Abstract
The genomic fine-typing of strains of Mycobacterium ulcerans, the causative agent of the emerging human disease Buruli ulcer, is difficult due to the clonal population structure of geographical lineages. Although large sequence polymorphisms (LSPs) resulted in the clustering of patient isolates originating from across the globe, differentiation of strains within continents using conventional typing methods is very limited. In this study, we analyzed M. ulcerans LSP haplotype-specific insertion sequence elements among 83 M. ulcerans strains and identified single nucleotide polymorphisms (SNPs) that differentiate between regional strains. This is the first genetic discrimination based on SNPs of M. ulcerans strains from African countries where Buruli ulcer is endemic, resulting in the highest geographic resolution of genotyping so far. The findings support the concept of genome-wide SNP analyses as tools to study the epidemiology and evolution of M. ulcerans at a local level.
Mycobacterium ulcerans causes the devastating cutaneous disease Buruli ulcer (BU). More than 30 countries worldwide have reported this emerging disease, reaching epidemic proportions in some areas, and children between the ages of 5 and 15 in the rural wetlands of West Africa are most affected (38). Although proximity to marshes and wetlands is a risk factor, the mode of transmission remains an enigma (10, 26, 27, 36, 37). Discrimination of genetic variants has become an indispensable tool to unravel the evolution, epidemiology, and transmission of pathogenic organisms and to gain insight into host-pathogen interactions (6, 13, 24). In M. ulcerans, such elucidation is impossible due to a remarkable lack of genetic diversity on a local geographic scale (22). Conventional genetic differentiation tools commonly used for phylogenetic profiling in Mycobacterium tuberculosis, such as restriction fragment length polymorphism, amplified fragment length polymorphism, variable-number tandem repeats (VNTR), and multilocus sequence typing, could distinguish between continental lineages only when applied to M. ulcerans (1-4, 7, 8, 15, 18, 29, 31, 34, 35). However, two publications using VNTRs reported the first discrimination of strains between and within African countries (16, 33). The identification of regions of difference (RDs) in a worldwide collection of M. ulcerans isolates led to an evolutionary scheme on the continental level, with two distinct genetic lineages that can be subgrouped into six haplotypes (20, 28). Strains of the “ancestral” lineage are genetically closer to Mycobacterium marinum, the progenitor of M. ulcerans, whereas the “classical” lineage accounts for the majority of BU cases and represents the most virulent genotype. Characterization of the large sequence polymorphisms (LSPs) showed that insertion sequence (IS) element (ISE) expansion is associated with the observed genome instability (17, 19, 40). ISs are compact mobile DNA segments capable of inserting at multiple sites in a target molecule, usually by a recombinase that is encoded by a coding sequence (CDS) contained within the ISE itself (23). Thus, uncontrolled duplications and insertions of ISEs occur at relatively high frequency in replicating bacteria, leading to genomic insertions, deletions, and rearrangements that have the potential for molecular epidemiological applications. In M. tuberculosis, until recently, IS-mediated insertions/deletions (InDels) used to be the principal source of genome plasticity (6) and are widely used as evolutionary markers in epidemiological studies. For M. ulcerans, two ISEs were defined, IS2404 and IS2606 (30, 32). Earlier, site-specific IS2404 elements were identified to be unique for and confined to distinct M. ulcerans haplotypes (19). Here, we specifically amplified such unique ISEs and compared their sequences for a collection of 83 M. ulcerans isolates including 67 derived from Africa. We aimed at the detection of single nucleotide polymorphisms (SNPs) in these ISEs that made genetic distinction within haplotypes and on a regional level possible.
MATERIALS AND METHODS
Bacterial strains.
Isolates used for SNP identification with their country origin are listed in Table 1 .
TABLE 1.
