Abstract
The prevalence, among human clinical cases, of Salmonella enterica serotype 4,5,12:i:−, a serotype antigenically similar to Salmonella enterica serotype Typhimurium but lacking second-phase flagellar antigens, has increased considerably over the last 10 years. To probe the evolution and ecology of this emerging serotype, we characterized 190 Salmonella isolates initially classified as Salmonella serotypes 4,5,12:i:− (n = 90) and Typhimurium (n = 100) and obtained from various sources in the United States and Spain. These isolates were characterized into six sequence types (determined by multilocus sequence typing [MLST]) and 79 pulsed-field gel electrophoresis types. The majority of Salmonella serotype 4,5,12:i:− and Typhimurium isolates (85 and 84 isolates, respectively) represented a single MLST type. Existing genome information revealed different genome deletions (which included genes responsible for phase 2 flagellum expression) in four Spanish Salmonella serotype 4,5,12:i:− isolates and one U.S. Salmonella serotype 4,5,12:i:− isolate. Fifty-nine isolates of both serotypes, representing different sources and geographical locations as well as different molecular subtypes, were thus screened for the presence of six genes and one specific region, all of which were previously found to show variable presence among Salmonella serotype 4,5,12:i:− and Typhimurium strains. All Salmonella serotype 4,5,12:i:− isolates lacked the phase 2 flagella genes fljA and fljB, which were present in all Salmonella serotype Typhimurium isolates. While all Spanish Salmonella serotype 4,5,12:i:− isolates carried the same deletion surrounding fljAB, all but two U.S. isolates showed a different genomic deletion; the two atypical U.S. isolates represented the “Spanish” deletion genotype and a unique deletion genotype. Salmonella serotype 4,5,12:i:− thus appears to represent at least two common clones, which cannot easily be differentiated with standard diagnostic procedures.
Salmonella spp. are one of the most common causes of bacterial food-borne diseases worldwide (34). In the United States nontyphoidal Salmonella serotypes cause an estimated 1.4 million human salmonellosis cases, including approximately 550 deaths annually (27). Serotyping with the Kaufmann-White scheme is used commonly as a first step to differentiate Salmonella isolates. Serotyping of Salmonella isolates is based on lipopolysaccharide moieties on the cell surface (O antigens) and the flagellar proteins (H antigens), as well as capsular protein antigens (Vi antigen), which are only found in a few Salmonella serotypes (e.g., Salmonella enterica serotype Typhi). According to the Kaufmann-White scheme, Salmonella includes over 2,500 recognized serotypes (20). Many Salmonella bacteria are motile due to peritrichous flagella (28), which include a basal body, a propeller, and a hook. The motility of Salmonella depends on the rotation of the flagellar propeller (i.e., the filament), which includes either FliC (phase 1 antigen) or FljB (phase 2 antigen) flagellin (11). Most Salmonella serotypes, including Salmonella enterica serotype Typhimurium, are biphasic, meaning that they can express two distinct flagellar antigens (i.e., phase 1 and phase 2 antigens). Regulation of phase 1 and 2 antigen expression is under the control of the recombinase Hin. This recombinase facilitates inversion of a promoter element so that it either (i) transcribes fljB (which encodes the phase 2 antigen FljB) and fljA (which encodes a repressor of fliC, the gene encoding the phase 1 antigen FliC) (4, 37) or (ii) does not transcribe either of these genes. If this promoter is located in an orientation that does not allow for transcription of fljB and fljA, the lack of a repression of fliC transcription leads to expression of phase 1 flagellar antigens.
Salmonella enterica serotype 4,5,12:i:− is a serotype that appears to be antigenically similar and genetically closely related to Salmonella serotype Typhimurium (which has the antigenic formula 4,5,12:i:1,2) but lacks expression of the second-phase flagellar antigen, which is 1,2 in Salmonella serotype Typhimurium (28). Salmonella serotype 4,5,12:i:− was the sixth most common Salmonella serotype among cases of human disease in the United States in 2006 (10) and the fourth most common serotype among human isolates in Spain in 1998 (18). Overall, the prevalence of Salmonella serotype 4,5,12:i:− among human cases has increased considerably in many countries in the world over the last 10 years (9, 10, 18, 29, 36). This Salmonella serotype has also been responsible for a number of human salmonellosis outbreaks over the last decades, including in Spain (1998), the United States (2004 and 2007), and Luxemburg (2006). Salmonella serotype 4,5,12:i:− has been isolated, particularly over the last decade, from a number of different foods and animals (1, 6, 13, 29, 38). While a number of separate studies, using molecular subtyping and characterization tools (e.g., genomic microarrays and PCR assays to test for gene presence/absence), have shown that Salmonella serotype 4,5,12:i:− isolates from Spain (15, 18) and the United States (1, 2, 38) are genetically closely related to Salmonella serotype Typhimurium, we are not aware of any comparative studies of Salmonella serotype 4,5,12:i:− isolates from Europe and the United States that have been published to date. In order to provide a better understanding of the transmission, ecology, and evolution of Salmonella serotype 4,5,12:i:−, we have assembled a collection of 190 Salmonella serotype 4,5,12:i:− and Typhimurium isolates from various sources and from two countries, the United States and Spain. These isolates were characterized by different molecular subtyping methods (i.e., multilocus sequence typing [MLST] and pulsed-field gel electrophoresis [PFGE]), followed by characterization of selected isolates for genomic deletions that may be responsible for the lack of phase 2 flagellum expression.
MATERIALS AND METHODS
Salmonella isolates.
A total of 190 Salmonella isolates initially identified as Salmonella serotypes Typhimurium (n = 100) and 4,5,12:i:− (n = 90) were used in this study (Table 1). These isolates were obtained from different states in the United States, including New York (69 isolates), Washington (52 isolates), and Georgia (26 isolates), and in Spain (43 isolates), as well as from different sources, including human clinical cases, foods, and cattle, poultry, and other warm-blooded animals (Table 1). Human isolates from New York State and Washington State were obtained from the New York State Department of Health and the Washington State Department of Health, respectively. Bovine isolates from New York State and Washington State were obtained from the Animal Health Diagnostic Center at Cornell University and the Washington Animal Disease Diagnostic Laboratory, Pullman, respectively; isolates from foods collected in New York State were obtained from the Food and Drug Administration (FDA) (see Table S1 in the supplemental material). Salmonella serotype Typhimurium and 4,5,12:i:− isolates from Georgia have previously been described (38). Salmonella serotype Typhimurium and 4,5,12:i:− isolates from Spain have also been described previously (5, 18) and were provided by J. Garaizar, University of the Basque Country, Vitoria-Gasteiz, Spain. Detailed information for all isolates (see Table S1 in the supplemental material), including serotype, source, gene sequence data, allelic types (ATs), and PFGE patterns are available via the Pathogen Tracker website (http://www.pathogentracker.net).
TABLE 1.
Salmonella serotype 4,5,12:i:− and Typhimurium isolates used in this study
| Serotype and state or country | No. of isolates froma |
||||||
|---|---|---|---|---|---|---|---|
| Bovine | Human | Poultry | Food | Others | Nondomestic birds | Total | |
| Salmonella serotype 4,5,12:i:− | |||||||
| Georgia | 2 (2) | 0 | 10 (7) | 0 | 0 | 1 (1) | 13 (10) |
| New York | 7 (5) | 9 (4) | 0 | 2 (1) | 0 | 0 | 18 (10) |
| Washington | 0 | 40 (10) | 0 | 0 | 2 (1) | 0 | 42 (11) |
| Spain | 0 | 11 | 0 | 2 | 0 | 0 | 13 (10) |
| Total | 86 (41) | ||||||
| Salmonella serotype Typhimurium | |||||||
| Georgia | 6 | 0 | 5 (2) | 0 | 0 | 1 (1) | 12 (3) |
| New York | 22 | 29 (4) | 0 | 0 | 0 | 0 | 51 (4) |
| Washington | 1 | 6 (2) | 0 | 0 | 0 | 0 | 7 (2) |
| Spainb | 0 | 5 | 0 | 8 | 17 | 0 | 30 (5) |
| Total | 100 (14) | ||||||
| Inconsistent (Salmonella serotype Typhimurium or 4,5,12:i:−)c | |||||||
| Georgia | 0 | 0 | 0 | 0 | 0 | 1 | 1 (1) |
| Washington | 3 (1) | 0 | 0 | 0 | 0 | 0 | 3 (3) |
| Total | 4 (4) | ||||||
| Total | 190 (59) | ||||||
Numbers in parentheses represent numbers of isolates that were used for further PCR screens to determine the presence/absence of selected genes and one specific region.
