. DNA was then digested in 200 μl 1× SmaI restriction buffer containing 40 U SmaI for 2 h at 25°C. Plugs were left at 4°C overnight. A gel was run (1% PFGE agarose in 0.5× TBE) with an initial switch time of 6.7 s and a final switch time of 38.3 s, with a total run time of 16 h. The gel was stained in ethidium bromide solution and examined under UV illumination.
PFGE gels were analyzed using the BioNumerics v. 4.01 software program (Applied Maths, Kortrijk, Belgium) using the Dice similarity coefficient, with 0.5% optimization and 1% tolerance, and dendrograms were done using the unweighted-pair group method with average linkages.
RESULTS
MLST.
Thirty-three isolates were assigned to nine different clonal complexes (Table 1). Sequence type 2772 was isolated four times but to date has not been assigned to a clonal complex. Overall, ST-45 was the most common clonal complex (11 isolates) identified in the dogs, followed by clonal complexes ST-21 (4 isolates), ST-508 (4 isolates), and ST-403 (3 isolates).
Sequence type 45, the central genotype of clonal complex ST-45, was the most common, being isolated five times. Although numbers were small, rescue dogs appeared to carry ST-45 more than other populations of dogs, while the three isolates belonging to ST-334 were exclusive to one hunt kennel within this study (Table 1).
All isolates belonging to clonal complexes ST-21 and ST-22 were obtained from hunt kennel dogs, as were the four ST-2772 isolates that were not assigned to a clonal complex (Table 1).
PFGE.
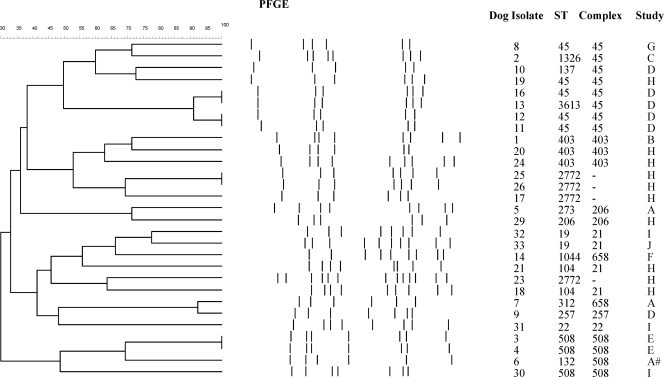
In general, the PFGE profiles agreed with the MLST results (Fig. 1). The dendrogram produced using SmaI clustered together all the isolates typed as clonal complexes ST-45, ST-206, ST-508, and ST-403. Unlike MLST, the PFGE method did not differentiate between dog isolates 13 (ST-3613; pubMLST identification no. 5297) and 16. However, the bottom fragment of isolate 13 did appear to have a slightly different weight from that of isolate 16, and the sequence types varied by only one base change (http://pubmlst.org/campylobacter), indicating that these isolates were closely related. PFGE demonstrated distinguishable profiles between the ST-2772 isolates, the ST-403 isolates, the ST-658 isolates, and the ST-21 isolates, whereas the MLST results did not (Fig. 1).
FIG. 1.
Results from a dendrogram constructed using the Dice (tolerance, 1.0% to 1.0%) (minimum height > 0.0%; minimum surface > 0.0%) (0.0% to 100.0%) coefficient for PFGE using SmaI digestion; #, rescue dog visiting a veterinary practice; A, national cross-sectional study of vet-visiting dogs (30); B, community-based study of household dogs (43); C, member of staff's dog; D, longitudinal study of dogs in a rescue shelter; E, longitudinal study of dogs in a boarding kennel; F, rescue shelter; G, stray block within a boarding kennel; H, hunt kennel 1; I, hunt kennel 2; J, hunt kennel 3.
Identical or near-identical PFGE patterns were observed between isolates from dogs from within the same population, i.e., isolates 3 and 4, obtained from the same boarding kennel, isolates 25 and 26, from the same hunt kennel, and isolates 11, 12, 13, and 16, obtained from a rescue kennel (Fig. 1). With the exception of isolates 13 and 16, which belonged to ST-3613 and ST-45, respectively, all these pairs of isolates were the same sequence type (Fig. 1). Four isolates (15, 22, 27, and 28) could not be digested with SmaI. Three of these isolates (22, 27, and 28) belonged to ST-334, while isolate 15 belonged to ST-267.
