Abstract
The eradication rate of Helicobacter pylori by standard therapy is decreasing due to antibiotic resistance, mainly to clarithromycin. Our aim was to provide a new molecular test to guide the treatment of new and relapsed cases. We first studied 126 H. pylori strains for phenotypic (MIC) and genotypic resistance to clarithromycin (rrl mutation) and levofloxacin (gyrA mutation) and then developed a DNA strip genotyping test on the basis of the correlation results and literature data. Clinical strains (n = 92) and gastric biopsy specimens containing H. pylori (n = 105) were tested blindly with the new molecular test GenoType HelicoDR. The presence of mutations or the absence of hybridization with wild-type sequences was predictive, in rrl for clarithromycin resistance in 91 cases (mostly the A2147G mutation) and in gyrA for levofloxacin resistance in 58 cases (mutations at codon 87 or 91). Genotyping revealed a mix of genotypes in 33% of the cases, reflecting a coinfection or selection for resistant mutants. The sensitivity and specificity of detecting resistance were 94% and 99% for clarithromycin and 87% and 98.5% for levofloxacin, respectively. The concordance scores were 0.96 for clarithromycin and 0.94 for levofloxacin. With global resistance rates of 46% for clarithromycin and 25% for levofloxacin, which were observed for consecutive positive biopsy specimens from 2007 and 2008, the positive and negative predictive values for detecting resistance were 99% and 94% for clarithromycin and 96% and 96% for fluoroquinolone. GenoType HelicoDR is efficient at detecting mutations predictive of antibiotic resistance in H. pylori when applied to strains or directly to gastric biopsy specimens.
Helicobacter pylori infection is a common chronic gastric infection worldwide with one-third prevalence (6). About 1 out of 10 humans infected with H. pylori suffers from various digestive diseases, such as duodenal and gastric ulcer and nonulcer dyspepsia; 1 out of 100 develops gastric adenocarcinoma; and ≤1 out of 1,000 may develop gastric mucosa-associated lymphoid tissue lymphoma. All consensus guidelines recommend eradication of H. pylori (6, 20) in symptomatic patients. Standard therapy combines a proton pump inhibitor, such as omeprazole, and two antibiotics, chosen from among amoxicillin, clarithromycin, and metronidazole (20). This therapy was assessed in studies in the early 1990s and demonstrated an eradication rate of H. pylori of over 80%. However, the eradication rate is decreasing, with as low as 60% success in some countries, and this is related to the increase in clarithromycin and metronidazole resistance reported worldwide (9, 10, 17). Fluoroquinolones, such as levofloxacin and moxifloxacin, are often used for rescue therapy in a third- or fourth-line treatment (20, 31).
Antibiotics used for the treatment of H. pylori infection are usually not chosen on the basis of routine susceptibility testing, because H. pylori is a fastidious microorganism requiring 3 to 10 days in a microaerobic atmosphere, and susceptibility results are not reliable for all antibiotics (17, 22). Indeed, susceptibility breakpoints have been difficult to set due to the lack of standard methods for susceptibility testing and difficulties in assessing the correlation between susceptibility results and clinical outcomes. Phenotypic resistance is correlated with clinical and microbiological failure for clarithromycin, but not for metronidazole (21). The eradication rate drops from 88% in the case of a clarithromycin-susceptible strain to less than 20% in the case of clarithromycin resistance (7, 21). Fluoroquinolone resistance was also shown to be correlated with treatment failure (24). Because resistance rates vary according to the country and patient characteristics, the choice of antibiotics on the basis of susceptibility results might be an effective strategy to improve H. pylori eradication. Since susceptibility testing is cumbersome, molecular methods for detection of resistance may be cost-effective.
