Abstract
The human T-lymphotropic virus (HTLV) proviral load remains the best surrogate marker for disease progression. Real-time PCR techniques have been developed for detection and quantification of cosmopolitan HTLV type 1a (HTLV-1a) and HTLV-2. Since a growing level of diversity in subtypes and genotypes is observed, we developed a multiplex quantitative PCR for simultaneous detection, genotyping, and quantification of proviral loads of HTLV-1, 2, and 3. Our assay uses tax type-specific primers and dually labeled probes and has a dynamic range of 105 to 10 HTLV copies. One hundred sixty-three samples were analyzed, among which all of the different subtypes within each HTLV genotype could be detected. The performance of proviral load determination of our multiplex assay was compared with that of a previously published HTLV-1 singleplex quantitative PCR based on SYBR green detection, developed at a different institute. Linear regression analysis showed a statistically significant (P < 0.0001) and strong (r2 = 0.87) correlation between proviral load values measured with the two distinct real-time PCR assays. In conclusion, our novel assay offers an accurate molecular diagnosis and genotyping, together with the determination of the proviral load of HTLV-infected individuals, in a single amplification reaction. Moreover, our molecular assay could offer an alternative when current available serological assays are insufficient.
Since the discovery of human T-lymphotropic virus type 1 (HTLV-1) in 1980 (16, 40), three other genotypes and 10 subtypes have been recognized. The precise geographical distribution and the clinical consequences of these infections are still a matter of debate. This can be attributed at least in part to the fact that there are insufficient accurate tools for HTLV diagnosis, genotyping, and measurement of viral burden.
HTLV-1 is endemic in several geographical areas, including sub-Saharan Africa, South America, the Caribbean Islands, Japan, and Melanesia. It has been estimated that worldwide 10 to 25 million people are infected with this retrovirus (41, 53). Most HTLV-1-infected individuals remain asymptomatic throughout their lifetimes. However, 5 to 10% of infected people develop clinical complications, among which adult T-cell leukemia/lymphoma (ATLL) and HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) are the most severe. Other manifestations of HTLV-1 infection include infective dermatitis (25), uveitis (34), arthritis (38), and Strongyloides stercoralis infection (53). Some of these manifestations could accelerate disease development and/or progression (12, 16). For HTLV-1, a distinction is made between seven subtypes: the worldwide, cosmopolitan subtype HTLV-1a; the Central African subtypes HTLV-1b, -d, -e, -f, and -g; and the Australo-Melanesic subtype HTLV-1c (8, 23, 41, 52).
HTLV-2 was discovered in 1982. This retrovirus is endemic in Amerindian and pygmy populations and epidemic in intravenous drug users (16, 49). In contrast to the case for HTLV-1, convincing epidemiological demonstrations of a definitive etiological role of HTLV-2 in human disease are limited. Nevertheless, HTLV-2 has been linked with the development of neurological disorders similar to HAM/TSP, with arthritis, with pulmonary disorders, and with increased mortality (2, 16, 42). HTLV-2 is divided into three subtypes, namely, HTLV-2a and HTLV-2b, mostly found on the American continent, and HTLV-2d, mostly found in Africa (10, 41, 44, 52).
In 2005, two more genotypes, HTLV-3 and -4, were discovered in asymptomatic individuals from Cameroon (6, 7, 47, 56). To date, no diseases have been reported in association with HTLV-3 or -4. Further research is needed to determine the distribution and prevalence as well as the pathogenicity of these two new genotypes.
The routine diagnosis of HTLV infections is based on conventional serological techniques such as enzyme-linked immunosorbent assay and Western blotting. However, among samples infected with HTLV-1 or HTLV-2, the proportion of seroindeterminate results is high (20, 21, 28, 57). Moreover, in the cases of HTLV-3 and HTLV-4, an indeterminate Western blot pattern appears to be the rule rather than the exception (6, 29). To confirm and/or support serological assays, diagnostic HTLV PCR techniques were created (51, 54). In the next phase, real-time or quantitative PCR (qPCR) assays were developed that confirm the diagnosis and at the same time quantify the HTLV proviral load (PVL). The majority of the published HTLV qPCR assays are singleplex assays, which detect one HTLV genotype per amplification reaction and hereby were developed for the most prevalent variant of HTLV-1, the cosmopolitan HTLV-1a, or for HTLV-2 infection (11, 22, 26, 32, 55). Multiplex qPCR allows the simultaneous detection and amplification of two or more target DNA sequences in only one amplification reaction. To our knowledge, one specific and one generic biplex qPCR for HTLV-1 and -2 (13, 26) and, just recently, one triplex qPCR for HTLV-1, -2, and -3 have been described (3).
To address the current problems with HTLV diagnosis and quantitation, taking into account the diversity in HTLV genotypes and subtypes, we developed a novel triplex qPCR assay for simultaneous detection, genotyping, and quantification of PVL of HTLV-1, -2, and -3 infections. In the future, HTLV-4 can be incorporated into our qPCR technique, provided that viral cell culture is possible. Furthermore, considering the increasing number of HTLV qPCR techniques available at present, together with the lack of validation, we performed the first comparative analysis between two qPCR assays developed at different institutions.
MATERIALS AND METHODS
Clinical specimens.
We analyzed 163 HTLV- or simian T-lymphotropic virus (STLV)-infected samples, including patient samples, cell lines, and plasmid DNA, with the samples comprising the HTLV-1, -2, and -3 genotypes and all known subtypes of HTLV-1 and HTLV-2. Samples were provided by the Instituto de Medicina Tropical Alexander von Humboldt (Lima, Peru [IMTAvH]; 116 patient samples), the Pasteur Institute (Paris, France; 35 patient samples and one STLV-3 [Tax 3] plasmid), and the Rega Institute (Leuven, Belgium; 8 patient samples and three cell lines) (9, 15, 27, 43, 50, 58) (Table 1). Genomic DNA (gDNA) was extracted from cell lines and peripheral blood mononuclear cells (PBMC) by use of a QIAamp DNA Blood Mini kit (Qiagen Benelux B.V., Venlo, The Netherlands). Plasmid DNA was isolated using a Qiagen Plasmid Midi kit (Qiagen Benelux B.V., Venlo, The Netherlands). We measured the concentration of gDNA or plasmid DNA by means of spectrophotometry at 260 nm with an Eppendorf BioPhotometer or a Qubit fluorometer (Invitrogen, Merelbeke, Belgium) and adjusted this concentration to 20 ng/μl or 10 ng/μl for qPCR analysis. We analyzed all samples in triplicate.
TABLE 1.
Overview of infected specimens tested in triplicate with our novel triplex qPCRa
| Sample group and HTLV subtype | Origin | No. of specimens | Disease manifestationb |
|---|---|---|---|
| Samples donated by IMTAvH | |||
| HTLV-1a | PBMC | 45 | AC |
| PBMC | 16 | HAM/TSP | |
| PBMC | 33 | OC | |
| Samples donated by Pasteur Institute | |||
| HTLV-1a | Human gDNA | 10 | ATLL |
| HTLV-1b | Human gDNA | 5 | ND |
| HTLV-1c | Human gDNA | 15 | ND |
| HTLV-1d | Human gDNA | 4 | ND |
| HTLV-2 | Human gDNA | 1 | ND |
| STLV-3 | Plasmid PSG5M-Tax3-pyl43 | 1 | ND |
| Samples from the Rega Institute | |||
| HTLV-1b | PBMC | 5 | ND |
| HTLV-1e | PBMC | 1 | ND |
| HTLV-1f | PBMC | 1 | ND |
| HTLV-2b | Gu cell line | 1 | ND |
| HTLV-2d | PBMC | 1 | ND |
| HTLV-2a | Cl19 cell line | 1 | ND |
| STLV-3 | PH969 cell line | 1 | ND |
| Samples donated by IMTAvH | |||
| HTLV-1a | PBMC | 8 | AC |
| PBMC | 6 | OC | |
| PBMC | 8 | HAM/TSP |
A total of 141 HTLV/STLV-infected specimens were used for assay performance validation of genotype and subtype detection, and 22 HTLV-1a-infected patient samples donated by IMTAvH were used for assay performance validation of PVL determination.