M. ulcerans strains used in this study
| Geographic regiona | Strain identifierb | Year of isolationa |
|---|---|---|
| Ghana, Greater Accra region | Agy99 | 1999 |
| Ghana, Greater Accra region | NM14/02 | 2002 |
| Ghana, Greater Accra region | NM18/02 | 2002 |
| Ghana, Greater Accra region | NM19/02 | 2002 |
| Ghana, Greater Accra region | NM20/02 | 2002 |
| Ghana, Greater Accra region | NM21/02 | 2002 |
| Ghana, Greater Accra region | NM22/02 | 2002 |
| Ghana, Greater Accra region | NM23/02 | 2002 |
| Ghana, Greater Accra region | NM27/02 | 2002 |
| Ghana, Greater Accra region | NM28/02 | 2002 |
| Ghana, Greater Accra region | NM30/02 | 2002 |
| Ghana, Greater Accra region | NM32/02 | 2002 |
| Ghana, Greater Accra region | NM33/02 | 2002 |
| Ghana, Greater Accra region | NM34/02 | 2002 |
| Ghana, Greater Accra region | NM37/02 | 2002 |
| Ghana, Greater Accra region | NM38/02 | 2002 |
| Ghana, Greater Accra region | NM40/02 | 2002 |
| Ghana, Greater Accra region | NM41/02 | 2002 |
| Ghana, Greater Accra region | NM43/02 | 2002 |
| Ghana, Greater Accra region | NM44/02 | 2002 |
| Ghana, Greater Accra region | NM46/02 | 2002 |
| Ghana, Greater Accra region | NM47/02 | 2002 |
| Ghana, Greater Accra region | NM48/02 | 2002 |
| Ghana, Greater Accra region | NM49/02 | 2002 |
| Ghana, Greater Accra region | NM50/02 | 2002 |
| Ghana, Greater Accra region | NM51/02 | 2002 |
| Ghana, Greater Accra region | NM52/02 | 2002 |
| Ghana, Greater Accra region | NM54/02 | 2002 |
| Ghana, Greater Accra region | NM56/02 | 2002 |
| Ghana, Greater Accra region | NM59/02 | 2002 |
| Ghana, Greater Accra region | NM60/02 | 2002 |
| Ghana, Greater Accra region | NM61/02 | 2002 |
| Ghana, Greater Accra region | NM62/02 | 2002 |
| Ghana, Greater Accra region | NM68/02 | 2002 |
| Ghana, Greater Accra region | NM69/02 | 2002 |
| Ghana, Greater Accra region | NM72/03 | 2003 |
| Ghana, Greater Accra region | NM74/03 | 2003 |
| Ghana, Greater Accra region | NM76/03 | 2003 |
| Ghana, Greater Accra region | NM77/03 | 2003 |
| Ghana, Greater Accra region | NM78/03 | 2003 |
| Ghana, Greater Accra region | NM89/03 | 2003 |
| Ghana, Greater Accra region | NM90/03 | 2003 |
| Ghana, Greater Accra region | NM97/03 | 2003 |
| Ghana, Greater Accra region | NM102/03 | 2003 |
| Ghana, Greater Accra region | NM103/03 | 2003 |
| Ghana, Ashanti region | ITM GH970321 | 1997 |
| Ghana, Ashanti region | ITM GH970359 | 1997 |
| Ghana, Eastern region | NM53/02 | 2002 |
| Ghana, region ND | NM01/03 | 2003 |
| Ghana, region ND | NM06/03 | 2003 |
| Ghana, region ND | NM94/03 | 2003 |
| Ghana, region ND | NM95/03 | 2003 |
| Ghana, region ND | NM98/03 | 2003 |
| Ghana, region ND | ITM GH970483 | 1997 |
| Angola | ITM AN960658 | 1996 |
| Ivory Coast | ITM IC940511 | 1994 |
| Ivory Coast | ITM IC940815 | 1994 |
| Ivory Coast | ITM IC940662 | 1994 |
| Democratic Republic of Congo | ITM DRC5150 | 1962 |
| Democratic Republic of Congo | ITM DRC5151 | ND |
| Democratic Republic of Congo | ITM DRC5155 | 1976 |
| Togo | ITM TOGO970680 | 1997 |
| Benin | ITM BEN970104 | 1997 |
| Benin | ITM BEN970111 | 1997 |
| Benin | ITM BEN940512 | 1994 |
| Benin | ITM BEN940886 | 1994 |
| Benin | ITM BEN001441 | 2000 |
| Papua New Guinea | ITM PNG9357 | ND |
| Papua New Guinea | ITM PNG941331 | 1994 |
| Malaysia | ITM MALAY941328 | 1994 |
| Australia, Queensland | ITM AU941324 | 1994 |
| Australia, Victoria | ITM AU5142 | 1967 |
| Australia, Victoria | ITM AU5147 | ND |
| Australia, Victoria | ITM AU9550 | ND |
| Australia, Victoria | ITM AU940339 | 1994 |
| Australia, region ND | ITM AU8849 | ND |
| Australia, region ND | ITM AU9549 | ND |
| Australia, region ND | ITM AU941325 | 1994 |
| Australia, region ND | ITM AU941327 | 1994 |
| Surinam | ITM SU842 | 1986 |
| French Guiana | ITM FG7922 | 1990 |
| China | ITM CH980912 | 1998 |
| Japan | ITM JP9756 | 1998 |
ND, not defined.