The mutations leading to resistance are now well known for macrolides and fluoroquinolones, although they are still unclear for metronidazole and amoxicillin. Clarithromycin resistance in H. pylori is due to point mutations in the rrl gene encoding the 23S rRNA, with three major mutations described: A2146C, A2146G, and A2147G (the numeration is from genome sequencing of NC000921 and NC000915, positions 2146 and 2147, formerly described as 2142 and 2143 [reviewed in references 21 and 22]). The resistance of H. pylori to quinolones is due to point mutations in the so-called quinolone resistance-determining region of the gyrA gene coding for the A subunit of the DNA gyrase, mainly at codons 87 and 91 (corresponding to 83 and 87 in Escherichia coli numbering) (1, 4, 23, 30).
Our objective was to develop and implement a molecular method to easily detect mutations predictive of clarithromycin and fluoroquinolone resistance in H. pylori. We based our test on the DNA strip methodology used with success for other pathogens, such as Mycobacterium tuberculosis (13). We first designed a prototype test using a panel test of 126 H. pylori strains for which the MICs of clarithromycin and fluoroquinolones and the rrl and gyrA genotypes had been determined. Then, the new test was applied blindly to clinical strains and gastric biopsy specimens containing H. pylori, and the results were compared to those of susceptibility testing done routinely. The specificity of the new test for H. pylori was evaluated by testing strains of Helicobacter species other than H. pylori, as well as negative biopsy specimens. The new test was concordant with reference tests for 94 to 98% of the samples, either performed on isolated strains or directly on gastric biopsy specimens containing H. pylori, and was easy to perform.
MATERIALS AND METHODS
H. pylori strains and gastric biopsy specimens containing H. pylori.
The first phase of the study compared phenotypic and genotypic profiles of resistance to clarithromycin and to fluoroquinolones (levofloxacin and ciprofloxacin) for 126 strains isolated from March 2004 to September 2005 at Henri Mondor Hospital, Créteil, France. The second phase studied 92 H. pylori strains and 105 gastric biopsy specimens positive for H. pylori. They were consecutive positive samples obtained either at Henri Mondor Hospital (67 strains and 74 biopsy specimens) or at the National Reference Center at Bordeaux Hospital (25 strains and 31 biopsy specimens) in 2006, 2007, and 2008. In the two hospitals, gastric biopsy specimens were routinely cultured and subjected to real-time PCR for H. pylori detection, and H. pylori-positive biopsy specimens were subjected to rrl mutation detection as described previously (18, 27).
Phenotypic susceptibility testing.
Susceptibility to clarithromycin and to fluoroquinolones (levofloxacin or ciprofloxacin, since we had previously shown that the results were concordant [4]) was assessed using the Etest method (AB Biodisk, Solna, Sweden) performed as previously described (4, 9) and using susceptibility breakpoints of ≤0.5 μg/ml for clarithromycin and <1 μg/ml for ciprofloxacin or levofloxacin (http://www.clsi.org and http://www.eucast.org). H. pylori NCTC11637 was used as a control. Briefly, a bacterial inoculum was prepared in brain heart infusion broth from subcultures grown on Pylori agar (bioMérieux, Marcy l'Etoile, France) so that the turbidity was equal to that of a 3 McFarland standard. Mueller-Hinton agar supplemented with 10% sheep blood and prepared extemporaneously was inoculated and incubated at 37°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) for 72 h.
DNA isolation and PCR sequencing of H. pylori strains and gastric biopsy specimens.
Genomic DNA of strains was extracted using the QIAmp DNA Mini Kit (Qiagen, Courtaboeuf, France). Total DNA from biopsy specimens was extracted using the High Pure PCR template kit after following the protocol for tissue lysis (Roche Diagnostics, Switzerland). Before DNA extraction, gastric biopsy specimens (from the antrum and corpus) were crushed or cut into pieces and mixed with 0.5 ml of brain heart infusion broth.