OC, other complications (uveitis, Strongyloides stercoralis infection, ID, other [scabies, genital warts]); ND, not determined.
Standards.
To detect and quantify HTLV copies integrated into the cellular genome, we created standard dilution series of known HTLV copies, using gDNA of HTLV- or STLV-infected cell lines. For HTLV-1, we used the chronically infected cell line MT-4, for HTLV-2 the Gu cell line, and for HTLV-3 the STLV-3-infected cell line VULy1824/VXLy6369-6397, derived from the PH969 cell line (17, 33, 45, 48, 58) (Table 2). Aliquots of 20 μl containing 800 ng gDNA of each of these cell lines were stored in siliconized tubes at −20°C. At the time of analysis, we prepared 10-fold serial dilutions of these aliquots in the gDNA background of an HTLV-negative human T-lymphocytic cell line, CEM (14) (Table 2). Total gDNA was equalized at 2,400 ng by the addition of gDNA of CEM at 120 ng/μl from aliquots stored at −20°C.
TABLE 2.
Overview of cell lines used to create standard dilution series to quantify HTLV copy number and β-globin copy number
| Genotype | Standard cell line | Dynamic range (copies/reaction) |
|---|---|---|
| HTLV-1 | MT-4 | ±4.6 × 104 to 4.6 |
| HTLV-2 | Gu | ±2.7 × 105 to 27 |
| STLV-3a | VULy1824/VXLy6369-6397 | ±1.1 × 105 to 11 |
| Human β-globin | CEM | ±2 × 105 to 2 × 101 |
PH969-derived cell line.
For analysis, we added 5 μl of each of the serial dilutions to the PCR mix, corresponding to 200 ng, 20 ng, 2 ng, 200 pg, and 20 pg of gDNA of each infected cell line.
To determine PVL, the HTLV copy number was normalized to the amount of cellular DNA by quantitation of the human β-globin gene via serial 10-fold dilutions of CEM gDNA, ranging from 2 × 105 to 2 × 101 β-globin copies. For this purpose, aliquots of CEM gDNA at 100 ng/μl were stored at −20°C.
To define the detection limit of the triplex qPCR assay and/or to assess the multiple integrations in the HTLV/STLV-infected cell lines, we performed singleplex qPCR assays based on HTLV/STLV plasmid DNA. We enclosed plasmid DNA of PSG5M-Tax1 for HTLV-1 (assuming a length of 5,106 bp and a molecular weight of 3,369,969 g/mol), PMOIA for HTLV-2 (assuming a length of 13,392 bp and a molecular weight of 8,838,720 g/mol), and PSG5M-Tax3-pyl43 for HTLV-3 (assuming a length of 5,184 bp and a molecular weight of 3,421,440 g/mol), with all ranging from 105 to 10 copies (9, 31).
Primers and probes.
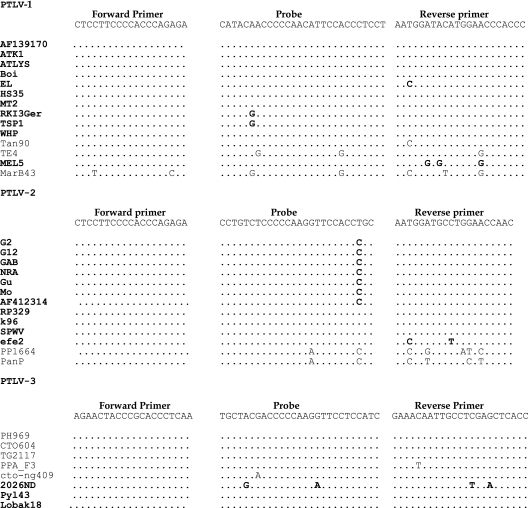
By means of a combination of Primer Express 2.0 and 3.0, we developed HTLV genotype-specific and subtype-generic primers and probes for the most conserved HTLV tax region, based on a full genome alignment of all known HTLV and STLV genotypes and subtypes (Fig. 1). For HTLV-1, the primers were the forward primer HTLV-1F (5′-CTCCTTCCCCACCCAGAGA-3′) and the reverse primer HTLV-1R (5′-GGGTGGGTTCCATGTATCCATT-3′); for HTLV-2, the primers were HTLV-2F (5′-CTCCTTCCCCACCCAGAGA-3′) and HTLV-2R (5′-GTTGGTTCCAGGCATCCATT-3′); and for HTLV-3, the primers were HTLV-3F (5′-AGAACTACCCGCACCCTCAA-3′) and HTLV-3R (5′-GGTGAGCTCGAGGCAATTGTTTC-3′). To obtain specific amplification signals, we used dually labeled fluorogenic probes, including the antisense probe 6-carboxyfluorescein-AGGAGGGTGGAATGTTGGGGGTTGTATG-BHQ1 for HTLV-1, the sense probe JOE-CCTGTCTCCCCCAAGGTTCCACCTGC-BHQ1 for HTLV-2, and the sense probe Cy5-TGCTACGACCCCCAAGGTTCCTCCATC-BHQ2 for HTLV-3.
FIG. 1.
Sequence alignments of oligonucleotides used for HTLV amplification in the tax region. For each genotype, several HTLV (in bold) and STLV strains are reported (GenBank accession no. AF139170, J02029, U19949, L36905, S74562, D13784, Z31660, AF042071, M86840, AF259264, AF074966, Z46900, L02534, AY590142, AF074965, L11456, Y13051, L20734, X89270, M10060, AF412314, AF326583, AF326584, AF139382, Y14365, NC_001815, U90557, Y07616, AF391797, AY217650, AF517775, AY222339, DQ093792, DQ462191, and EU649782.1). Polymorphisms compared to the sequences of the HTLV primers and probes are indicated.
For amplification of the human β-globin gene, the primers were β-glob F (5′-GCAAGAAAGTGCTCGGTG-3′) and β-glob R (5′-CTACTCAGTGTGGCAAAGGTG-3′), with the minor groove binder probe NED-TAGTGATGGCCTGGCTC-NFQ (GenBank accession no. NG_000007).
PCR amplification.
Triplex HTLV amplification reaction mixtures consisted of 5 μl gDNA (20 ng/μl patient sample or standard template), 12.5 μl 2× TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA), 400 nM (each) forward and reverse primers, and 200 nM of the probe for each of the three HTLV genotypes, brought up to a 25-μl reaction volume with water.
Singleplex β-globin amplification reaction mixtures consisted of 5 μl gDNA (10 ng/μl patient sample or standard template), 12.5 μl 2× TaqMan universal PCR master mix, 400 nM (each) forward and reverse primers, and 200 nM of the β-globin probe, brought up to a 25-μl reaction volume with water.
For both the HTLV and β-globin reactions, amplification and data acquisition were carried out using an ABI 7500 Fast real-time PCR system (Applied Biosystems, Foster City, CA). Thermal cycling conditions consisted of an initial activation step at 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min.
For all standard dilutions, controls, and infected specimens, we performed triplicate analysis for each gene (tax and β-globin) in an optical 96-well plate. We implemented the average value of the copy number to quantify both HTLV and β-globin DNAs. The PVL was calculated as the ratio (mean HTLV copy number/mean globin copy number/2) × 104 and then expressed as the number of HTLV copies per 104 lymphocytes.