Spanish Salmonella serotype Typhimurium isolates in the category “Other” were obtained from water and other environments; detailed information for Spanish isolates is in Table S1 in the supplemental material.
These isolates were serotyped as Salmonella serotype 4,5,12:i:− in one replicate and Salmonella serotype Typhimurium in another replicate (including one isolate that was classified as Salmonella serotype 4,5,12:i:− in two replicates and Salmonella serotype Typhimurium in one replicate) and were thus designated “inconsistent.”
While serotype data were provided for all isolates, isolates that were initially classified as Salmonella serotype 4,5,12:i:− but contained an intact copy of the phase 2 flagellum gene fljB were resubmitted for serotyping at the National Veterinary Service Laboratories (USDA APHIS VS, Ames, IA). Isolates that were classified as Salmonella serotype 4,5,12:i:− in one replicate and Salmonella serotype Typhimurium in another replicate (including one isolate that was classified as Salmonella serotype 4,5,12:i:− in two replicates and Salmonella serotype Typhimurium in one replicate) were designated “inconsistent serotype” isolates.
MLST.
While traditional MLST schemes target seven housekeeping genes (24), we initially used a previously reported MLST scheme targeting three genes (i.e., manB, mdh, and fimA) (2, 33) to characterize all isolates used in this study. In addition to these three genes, we also sequenced an 826-nucleotide fragment of a fourth gene (aroC) in all isolates to determine whether the use of additional genes would increase discriminatory power. aroC was chosen as an additional gene because it was found to represent the greatest number of different ATs among all isolates in the seven-gene MLST database for Salmonella in July 2007 (http://web.mpiib-berlin.mpg.de/mlst/dbs/Senterica). ATs for fimA, mdh, and manB and three-gene sequence types (STs) were assigned to be consistent with previous studies published by our group (2, 3, 33). ATs for aroC were also assigned to be consistent with the seven-gene MLST Max Planck Institute database (http://web.mpiib-berlin.mpg.de/mlst/dbs/Senterica). For example, three-gene ST6 includes the same AT combination for three genes as reported in two studies by Alcaine et al. (2, 3), while aroC AT 18 is identical to AT AROC18 in the seven-gene MLST database (http://web.mpiib-berlin.mpg.de/mlst/dbs/Senterica). STs were also determined based on ATs for all four genes; these STs do not correspond to any previously reported STs.
Salmonella DNA used as a template for PCRs performed for MLST was purified using the QIAmp DNA mini kit (Qiagen Inc., Chatsworth, CA). PCR primers for manB, mdh, and fimA have been previously reported (2, 33); PCR primers for aroC amplification were obtained from the Salmonella enterica MLST database at Max Plank Institute (http://web.mpiib-berlin.mpg.de/mlst/dbs/Senterica). All primers used are summarized in Table S2 in the supplemental material. PCR products were purified using exonuclease I (USB, Cleveland, OH) and shrimp alkaline phosphatase (USB, Cleveland, OH). Purified PCR products were sequenced using Big Dye Terminator chemistry and AmpliTaq-FS DNA polymerase, and sequencing reactions were analyzed using the Applied Biosystems automated 3730 DNA analyzer at the Cornell University Life Sciences Core Laboratories Center. Sequences were assembled and proofread using SeqMan and aligned using the Clustal W algorithm implanted in MegAlign (DNAStar Inc., Madison, WI).
Phylogenetic analysis.
Phylogenetic analysis was performed using all three-gene STs found among Salmonella serotype Typhimurium and 4,5,12:i:− isolates as well as three-gene STs available for other serotypes (e.g., STs reported by Alcaine et al. ([2]). As manB is duplicated in some isolates, thus yielding sequence data not suitable for phylogenetic analyses (2), STs representing sequences with manB duplications were not included in the phylogenetic analyses. For each unique ST, the sequences of the three genes were concatenated. Concatenated sequences were aligned using MegAlign (DNAStar Inc., Madison, WI) and MACCLADE version 4.08 (Sinauer Associates Inc., Sunderland, MA). MODELTEST (30) was used to determine the best-fitting model of evolution (i.e., TrN+I+G), which was used for construction of a maximum-likelihood (ML) tree. The ML tree was constructed, using the concatenated three-gene MLST sequences, using PAUP* Portable version 4.0b10 for Unix (35). No phylogenetic analyses were performed on the four-gene MLST data, as insufficient ST data are available for serotypes other than Salmonella serotypes Typhimurium and 4,5,12:i:−.
PFGE.
XbaI PFGE was performed according to the Centers for Disease Control and Prevention PulseNet protocol (31). Analysis of PFGE types was performed using the BioNumerics Software package (Applied Maths, Austin, TX). Similarity analysis was performed by using the Dice coefficient, and clustering was performed using the unweighted pair group method with arithmetic mean.
SID.
Simpson's index of diversity (SID) was calculated as previously described (23).
Analysis of microarray and genome sequence data to identify gene deletions in Salmonella serotype 4,5,12:i:−.
In order to identify gene deletions and other genomic differences between Salmonella serotypes 4,5,12:i:− and Typhimurium, we used (i) comparative genomic microarray data on gene presence/absence patterns in four Spanish Salmonella serotype 4,5,12:i:− isolates (reported by Garaizar et al. [18]) and (ii) the full genome sequence data for the U.S. Salmonella serotype 4,5,12:i:− isolate CVM23701 (GenBank accession no. NZ_ABAO00000000; http://msc.jcvi.org/salmonella/salmonella_enterica_subsp__enterica_serovar_4__5__12_i___str__cvm23701/index.shtml) (32) and Salmonella serotype Typhimurium LT2 (AE006468). Genomic microarray data reported by Garaizar et al. (18) revealed one genomic deletion (termed cluster V) in Salmonella serotype 4,5,12,:i:−, which included deletion of fljB (encoding phase 2 flagella, thus providing a functional explanation for the absence of phase 2 flagellar expression observed in Salmonella serotype 4,5,12:i:−) as well as a second deletion (termed cluster IV) located approximately 16 kb 5′ of cluster V. BLAST searches were used to determine whether genes in cluster IV (genes STM2694 to STM2740) and cluster V (genes STM2758 to STM2773) as well as genes in the intervening regions and upstream and downstream were present in the Salmonella serotype 4,5,12:i:− isolate CVM23701 genome. Specifically, BLAST searches were used to determine whether Salmonella serotype Typhimurium LT2 genes STM2691 through STM2775 were present in the CVM23701 genome. BLAST searches were performed using the National Center for Biotechnology Information (NCBI) BLAST tools and gene sequences downloaded from the J. Craig Venter Institute Comprehensive Microbial Resource. BLAST searches were also used to determine whether genes in three other clusters (I, II, and III), which were previously reported to be present in Salmonella serotype Typhimurium LT2, but absent in Spanish Salmonella serotype 4,5,12:i:− isolates, were present in the genome sequence for the U.S. Salmonella serotype 4,5,12:i:− isolate CVM23701.
PCR-based characterization of gene deletion patterns in representative Salmonella serotype 4,5,12:i:− and Typhimurium isolates.