Genomic DNA preparation.
Genomic DNA from clinical and environmental isolates was extracted from bacterial pellets using an optimized method for mycobacterial DNA preparation (21). Bacterial pellets of about 20 mg (wet weight) were heat inactivated for 1 h at 95°C, followed by cell wall disruption and digestion. DNA was extracted from the supernatants by phenol-chloroform (Fluka, Buchs, Switzerland) extraction and subjected to ethanol precipitation as described previously (21). DNA was measured by the optical density at 260 nm using a NanoDrop 1000 spectrophotometer (Thermo Fisher, Waltham, MA).
Amplification, sequencing, and alignment of haplotype-specific IS2404 elements.
Versions of IS2404 were selected that are haplotype specific. For the African/Australian haplotypes, the primer pair MK323 (GCGGTACAAGCTTCCCAAAG) and MK814 (AGCCAGAGCTTTGGATTTGA) was applied to yield a PCR product of 2 kb comprising IS2404 (MUL_3871) in RD12, and the pair MK809 (GGTGCTTAACGAAACGTGCT) and MK808 (ACGAAATCGAATTCCTCGTG) was used to yield a PCR product of 2 kb comprising IS2404 (MUL_2990) in RD1. Primers MK808 and MK809 amplified a 360-bp PCR fragment of glnA3 lacking IS2404 in the South American and Asian haplotypes. The primer pair MK382 (GATCCTCGATCCGGTGTTC) and MK410 (GGATCTCCACCTTCGTCAAC) amplified a specific IS2404 element within RD9 confined to the South American haplotype, and primer pair MK892 (GCAATGTGATGCACAACCTC) and MK650 (CGTTCGATTTCACCTCACC) amplified a specific IS2404 element within RD11 unique for the Asian haplotype. Sequencing of the respective PCR products was done using the primers used for the PCR and the IS2404-specific internal primers MK661 (GATTGGTGCTCGGTCAACTC), MK662 (TCAGGTAGTGCGACTTCAAGG), MK663 CAGCGTGGAGGTGGTCTATG), and SR685 (AGGCCAACACATCGAGAAAC) to cover the entire amplicon. PCR was performed using the FirePol 10× BD buffer and 0.5 μl of FirePol Taq polymerase (Solis BioDyne, Tartu, Estonia) with 5 ng of genomic DNA, 0.6 μM (each) of forward and reverse primer, 1.7 mM MgCl2, and a 0.3 mM concentration of each deoxynucleoside triphosphate in a total volume of 30 μl. PCRs were run in a GeneAmp PCR System 9700 PCR machine. The thermal profile for PCR amplification of Escherichia coli plasmids and M. ulcerans genomic DNA included an initial denaturation step of 95 to 98°C for 3 min, followed by 32 cycles of 95°C for 20 s, annealing at 58 to 65°C for 20 s, and elongation at 72°C for 30 s up to 2 min. The PCRs were finalized by an extension step at 72°C for 10 min. PCR products were analyzed on 1% agarose gels by gel electrophoresis using ethidium bromide staining and an AlphaImager illuminator (Alpha Innotech, San Leandro, CA). PCR amplicons were purified using a NucleoSpin purification kit (Machery-Nagel, Düren, Germany) and subjected to direct sequencing by Macrogen, Seoul, South Korea. Primers (Sigma-Aldrich, Steinheim, Germany) were designed using Primer3 software, version 0.4.0 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). All sequences were subjected to multiple sequence alignments using the ClustalW2 tool of the European Molecular Biology Library-European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/clustalw2/index.html) for phylogenetic analysis.