Statistical analysis.
To evaluate intra-assay and interassay reproducibility, we calculated the coefficient of variation (CV) of the threshold cycle (CT) values and the PVL results as follows: %CV = (standard deviation/mean) × 100. In the literature, CVs of CT values of ≤3% for intra-assay reproducibility and ≤10% for interassay reproducibility, based on standard dilutions, are acceptable (11, 13). For comparison of the HTLV-1 PVL results measured with two different assays, we used GraphPad Prism 5 software to perform linear regression. For evaluation of the effect of clinical status on HTLV-1 PVL, we checked normality and used GraphPad Prism 5 software to perform the nonparametric Mann-Whitney U test.
RESULTS
Specificity, sensitivity, and dynamic range of qPCR.
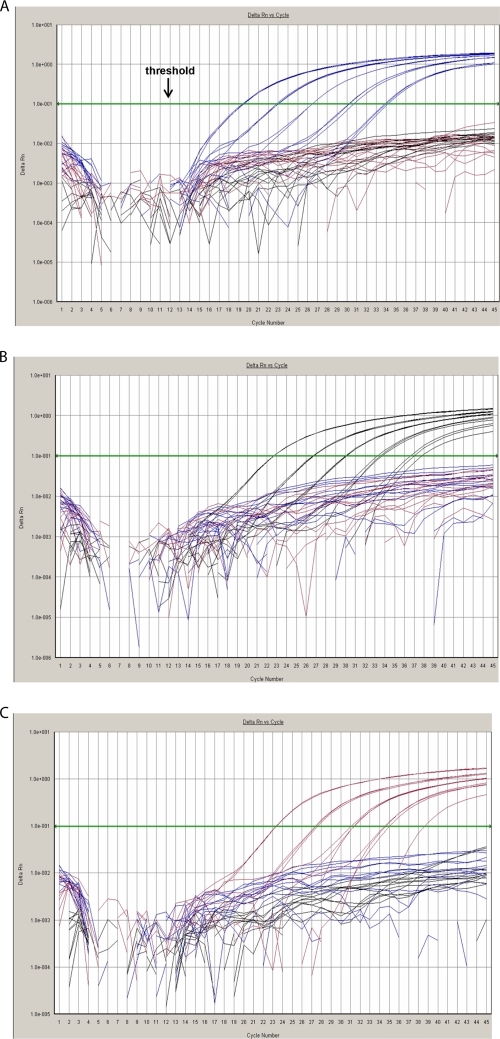
Figure 1 shows the alignments of the sequences from several HTLV and STLV strains, corresponding to the primer and probe regions. We verified the specificity of the HTLV primers and probes through a BLAST search in GenBank (www.ncbi.nih.gov). Given that all HTLV genotype-specific primer-probe sets are included in one triplex amplification reaction, accurate genotyping is conceivable only with highly specific probes. Therefore, we evaluated the three HTLV primer-probe sets in the presence of each of the individual standard templates, corresponding to one specific genotype, separately. None of the individual probes showed cross-reaction with other HTLV types, as demonstrated in Fig. 2. We confirmed the specificity by analyzing PBMC from healthy donors together with HTLV-negative cell lines and found no nonspecific fluorescence signals.
FIG. 2.
HTLV amplification results in order to verify specificity of HTLV probes. Fluorescence signals are plotted against the cycle number at each extension step. The x axis shows the PCR cycle number, and the y axis shows the baseline corrected and normalized fluorescence intensity (ΔRn). The qPCR cycle at which the threshold, placed in the exponential or geometric phase of the plot (A), crosses the fluorescence signal represents the CT, where actual quantification takes place. Amplification plots were obtained based on 10-fold dilutions of each standard template in combination with all genotype-specific primer-probe sets. Each HTLV genotype is color coded to show the fluorescence emitted by each genotype-specific probe, with HTLV-1 in blue, HTLV-2 in black, and HTLV-3 in red. (A) For HTLV-1, amplification plots were obtained based on 10-fold dilutions of MT-4 cells in combination with all genotype-specific primer-probe sets. (B) For HTLV-2, amplification plots were obtained based on 10-fold dilutions of Gu cells in combination with all genotype-specific primer-probe sets. (C) For HTLV-3, amplification plots were obtained based on 10-fold dilutions of VULy1824/VXLy6369-6397 cells in combination with all genotype-specific primer-probe sets. No cross-reaction of the individual probes with the other HTLV genotypes was detected, as fluorescence signals were all below the threshold for the two other genotypes.
Singleplex assays based on standard dilution series of plasmid DNAs of PSG5M-Tax1 for HTLV-1, pMOIA for HTLV-2, and PSG5M-Tax3-pyl43 for HTLV-3 revealed a dynamic range encompassing at least 105 to 10 HTLV copies per amplification reaction.
To mimic the in vivo situation, in which the HTLV provirus is integrated into the cellular genome, we studied the effect of background gDNA of a noninfected cell line, human cutaneous T-lymphocyte HUT-78 (American Type Culture Collection, Manassas, VA), on HTLV detection. This revealed no significant effect, with the condition that background gDNA input did not surpass 105 cells.
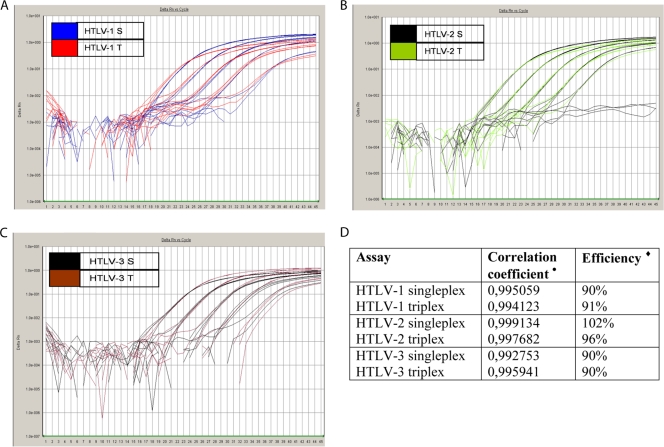
We validated the efficiency and accuracy of multiplex qPCR amplification by comparing CT values, ΔRn values, correlation coefficients (r2), and slopes of standard curves of triplex reactions with corresponding singleplex amplification parameters. We observed little or no variation between CT values of triplex and singleplex reactions, while triplex ΔRn values exhibited a small but acceptable decline in comparison to those of the singleplex reactions (Fig. 3A to C). However, with the exception of the minimal standard dilutions (corresponding to 10 to 100 HTLV copies), the triplex reactions achieved the postulated limit of ΔRn values of ≥0.8. Furthermore, qPCR efficiency and correlation coefficients (r2) were all acceptable for the singleplex as well as the triplex reactions, as illustrated in Fig. 3D.
FIG. 3.
Amplification plots of singleplex (S) and triplex (T) assays for HTLV-1 (A), HTLV-2 (B), and HTLV-3 (C). In singleplex qPCR mixtures, in which only one standard DNA sequence was present, gDNA of MT-4 (HTLV-1), Gu (HTLV-2), or VULy1824/VXLy6369-6397 (HTLV-3) cells was used and amplified. In triplex qPCR mixtures, in which three standard DNA sequences were present, gDNAs of MT-4, Gu, and VULy1824/VXLy6369-6397 cells were combined in one amplification tube, creating a 10-fold dilution series. Fluorescence signals were recorded at each extension step and plotted against cycle number. The ABI software then plots the CT for each dilution against the dilution's known quantity, after which linear regression is performed, creating a standard curve. (D) Correlation coefficients (r2) and PCR efficiency, provided by the standard curves of singleplex and triplex reactions, are represented for the three genotypes. •, correlation coefficient r2, a measurement of the correlation between log copy number of HTLV provirus and the CT. We were aiming at r2 values of ≥0.99. ⧫, PCR efficiency, calculated as (10−1/slope − 1) × 100. We were aiming at 90% ≤ efficiency ≤ 110%.