Based on our analyses of (i) the genomic microarray data reported by Garaizar et al. (18) and (ii) the Salmonella serotype 4,5,12:i:− isolate CVM23701 genome, we designed PCR primers to test for the presence of selected genes in clusters IV and V and adjoining regions (Table 2). We initially designed eight primer sets for genes that are at the junctions of clusters IV and V (as reported by Garaizar et al. [18]); these primers target STM2692, STM2694, STM2740, STM2741, STM2757, STM2758, STM2773 (iroB), and STM2774 (Fig. 1 shows primer locations). In addition, we designed primer sets for (i) two genes (fljA and fljB) absent from both the Salmonella serotype 4,5,12:i:− isolates from Spain (based on the genomic microarray data reported by Garaizar et al. [18]) and the CVM23701 genome and for (ii) one gene (hin) present in CVM23701 and absent in the Salmonella serotype 4,5,12:i:− isolates from Spain. We also designed one set of primers targeting a region found upstream of hin in only the CVM23701 genome; this region was designated the STM1053-1997 region, as the primers designed are located in genes with homology to STM1053 (forward primer) and STM1997 (reverse primer) (Fig. 1). PCR was performed on DNA purified using the QIAmp DNA mini kit (Qiagen Inc., Chartsworth) as detailed below, using either Ampli Taq Gold (Applied Biosystems, Foster City, CA) or Go Taq (Promega, Madison, WI).
TABLE 2.
PCR conditions and primers for six genes and one region that show variable presence among Salmonella serotype 4,5,12:i:− isolates from the United States and Spain
| Gene | Functiona | Amplicon size (bp) | Primers (5′ to 3′)b | Temp, °C (time) for: |
||
|---|---|---|---|---|---|---|
| Denaturation | Annealing | Extension | ||||
| fljA | Repressor of phase 1 flagellin gene | 642 | F, TTC ATT AGG TCC CCT CCG G; R, ATT CAG CCC CGT GAA TTC GGG | 95 (10 min) | 55 (45 s) | 72 (1 min) |
| fljB | Phase 2 flagellin structural protein | 561 | F, TTTACCGTCTACGCCACCC; R, GGTACTACACTGGATGTATCGGG | 95 (10 min) | 52 (45 s) | 72 (1 min) |
| hin | H inversion: regulation of flagellar gene expression | 570 | F, TGG CTA CTA TTG GGT ATA TTC GGG; R, AAT TCA TTC GTT TTT TTA TGC GGC | 95 (10 min) | 52 (45 s) | 72 (1 min) |
| STM1053-1997 | 614 | F, CCA TTT TTA TAC TGC CAG TCG CC; R, CAG CGA AAT ACT GAT GGC GG | 95 (10 min) | 55 (45 s) | 72 (1 min) | |
| STM2740 | Integrase, phage family | 980 | F, AAT GTG GAG ATC GCT GGC GCG; R, AGT TCG CCG CCG AAC CCC | 95 (2 min) | 55 (45 s) | 72 (1.5 min) |
| STM2757 | Putative cytoplasmic protein | 717 | F, ATG ATG ATG GCG TAA TGG CGC; R, AAA ACG TTC CGG TGC GGC G | 95 (10 min) | 55 (45 s) | 72 (1 min) |
| iroB | Glucosyltransferase homolog | 858 | F, TTC GAT TCG GAA GCG GGT TAT CGC CG; R, CTC GCG AAG CGC GCG | 95 (2 min) | 65-55 TDc (45 s) | 72 (1.5 min) |
Gene functions for Salmonella serotype Typhimurium LT2 were obtained from the J. Craig Venter Institute Comprehensive Microbial Resource website.
R, reverse primer; F, forward: primer.
TD, touchdown PCR; annealing temperatures decreased 0.5°C/cycle during the first 20 cycles, followed by 20 cycles at 55°C.
FIG. 1.
Deduced genome structure for the genomic region between STM2691 and STM2774 for four Salmonella serotype 4,5,12:i:− isolates from Spain (based on genomic microarray data reported by Garaizar et al. [18]) (A) and for the U.S. Salmonella serotype 4,5,12:i:− isolate CVM 23701 (based on an unfinished genome sequence reported by Rosovitz et al. [http://msc.jcvi.org/salmonella/salmonella_enterica_subsp__enterica_serovar_4__5__12_i___str__cvm23701/index.shtml]) (B). Genes are represented as open arrows or boxes; gene numbers (e.g., 2691) represent locus numbers based on primary annotation of Salmonella serotype Typhimurium LT2 (with the prefix “STM”). White represents the genes present in Salmonella serotype Typhimurium LT2 and all Salmonella serotype 4,5,12:i:− isolates, gray represents genes present in Salmonella serotype Typhimurium LT2 and absent from both U.S. and Spanish Salmonella serotype 4,5,12:i:− isolates, and a halftone pattern represents genes present in Salmonella serotype Typhimurium LT2 and Spanish Salmonella serotype 4,5,12:i:− isolates but absent from the U.S. Salmonella serotype 4,5,12:i:− isolate. Black represents a unique insertion in the U.S. Salmonella serotype 4,5,12:i:− isolate CVM 23701, which includes genes with full or partial homology with the Salmonella serotype Typhimurium LT2 genes STM1054 (94% homology with LT2 over 79% gene length), STM1053 (93% homology with LT2 over 85% gene length), STM1997 (92% homology with LT2 over 42% gene length), STM2704 (87% homology with LT2 over 100% gene length), and STM2706 (87% homology with LT2 over 18% gene length). hin and iroB, which are present in LT2 and the U.S. Salmonella serotype 4,5,12:i:− isolate, are also shown in black. Small arrows represent PCR primers, including five primer sets (see Table S2 in the supplemental material) used only for an initial screen of six isolates (two Salmonella serotype 4,5,12:i:− isolates each from Spain and the United States and one Salmonella serotype Typhimurium isolate each from Spain and the United States) (shown as thin black arrows) and seven primer sets (Table 2) used to screen a total of 59 isolates (shown as thick arrows).
All PCR primers were used initially to screen for gene presence/absence among four Salmonella serotype 4,5,12:i:− isolates (two each from Spain and the United States) as well as two Salmonella serotype Typhimurium isolates (one each from Spain and the United States). Subsequently, primers targeting six genes (i.e., STM2740, STM2757, fljA, fljB, hin, and iroB) and the STM1053-1997 region (Table 2 lists all primers) were used to screen for presence/absence of the selected genes among 59 representative isolates, representing Salmonella serotypes 4,5,12:i:− (41 isolates) and Typhimurium (14 isolates) as well as all four isolates with inconsistent serotype data (i.e., serotyped as Salmonella serotypes 4,5,12:i:− and Typhimurium). These isolates were selected to represent all PFGE types and STs found among the Spanish isolates. Isolates obtained in the United States were selected to represent the most common PFGE types found among different isolate sources (e.g., human, food, cattle, poultry, and nondomestic birds); for Salmonella serotype 4,5,12:i:−, isolates from the United States were selected to ensure inclusion of at least one representative of each ST and PFGE.
Nucleotide sequence accession number.
The representative sequence for Salmonella isolate FSL S9-102 has been deposited in GenBank with accession no. FJ763347.
RESULTS AND DISCUSSION
In order to better understand the evolution, ecology, and genetic characteristics of Salmonella serotype 4,5,12:i:−, we characterized 190 Salmonella serotype 4,5,12:i:− and Typhimurium isolates from the United States and Spain using a variety of molecular methods. Overall, our data indicate that (i) Salmonella serotypes 4,5,12:i:− and Typhimurium represent a highly clonal group, which can be differentiated by PFGE; (ii) U.S. and Spanish Salmonella serotype 4,5,12:i:− isolates show different patterns of gene deletion in the regions encoding phase 2 flagella and represent distinct PFGE patterns; and (iii) in addition to two common Salmonella serotype 4,5,12:i:− genotypes (designated here the “Spanish” and the “U.S.” Salmonella serotype 4,5,12:i:− clones), other 4,5,12:i:− genotypes exist. We thus conclude that Salmonella serotype 4,5,12:i:− most likely represents multiple clones that emerged through independent deletion events.
Salmonella serotypes 4,5,12:i:− and Typhimurium represent a highly clonal group, which can be differentiated by PFGE.