DNA regions involved in clarithromycin (rrl gene) and fluoroquinolone (gyrA gene) resistance were amplified by PCR as previously described (4, 22). Typical reaction mixtures (50 μl) contained 1× reaction buffer, 1.5 mM of MgCl2, 200 μM of deoxynucleotide triphosphates, 1 μM of each primer (Proligo, France), 1.25 U of Taq polymerase (Q-Biogene, Illkirch, France), and 150 ng of DNA template. PCR-amplified fragments were purified by using Montage PCR centrifugal filter devices (Millipore, Molsheim, France) and sequenced by the dideoxy chain termination method with the ABI Prism BigDye Terminator v3.1 cycle-sequencing kit (Applied Biosystems, Courtaboeuf, France). The oligonucleotide primers used for DNA sequencing were those used for PCR. The nucleotide and deduced amino acid sequences were analyzed with Seqscape v2.0 software (Applied Biosystems).
GenoType HelicoDR testing.
Genomic DNA of the H. pylori strains of the phase 2 panel and total DNA extracted from the gastric biopsy specimens were subjected blindly to DNA strip testing. The strips were coated at the Hain Lifescience factory (Nehren, Germany) with different specific oligonucleotides (DNA probes) using the DNA strip technology. The probes were designed to hybridize with the sequences of the wild-type alleles (WT probes) or the mutated alleles (MUT probes).
Amplification, hybridization, and interpretation were performed in a procedure similar to those for other GenoType tests (12). Briefly, 35 μl of 5′-biotinylated primers and nucleotide mixture, 5 μl of polymerase buffer, 2 μl of 25 mM MgCl2 stock solution, 3 μl of water, and 5 μl of total DNA (20 to 100 ng) were mixed with 1 U of Hot Star Taq polymerase (Qiagen) per reaction mixture. The PCR run comprised 30 cycles for strains and 35 cycles for biopsy specimens. In both protocols, the denaturation cycle was 1 cycle at 95°C for 15 min, followed by 10 cycles at 95°C for 30 s and at 58°C for 2 min. Then, 20 cycles (strain) or 25 cycles (biopsy specimen) were composed of a first step at 95°C for 25 s, a second step at 53°C for 40 s, and a third step at 70°C for 40 s. The PCR ended with 8 min at 70°C. Hybridization was performed using the TwinCubator at a temperature of 45°C. The denaturation solution was mixed with 20 μl of the amplified sample and submitted to the usual protocol for hybridization (12, 13).
In order to assess positive and negative bands, the DNA strips were stuck on an evaluation sheet after the hybridization, and a template was aligned side by side with the conjugate control band of the respective strip. Control bands that should appear positive to validate the test were bands corresponding to the conjugate control and the amplification control, an identification band for H. pylori (the PCR fragment was from the rrl gene, which codes for the 23S rRNA, as stated above), and amplification controls of the rrl and gyrA genes.
The analytical sensitivity of the GenoType HelicoDR test for the detection of H. pylori was tested by comparing the positivity of the H. pylori detection probe and dilutions of bacterial suspensions. The analytical specificity of the GenoType HelicoDR test was determined by subjecting to the test DNA extracted from reference strains of 20 Helicobacter species other than H. pylori. In addition, 23 biopsy specimens known to be negative for H. pylori were also tested for specificity.
RESULTS
Concordance between phenotypes and genotypes.
The clarithromycin, levofloxacin, and ciprofloxacin MICs for all 126 strains included in the phase 1 study were determined, and rrl and gyrA resistance-determining regions were sequenced. The results are detailed in Table 1. For the strains and biopsy specimens included in the phase 2 study, clarithromycin susceptibility was compared to the rrl mutation detection by real-time PCR, since the molecular detection is done routinely. Determination of gyrA mutations was not done routinely. PCR sequencing of rrl or gyrA was done in cases of discord between the susceptibility data and the results of the GenoType HelicoDR test.
TABLE 1.