Determination of the dynamic range of the HTLV triplex qPCR requires quantification of the HTLV copy number per cell, as cell lines can harbor multiple integrations. Singleplex qPCR assays based on 10-fold serial plasmid DNA dilutions revealed 1.4 HTLV-1 copies/cell for MT-4 cells, 8.3 HTLV-2 copies/cell for Gu cells, and 3.4 STLV-3 copies/cell for VULy1824/VXLy6369-6397 cells. Assuming a genome size of 3 × 109 base pairs and a molecular weight of 1.85 × 1012 g/mol, the dynamic range changes to ±4.6 × 104 to 4.6 HTLV-1 copies, ±2.7 × 105 to 27 HTLV-2 copies, and ±1.1 × 105 to 11 HTLV-3 copies per amplification reaction.
To establish PVL determination, quantitation and standardization for the input amount of cellular gDNA has to be implemented in our qPCR assay. This is referred to as quantitation of the endogenous control (or reference), used to rectify variations in the input of a patient's gDNA amount due to differences in PBMC counts or DNA extraction. For this purpose, we performed singleplex qPCR assays based on gDNA standard dilutions of noninfected cell lines, using primer-probe sets for different human cellular genes (ERV-3, β-actin, albumin, and β-globin). We verified the optimal conditions based on CT, ΔRn, and r2 values and qPCR efficiency. The primers and probe for the human β-globin gene, using CEM gDNA standard dilutions, provided the best results. However, when the β-globin reaction was included in the multiplex qPCR, the sensitivity for the detection of HTLV decreased considerably. Therefore, we decided to exclude the amplification of β-globin from the triplex HTLV qPCR assay. Our molecular assay comprises, in addition to the triplex qPCR for the detection of HTLV, a separate singleplex qPCR assay for the quantitation of β-globin, with 100% detection of 105 to 10 cells (assuming a genome size of 3 × 109 base pairs and a molecular weight of 1.85 × 1012 g/mol).
Intra- and interassay reproducibility.
We measured intra-assay reproducibility by running 12 replicates of standard DNA dilutions in one plate, followed by evaluation of the average and standard deviation of CT values together with the CV for each dilution. Using HTLV standard gDNA dilutions for the triplex assay and standard CEM gDNA dilutions for the β-globin singleplex assay, CVs for CT values were all ≤2.18% and thus within the control limit of 3% (Table 3). Repeating the intra-assay reproducibility analysis of the HTLV standard templates based on different gDNA extractions, CT values ranged from 0.5% to 3.5%, indicating a slight increase in CV for the CT values (data not shown).
TABLE 3.
Intra-assay and interassay reproducibility analysis of triplex HTLV and singleplex β-globin qPCR assays, based on standard templates
| Analysis group, HTLV type, and dilution | μ ± σ | %CV |
|---|---|---|
| Intra-assay reproducibility | ||
| HTLV-1 | ||
| 105 | 22.02 ± 0.08 | 0.38 |
| 104 | 25.31 ± 0.07 | 0.26 |
| 103 | 28.84 ± 0.14 | 0.47 |
| 102 | 32.33 ± 0.17 | 0.53 |
| 10 | 36.63 ± 0.66 | 1.81 |
| HTLV-2 | ||
| 105 | 21.94 ± 0.08 | 0.35 |
| 104 | 25.28 ± 0.06 | 0.24 |
| 103 | 28.70 ± 0.09 | 0.31 |
| 102 | 32.13 ± 0.19 | 0.58 |
| 10 | 35.63 ± 0.45 | 1.27 |
| HTLV-3 | ||
| 105 | 22.15 ± 0.06 | 0.27 |
| 104 | 25.59 ± 0.09 | 0.37 |
| 103 | 29.12 ± 0.06 | 0.21 |
| 102 | 32.67 ± 0.26 | 0.80 |
| 10 | 35.37 ± 0.46 | 1.29 |
| β-Globin | ||
| 105 | 23.17 ± 0.05 | 0.23 |
| 104 | 26.51 ± 0.06 | 0.22 |
| 103 | 29.97 ± 0.12 | 0.41 |
| 102 | 33.46 ± 0.23 | 0.68 |
| 10 | 36.85 ± 0.80 | 2.18 |
| Interassay reproducibility | ||
| HTLV-1 | ||
| 105 | 22.72 ± 0.26 | 1.13 |
| 104 | 26.94 ± 0.47 | 1.75 |
| 103 | 30.52 ± 0.52 | 1.70 |
| 102 | 34.08 ± 0.56 | 1.65 |
| 10 | 37.70 ± 1.60 | 4.25 |
| HTLV-2 | ||
| 105 | 23.70 ± 0.14 | 0.58 |
| 104 | 27.30 ± 0.21 | 0.75 |
| 103 | 30.83 ± 0.22 | 0.70 |
| 102 | 34.28 ± 0.27 | 0.77 |
| 10 | 38.38 ± 1.45 | 3.79 |
| HTLV-3 | ||
| 105 | 24.33 ± 0.16 | 0.65 |
| 104 | 28.11 ± 0.21 | 0.74 |
| 103 | 31.66 ± 0.31 | 0.98 |
| 102 | 35.23 ± 0.56 | 1.56 |
| 10 | 39.52 ± 2.00 | 5.00 |
| β-Globin | ||
| 105 | 21.58 ± 0.18 | 0.82 |
| 104 | 24.88 ± 0.18 | 0.74 |
| 103 | 28.37 ± 0.19 | 0.68 |
| 102 | 31.84 ± 0.25 | 0.77 |
| 10 | 35.61 ± 0.69 | 1.95 |
Next, we evaluated interassay reproducibility via analysis of different aliquots from the same stocks of HTLV and β-globin standards in 10 independent experiments. CVs for CT values were all within the postulated limit of 5% (Table 3). In addition, the r2 values were all ≥0.99 and the slopes of the standard curves ranged between −3.1 and −3.6, demonstrating high amplification efficiency. Furthermore, intra- and interassay reproducibility of the HTLV-1 PVL determinations was measured in HTLV-1-infected patient samples (n = 4). For intra-assay reproducibility, we performed nine replicate amplifications for each of the patient samples in one plate; analogously, the same patient samples were amplified in three different plates to assess the interassay reproducibility (for both HTLV and β-globin). We observed variations for both the HTLV and β-globin reactions as well as in the intra- and interassay analyses for all patient samples (Table 4). The intra-assay variation for HTLV copies was reduced for two of four samples when the HTLV-1 copy number was normalized to the β-globin copy number (Table 4). The interassay variation for the HTLV-1 copy number was reduced for all samples when data were normalized to the β-globin copy number. Such observed variability is in agreement with the previously reported reproducibility of qPCR assays based on patient samples (11, 32).
TABLE 4.