RESULTS
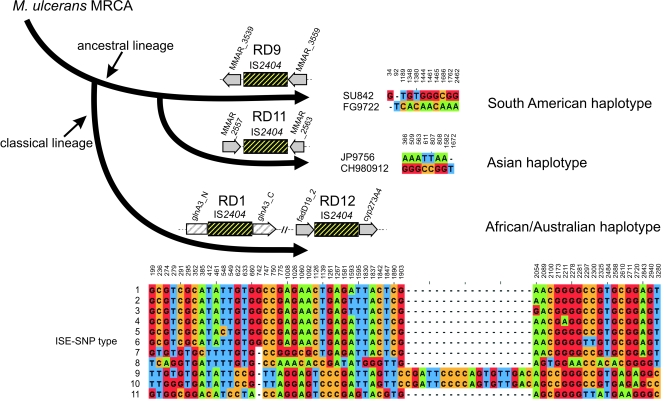
We selected copies of four IS2404 elements that were earlier identified to be confined to one specific haplotype only (19). In RD9, an IS2404 element was inserted in between orthologues of the partial CDSs, MMAR_3539 and MMAR_3559, in only the South American haplotype (Fig. 1). In RD11, an IS2404 element was inserted in between the orthologues of the CDSs, MMAR_2557 and MMAR_2563 in only the Asian haplotype (Fig. 1). The two IS2404 elements in RD1 (MUL_2990) and RD12 (MUL_3871) were confined to the classical lineage (Fig. 1), represented by the African/Australian isolates. Primers flanking the respective ISEs and specific for the haplotype-specific constellation of the respective RDs were used to specifically amplify these four IS2404 element-containing loci without contamination by the many other copies of ISEs present in the M. ulcerans genomes. The RD9-associated IS2404 was amplified for two South American strains, and the RD11-associated IS2404 was amplified for two Asian strains, each belonging to the ancestral lineage. The two IS2404 versions associated with RD1 and RD12 were amplified for 79 M. ulcerans strains belonging to the classical lineage (Fig. 1). Fifty-four of these strains originated from Ghana, and 13 more were from other West and Central African countries; 12 strains derived from Australia, Malaysia, and Papua New Guinea. The chromosomal context within each haplotype was identical for all strains (data not shown), i.e., the nucleotide composition at the breakpoints, the adjacent regions, and the deleted DNA stretches associated with the ISE insertion (in RD9 and RD11). The IS2404 elements were between 1,362 and 1,367 bp long.
FIG. 1.
Sequence variation in haplotype-specific IS2404 elements. IS2404 elements that are unique for, and confined to, the shown haplotypes were amplified with site-specific primers located outside of the ISEs, and SNPs were determined by sequencing. Only sites that differ in the aligned nucleotides (European Bioinformatics Institute ClustalW2) are shown in the sequence panels (JalView, version 2.4) (9) from the 1,362-bp (RD9), 1,366-bp, and adjacent regions (RD11) and from 1,497 bp (RD1) combined with 1,822 bp (RD12) to yield 3,319 bp of concatenated chromosomal ISE fragments, including their flanking regions. SNP position numbers in RD1 and RD12 are given according to the M. ulcerans Agy99 sequence, with 1 corresponding to position 3313231 in RD1 and 1498 to position 4326896 in RD12. SNP position numbers in RD9 and RD11 are given according to the M. marinum M sequence, with 1 corresponding to position 4348015 in RD9 and 1 corresponding to position 3108032 in RD11. For the 79 sequenced M. ulcerans isolates of the African/Australian haplotype, 11 various ISE-SNP types (1 to 11) were identified, as indicated in the sequence panel. MRCA, most recent common ancestor.