DISCUSSION
MLST demonstrated considerable diversity among the C. jejuni sequence types and clonal complexes isolated from the dogs in this study; the PFGE profiles were largely in concordance with these results and showed a similar amount of genetic diversity. The majority of clonal complexes found in isolates from dogs were the same as those reported for humans, including some of the complexes most frequently isolated from humans, i.e., ST-45, ST-21, ST-22, ST-257, and ST-206 (12, 15, 26, 31, 35, 36; http://pubmlst.org/campylobacter). The main exception was ST-2772, which was isolated from four samples and could not be assigned to a clonal complex. The most commonly identified clonal complexes for dogs were ST-45, ST-21, ST-508, and ST-403.
There are various possible sources from which dogs might acquire clonal complex ST-45. This clonal complex has been isolated from a range of sources, such as water, wild birds, cattle, sheep, rabbits, badgers, turkey chicks, broiler chicks, and soil, as well as human disease (7, 18, 31). Sopwith et al. found that ST-45 was the sequence type most commonly isolated from water and suggested that it might be better adapted to survive outside a host and thus is crucial in the transmission of C. jejuni throughout the environment (36). Interestingly, open drains and possibly lakes have been associated with Campylobacter spp. carriage in dogs (3, 45), and the ST-45 complex isolated from humans has also been significantly associated with contact with pet cats and dogs (22). This may indicate common sources of infection for humans and dogs or possibly that dogs may act as conduits of infection from the environment to humans.
Clonal complexes ST-508 and ST-403 were isolated from several dogs within this study. Both complexes have also been isolated from humans, and interestingly, both dominated in human C. jejuni isolates obtained from a study in Curacao (12, 14, 15). Possible origins of these complexes are unclear, although ST-508 has been isolated from sources such as wild birds and cattle, while ST-403 has been found in porcine isolates and also occasionally in cattle (18, 25, 46; http://pubmlst.org/campylobacter).
Isolates from the rescue dogs in this study had the lowest diversity of clonal complexes. Clonal complex ST-45 was the most common among the rescue dogs, and within this complex the central genotype, ST-45, dominated. In contrast, dogs from hunt kennels had the greatest diversity of clonal complexes, which could be a result of frequent exposure to possible sources of infection since dogs from hunt kennels 1 and 2 were exercised daily through fields grazed by livestock and dogs in hunt kennel 1 were fed a diet of raw meat and bone. Cattle feces, carcasses, wildlife, birds, soil, and water have all been shown to carry C. jejuni (4, 9, 23, 41, 48). Although a variety of sequence types were isolated from hunt kennels, several dogs had ST-2772, ST-334, or complex ST-21, none of which were observed for any of the other dog populations within this study. The only other instance of ST-2772 found to date was in isolation from cattle (http://pubmlst.org/campylobacter), whereas clonal complex ST-21 has been isolated in poultry, cattle, and human disease (12, 31).
Although many of the clonal complexes present in dogs are also found in humans, it is not clear if there are common sources of infection or whether on occasion dog-human or indeed human-dog transmission may occur. Nevertheless, such potential zoonotic risk should be put into context. The prevalence of C. jejuni in dog populations varies between different studies. High prevalences, such as those found in our study of kennelled and hunt dogs, have also been found in various study populations and particular demographic groups in other countries (19, 20, 24, 40, 48). However, in other studies, including our previous work with United Kingdom vet-visiting dogs and healthy pet dogs in the community, the prevalence of C. jejuni appears to be relatively low (30, 33, 34, 43, 45). In such populations, it is therefore likely that the overall zoonotic risk is low, although where an individual dog does carry C. jejuni, the risk may be high, depending on factors such as the level of contact between the dogs and humans (42). Such instances may account for case reports of dog-human transmission (8, 47). In addition, with kennelled and hunt dogs, the overall zoonotic risk may be greater than that with household pets. Standard hygiene measures, such as washing hands after contact with dog and/or dog feces before mouth-to-hand contact, should greatly reduce the likelihood of transmission.