Intra-assay and interassay reproducibility analysis of triplex HTLV and singleplex β-globin qPCR assays, based on four infected patient samples
| Analysis group and patient sample no. | HTLV-1 copies |
β-Globin copies |
PVL |
|||
|---|---|---|---|---|---|---|
| Mean ± SD | %CV | Mean ± SD | %CV | Mean ± SD | %CV | |
| Intra-assay reproducibility | ||||||
| 1 | 12.0 ± 1.0 | 8.2 | 6,829.4 ± 416.1 | 6.1 | 24.7 ± 1.9 | 7.5 |
| 2 | 21.1 ± 3.5 | 16.5 | 7,888.0 ± 188.6 | 2.4 | 37.5 ± 6.5 | 17.4 |
| 3 | 1.4 ± 0.2 | 14.5 | 13,204.1 ± 503.2 | 3.8 | 1.5 ± 0.2 | 15.6 |
| 4 | 57.2 ± 8.6 | 15.0 | 6,229.7 ± 47.4 | 0.8 | 128.6 ± 18.4 | 14.3 |
| Interassay reproducibility | ||||||
| 1 | 9.1 ± 2.7 | 29.4 | 6,595.0 ± 585.3 | 8.9 | 19.3 ± 5.3 | 27.6 |
| 2 | 15.8 ± 5.0 | 31.4 | 7,029.5 ± 1,591.6 | 22.6 | 31.5 ± 7.5 | 23.7 |
| 3 | 1.8 ± 0.4 | 24.4 | 13,518.8 ± 454.8 | 3.4 | 1.9 ± 0.4 | 22.6 |
| 4 | 61.6 ± 7.9 | 12.7 | 6,142.3 ± 531.0 | 8.6 | 140.7 ± 15.5 | 11.0 |
Assay performance validation of HTLV genotype and subtype detection.
Next, we tested a set of previously characterized HTLV/STLV-infected specimens (n = 141) with our qPCR assay. gDNAs of HTLV-1a-, HTLV-1b-, HTLV-1c-, HTLV-1d-, HTLV-1e-, HTLV-1f-, and HTLV-2d-infected patient samples and HTLV-2a-, HTLV-2b-, and STLV-3-infected cell lines were analyzed together with PSG5M-Tax3-pyl43 plasmid DNA (Table 1). As presented in Table 5, our novel assay correctly identified 137 of 141 infected samples. One HTLV-1b strain, one HTLV-1c strain, and two HTLV-1d strains were not detected.
TABLE 5.
Detection of different HTLV genotypes and subtypes through analysis of a set of 141 HTLV/STLV-infected specimens
| HTLV subtype | No. of specimens for which subtype was detected/total no. of specimens |
|---|---|
| HTLV-1a | 104/104 |
| HTLV-1b | 9/10 |
| HTLV-1c | 14/15 |
| HTLV-1d | 2/4 |
| HTLV-1e | 1/1 |
| HTLV-1f | 1/1 |
| HTLV-2 | 1/1 |
| HTLV-2a | 1/1 |
| HTLV-2b | 1/1 |
| HTLV-2d | 1/1 |
| STLV-3 | 2/2 |
Assay performance validation of PVL determination.
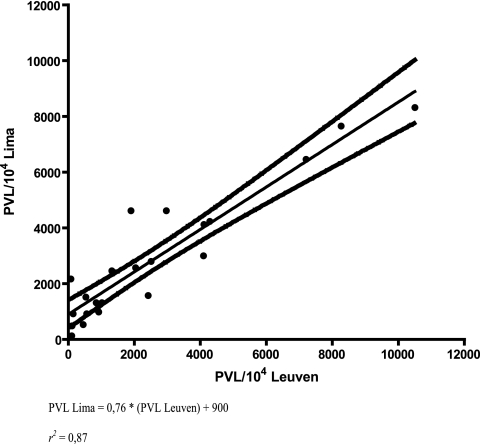
In order to validate PVL determination, we compared two diverse qPCR assays developed at different institutions. The same set of 22 HTLV-1a-infected patient samples (Table 1) was quantified in triplicate by both our triplex assay, performed in Leuven, Belgium, and the singleplex qPCR assay developed at IMTAvH, Lima, Peru, based on SYBR green detection, HTLV-1 plasmid, and ERV-3 standardization, performed in Lima by a different technician (1). To evaluate reproducibility, we divided the 22 HTLV-1 patient samples into two sets of samples, with 15 and 7 gDNA samples. Next, both sets of samples were analyzed by both triplex and SYBR green singleplex qPCR in two separate runs at different times (time interval of ±2 months). For each patient sample, 100 ng of gDNA was analyzed in the HTLV triplex assay and 50 ng of gDNA was analyzed in the β-globin singleplex qPCR, while 40 ng of gDNA was analyzed in the SYBR green singleplex qPCR. Subsequently, we examined the correlation between PVL values obtained with both assays, pooling all data by performing linear regression. The latter revealed a significant (P < 0.0001) and strong (r2 = 0.87) correlation between the two sets of PVL values (Fig. 4).
FIG. 4.
Correlation between PVL quantified with a triplex assay developed at Rega Institute, Leuven, Belgium, and a singleplex assay developed at IMTAvH, Lima, Peru.
Evaluation of HTLV-1 PVL in relation to clinical status.
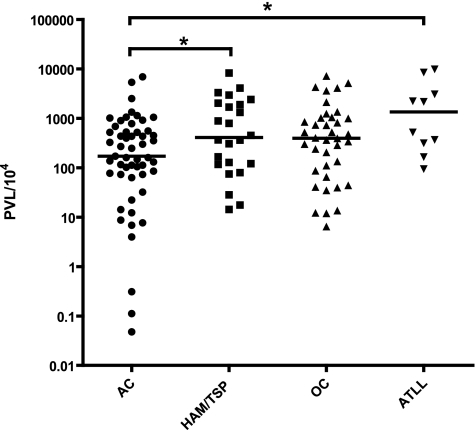
In addition, we evaluated the effect of clinical status on PVL with a set of 126 HTLV-1a-infected samples analyzed during validation of our assay (Table 1). Proviral DNA was detected in the samples of all HTLV-1a-infected individuals, but one sample was excluded because the HTLV copy number was not within the detection limit. We studied a total of 53 asymptomatic carriers (ACs), 24 HAM/TSP patients, 10 ATLL patients, and 38 patients presenting with other complications. The latter included infectious and inflammatory complications such as infective dermatitis (ID), uveitis, Strongyloides stercoralis infection, and other manifestations (scabies, genital warts). In the case of ID and uveitis, the etiological role of HTLV-1 infection is well established (25, 35). HTLV-1 has also been reported in association with S. stercoralis infection (19), but for other manifestations, such as scabies, a definitive association has not been demonstrated (4, 5, 19). The PVL, expressed as the number of HTLV copies per 104 lymphocytes, varied from 0.05 to 6,923 in ACs, from 14.31 to 8,264 in HAM/TSP patients, from 6.43 to 7,201 in patients with other complications, and from 94.24 to 9,895 in ATLL patients. The median PVL in ATLL patients was 1,348 copies per 104 PBMC, compared to 171 copies in ACs, indicating a significant difference in PVL between ATLL patients and ACs (Mann-Whitney U test; P = 0.01) (Fig. 5). In addition, the median PVL in HAM/TSP patients was 409 copies per 104 PBMC, compared to 171 copies in ACs, also indicating a significant difference in PVL between HAM/TSP patients and ACs (Mann-Whitney U test; P = 0.04) (Fig. 5).
FIG. 5.
HTLV-1a PVL according to clinical status. The PVL is expressed as the number of HTLV copies per 104 lymphocytes. OC, other complications.