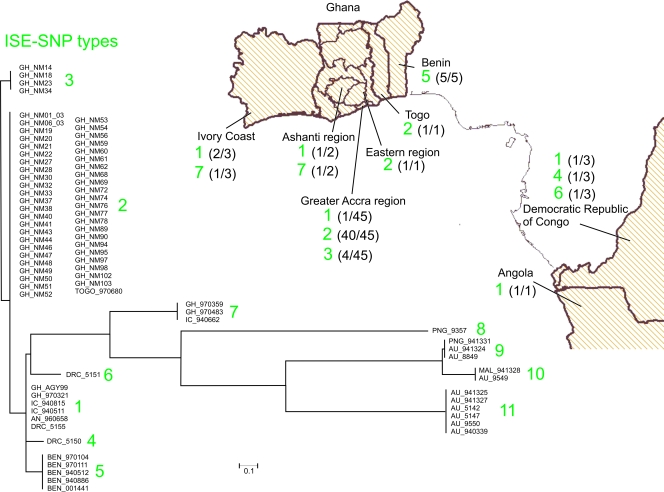
Within the South American haplotype, the patient isolate originating from Surinam could be distinguished by 11 SNPs (including one gap) from the one from French Guiana (Fig. 1). The two strains from Japan and China, belonging to the Asian haplotype, could be distinguished by eight SNPs (including one gap) from each other (Fig. 1). Within the African/Australian haplotype for the two RD9- and RD11-associated ISEs, altogether 72 variable nucleotide positions were found that cluster the 79 strains into 11 groups that we designated ISE-SNP types (Fig. 1). When these SNPs are linked to epidemiological data (Fig. 2), we found that the West African region from the Ivory Coast to Togo harbors a mixture of ISE-SNP types. All four patient isolates from Benin showed identical nucleotide sequences defined by one common SNP (T548C). This polymorphism is shared by the environmental strain BEN001441 (25). We found four genotypes represented within southern Ghana, with a majority having ISE-SNP type 2. The ISE-SNP type 1 cluster contains the sequenced reference strain Agy99 along with another strain from Ghana as well as isolates from the Democratic Republic of Congo, the Ivory Coast, and Angola. ISE-SNP type 3 from within the Greater Accra region in Ghana comprises four identical strains that differ in one SNP (T-A) from other M. ulcerans isolates of the same area. Two strains from Ghana and one from the Ivory Coast (forming ISE-SNP type 7) are, with respect to the ISE-SNP type, quite distinct from the remainder of the African isolates but were found in closer genetic proximity to strains from Southeast Asia and Australia (ISE-SNP types 8 to 11) (Fig. 2). Within the Australian isolates, the insertion of 18 nucleotides in five strains (Fig. 1, ISE-SNP types 9 and 10) has probably emerged through homologous recombination of an internal part of another IS2404 fragment in a common progenitor, grouping these strains originating from Papua New Guinea, Australia, and Malaysia together. However, other M. ulcerans isolates from southern Australia, i.e., Victoria (ISE-SNP type 11), do not have this small insert, resulting in two different clusters within Australia.
FIG. 2.
Phylogenetic and epidemiological patterns of the ISE-SNP types in the classical lineage. Identified SNPs were used to create a neighbor-joining tree (using percent identity) with JalView, version 2.4 (9), for visualization of their phylogenetic relationships. Clusters were assigned ISE-SNP type numbers (1 to 11, corresponding to Fig. 1). The geographic origins of the African strains are shown on the map (created using HealthMapper, version 4.3.1, software [http://www.who.int/health_mappping/tools/healthmapper/en/index html] with the permission of the WHO Geographic Information Systems department). Frequencies of the respective ISE-SNP type per geographic sample size are indicated in brackets for each country or region. Genetic distances (legend) are reflected by the lengths of the branches. GH, Ghana; IC, Ivory Coast; DRC, Democratic Republic of Congo; AN, Angola; BEN, Benin; PNG, Papua New Guinea; AU, Australia; MAL, Malaysia.
Here, we defined ISE-SNP types in Africa that seem to be either geographically clustered (e.g., ISE-SNP types 5 and 6) or more widespread (e.g., ISE-SNP types 1 and 7). These genotypes unveil a clearer picture of M. ulcerans dispersal and epidemiology in Africa and on a worldwide scale.