Among the 190 Salmonella isolates initially characterized as Salmonella serotypes Typhimurium (100 isolates) and 4,5,12:i:−, we identified six distinct STs based on a four-gene MLST scheme (Table 3). While the three-gene MLST scheme initially used had previously been shown to provide discriminatory power similar to that for a seven-gene MLST scheme (33), sequencing of a fourth gene (aroC) was included because the three-gene MLST allowed for only limited discrimination among the isolates used. Even with a four-gene MLST, a single ST (ST1) represented the vast majority of Salmonella serotype Typhimurium and 4,5,12:i:− isolates; 84 out of 100 Salmonella serotype Typhimurium and 85 out of 86 Salmonella serotype 4,5,12:i:− isolates were classified as ST1. Analyses of the relevant genes in the genomes of Salmonella strain LT2 and the U.S. Salmonella serotype 4,5,12:i:− isolate CVM23701 showed that these two strains also represent ST1. One Salmonella serotype 4,5,12:i:− isolate from Spain represented ST3; ST3 also represented seven U.S. Salmonella serotype Typhimurium isolates and one U.S. isolate with inconsistent serotype data (i.e., serotyped as Salmonella serotypes 4,5,12:i:− and Typhimurium in replicate experiments). ST3 differs from ST1 by only one nucleotide difference in manB. While Salmonella serotype 4,5,12:i:− represented only two STs (SID = 0.02), Salmonella serotype Typhimurium isolates represented six STs (SID = 0.29), indicating considerably higher ST diversity among the Salmonella serotype Typhimurium isolates characterized. Guerra et al. (21) previously also proposed that Salmonella serotype 4,5,12:i:− represents a lower diversity than Salmonella serotype Typhimurium, even though their molecular subtype study only used 16 Salmonella serotype 4,5,12:i:− and 2 Salmonella serotype Typhimurium isolates from Spain.
TABLE 3.
Distribution of four-gene STs among Salmonella isolates
| Four-gene STa | No. of isolates |
|||
|---|---|---|---|---|
| Salmonella serotype 4,5,12:i:− | Salmonella serotype Typhimurium | Inconsistent Salmonella serotypeb | Total | |
| 1 | 85 | 84 | 3 | 172 |
| 2 | 0 | 1 | 0 | 1 |
| 3 | 1 | 7 | 1 | 9 |
| 7 | 0 | 5 | 0 | 5 |
| 8 | 0 | 1 | 0 | 1 |
| 9 | 0 | 2 | 0 | 2 |
STs were based on ATs for partial fimA, mdh, manB, and aroC sequences.
These isolates were serotyped as Salmonella serotype 4,5,12:i:− in one replicate and Salmonella serotype Typhimurium in another replicate (including one isolate that was classified as Salmonella serotype 4,5,12:i:− in two replicates and Salmonella serotype Typhimurium in one replicate) and were thus designated “inconsistent.”
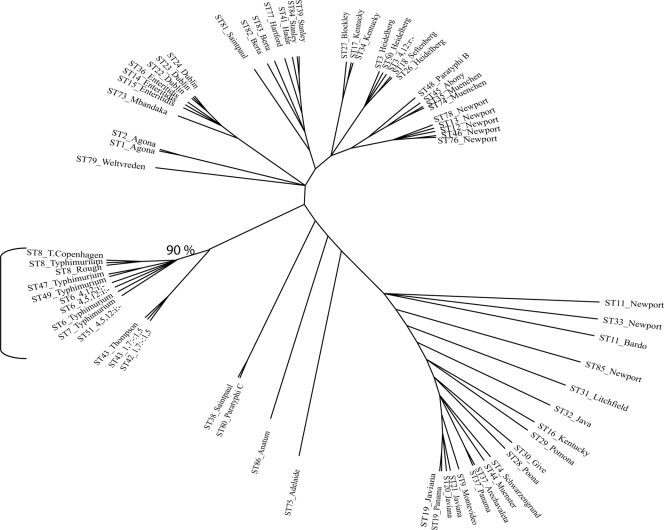
Phylogenetic analyses of three-gene MLST data (Fig. 2) also supported that Salmonella serotypes 4,5,12:i:− and Typhimurium are genetically closely related and highly clonal, as shown by the fact that all Salmonella serotype 4,5,12:i:− and Typhimurium STs form a single branch with strong bootstrap support. This observation is consistent with a number of studies (2, 14) that have shown that Salmonella serotype Typhimurium is highly clonal. The observation that all Salmonella serotype 4,5,12:i:− isolates characterized here share identical STs with Typhimurium isolates is consistent with a number of studies (see, e.g., references 1, 6, 13, 14, 16, and 38) that have shown, using different molecular subtyping methods (e.g., PFGE and MLST), that Salmonella serotype 4,5,12:i:− isolates are genetically and phenotypically closely related to Salmonella serotype Typhimurium. While according to serological characterization, Salmonella serotype 4,5,12:i:− is also closely related to Salmonella serotypes Lagos (4,5,12:i:1,5), Agama (4,12:i:1,6), Farsta (4,12:i:e,n,x), Tsevie (4,12:i:e,n,z15), Cloucester (1,5,12,27:i:l,w), and Tumodi (1,4,12:i:z6) and to an unnamed subspecies II serotype (4,5,27:i:z35) (28), we are not aware of any data indicating that any of these serotypes might be an ancestor of a Salmonella serotype 4,5,12:i:− strain. Echeita et al. (16) specifically reported that two genomic regions, i.e., a 1,000-bp fliB-fliA intergenic region and a 162-bp region specific for DT104 and DT U302 phage types, were absent in Salmonella serotype Lagos but present in Salmonella serotype Typhimurium phage types DT104 and DT U302, as well as in Spanish Salmonella serotype 4,5,12:i:− isolates; these data suggest that Spanish Salmonella serotype 4,5,12:i:− isolates are genetically closely related to Salmonella serotype Typhimurium DT104 and DT U302 and are unlikely to have originated from a Salmonella serotype Lagos ancestor. In our analysis of aroC ATs (including aroC ATs obtained from the Max Planck Institute MLST website at http://web.mpiib-berlin.mpg.de/mlst/dbs/Senterica), we also found that the aroC AT (i.e., AROC146) for the only Salmonella serotype Lagos isolate in this database was distinct from the aroC AT found among all Salmonella serotype 4,5,12:i:− isolates and all but one Salmonella serotype Typhimurium isolate characterized here (i.e., AT 10, which differs by four nucleotides from AT AROC146), further supporting that Salmonella serotype Lagos is unlikely to be the ancestor of Salmonella serotype 4,5,12:i:−. Similarly, the aroC AT for the one Salmonella serotype Agama isolate represented in the Max Planck Institute MLST database represents an aroC AT (AROC136), which is distinct from AT 10 (four nucleotide differences between AT 10 and AT AROC136). These data indicate that Salmonella serotype Agama is unlikely to be the ancestor of Salmonella serotype 4,5,12:i:−. While, overall, these data suggest that Salmonella serotype 4,5,12:i:− is a monophasic variant of Salmonella serotype Typhimurium, the rare Salmonella serotypes Farsta, Tsevie, Cloucester, Tumodi, and subspecies II serotype 4,5,27:i:z35 cannot be definitively excluded as ancestors of Salmonella serotype 4,5,12:i:− until isolates representing these serotypes have been characterized by molecular methods and compared to Salmonella serotype Typhimurium and 4,5,12:i:− isolates.
FIG. 2.
Phylogenetic tree for all three-gene (fimA, manB, and mdh) STs identified among Salmonella serotype 4,5,12:i:− and Typhimurium isolates as well as selected isolates representing other Salmonella serotypes (these STs were taken from reference 2). For each unique ST, fimA, manB, and mdh sequences were concatenated and aligned, followed by construction of an ML tree (100 bootstrap replicates), using the TrN+I+G model of evolution (identified by MODELTEST as the appropriate model for the data set used). While a number of nodes in this tree were supported by high bootstrap values, bootstrap support is shown only for the clade containing Salmonella serotype 4,5,12:i:− and Typhimurium isolates. Branch lengths do not represent phylogenetic distances.
As PFGE has been shown to be a highly discriminatory subtyping method for a number of Salmonella serotypes (1, 14, 38), we further characterized all 190 Salmonella isolates using PFGE with the enzyme XbaI. Overall, we identified 79 PFGE patterns (SID = 0.96) among all 190 isolates. A total of 29 and 50 PFGE types were differentiated among the 86 and 100 Salmonella serotype 4,5,12:i:− and Typhimurium isolates (SID = 0.92 and 0.93, respectively); the four isolates with inconsistent serotypes represented three different PFGE patterns. Overall, these data support previous studies which have shown that Salmonella serotype 4,5,12:i:− isolates represent considerable PFGE diversity (1, 21, 38) and that PFGE, in general, allows for more sensitive subtype discrimination among Salmonella isolates than MLST (12, 17, 22).