DISCUSSION
We have demonstrated the broad dynamic range, specificity, efficiency, and performance in genotyping, subtyping, and PVL determination of our triplex assay with 163 infected specimens. Assay performance validation of HTLV genotype and subtype detection pointed out four HTLV-1 strains which could not be detected. Possible explanations are a PVL beneath the assay detection limit or conceivable polymorphisms in the viral strains with regard to the implemented HTLV-1 primers and probe. Due to the limited amount of gDNA, sequence analysis of these samples could not be performed, and therefore the latter hypothesis could not be verified. Nevertheless, our assay detects a great degree of diversity in subtypes within the different genotypes. Moreover, comparison of two HTLV qPCR techniques developed at independent institutes revealed an excellent correlation between HTLV-1 PVL values determined with both assays, indicating the reliability and relevance of both techniques. Consequently, both real-time PCR techniques allow solid and reproducible quantification and therefore enable the determination of the PVL of HTLV-1-infected patient samples. Nonetheless, both qPCR assays differ in primer and probe sequences and in utilized fluorescent reporters and standard templates, that is, probes (Leuven) versus dye (Lima) and cell lines (Leuven) versus plasmid (Lima). Such validation offers excellent quality control of currently implemented qPCR assays for the analysis of HTLV infections. Furthermore, a multiplex qPCR assay is labor-, time-, and cost-effective in comparison to individual singleplex assays when the HTLV type of the sample is unclear.
Although a similar triplex qPCR assay was recently developed for HTLV-1, -2, and -3 genotypes, with a dynamic range of 106 to 100 copies per assay (3), the authors did not present an evaluation of intra- and interassay reproducibility. In addition, our triplex assay was validated with a much broader range of HTLV-infected samples, including a wide variety of HTLV genotypes and subtypes together with PBMC samples obtained from individuals with diverse HTLV-1-associated clinical manifestations. We were thus able to assess the difference in PVL according to disease manifestation: the median PVL in ATLL and HAM/TSP patients was significantly higher than that in ACs, a finding consistent with previous publications (11, 18, 22, 30, 36, 39).
When developing our triplex qPCR assay, we aimed at detecting the diversity in the HTLV genotypes and subtypes. Within the HTLV-1 and HTLV-2 genotypes, a variety of subtypes, arisen from interspecies transmissions and human migration, can be distinguished. Given that HTLV-3 infection was reported only for three Cameroonese asymptomatic individuals, one can doubt the usefulness of including this genotype in our assay. However, analysis of the molecular properties of the HTLV-3 Tax protein revealed that this protein is functionally closely related to the transforming HTLV-1 Tax protein, suggesting that HTLV-3, like HTLV-1, could be pathogenic in vivo (9). Furthermore, phylogenetic analysis based on the complete HTLV-3 sequence isolated from an HTLV-3-infected hunter shows a strong relationship between HTLV-3 and STLV-3, suggesting a primate origin (46). Based upon this plausible interspecies transmission, HTLV-3 can be present throughout the whole African continent. Consequently, the availability of our triplex qPCR including the HTLV-3 genotype is essential.
Moreover, our assay may contribute to currently implemented serodiagnostic assays for HTLV infections. Commercially available enzyme-linked immunosorbent assays and Western blot assays are characterized by a large proportion of seroindeterminate results and are not able to detect early infections when an immune response is lacking. Nevertheless, these conventional serodiagnostic techniques are indispensable for screening for HTLV infections. Therefore, the significance of our multiplex assay mainly concerns ratification of diagnosis, with occasional typing of the virus and determination of PVL, which is still the most accepted surrogate marker for HTLV-associated disease progression (22, 24, 37).
In conclusion, our technique provides an accurate molecular diagnosis, genotyping, and measure of PVL in a single amplification reaction. We stress here some of the difficulties in developing a multiplex assay. Although various parameters can be investigated simultaneously, the combination of multiple standard dilution series negatively affects the intensity and stability of generated fluorescence signals. As a result, we selected HTLV/STLV-infected cell lines instead of HTLV plasmids in order to avoid contamination and instability. Furthermore, we emphasize the necessity to validate currently implemented real-time PCR assays for HTLV detection and quantification. In order to enable a reliable, solid comparison between published results, broader comparisons between published PVL assays are necessary.
Acknowledgments
Britta Moens is supported by a Ph.D. grant from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen). This work was sponsored by Flemish Interuniversity Council (VLIR) Own Initiative grant no. 1179 and by Leerstoel voor Wetenschappelijk onderzoek over infectieziekten in ontwikkelingslanden.
We thank Jo Martens for excellent technical assistance. We thank Kristel Van Laethem and Johan Van Weyenbergh for helpful discussions and Renaud Mahieux and Sébastien Chevalier for the donation of HTLV plasmids.
Footnotes
Published ahead of print on 9 September 2009.
REFERENCES
- 1.Adaui, V., K. Verdonck, I. Best, E. Gonzalez, M. Tipismana, J. Arevalo, G. Vanham, M. Campos, M. Zimic, and E. Gotuzzo. 2006. SYBR green-based quantitation of human T-lymphotropic virus type 1 proviral load in Peruvian patients with neurological disease and asymptomatic carriers: influence of clinical status, sex, and familial relatedness. J. Neurovirol. 12:456-465. [DOI] [PubMed] [Google Scholar]
- 2.Araujo, A., and W. W. Hall. 2004. Human T-lymphotropic virus type II and neurological disease. Ann. Neurol. 56:10-19. [DOI] [PubMed] [Google Scholar]
- 3.Besson, G., and M. Kazanji. 2009. One-step, multiplex, real-time PCR assay with molecular beacon probes for simultaneous detection, differentiation and quantification of human T-cell leukemia virus types 1, 2, and 3. J. Clin. Microbiol. 47:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blas, M., F. Bravo, W. Castillo, W. J. Castillo, R. Ballona, P. Navarro, J. Catacora, R. Cairampoma, and E. Gotuzzo. 2005. Norwegian scabies in Peru: the impact of human T cell lymphotropic virus type I infection. Am. J. Trop. Med. Hyg. 72:855-857. [PubMed] [Google Scholar]
- 5.Brites, C., M. Weyll, C. Pedroso, and R. Badaro. 2002. Severe and Norwegian scabies are strongly associated with retroviral (HIV-1/HTLV-1) infection in Bahia, Brazil. AIDS 16:1292-1293. [DOI] [PubMed] [Google Scholar]