DISCUSSION
Characterization of InDel diversity among a worldwide collection of M. ulcerans strains by comparative genomic hybridization analysis (20) has yielded markers for the investigation of the phylogeography of M. ulcerans patient isolates on a global scale. Continental haplotypes with unique constellations in particular RDs were defined (19, 20). Here, we combine the strength of lineage-specific unequivocal genetic InDel markers with the high-resolution power of SNPs. We now determined the nucleotide sequence of RD-associated haplotype-specific copies of IS2404 and identified SNPs, allowing further subdivision of continental lineages. In particular, within the classical lineage, sequence analysis of the RD9- and RD11-associated IS2404 elements yielded 11 SNP types (ISE-SNP types) across a panel of 79 M. ulcerans strains. Since the two selected ISEs are identical in their chromosomal context across the tested classical lineage strains, the haplotype-specific insertions in RD9 and RD11 must have occurred in a common ancestor, and accumulation of SNPs represents secondary events. Since IS2404 is highly redundant in M. ulcerans, the occurrence of point mutations, whether synonymous or nonsynonymous in nature, is irrelevant for the microbe's biology.
The resolution of ISE-SNP typing is higher than that achieved with other DNA fingerprinting techniques: ISE-SNP types correlated with the more limited VNTR/ mycobacterial interspersed repetitive unit-VNTR fine-typing and, in particular, enhanced the resolution within the Atlantic African genotype (16, 33). Some ISE-SNP types seem to be widespread across West African countries (e.g., ISE-SNP types 1, 2, 3, and 7 in the Ivory Coast, Ghana, Togo, the Democratic Republic of the Congo, and Angola). Others appear more delimited (such as ISE-SNP type 5 in Benin and types 4 and 6 in the Democratic Republic of the Congo). Among M. ulcerans isolates from Ghana, four ISE-SNP types (1, 2, 3, and 7) were identified. The retrieved phylogenetic tree (Fig. 2) depicts the highest resolution of M. ulcerans phylogeny within and between continents. The ISE-SNP type analysis revealed genetic relatedness of a subgroup of African strains (ISE-SNP type 7 from Ghana and the Ivory Coast) to the Southeast Asian/Australian clusters (ISE-SNP types 8 through 11). The latter indicates the possible link of an origin from common ancestors of ISE-SNP type 7 to Australian M. ulcerans strains. Interestingly, the only M. ulcerans isolate ever cultivated from the environment and originating from Benin (25) showed the same ISE-SNP type as the patient isolates coming from the same country, supporting the current hypothesis that infection with BU disease results from environmental exposure.
Within the M. tuberculosis complex, and even within M. ulcerans-related mycolactone-producing mycobacteria, analysis of LSPs represents a valuable approach for genetic fingerprinting (5, 6, 12, 18). However, with the increasing availability of multiple whole-genome sequences, SNP identification adds considerably to phylogeographic analyses (11, 13, 14). We conclude that also for M. ulcerans, SNP typing rather than analysis of LSPs will yield sufficient resolution for microepidemiological studies. The resolution obtained here for the classical lineage is thus far based on only two copies of IS2404. Analysis of a larger number of ISE copies or of the entire genome of a collection of isolates may yield a large enough number of SNPs to resolve the spatial and temporal dispersal of genetic M. ulcerans variants on the regional level.
ADDENDUM IN PROOF
A subsequent study that applied next-generation sequencing to two additional genomes of M. ulcerans strains from Ghana confirmed our conclusion in revealing 68 SNP loci that led to the differentiation of a collection of 54 strains from this region of endimicity into 13 SNP haplotypes (W. Qi, M. Käser, K. Röltgen, D. Yeboah-Manu, and G. Pluschke, PLoS Pathog. 5:e1000580, 2009).
Acknowledgments
This research activity was part of the Stop Buruli initiative funded by the UBS Optimus Foundation, Switzerland.