Interestingly, two PFGE patterns (P1 and P71) (Fig. 3) were shared by Salmonella serotype 4,5,12:i:− and Typhimurium isolates. PFGE type P1 was found in three Salmonella serotype 4,5,12:i:− and four Salmonella serotype Typhimurium isolates from the United States, while P71 represented four Salmonella serotype 4,5,12:i:− isolates and one Salmonella serotype Typhimurium isolate as well as one isolate with an inconsistent serotype, all isolated in the United States. These observations extend previous observations by de la Torre et al. (13) and Zamperini et al. (38). de la Torre et al. (13) showed that at least one XbaI type and one BlnI PFGE type were shared between Spanish Salmonella serotype 4,5,12:i:− and Typhimurium isolates, even though these two serotypes never shared the same combined XbaI/BlnI PFGE type (13). However, Zamperini et al. found one combined XbaI/BlnI PFGE type shared by one Salmonella serotype 4,5,12:i:− and one Salmonella serotype Typhimurium isolate, both isolated from poultry in the United States (38). In general, these observations further support that Salmonella serotype 4,5,12:i:− has evolved from a Salmonella serotype Typhimurium ancestor.
FIG. 3.
Representative XbaI PFGE patterns for Salmonella serotype 4,5,12:i:− and Typhimurium isolates as well as four isolates with inconsistent serotype data (i.e., isolates that were initially identified as Salmonella serotype 4,5,12:i:− but were classified as Salmonella serotype Typhimurium when they were resubmitted for serotyping). The PFGE types shown represent all 79 unique types found among the 190 isolates characterized. If identical PFGE types were found among isolates representing two serotypes, different sources (e.g., human and bovine), or different countries, one representative from each group was included in this figure; solid vertical lines indicate multiple isolates with identical PFGE patterns. For example PFGE pattern 28 (P28) was identified in five Salmonella serotype 4,5,12:i:− isolates from Spain and one Salmonella serotype 4,5,12:i:− isolate from a nondomestic bird in the United States. The number of isolates with a given PFGE type is indicated in parentheses after the PFGE type designation.
A total of four PFGE patterns (P12, P19, P35, and P45) were found among both Spanish and U.S. Salmonella serotype Typhimurium isolates. P12 represented 14 and 10 Salmonella serotype Typhimurium isolates from Spain and the United States, respectively. P35 represented two Salmonella serotype Typhimurium isolates from the United States and one Salmonella serotype Typhimurium isolate from Spain. PFGE patterns P19 and P45 each represented one Salmonella serotype Typhimurium isolate from the United States and one isolate from Spain. Identification of identical XbaI PFGE patterns among Salmonella serotype Typhimurium isolates from different continents is consistent with previous studies which have shown that some genetically closely related Salmonella strains are distributed worldwide (7, 19), including some other studies that have found Salmonella serotype Typhimurium isolates with identical PFGE types in different countries and continents (7). While PFGE patterns for most Spanish Salmonella serotype 4,5,12:i:− isolates were similar to each other and different from the patterns of U.S. Salmonella serotype 4,5,12:i:− isolates, one PFGE pattern (PFGE type P28) was shared among five Spanish Salmonella serotype 4,5,12:i:− isolates and one Salmonella serotype 4,5,12:i:− isolate from the United States. This U.S. isolate was obtained from a free-ranging owl in Georgia (38). As most owls are nonmigratory, there is no apparent hypothesis as to the source of an infection with a “Spanish clone” Salmonella serotype 4,5,12:i:− isolate in this animal.
U.S. and Spanish Salmonella serotype 4,5,12:i:− isolates show different patterns of gene deletion in the regions encoding phase 2 flagella and have different PFGE patterns.
A previous study of four multidrug-resistant Spanish Salmonella serotype 4,5,12:i:− isolates and Salmonella serotype Typhimurium LT2 identified five genomic regions (clusters) that were absent in all four Spanish Salmonella serotype 4,5,12:i:− isolates, but present in Salmonella serotype Typhimurium LT2. As our initial PFGE data indicated that Spanish Salmonella serotype 4,5,12:i:− isolates are genetically distinct from most U.S. Salmonella serotype 4,5,12:i:− isolates, we analyzed an available genome sequence for a U.S. Salmonella serotype 4,5,12:i:− isolate (strain CVM23701 [32]) for the presence of these five clusters (i.e., clusters I to V). BLAST searches against the CVM23701 genome sequences showed that cluster I (STM0517 to STM0529), which includes 13 genes, most of which are involved in allantoin-glyoxylate pathway-related functions, is present in the genome of this U.S. Salmonella serotype 4,5,12:i:− isolate, even though this cluster appears to be absent from the four Spanish Salmonella serotype 4,5,12:i:− isolates previously characterized by genomic microarrays (18). Cluster II (STM0893 to STM0929), which includes 35 Fels-1 prophage genes and two adjacent genes, was reported to be absent from the four Spanish Salmonella serotype 4,5,12:i:− isolates and was not identified in the available unfinished genome of the U.S. Salmonella serotype 4,5,12:i:− isolate. As the genes upstream and downstream of cluster II were located on a single contig, we conclude that this cluster is likely absent from the CVM23701 genome. These findings are consistent with the observation that the Fels-1 prophage is present in LT2 but typically is absent in other Salmonella serotype Typhimurium isolates (25). Cluster III (STM2616 to STM2617) encodes the Gifsy-1 prophage and was found in the genome of the U.S. Salmonella serotype 4,5,12:i:− isolate CVM23701, but it was reported to be absent from the four Spanish Salmonella serotype 4,5,12:i:− isolates previously characterized (18). The findings that clusters I to III were all absent from these four Spanish Salmonella serotype 4,5,12:i:− isolates while only cluster II was absent from the U.S. Salmonella serotype 4,5,12:i:− isolate CVM23701 provide initial evidence that U.S. and Spanish Salmonella serotype 4,5,12:i:− isolates may represent distinct genotypes.
Clusters IV (STM2694 to STM2740) and V (STM2758 to STM2773), which both were reported to be absent in the four Spanish Salmonella serotype 4,5,12:i:− isolates (Fig. 1A), are located in close proximity to each other. While cluster IV contains 47 Fels-2 prophage genes, cluster V contains a number of genes associated with different functions, including the fljAB operon (18). Notably, deletion of fljAB provides a functional explanation for the absence of phase 2 flagellar expression observed in Salmonella serotype 4,5,12:i:−, as fljB encodes the phase 2 flagellar protein and fljA encodes a repressor of fliC transcription (which encodes the phase 1 flagellar protein). Initial BLAST searches against the genome sequence for the U.S. Salmonella serotype 4,5,12:i:− isolate CVM23701 showed that both clusters IV and V were absent from the CVM23701 genome (cluster IV and V as well as intervening genes were located on a single CVM23701 contig, NZ_ABA001000014.1). Two genes located in the 3′ end of cluster V, including STM2772 (hin, encodes a recombinase that regulates the regulation of flagellar gene expression) and STM2773 (iroB, encodes glucosyltransferase homolog protein), were present in the CVM23701 genome, even though they were reported to be absent from the four Spanish Salmonella serotype 4,5,12:i:− isolates, based on genomic microarray data (18). Further analysis of the CVM23701 genome sequence indicated that the region between clusters IV and V (STM2739 to STM2757) was also absent from the CVM23701 genome, indicating that this strain contains a larger deletion than the four Spanish Salmonella serotype 4,5,12:i:− isolates; this deletion spans cluster IV and most of cluster V (except for two genes at the 3′ end) as well as the region between these two clusters. Interestingly, in the genome sequence of the Salmonella serotype 4,5,12:i:− isolate CVM23701, an approximately 7-kb region is inserted into this deleted section of the genome. This insertion includes two partial Fels-2 genes (STM2704 and STM2706) and three genes homologous to STM1054, STM1053, and STM1997 (umuC), which encode two Gifsy-2 prophage genes and a component of DNA polymerase V (umuC) (Fig. 1B). We will refer to this insertion as the “STM1053-1997” region; this region is not found in LT2. The presence of this region in CVM23701 suggests the intriguing hypothesis that deletion, in the U.S. Salmonella serotype 4,5,12:i:− clone, of clusters IV and V and the intervening region may have been caused by abortive, imprecise excision of a prophage.