- 6.Calattini, S., E. Betsem, S. Bassot, S. A. Chevalier, R. Mahieux, A. Froment, and A. Gessain. 2009. New strain of human T lymphotropic virus (HTLV) type 3 in a pygmy from Cameroon with peculiar HTLV serologic results. J. Infect. Dis. 199:561-564. [DOI] [PubMed] [Google Scholar]
- 7.Calattini, S., S. A. Chevalier, R. Duprez, S. Bassot, A. Froment, R. Mahieux, and A. Gessain. 2005. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology 2:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassar, O., C. Capuano, S. Bassot, F. Charavay, R. Duprez, P. V. Afonso, M. Abel, H. Walter, W. Mera, P. M. Martin, E. Chungue, and A. Gessain. 2007. Human T lymphotropic virus type 1 subtype C Melanesian genetic variants of the Vanuatu Archipelago and Solomon Islands share a common ancestor. J. Infect. Dis. 196:510-521. [DOI] [PubMed] [Google Scholar]
- 9.Chevalier, S. A., L. Meertens, C. Pise-Masison, S. Calattini, H. Park, A. A. Alhaj, M. Zhou, A. Gessain, F. Kashanchi, J. N. Brady, and R. Mahieux. 2006. The Tax protein from the primate T-cell lymphotropic virus type 3 is expressed in vivo and is functionally related to HTLV-1 Tax rather than HTLV-2 Tax. Oncogene 25:4470-4482. [DOI] [PubMed] [Google Scholar]
- 10.Covas, D. T., and S. Kashima. 2003. Complete nucleotide sequences of the genomes of two Brazilian specimens of human T lymphotropic virus type 2 (HTLV-2). AIDS Res. Hum. Retrovir. 19:689-697. [DOI] [PubMed] [Google Scholar]
- 11.Dehee, A., R. Cesaire, N. Desire, A. Lezin, O. Bourdonne, O. Bera, Y. Plumelle, D. Smadja, and J. C. Nicolas. 2002. Quantitation of HTLV-I proviral load by a TaqMan real-time PCR assay. J. Virol. Methods 102:37-51. [DOI] [PubMed] [Google Scholar]
- 12.Edlich, R. F., L. G. Hill, and F. M. Williams. 2003. Global epidemic of human T-cell lymphotrophic virus type-I (HTLV-I): an update. J. Long Term Eff. Med. Implants 13:127-140. [DOI] [PubMed] [Google Scholar]
- 13.Estes, M. C., and J. S. Sevall. 2003. Multiplex PCR using real time DNA amplification for the rapid detection and quantitation of HTLV I or II. Mol. Cell. Probes 17:59-68. [DOI] [PubMed] [Google Scholar]
- 14.Foley, G. E., H. Lazarus, S. Farber, B. G. Uzman, B. A. Boone, and R. E. McCarthy. 1965. Continuous culture of human lymphoblasts from peripheral blood of a child with acute leukemia. Cancer 18:522-529. [DOI] [PubMed] [Google Scholar]
- 15.Gallo, D., L. M. Penning, and C. V. Hanson. 1991. Detection and differentiation of antibodies to human T-cell lymphotropic virus types I and II by the immunofluorescence method. J. Clin. Microbiol. 29:2345-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallo, R. C. 2002. Human retroviruses after 20 years: a perspective from the past and prospects for their future control. Immunol. Rev. 185:236-265. [DOI] [PubMed] [Google Scholar]
- 17.Goubau, P., M. Van Brussel, A. M. Vandamme, H. F. Liu, and J. Desmyter. 1994. A primate T-lymphotropic virus, PTLV-L, different from human T-lymphotropic viruses types I and II, in a wild-caught baboon (Papio hamadryas). Proc. Natl. Acad. Sci. USA 91:2848-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashimoto, K., I. Higuchi, M. Osame, and S. Izumo. 1998. Quantitative in situ PCR assay of HTLV-1 infected cells in peripheral blood lymphocytes of patients with ATL, HAM/TSP and asymptomatic carriers. J. Neurol. Sci. 159:67-72. [DOI] [PubMed] [Google Scholar]
- 19.Hirata, T., N. Uchima, K. Kishimoto, O. Zaha, N. Kinjo, A. Hokama, H. Sakugawa, F. Kinjo, and J. Fujita. 2006. Impairment of host immune response against Strongyloides stercoralis by human T cell lymphotropic virus type 1 infection. Am. J. Trop. Med. Hyg. 74:246-249. [PubMed] [Google Scholar]
- 20.Ishak, R., A. C. Vallinoto, V. N. Azevedo, A. C. Vicente, W. W. Hall, and M. O. Ishak. 2007. Molecular evidence for infection by HTLV-2 among individuals with negative serological screening tests for HTLV antibodies. Epidemiol. Infect. 135:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob, F., E. Santos-Fortuna, R. S. Azevedo, and A. Caterino-de-Araujo. 2008. Serological patterns and temporal trends of HTLV-1/2 infection in high-risk populations attending public health units in Sao Paulo, Brazil. J. Clin. Virol. 42:149-155. [DOI] [PubMed] [Google Scholar]
- 22.Kamihira, S., N. Dateki, K. Sugahara, T. Hayashi, H. Harasawa, S. Minami, Y. Hirakata, and Y. Yamada. 2003. Significance of HTLV-1 proviral load quantification by real-time PCR as a surrogate marker for HTLV-1-infected cell count. Clin. Lab. Haematol. 25:111-117. [DOI] [PubMed] [Google Scholar]
- 23.Kazanji, M., and A. Gessain. 2003. Human T-cell lymphotropic virus types I and II (HTLV-I/II) in French Guiana: clinical and molecular epidemiology. Cad. Saude Publica 19:1227-1240. [DOI] [PubMed] [Google Scholar]
- 24.Komori, K., A. Hasegawa, K. Kurihara, T. Honda, H. Yokozeki, T. Masuda, and M. Kannagi. 2006. Reduction of human T-cell leukemia virus type 1 (HTLV-1) proviral loads in rats orally infected with HTLV-1 by reimmunization with HTLV-1-infected cells. J. Virol. 80:7375-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaGrenade, L., B. Hanchard, V. Fletcher, B. Cranston, and W. Blattner. 1990. Infective dermatitis of Jamaican children: a marker for HTLV-I infection. Lancet 336:1345-1347. [DOI] [PubMed] [Google Scholar]
- 26.Lee, T. H., D. M. Chafets, M. P. Busch, and E. L. Murphy. 2004. Quantitation of HTLV-I and II proviral load using real-time quantitative PCR with SYBR green chemistry. J. Clin. Virol. 31:275-282. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H. F., A. M. Vandamme, K. Kazadi, H. Carton, J. Desmyter, and P. Goubau. 1994. Familial transmission and minimal sequence variability of human T-lymphotropic virus type I (HTLV-I) in Zaire. AIDS Res. Hum. Retrovir. 10:1135-1142. [DOI] [PubMed] [Google Scholar]
- 28.Lu, S. C., and B. H. Chen. 2003. Seroindeterminate HTLV-1 prevalence and characteristics in blood donors in Taiwan. Int. J. Hematol. 77:412-413. [DOI] [PubMed] [Google Scholar]
- 29.Mahieux, R., and A. Gessain. 2009. The human HTLV-3 and HTLV-4 retroviruses: new members of the HTLV family. Pathol. Biol. (Paris) 57:161-166. [DOI] [PubMed] [Google Scholar]
- 30.Manns, A., W. J. Miley, R. J. Wilks, O. S. Morgan, B. Hanchard, G. Wharfe, B. Cranston, E. Maloney, S. L. Welles, W. A. Blattner, and D. Waters. 1999. Quantitative proviral DNA and antibody levels in the natural history of HTLV-I infection. J. Infect. Dis. 180:1487-1493. [DOI] [PubMed] [Google Scholar]
- 31.Meertens, L., S. Chevalier, R. Weil, A. Gessain, and R. Mahieux. 2004. A 10-amino acid domain within human T-cell leukemia virus type 1 and type 2 Tax protein sequences is responsible for their divergent subcellular distribution. J. Biol. Chem. 279:43307-43320. [DOI] [PubMed] [Google Scholar]
- 32.Miley, W. J., K. Suryanarayana, A. Manns, R. Kubota, S. Jacobson, J. D. Lifson, and D. Waters. 2000. Real-time polymerase chain reaction assay for cell-associated HTLV type I DNA viral load. AIDS Res. Hum. Retrovir. 16:665-675. [DOI] [PubMed] [Google Scholar]