We thank Konstantina Boutsika for technical assistance.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Ablordey, A., P. A. Fonteyne, P. Stragier, P. Vandamme, and F. Portaels. 2007. Identification of a new variable number tandem repeat locus in Mycobacterium ulcerans for potential strain discrimination among African isolates. Clin. Microbiol. Infect. 13:734-736. [DOI] [PubMed] [Google Scholar]
- 2.Ablordey, A., M. Hilty, P. Stragier, J. Swings, and F. Portaels. 2005. Comparative nucleotide sequence analysis of polymorphic variable-number tandem-repeat loci in Mycobacterium ulcerans. J. Clin. Microbiol. 43:5281-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ablordey, A., R. Kotlowski, J. Swings, and F. Portaels. 2005. PCR amplification with primers based on IS2404 and GC-rich repeated sequence reveals polymorphism in Mycobacterium ulcerans. J. Clin. Microbiol. 43:448-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ablordey, A., J. Swings, C. Hubans, K. Chemlal, C. Locht, F. Portaels, and P. Supply. 2005. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J. Clin. Microbiol. 43:1546-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosch, R., A. S. Pym, S. V. Gordon, and S. T. Cole. 2001. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 9:452-458. [DOI] [PubMed] [Google Scholar]
- 7.Chemlal, K., K. De Ridder, P. A. Fonteyne, W. M. Meyers, J. Swings, and F. Portaels. 2001. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographical areas. Am. J. Trop. Med. Hyg. 64:270-273. [DOI] [PubMed] [Google Scholar]
- 8.Chemlal, K., G. Huys, P. A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 10.Debacker, M., F. Portaels, J. Aguiar, C. Steunou, C. Zinsou, W. Meyers, and M. Dramaix. 2006. Risk factors for Buruli ulcer, Benin. Emerg. Infect. Dis. 12:1325-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbon, d. Bobadilla, V., J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagneux, S., K. DeRiemer, T. Van, M. Kato-Maeda, B. C. de Jong, S. Narayanan, M. Nicol, S. Niemann, K. Kremer, M. C. Gutierrez, M. Hilty, P. C. Hopewell, and P. M. Small. 2006. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 103:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagneux, S., and P. M. Small. 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7:328-337. [DOI] [PubMed] [Google Scholar]
- 14.Gutacker, M. M., B. Mathema, H. Soini, E. Shashkina, B. N. Kreiswirth, E. A. Graviss, and J. M. Musser. 2006. Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J. Infect. Dis. 193:121-128. [DOI] [PubMed] [Google Scholar]
- 15.Hilty, M., M. Kaser, J. Zinsstag, T. Stinear, and G. Pluschke. 2007. Analysis of the Mycobacterium ulcerans genome sequence reveals new loci for variable number tandem repeats (VNTR) typing. Microbiology 153:1483-1487. [DOI] [PubMed] [Google Scholar]
- 16.Hilty, M., D. Yeboah-Manu, D. Boakye, E. Mensah-Quainoo, S. Rondini, E. Schelling, D. Ofori-Adjei, F. Portaels, J. Zinsstag, and G. Pluschke. 2006. Genetic diversity in Mycobacterium ulcerans isolates from Ghana revealed by a newly identified locus containing a variable number of tandem repeats. J. Bacteriol. 188:1462-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huber, C. A., M. T. Ruf, G. Pluschke, and M. Kaser. 2008. Independent loss of immunogenic proteins in Mycobacterium ulcerans suggests immune evasion. Clin. Vaccine Immunol. 15:598-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaser, M., J. Hauser, P. Small, and G. Pluschke. 2009. Large sequence polymorphisms unveil phylogenetic relationship of environmental and pathogenic mycobacteria related to Mycobacterium ulcerans. Appl. Environ. Microbiol. 75:5667-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaser, M., and G. Pluschke. 2008. Differential gene repertoire in Mycobacterium ulcerans identifies candidate genes for patho-adaptation. PLoS Negl. Trop. Dis. 2:e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaser, M., S. Rondini, M. Naegeli, T. Stinear, F. Portaels, U. Certa, and G. Pluschke. 2007. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol. Biol. 7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaser, M., M. T. Ruf, J. Hauser, L. Marsollier, and G. Pluschke. 2009. Optimized method for preparation of DNA from pathogenic and environmental mycobacteria. Appl. Environ. Microbiol. 75:414-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Käser, M., O. Gutmann, J. Hauser, T. Stinear, S. Cole, D. Yeboah-Manu, G. Dernick, U. Certa, and G. Pluschke. 