As our analyses detailed above indicated that the U.S. Salmonella serotype 4,5,12:i:− isolate CVM23701 shows distinct genomic gene presence/absence patterns compared to four Spanish Salmonella serotype 4,5,12:i:− isolates previously characterized (18), we designed PCR primers to determine the absence/presence of eight genes that are at the junctions of clusters IV and V (i.e., STM2692, STM2694, STM2740, STM2741, STM2757, STM2758, STM2773 [iroB], and STM2774; Fig. 1 shows primer locations) and three genes (i.e., fljA, fljB, and hin) that are responsible for expression of phase 2 flagellar antigen. In addition, a set of primers was designed to allow for the detection of the “STM1053-1997” region, which was found in the CVM23701 genome sequence. While negative PCR results may indicate the absence of a gene or the presence of a distinct allelic variant of a gene, which does not allow for PCR amplification, we surmised that in this study, negative PCR results for Salmonella serotype 4,5,12:i:− due to gene diversification (rather than gene absence) are extremely unlikely due to the high genetic similarity between Salmonella serotype 4,5,12:i:− and Typhimurium isolates (e.g., as indicated by identical or highly similar MLST types for these two serotypes). Characterization of an initial six isolates (two Salmonella serotype 4,5,12:i:− isolates each from Spain and the United States and one Salmonella serotype Typhimurium isolate each from Spain and the United States) showed that the Spanish Salmonella serotype 4,5,12:i:− isolates had STM2692, STM2740, STM2741, STM2757, and STM2774 but lacked STM2694, STM2758, STM2773 (iroB), fljA, fljB, hin, and the STM1053-1997 region. These results confirmed the gene presence/absence patterns previously reported for four Spanish Salmonella serotype 4,5,12:i:− isolates (18), except for the fact that the PCR primers for STM2740, which was previously reported as absent in Spanish Salmonella serotype 4,5,12:i:−, yielded positive results, indicating the presence of at least part of this gene. The PCR results for the two U.S. Salmonella serotype 4,5,12:i:− isolates were consistent with the observations based on our analysis of the CVM23701 genome (representing a U.S. Salmonella serotype 4,5,12:i:− isolate). Specifically, the PCR data indicated that the two U.S. Salmonella serotype 4,5,12:i:− isolates (i) lack clusters IV and V as well as the intervening region (as supported by negative PCR results for STM2694, STM2740, STM2741, STM2757, STM2758, fljA, and fljB), (ii) contain hin and iroB (which are located in the 3′ end of cluster V and absent in the Spanish isolates), and (iii) contain an insertion upstream of the hin gene (i.e., the STM1053-1997 region), which is absent in the Spanish Salmonella serotype 4,5,12:i:− isolates. These data provided further support that Spanish Salmonella serotype 4,5,12:i:− isolates may be distinct from U.S. Salmonella serotype 4,5,12:i:− isolates.
To further test the hypothesis that Spanish and U.S. Salmonella serotype 4,5,12:i:− isolates represent different clonal groups with distinct genome deletion patterns, we screened 59 Salmonella serotype 4,5,12:i:− and Typhimurium isolates from these two countries (representing all PFGE patterns represented among 4,5,12:i:− isolates) for the presence/absence of six genes (i.e., STM2740, STM2757, fljA, fljB, hin, and iroB) and the STM1053-1997 region (Table 2). These PCR targets were selected because they (i) allow for clear differentiation of Salmonella serotype Typhimurium and 4,5,12:i:− genotypes and (ii) allow for differentiation of the “Spanish” and “U.S.” genomic deletion patterns in the cluster IV and V region of Salmonella serotype 4,5,12:i:− isolates. The PCR data generated clearly indicated that (i) all Spanish Salmonella serotype 4,5,12:i:− isolates show a deletion of clusters IV and V but presence of the intervening region (STM2740 to STM2757) and (ii) all but two U.S. Salmonella serotype 4,5,12:i:− isolates show a deletion of clusters IV and V, including a deletion of the intervening region between clusters IV and V, as well as presence of hin and iroB (which are absent in the Spanish Salmonella serotype 4,5,12:i:− isolates) and presence of the STM1053-1997 region. We thus propose that Salmonella serotype 4,5,12:i:− isolates from the United States and Spain represent two distinct clones (i.e., the “Spanish” and the “U.S.” clones). These findings are consistent with our observations that XbaI PFGE types of Spanish Salmonella serotype 4,5,12:i:− isolates generally are clearly distinct from the PFGE patterns for U.S. Salmonella serotype 4,5,12:i:− isolates. Interestingly, one Salmonella serotype 4,5,12:i:− isolate from the United States (isolated from a free-ranging owl in Georgia) had the same deletion pattern as Spanish Salmonella serotype 4,5,12:i:− isolates. As this isolate also shared an identical PFGE pattern (P28 [Fig. 3]) with five Spanish Salmonella serotype 4,5,12:i:− isolates, we also provide initial evidence for intercontinental spread of the “Spanish” Salmonella serotype 4,5,12:i:− clone.
Interestingly, Matiasovicova et al. (25) suggested that multidrug-resistant Salmonella serotype Typhimurium might have evolved from a Salmonella serotype Typhimurium ancestor that first lost the region including STM0517-0529 (designated cluster I by Garaizar et al. [18]), allowing the utilization of allantoin as a sole nitrogen source, followed by acquisition of the Salmonella genomic island 1, which includes genes responsible for multidrug resistance. Since multidrug-resistant Spanish Salmonella serotype 4,5,12:i:− isolates lack cluster I (18), while the U.S. Salmonella serotype 4,5,12:i:− (CVM23701) contains this cluster, one might hypothesize that Spanish Salmonella serotype 4,5,12:i:− strains might have emerged from multidrug-resistant Salmonella serotype Typhimurium, while U.S. Salmonella serotype 4,5,12:i:− might have emerged from non-drug-resistant Salmonella serotype Typhimurium through independent events. Future studies of larger sets of multidrug-resistant and pansusceptible Salmonella serotype Typhimurium and 4,5,12:i:− isolates from different countries will be needed, though, to test this hypothesis.
In addition to two common Salmonella serotype 4,5,12:i:− clones (i.e., the “Spanish” and the “U.S.” clones), we identified one rare Salmonella serotype 4,5,12:i:− genotype in North America.
In addition to the common “Spanish” and “U.S.” Salmonella serotype 4,5,12:i:− clones described above, we also identified one rare Salmonella serotype 4,5,12:i:− genotype in North America. Specifically, a human Salmonella serotype 4,5,12:i:− isolate from New York State (isolate FSL S5-635) (Table 4) was found to lack hin and the STM1053-1997 region, which are both present in the typical U.S. Salmonella serotype 4,5,12:i:− isolates, but contained iroB, which is typically absent in the Spanish clone. This isolate also was positive in the PCR assays targeting STM2741 and 2757, suggesting that this isolate did maintain the genomic region between clusters IV and V, which is present in the Spanish but absent in the U.S. Salmonella serotype 4,5,12:i:− clone. This isolate thus seems to be similar to the Spanish clone but shows a deletion pattern different from that of Spanish clone isolates at the 3′ end of cluster V (Table 4). Further characterization of this isolate will be needed to determine whether it represents a third emergence event, independent of both the emergence of the “Spanish” and the “U.S.” clones of Salmonella serotype 4,5,12:i:−, or whether it represents an evolutionary intermediate related to the Spanish Salmonella serotype 4,5,12:i:− clone. While this isolate represents a unique PFGE pattern not found among any other Salmonella serotype 4,5,12:i:− or Typhimurium isolates, it was classified as ST1, the same ST that represented the majority of Salmonella serotype Typhimurium isolates (84/100) as well as the majority of Spanish and U.S. clone Salmonella serotype 4,5,12:i:− isolates (12/13 and 73/73, respectively). This indicates that this strain most likely also emerged from a Salmonella serotype Typhimurium ancestor. Overall, our findings suggest that Salmonella serotype 4,5,12:i:− represents multiple genotypes, possibly indicating a strong selective pressure for loss of phase 2 flagellum expression.