- 33.Miyoshi, I., S. Yoshimoto, I. Kubonishi, H. Taguchi, Y. Shiraishi, Y. Ohtsuki, and T. Akagi. 1981. Transformation of normal human cord lymphocytes by co-cultivation with a lethally irradiated human T-cell line carrying type C virus particles. Gann 72:997-998. [PubMed] [Google Scholar]
- 34.Mochizuki, M., T. Watanabe, K. Yamaguchi, K. Takatsuki, K. Yoshimura, M. Shirao, S. Nakashima, S. Mori, S. Araki, and N. Miyata. 1992. HTLV-I uveitis: a distinct clinical entity caused by HTLV-I. Jpn. J. Cancer Res. 83:236-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mochizuki, M., T. Watanabe, K. Yamaguchi, K. Yoshimura, S. Nakashima, M. Shirao, S. Araki, K. Takatsuki, S. Mori, and N. Miyata. 1992. Uveitis associated with human T-cell lymphotropic virus type I. Am. J. Ophthalmol. 114:123-129. [DOI] [PubMed] [Google Scholar]
- 36.Montanheiro, P. A., A. C. Oliveira, M. P. Posada-Vergara, A. C. Milagres, C. Tauil, P. E. Marchiori, A. J. Duarte, and J. Casseb. 2005. Human T-cell lymphotropic virus type I (HTLV-I) proviral DNA viral load among asymptomatic patients and patients with HTLV-I-associated myelopathy/tropical spastic paraparesis. Braz. J. Med. Biol. Res. 38:1643-1647. [DOI] [PubMed] [Google Scholar]
- 37.Nagai, M., K. Usuku, W. Matsumoto, D. Kodama, N. Takenouchi, T. Moritoyo, S. Hashiguchi, M. Ichinose, C. R. Bangham, S. Izumo, and M. Osame. 1998. Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J. Neurovirol. 4:586-593. [DOI] [PubMed] [Google Scholar]
- 38.Nishioka, K., I. Maruyama, K. Sato, I. Kitajima, Y. Nakajima, and M. Osame. 1989. Chronic inflammatory arthropathy associated with HTLV-I. Lancet i:441. [DOI] [PubMed] [Google Scholar]
- 39.Olindo, S., A. Lezin, P. Cabre, H. Merle, M. Saint-Vil, M. E. Kaptue, A. Signate, R. Cesaire, and D. Smadja. 2005. HTLV-1 proviral load in peripheral blood mononuclear cells quantified in 100 HAM/TSP patients: a marker of disease progression. J. Neurol. Sci. 237:53-59. [DOI] [PubMed] [Google Scholar]
- 40.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proietti, F. A., A. B. Carneiro-Proietti, B. C. Catalan-Soares, and E. L. Murphy. 2005. Global epidemiology of HTLV-I infection and associated diseases. Oncogene 24:6058-6068. [DOI] [PubMed] [Google Scholar]
- 42.Roucoux, D. F., and E. L. Murphy. 2004. The epidemiology and disease outcomes of human T-lymphotropic virus type II. AIDS Rev. 6:144-154. [PubMed] [Google Scholar]
- 43.Salemi, M., S. Van Dooren, E. Audenaert, E. Delaporte, P. Goubau, J. Desmyter, and A. M. Vandamme. 1998. Two new human T-lymphotropic virus type I phylogenetic subtypes in seroindeterminates, a Mbuti pygmy and a Gabonese, have closest relatives among African STLV-I strains. Virology 246:277-287. [DOI] [PubMed] [Google Scholar]
- 44.Salemi, M., S. Van Dooren, and A.-M. Vandamme. 1999. Origin and evolution of human and simian T-cell lymphotropic viruses. AIDS Rev. 1:131-139. [Google Scholar]
- 45.Salemi, M., A. M. Vandamme, F. Guano, C. Gradozzi, E. Cattaneo, C. Casoli, and U. Bertazzoni. 1996. Complete nucleotide sequence of the Italian human T-cell lymphotropic virus type II isolate Gu and phylogenetic identification of a possible origin of South European epidemics. J. Gen. Virol. 77:1193-1201. [DOI] [PubMed] [Google Scholar]
- 46.Switzer, W. M., S. H. Qari, N. D. Wolfe, D. S. Burke, T. M. Folks, and W. Heneine. 2006. Ancient origin and molecular features of the novel human T-lymphotropic virus type 3 revealed by complete genome analysis. J. Virol. 80:7427-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Switzer, W. M., M. Salemi, S. H. Qari, H. Jia, R. R. Gray, A. Katzourakis, S. J. Marriott, K. N. Pryor, N. D. Wolfe, D. S. Burke, T. M. Folks, and W. Heneine. 2009. Ancient, independent evolution and distinct molecular features of the novel human T-lymphotropic virus type 4. Retrovirology 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Brussel, M., P. Goubau, R. Rousseau, J. Desmyter, and A. M. Vandamme. 1996. The genomic structure of a new simian T-lymphotropic virus, STLV-PH969, differs from that of human T-lymphotropic virus types I and II. J. Gen. Virol. 77:347-358. [DOI] [PubMed] [Google Scholar]
- 49.Vandamme, A. M., U. Bertazzoni, and M. Salemi. 2000. Evolutionary strategies of human T-cell lymphotropic virus type II. Gene 261:171-180. [DOI] [PubMed] [Google Scholar]
- 50.Vandamme, A. M., H. F. Liu, M. Van Brussel, W. De Meurichy, J. Desmyter, and P. Goubau. 1996. The presence of a divergent T-lymphotropic virus in a wild-caught pygmy chimpanzee (Pan paniscus) supports an African origin for the human T-lymphotropic/simian T-lymphotropic group of viruses. J. Gen. Virol. 77:1089-1099. [DOI] [PubMed] [Google Scholar]
- 51.Vandamme, A. M., K. Van Laethem, H. F. Liu, M. Van Brussel, E. Delaporte, C. M. Castro Costa, C. Fleischer, G. Taylor, U. Bertazzoni, J. Desmyter, and P. Goubau. 1997. Use of a generic polymerase chain reaction assay detecting human T-lymphotropic virus (HTLV) types I, II and divergent simian strains in the evaluation of individuals with indeterminate HTLV serology. J. Med. Virol. 52:1-7. [PubMed] [Google Scholar]
- 52.Van Dooren, S., M. Salemi, and A. M. Vandamme. 2001. Dating the origin of the African human T-cell lymphotropic virus type I (HTLV-I) subtypes. Mol. Biol. Evol. 18:661-671. [DOI] [PubMed] [Google Scholar]
- 53.Verdonck, K., E. Gonzalez, S. Van Dooren, A. M. Vandamme, G. Vanham, and E. Gotuzzo. 2007. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect. Dis. 7:266-281. [DOI] [PubMed] [Google Scholar]
- 54.Vet, J. A., A. R. Majithia, S. A. Marras, S. Tyagi, S. Dube, B. J. Poiesz, and F. R. Kramer. 1999. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA 96:6394-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vitone, F., D. Gibellini, P. Schiavone, A. D'Antuono, L. Gianni, I. Bon, and M. C. Re. 2006. Human T-lymphotropic virus type 1 (HTLV-1) prevalence and quantitative detection of DNA proviral load in individuals with indeterminate/positive serological results. BMC Infect. Dis. 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfe, N. D., W. Heneine, J. K. Carr, A. D. Garcia, V. Shanmugam, U. Tamoufe, J. N. Torimiro, A. T. Prosser, M. Lebreton, E. Mpoudi-Ngole, F. E. McCutchan, D. L. Birx, T. M. Folks, D. S. Burke, and W. M. Switzer. 2005. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc. Natl. Acad. Sci. USA 102:7994-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao, K., M. Hisada, E. Maloney, Y. Yamano, B. Hanchard, R. Wilks, M. Rios, and S. Jacobson. 2006. Human T lymphotropic virus types I and II Western blot seroindeterminate status and its association with exposure to prototype HTLV-I. J. Infect. Dis. 193:427-437. [DOI] [PubMed] [Google Scholar]
- 58.Zella, D., A. Cavicchini, M. Salemi, C. Casoli, F. Lori, G. Achilli, E. Cattaneo, V. Landini, and U. Bertazzoni. 1993. Molecular characterization of two isolates of human T cell leukaemia virus type II from Italian drug abusers and comparison of genome structure with other isolates. J. Gen. Virol. 74:437-444. [DOI] [PubMed] [Google Scholar]