2009. Lack of insertional-deletional polymorphism in a collection of Mycobacterium ulcerans isolates from Ghanaian Buruli ulcer patients. J. Clin. Microbiol. 47:3640-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathema, B., N. E. Kurepina, P. J. Bifani, and B. N. Kreiswirth. 2006. Molecular epidemiology of tuberculosis: current insights. Clin. Microbiol. Rev. 19:658-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Portaels, F., W. M. Meyers, A. Ablordey, A. G. Castro, K. Chemlal, P. de Rijk, P. Elsen, K. Fissette, A. G. Fraga, R. Lee, E. Mahrous, P. L. Small, P. Stragier, E. Torrado, A. Van Aerde, M. T. Silva, and J. Pedrosa. 2008. First cultivation and characterization of Mycobacterium ulcerans from the environment. PLoS Negl. Trop. Dis. 2:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pouillot, R., G. Matias, C. M. Wondje, F. Portaels, N. Valin, F. Ngos, A. Njikap, L. Marsollier, A. Fontanet, and S. Eyangoh. 2007. Risk factors for Buruli ulcer: a case control study in Cameroon. PLoS Negl. Trop. Dis. 1:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quek, T. Y., E. Athan, M. J. Henry, J. A. Pasco, J. Redden-Hoare, A. Hughes, and P. D. Johnson. 2007. Risk factors for Mycobacterium ulcerans infection, southeastern Australia. Emerg. Infect. Dis. 13:1661-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rondini, S., M. Käser, T. Stinear, M. Tessier, C. Mangold, G. Dernick, M. Naegeli, F. Portaels, U. Certa, and G. Pluschke. 2007. Ongoing genome reduction in Mycobacterium ulcerans. Emerg. Infect. Dis. 13:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stinear, T., J. K. Davies, G. A. Jenkin, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J. A. Hayman, and P. D. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stinear, T., B. C. Ross, J. K. Davies, L. Marino, R. M. Robins-Browne, F. Oppedisano, A. Sievers, and P. D. Johnson. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 37:1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stinear, T. P., G. A. Jenkin, P. D. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stinear, T. P., T. Seemann, S. Pidot, W. Frigui, G. Reysset, T. Garnier, G. Meurice, D. Simon, C. Bouchier, L. Ma, M. Tichit, J. L. Porter, J. Ryan, P. D. Johnson, J. K. Davies, G. A. Jenkin, P. L. Small, L. M. Jones, F. Tekaia, F. Laval, M. Daffe, J. Parkhill, and S. T. Cole. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stragier, P., A. Ablordey, L. M. Bayonne, Y. L. Lugor, I. S. Sindani, P. Suykerbuyk, H. Wabinga, W. M. Meyers, and F. Portaels. 2006. Heterogeneity among Mycobacterium ulcerans isolates from Africa. Emerg. Infect. Dis. 12:844-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stragier, P., A. Ablordey, L. Durnez, and F. Portaels. 2007. VNTR analysis differentiates Mycobacterium ulcerans and IS2404 positive mycobacteria. Syst. Appl. Microbiol. 30:525-530. [DOI] [PubMed] [Google Scholar]
- 35.Stragier, P., A. Ablordey, W. M. Meyers, and F. Portaels. 2005. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by using mycobacterial interspersed repetitive units. J. Bacteriol. 187:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner, T., M. E. Benbow, T. O. Brenden, J. Qi, and R. C. Johnson. 2008. Buruli ulcer disease prevalence in Benin, West Africa: associations with land use/cover and the identification of disease clusters. Int. J. Health Geogr. 7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williamson, H. R., M. E. Benbow, K. D. Nguyen, D. C. Beachboard, R. K. Kimbirauskas, M. D. McIntosh, C. Quaye, E. O. Ampadu, D. Boakye, R. W. Merritt, and P. L. Small. 2008. Distribution of Mycobacterium ulcerans in Buruli Ulcer Endemic and Non-Endemic Aquatic Sites in Ghana. PLoS Negl. Trop. Dis. 2:e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization. 2008. Buruli ulcer: progress report, 2004-2008. Wkly. Epidemiol. Rec. 83:145-156. [PubMed] [Google Scholar]
- 39.Yeboah-Manu, D., T. Bodmer, E. Mensah-Quainoo, S. Owusu, D. Ofori-Adjei, and G. Pluschke. 2004. Evaluation of decontamination methods and growth media for primary isolation of Mycobacterium ulcerans from surgical specimens. J. Clin. Microbiol. 42:5875-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yip, M. J., J. L. Porter, J. A. Fyfe, C. J. Lavender, F. Portaels, M. Rhodes, H. Kator, A. Colorni, G. A. Jenkin, and T. Stinear. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 189:2021-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]