PFGE was useful in identifying C. jejuni isolates from within a dog population, and in general the results agreed with MLST data. This was also observed in another study, where PFGE and MLST complexes were associated with each other (31). For example, two dogs (isolates 3 and 4) who had been negative for Campylobacter spp. for more than a week in a boarding kennel began to shed C. jejuni within a couple of days of each other. Results from PFGE indicated that the patterns were identical, suggesting possible transmission or that the two dogs may have shared a common source of infection. This was further supported by MLST data, which also indicated identical sequence types. A similar situation was found for identical isolates 11 and 12 from a rescue kennel.
Although the PFGE technique failed to digest four isolates overall, interestingly these four isolates all belonged to ST-334 or ST-267, both of which could not be digested using SmaI in another study (31). These two sequence types differ by only one base, so despite being undigested, PFGE demonstrated relative clonality for these isolates (http://pubmlst.org/campylobacter). These findings support the need for two independent molecular typing techniques, particularly when analyzing a bacterium with a potentially unstable genome, such as Campylobacter spp. (21, 37). This is important because a single base change can result in a different sequence type or the alteration of a restriction site, which can subsequently lead to a three-fragment difference in PFGE (39).
In conclusion, this study suggests that there is considerable genetic diversity between C. jejuni sequence types obtained from dogs from various sources, and on the whole, evidence to date suggests that dogs do not have strains of C. jejuni particular to them. The majority of sequence types found in dogs within this study have also been isolated from humans. These data may indicate that there are common sources of infection for both humans and dogs and that dogs remain a possible zoonotic risk of C. jejuni infection for humans. However, the exposure risk to dogs and thus possibly humans for certain sequence types may differ depending upon the circumstances of the dog (42).
Acknowledgments
This work was supported by funding from the Department for Environment, Food and Rural Affairs, research contract number OZ0612, and made use of the Internet-accessible databases located at http://pubmlst.org/campylobacter, the development, maintenance, and curation of which are supported by funding from the Wellcome Trust and DEFRA, contract numbers OZ0611 and OZ0615.
We thank Lynne Richardson of the University of Oxford Sequencing Facility and Intervet-Schering-Plough for funding Jenny Stavisky's Ph.D.
None of the authors of this article has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the article.
Footnotes
Published ahead of print on 30 September 2009.
REFERENCES
- 1.Adak, G. K., J. M. Cowden, S. Nicholas, and H. S. Evans. 1995. The Public Health Laboratory Service national case-control study of primary indigenous sporadic cases of campylobacter infection. Epidemiol. Infect. 115:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adak, G. K., S. M. Long, and S. J. O'Brien. 2002. Trends in indigenous foodborne disease and deaths, England and Wales: 1992 to 2000. Gut 51:832-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, J., M. D. Barton, and J. Lanser. 1999. Campylobacter species in cats and dogs in South Australia. Aust. Vet. J. 77:662-666. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. E., O. F. Christensen, H. E. Clough, P. J. Diggle, C. A. Hart, S. Hazel, R. Kemp, A. J. Leatherbarrow, A. Moore, J. Sutherst, J. Turner, N. J. Williams, E. J. Wright, and N. P. French. 2004. Frequency and spatial distribution of environmental Campylobacter spp. Appl. Environ. Microbiol. 70:6501-6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2008. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2007. MMWR Morb. Mortal. Wkly. Rep. 57:366-370. [PubMed] [Google Scholar]
- 6.Colles, F. M., K. E. Dingle, A. J. Cody, and M. C. Maiden. 2008. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl. Environ. Microbiol. 74:3583-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colles, F. M., K. Jones, R. M. Harding, and M. C. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damborg, P., K. E. Olsen, E. Moller Nielsen, and L. Guardabassi. 2004. Occurrence of Campylobacter jejuni in pets living with human patients infected with C. jejuni. J. Clin. Microbiol. 42:1363-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Cesare, A., A. Parisi, V. Bondioli, G. Normanno, and G. Manfreda. 2008. Genotypic and phenotypic diversity within three campylobacter populations isolated from broiler ceca and carcasses. Poult. Sci. 87:2152-2159. [DOI] [PubMed] [Google Scholar]
- 10.Department for Environmental Food and Rural Affairs. 2007. Zoonoses Report United Kingdom 2007. Department for Environmental Food and Rural Affairs, London, United Kingdom. http://www.defra.gov.uk/foodfarm/tarmanimal/diseases/atoz/zoonoses/documents/reports/zoonoses2007.pdf. Accessed 26 May 2009.