TABLE 4.
Presence/absence of selected genes in isolates representing the Spanish and U.S. Salmonella serotype 4,5,12:i:− clones as well as other Salmonella isolates
| Gene | Presence of genes ina: |
||||
|---|---|---|---|---|---|
| U.S. Salmonella serotype 4,5,12:i:− clone (n = 30) | Spanish Salmonella serotype 4,5,12:i:− clone (n = 10) | FSL S5-635 (Salmonella serotype 4,5,12:i:−; n = 1) | Isolates with inconsistent serotype results (n = 4)b | Salmonella serotype Typhimurium (n = 14) | |
| STM2740 | − | + | + | + | + |
| STM2757 | − | + | + | + | + |
| STM1053-1997 | + | − | − | − | − |
| fljA | − | − | − | + | + |
| fljB | − | − | − | + | + |
| hin | + | − | − | + | + |
| iroB | + | − | + | + | + |
+ and − signs designate positive and negative PCR results, indicating the presence or absence of a gene, respectively.
These isolates were serotyped as Salmonella serotype 4,5,12:i:− in one replicate and Salmonella serotype Typhimurium in another replicate (including one isolate that was classified as Salmonella serotype 4,5,12:i:− in two replicates and Salmonella serotype Typhimurium in one replicate) and were thus designated “inconsistent.”
We also identified four isolates from the United States (FSL S9-102, FSL S9-165, FSL S9-166, and FSL R6-084) that were initially determined to be Salmonella serotype 4,5,12:i:− but were found in the PCR screens to contain fljA, fljB, and hin, three genes critical for phase 2 flagellar expression (Table 4). PCR screens for other genes in clusters IV and V indicated that both of these clusters were present in these four isolates. As Zamperini et al. (38) suggested that mutations in fljB (the gene encoding phase 2 flagella) may also cause a Salmonella serotype 4,5,12:i:− phenotype, we sequenced the 1,521-nucleotide fljB open reading frame in three of these isolates (isolates FSL S9-165 and FSL S9-166 showed the same PFGE type [P23], and thus fljB was sequenced for only one of these isolates). All of these isolates had an identical fljB sequence, which showed one synonymous single-nucleotide polymorphism compared to Salmonella serotype Typhimurium LT2; these isolates did not show any nonsynonymous changes or other mutations that would explain a lack of phase 2 flagellum expression. These four isolates were thus submitted to National Veterinary Service Laboratories for serotype confirmation. While isolates FSL S9-166 and FSL R6-084 were reserotyped as Salmonella serotype Typhimurium, FSL S9-102 was reserotyped twice, once as Salmonella serotype 4,5,12:i:− and once as Salmonella serotype Typhimurium, and FSL S9-165 was reserotyped as Salmonella serotype Typhimurium twice. These results are consistent with previous reports (26) that serotyping of Salmonella may sometimes be difficult to reproduce and suggest that Salmonella serotype Typhimurium isolates may sometimes be misclassified as Salmonella serotype 4,5,12:i:− (and vice versa). While the four specific isolates with inconsistent serotype results characterized here appear to represent Salmonella serotype Typhimurium (based on genetic evidence for the presence of intact phase 2 genes and at least one serotype result characterizing them as Salmonella serotype Typhimurium), it is tempting to speculate that these isolates may show reduced phase 2 flagellum expression, which could be responsible for the inconsistent serotype data. This hypothesis would need to be tested further by expression analyses (e.g., quantitative reverse transcription-PCR analysis).
Conclusions.
Overall, our observations suggest that Salmonella serotype 4,5,12:i:− evolved through multiple independent emergence events, most likely from Salmonella serotype Typhimurium ancestors. Salmonella serotype 4,5,12:i:− isolates from Spain and the United States appear to represent two different clones with distinct geographical distributions. This hypothesis is supported by multiple independent pieces of evidence. First, different genome-wide deletion patterns were found in four Spanish Salmonella serotype 4,5,12:i:− isolates (as previously determined by genomic microarrays [18]) and one U.S. Salmonella serotype 4,5,12:i:− isolate (based on an available whole genome sequence [32]). In particular, clusters I and III were present in the U.S. Salmonella serotype 4,5,12:i:− isolate (CVM23701), even though these clusters were reported to be absent in Spanish Salmonella serotype 4,5,12:i:− isolates (18). Second, genome analyses and PCR-based mapping showed clearly distinct deletion patterns in the genome region up- and downstream of the genes encoding proteins critical for phase 2 flagella and phase variation (i.e., fljA, fljB, and hin) in all Spanish and all but two U.S. Salmonella serotype 4,5,12:i:− isolates. Specifically, the Spanish isolates showed two deletions (of clusters IV and V), while the majority of U.S. isolates showed a single larger deletion (encompassing both clusters IV and V as well as the intervening region) with a 3′ junction different from that observed in the Spanish isolates. These findings provide another example of a Salmonella serotype of considerable public health relevance that represents at least two independent genetic lineages. For example, Salmonella serotype Newport has previously been shown to represent two distinct genetic lineages, including one lineage that contains predominantly pansusceptible isolates and one that contains predominantly multidrug-resistant isolates (2, 8, 22). In addition, the multiple independent emergences of Salmonella serotype 4,5,12:i:− and subsequent ecological success of multiple lineages (as evidenced by common isolation from human clinical cases in both Spain and the United States) suggest a strong selective pressure for loss of phase 2 flagella or a closely linked genotype. Future efforts to define the possible selection for loss of phase 2 flagella and to understand the specific Salmonella serotype 4,5,12:i:− genotypes circulating in countries other than the United States and Spain will be critical for understanding of the ecology and evolution of human disease-associated nontyphoidal Salmonella.
Findings of particular clinical relevance include the following: (i) PFGE allows for sensitive subtype discrimination of the emerging Salmonella serotype 4,5,12:i:−; (ii) Salmonella serotype 4,5,12:i:− appears to represent at least two common clones, which cannot easily be differentiated with standard diagnostic procedures (but can easily be discriminated with the PCR primers described here); and (iii) serological misclassification of Salmonella isolates as Salmonella serotype 4,5,12:i:− or Typhimurium may occasionally occur.
Supplementary Material
Acknowledgments
We thank Javier Garaizar, University of the Basque Country, Vitoria-Gasteiz, Spain, for providing Salmonella isolates from Spain and David White, FDA, for providing Salmonella food isolates.
Support for this project was provided by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract N01-AI-30054-ZC-006-07 (to Lorin Warnick). Andrea Moreno was supported by National Research Initiative Competitive Grant 2006-55212-17250 from the USDA Cooperative State Research, Education, and Extension Service Epidemiological Approaches to Food Safety program awarded to Randall Singer.