Concordance between phenotype (MIC determined by Etest) and genotype (sequencing or real-time PCR) for H. pylori strains included in the study
| H. pyloria | n |
rrl genotype |
gyrA genotype |
||||
|---|---|---|---|---|---|---|---|
| Wild type | Missense mutation | No data | Wild type | Missense mutation | No data | ||
| Phase 1 | |||||||
| Development panel H. pylori strains | 126 | ||||||
| CLA-R | 40 | 3b | 37 | ||||
| CLA-S | 86 | 86 | 0 | ||||
| FQ-R | 21 | 0 | 21 | ||||
| FQ-S | 105 | 105c | 0 | ||||
| Phase 2 | |||||||
| Clinical H. pylori strains | 92 | ||||||
| CLA-R | 48 | 2 | 34 | 12 | |||
| CLA-S | 44 | 21 | 1 | 22 | |||
| FQ-R | 41 | 41 | |||||
| FQ-S | 51 | 51 | |||||
| Gastric biopsy specimens | 105 | ||||||
| CLA-R | 49 | 0 | 49 | ||||
| CLA-S | 56 | 56 | 10 | ||||
| FQ-R | 26 | 26 | |||||
| FQ-S | 79 | 79 | |||||
CLA, clarithromycin; FQ, fluoroquinolone; S, susceptible; R, resistant.
Clarithromycin MIC, 2 μg/ml.
Sixteen strains harbored a Thr87, which confers fluoroquinolone susceptibility.
GenoType HelicoDR probe design according to the sequencing of strains from phase 1.
The MUT and WT probes were designed from the mutations observed in the resistant strains from the phase 1 study; mutations in the rrl gene encoding the 23S rRNA for the clarithromycin-resistant strains and mutations in the gyrA gene for the fluoroquinolone-resistant strains. These results were gathered with those described in the literature (reviewed in reference 22). The probes are listed in Table 2. In rrl, the most prevalent mutations were A2147G (92% in our study and 54% in the literature), A2146G (8% in our study and 28% in the literature), and A2146C (no strains in our study but 4% in the literature). gyrA mutations have been previously described and compared to those in the literature (4). Overall, in gyrA, the mutations were distributed at codons 87 and 91 with the following prevalences: N87K, 27% in our study and 41% in the literature; T87Y, 9% and 10%; D91N, 36% and 30%; D91Y, 14% and 15%; D91G, 0% and 11%; and D86N, 4% and 2%. By sequencing the gyrA quinolone resistance-determining region in fluoroquinolone-susceptible strains, we observed an Asn (N)/Thr (T) polymorphism at codon 87, with 15% of the strains with T87. It was then necessary to include four wild-type probes corresponding to codon 87 (two for Asn87 and two for Thr87) in gyrA in the DNA strip, as shown in Table 2 and Fig. 1.
TABLE 2.
Probes hybridized on the DNA strip of the GenoType HelicoDR test for detection of mutations in the gyrA and the rrl genesa
| Probe | Codon | Nucleotides | Associated phenotypeb |
|---|---|---|---|
| gyr87-WT1 | N87 | AAC | FQ-S |
| gyr87-WT2 | N87 | AAT | FQ-S |
| gyr87-WT3 | T87 | ACC | FQ-S |
| gyr87-WT4 | T87 | ACT | FQ-S |
| gyr87-MUT | N87K | AAA | FQ-R |
| gyr91-WT | D91 | GAT | FQ-S |
| gyr91-MUT1 | D91N | AAT | FQ-R |
| gyr91-MUT2 | D91G | GGT | FQ-R |
| gyr91-MUT3 | D91Y | TAT | FQ-R |
| 23S-WT | 2146 and 2147 | AA | CLA-S |
| 23S-MUT1 | 2146 | A2146G | CLA-R |
| 23S-MUT2 | 2146 | A2146C | CLA-R |
| 23S-MUT3 | 2147 | A2147G | CLA-R |
Numbering system used in H. pylori J99 and 26695 (GenBank accession no. NC000921 and NC000915).
CLA, clarithromycin; FQ, fluoroquinolone; S, susceptible; R, resistant.
FIG. 1.