- 11.Dingle, K. E., F. M. Colles, D. Falush, and M. C. Maiden. 2005. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J. Clin. Microbiol. 43:340-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingle, K. E., N. D. McCarthy, A. J. Cody, T. E. Peto, and M. C. Maiden. 2008. Extended sequence typing of Campylobacter spp., United Kingdom. Emerg. Infect. Dis. 14:1620-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duim, B., P. C. Godschalk, N. van den Braak, K. E. Dingle, J. R. Dijkstra, E. Leyde, J. van der Plas, F. M. Colles, H. P. Endtz, J. A. Wagenaar, M. C. Maiden, and A. van Belkum. 2003. Molecular evidence for dissemination of unique Campylobacter jejuni clones in Curacao, Netherlands Antilles. J. Clin. Microbiol. 41:5593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan, R. C., J. M. Neal-McKinney, A. S. Dhillon, W. G. Miller, and M. E. Konkel. 2009. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 77:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Food Standards Agency. 2005. Advisory Committee on the Microbiological Safety of Food: second report on Campylobacter. Food Standards Agency, London, United Kingdom. http://www.food.gov.uk/news/newsarchive/2005/jul/campylobacter. Accessed 29 May 2008.
- 18.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherbarrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 19.Hald, B., and M. Madsen. 1997. Healthy puppies and kittens as carriers of Campylobacter spp., with special reference to Campylobacter upsaliensis. J. Clin. Microbiol. 35:3351-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hald, B., K. Pedersen, M. Waino, J. C. Jorgensen, and M. Madsen. 2004. Longitudinal study of the excretion patterns of thermophilic Campylobacter spp. in young pet dogs in Denmark. J. Clin. Microbiol. 42:2003-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karenlampi, R., H. Rautelin, D. Schonberg-Norio, L. Paulin, and M. L. Hanninen. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemp, R., A. J. Leatherbarrow, N. J. Williams, C. A. Hart, H. E. Clough, J. Turner, E. J. Wright, and N. P. French. 2005. Prevalence and genetic diversity of Campylobacter spp. in environmental water samples from a 100-square-kilometer predominantly dairy farming area. Appl. Environ. Microbiol. 71:1876-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koene, M. G., D. J. Houwers, J. R. Dijkstra, B. Duim, and J. A. Wagenaar. 2004. Simultaneous presence of multiple Campylobacter species in dogs. J. Clin. Microbiol. 42:819-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwan, P. S., A. Birtles, F. J. Bolton, N. P. French, S. E. Robinson, L. S. Newbold, M. Upton, and A. J. Fox. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74:3626-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levesque, S., E. Frost, R. D. Arbeit, and S. Michaud. 2008. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 46:3404-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogden, I. D., M. MacRae, M. Johnston, N. J. Strachan, A. J. Cody, K. E. Dingle, and D. G. Newell. 2007. Use of multilocus sequence typing to investigate the association between the presence of Campylobacter spp. in broiler drinking water and Campylobacter colonization in broilers. Appl. Environ. Microbiol. 73:5125-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen, R. J., K. Sutherland, C. Fitzgerald, J. Gibson, P. Borman, and J. Stanley. 1995. Molecular subtyping scheme for serotypes HS1 and HS4 of Campylobacter jejuni. J. Clin. Microbiol. 33:872-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons, B. N., C. J. Porter, R. Ryvar, J. Stavisky, N. J. Williams, G. L. Pinchbeck, R. J. Birtles, R. M. Christley, A. J. German, A. D. Radford, C. A. Hart, R. M. Gaskell, and S. Dawson. Prevalence of Campylobacter spp. in a cross-sectional study of dogs attending veterinary practices in the UK and risk indicators associated with shedding. Vet. J., in press. [DOI] [PubMed]