Footnotes
Published ahead of print on 9 September 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Agasan, A., J. Kornblum, G. Williams, C. C. Pratt, P. Fleckenstein, M. Wong, and A. Ramon. 2002. Profile of Salmonella enterica subsp. enterica (subspecies I) serotype 4,5,12:I:− strains causing food-borne infections in New York City. J. Clin. Microbiol. 40:1924-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaine, S. D., Y. Soyer, L. D. Warnick, W. L. Su, S. Sukhnanand, J. Richards, E. D. Fortes, P. McDonough, T. P. Root, N. B. Dumas, Y. Grohn, and M. Wiedmann. 2006. Multilocus sequence typing supports the hypothesis that cow- and human-associated Salmonella isolates represent distinct and overlapping populations. Appl. Environ. Microbiol. 72:7575-7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcaine, S. D., S. S. Sukhnanand, L. D. Warnick, W. L. Su, P. McGann, P. McDonough, and M. Wiedmann. 2005. Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob. Agents Chemother. 49:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldridge, P. D., C. Wu, J. Gnerer, J. E. Karlinsey, K. T. Hughes, and M. S. Sachs. 2006. Regulatory protein that inhibits both synthesis and use of the target protein controls flagellar phase variation in Salmonella enterica. Proc. Natl. Acad. Sci. USA 103:11340-11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez, J., M. Sota, A. B. Vivanco, I. Perales, R. Cisterna, A. Rementeria, and J. Garaizar. 2004. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 42:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amavisit, P., W. Boonyawiwat, and A. Bangtrakulnont. 2005. Characterization of Salmonella enterica serovar Typhimurium and monophasic Salmonella serovar 1,4,[5],12:I:− isolates in Thailand. J. Clin. Microbiol. 43:2736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baggesen, D. L., D. Sandvang, and F. M. Aarestrup. 2000. Characterization of Salmonella enterica serovar Typhimurium DT104 isolated from Denmark and comparison with isolates from Europe and the United States. J. Clin. Microbiol. 38:1581-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berge, A. C., J. M. Adaska, and W. M. Sischo. 2004. Use of antibiotic susceptibility patterns and pulsed-field gel electrophoresis to compare historic and contemporary isolates of multi-drug-resistant Salmonella enterica subsp. Enterica serovar Newport. Appl. Environ. Microbiol. 70:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boonmar, S., A. Bangtrakulnonth, S. Pornruangwong, S. Samosornsuk, K. Kaneko, and M. Ogawa. 1998. Significant increase in antibiotic resistance of Salmonella isolates from human beings and chicken meat in Thailand. Vet. Microbiol. 62:73-80. [DOI] [PubMed] [Google Scholar]
- 10.CDC. 2008. Salmonella surveillance: annual summary, 2006. CDC, Department of Health and Human Services, Atlanta, GA.
- 11.Chevance, F. F., and K. T. Hughes. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke, F. J., D. J. Brown, M. Fookes, D. Pickard, A. Ivens, J. Wain, M. Roberts, R. A. Kingsley, N. R. Thomson, and G. Dougan. 2008. Characterization of the genomes of a diverse collection of Salmonella enterica serovar Typhimurium definitive phage type 104. J. Bacteriol. 190:8155-8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Torre. E., D. Zapata, M. Tello, W. Mejia, N. Frias, F. J. Garcia Pena, E. M. Mateu, and E. Torre. 2003. Several Salmonella enterica subsp. enterica serotype 4,5,12:I:− phage types isolated from swine samples originate from serotype Typhimurium DT U302. J. Clin. Microbiol. 41:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado Ronda, N., J. L. Muñoz Bellido, M. I. García García, R. Ibáñez Pérez, S. Muñoz Criado, R. Serrano Heranz, M. C. Sáenz González, and J. A. García Rodríguez. 2006. Molecular epidemiology of drug-resistant Salmonella Typhimurium in Spain. Rev. Esp. Quimioterap. 19:152-160. [PubMed] [Google Scholar]
- 15.Echeita, M. A., A. Aladuena, S. Cruchaga, and M. A. Usera. 1999. Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:I:− strain in Spain. J. Clin. Microbiol. 37:3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Echeita, M. A., S. Herrera, and M. A. Usera. 2001. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:I:− appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 39:2981-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakhr, M. K., L. K. Nolan, and C. M. Logue. 2005. Multilocus sequence typing lacks the discriminatory ability of pulsed-field gel electrophoresis for typing Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 43:2215-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garaizar, J., S. Porwollik, A. Echeita, A. Rementeria, S. Herrera, R. M. Y. Wong, J. Frye, M. A. Usera, and M. McClelland. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatto, A. J., T. M. Peters, J. Green, I. S. Fisher, O. N. Gill, J. S. O'Brien, C. Maguire, C. Berghold, I. Lederer, P. Gerner-Smidt, M. Torpdahl, A. Siitonen, S. Lukinmaa, H. Tschape, R. Prager, I. Luzzi, A. M. Dionisi, W. K. van der Zwaluw, M. Heck, J. Coia, D. Brown, M. Usera, A. Echeita, and E. J. Threlfall. 2006. Distribution of molecular subtypes within Salmonella enterica serotype Enteritidis phage type 4 and S. Typhimurium definitive phage type 104 in nine European countries, 2000-2004: results of an international multi-centre study. Epidemiol. Infect. 134:729-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimont, P., and F. Weil. 2007. Antigenic formulae of the Salmonella servovars, 9th ed. World Health Organization Centre for Reference and Research on Salmonella, Pasteur Institute, Paris, France.
- 21.Guerra, B., I. Laconcha, S. M. Soto, M. A. Gonzalez-Hevia, and M. C. Mendoza. 2000. Molecular characterisation of emergent multiresistant Salmonella enterica serotype [4,5,12:I:−] organisms causing human salmonellosis. FEMS Microbiol. Lett. 190:341-347. [DOI] [PubMed] [Google Scholar]
- 22.Harbottle, H., D. G. White, P. F. McDermott, R. D. Walker, and S. Zhao. 2006. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J. Clin. Microbiol. 44:2449-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matiasovicova, J., P. Adams, P. A. Barrow, H. Hradecka, M. Malcova, R. Karpiskova, E. Budinska, L. Pilousova, and I. Rychlik. 2007. Identification of putative ancestors of the multidrug-resistant Salmonella enterica serovar Typhimurium DT104 clone harboring the Salmonella genomic island 1. Arch. Microbiol. 187:415-424. [DOI] [PubMed] [Google Scholar]
- 26.McQuiston, J. R., R. Parrenas, M. Ortiz-Rivera, L. Gheesling, F. Brenner, and P. I. Fields. 2004. Sequencing and comparative analysis of flagellin genes fliC, fljB, and flpA from Salmonella. J. Clin. Microbiol. 42:1923-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno Switt, A. I., Y. Soyer, L. D. Warnick, and M. Wiedmann. 2009. Emergence, distribution and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:I:−. Foodborne Pathog. Dis. 6:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossong, J., P. Marques, C. Ragimbeau, P. Huberty-Krau, S. Losch, G. Meyer, G. Moris, C. Strottner, W. Rabsch, and F. Schneider. 2007. Outbreaks of monophasic Salmonella enterica serovar 4,[5],12:I:− in Luxembourg, 2006. Euro. Surveill. 12:E11-E12. [DOI] [PubMed] [Google Scholar]
- 30.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 31.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 32.Rosovitz, M. J., P. McDermott, D. White, J. E. LeClerc, M. K. Mammel, T. A. Cebula, and J. Ravel. Annotation of Salmonella enterica subsp. enterica serovar 4,[5],12:I:− str. CVM23701. The Institute for Genomic Research, Rockville, MD. http://msc.jcvi.org/Salmonella/Salmonella_enterica_subsp__enterica_serovar_4__5__12_i___str__cvm23701/index.shtml.
- 33.Sukhnanand, S., S. Alcaine, L. D. Warnick, W. L. Su, J. Hof, M. P. Craver, P. McDonough, K. J. Boor, and M. Wiedmann. 2005. DNA sequence-based subtyping and evolutionary analysis of selected Salmonella enterica serotypes. J. Clin. Microbiol. 43:3688-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaminathan, B., P. Gerner-Smidt, and T. Barrett. 2006. Focus on Salmonella. Foodborne Pathog. Dis. 3:154-156. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 2002. Paup*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 36.Tavechio, A. T., A. C. Ghilardi, and S. A. Fernandes. 2004. “Multiplex PCR” identification of the atypical and monophasic Salmonella enterica subsp. enterica serotype 1,4,[5],12:I:− in Sao Paulo State. Brazil: frequency and antibiotic resistance patterns. Rev. Inst. Med. Trop. Sao Paulo 46:115-117. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto, S., and K. Kutsukake. 2006. fljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:958-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamperini, K., V. Soni, D. Waltman, S. Sanchez, E. C. Theriault, J. Bray, and J. J. Maurer. 2007. Molecular characterization reveals Salmonella enterica serovar 4,[5],12:I:− from poultry is a variant Typhimurium serovar. Avian Dis. 51:958-964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.