- 31.Ragimbeau, C., F. Schneider, S. Losch, J. Even, and J. Mossong. 2008. Multilocus sequence typing, pulsed-field gel electrophoresis, and fla short variable region typing of clonal complexes of Campylobacter jejuni strains of human, bovine, and poultry origins in Luxembourg. Appl. Environ. Microbiol. 74:7715-7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi, M., M. L. Hanninen, J. Revez, M. Hannula, and R. G. Zanoni. 2008. Occurrence and species level diagnostics of Campylobacter spp., enteric Helicobacter spp. and Anaerobiospirillum spp. in healthy and diarrheic dogs and cats. Vet. Microbiol. 129:304-314. [DOI] [PubMed] [Google Scholar]
- 34.Sandberg, M., B. Bergsjo, M. Hofshagen, E. Skjerve, and H. Kruse. 2002. Risk factors for Campylobacter infection in Norwegian cats and dogs. Prev. Vet. Med. 55:241-253. [DOI] [PubMed] [Google Scholar]
- 35.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2006. Campylobacter jejuni multilocus sequence types in humans, northwest England, 2003-2004. Emerg. Infect. Dis. 12:1500-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2008. Identification of potential environmentally adapted Campylobacter jejuni strain, United Kingdom. Emerg. Infect. Dis. 14:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steinbrueckner, B., F. Ruberg, and M. Kist. 2001. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human campylobacter enteritis? J. Clin. Microbiol. 39:4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenkate, T. D., and R. J. Stafford. 2001. Risk factors for campylobacter infection in infants and young children: a matched case-control study. Epidemiol. Infect. 127:399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, H. J., H. C. Huang, C. M. Lin, Y. Y. Lien, and C. H. Chou. 2007. Salmonellae and campylobacters in household and stray dogs in northern Taiwan. Vet. Res. Commun. 31:931-939. [DOI] [PubMed] [Google Scholar]
- 41.Waldenstrom, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenaar, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westgarth, C., G. L. Pinchbeck, J. W. Bradshaw, S. Dawson, R. M. Gaskell, and R. M. Christley. 2008. Dog-human and dog-dog interactions of 260 dog-owning households in a community in Cheshire. Vet. Rec. 162:436-442. [DOI] [PubMed] [Google Scholar]
- 43.Westgarth, C., C. J. Porter, L. Nicolson, R. J. Birtles, N. J. Williams, C. A. Hart, G. L. Pinchbeck, R. M. Gaskell, R. M. Christley, and S. Dawson. Risk factors for Campylobacter upsaliensis carriage in pet dogs in a UK community. Vet. Rec., in press. [DOI] [PubMed]
- 44.Westrell, T., N. Ciampa, F. Boelaert, B. Helwigh, H. Korsgaard, M. Chriel, A. Ammon, and P. Makela. 22 January 2009. Zoonotic infections in Europe in 2007: a summary of the EFSA-ECDC annual report. Euro Surveill. 14:pii=19100. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19100. [PubMed]
- 45.Wieland, B., G. Regula, J. Danuser, M. Wittwer, A. P. Burnens, T. M. Wassenaar, and K. D. Stark. 2005. Campylobacter spp. in dogs and cats in Switzerland: risk factor analysis and molecular characterization with AFLP. J. Vet. Med. B Infect. Dis. Vet. Public Health 52:183-189. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, D. J., E. Gabriel, A. J. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, P. Fearnhead, C. A. Hart, and P. J. Diggle. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfs, T. F., B. Duim, S. P. Geelen, A. Rigter, F. Thomson-Carter, A. Fleer, and J. A. Wagenaar. 2001. Neonatal sepsis by Campylobacter jejuni: genetically proven transmission from a household puppy. Clin. Infect. Dis. 32:E97-E99. [DOI] [PubMed] [Google Scholar]
- 48.Workman, S. N., G. E. Mathison, and M. C. Lavoie. 2005. Pet dogs and chicken meat as reservoirs of Campylobacter spp. in Barbados. J. Clin. Microbiol. 43:2642-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]