Prototype of the strip of the GenoType HelicoDR test. Lane 1, gyrA pattern with a wt1 codon at position 87, a wt codon at codon 91, and a wild-type rrl pattern; lane 2, gyrA pattern with a MUT codon at position 87, a wt codon at codon 91, and a wild-type rrl pattern; lane 3, gyrA pattern with a wt2 codon at position 87, a MUT2 codon at position 91, and a wild-type rrl pattern; lane 4, gyrA pattern with a wt3 codon at position 87, a wt codon at position 91, and a MUT1 rrl mutation; lane 5, double mutation in gyrA with mutation at both 87 (MUT) and 91 (MUT3) and a wild-type rrl pattern.
GenoType HelicoDR results for detection of clarithromycin resistance for the nondevelopmental strains and specimens (phase 2 study).
There were 197 results of testing by the GenoType HelicoDR test for the 92 clinical strains and the 105 gastric biopsy specimens containing H. pylori in phase 2. Representative strips are shown in Fig. 2. Several times (see Heterogeneity of the H. pylori population below), more than one mutation was detected per strain or biopsy specimen. One rrl mutation was observed in 91 strains or biopsy specimens, but 100 “cases” of rrl mutation were reported overall (for the observed genotypes, see Table 4). Overall, the most frequent mutation was A2147G (MUT3 profile), observed in 35 strains and 41 biopsy specimens (76% of the mutated alleles), followed by A2146G (MUT1 profile) in 12 strains and 8 biopsy specimens (20%) and A2146C (MUT2 profile) in 1 strain and 2 biopsy specimens (3%).
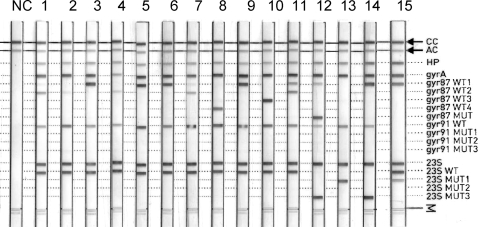
FIG. 2.
Representative GenoType HelicoDR DNA strip results obtained with H. pylori strains and biopsy specimens showing (i) rrl alleles that are wild type (lanes 1 to 11) or mutated (12 to 15), (ii) gyrA alleles that are wild type (lanes 1, 3 to 11, and 15) or mutated (a mutation at codon 87 in lane 12 and no hybridization with probes of codon 87 in lane 2), and (iii) heterogeneity of the H. pylori population (lanes 7, 11, 12, and 15). NC, negative control; CC, conjugate control; AC, amplification control.
TABLE 4.
Details of genotypes detected by the GenoType HelicoDR test for the rrl or gyrA gene in H. pylori strains and gastric biopsy specimens in the phase 2 study
| Genotype | No. in clinical strains (n = 92) | No. in biopsy specimens (n = 105) |
|---|---|---|
| 23S rRNA gene (rrl) | ||
| WT | 48 | 57 |
| MUT1 | 6 | 5 |
| MUT3 | 25 | 34 |
| WT + MUT1 | 2 | 1 |
| WT + MUT3 | 5 | 5 |
| WT + MUT1 + MUT3 | 1 | 0 |
| WT + MUT1 + MUT2 | 0 | 1 |
| WT + MUT2 + MUT3 | 1 | 1 |
| MUT1 + MUT3 | 3 | 1 |
| gyrA gene | ||
| Codon 87 | ||
| WT1 | 34 | 41 |
| WT2 | 8 | 11 |
| WT3 | 15 | 8 |
| WT4 | 2 | 6 |
| WT1+WT2 | 15 | 20 |
| WT2+WT4 | 1 | 0 |
| WT3+WT4 | 0 | 2 |
| WT1+WT2 + WT3 | 0 | 1 |
| WT 1 + MUT | 2 | 3 |
| MUT | 7 | 2 |
| WT1+WT2 + MUT | 1 | 0 |
| No WT or MUT band | 7 | 11 |
| Codon 91 | ||
| WT | 73 | 93 |
| MUT1 | 6 | 5 |
| MUT2 | 4 | 3 |
| MUT3 | 1 | 1 |
| WT + MUT1 | 4 | 0 |
| WT + MUT3 | 1 | 0 |
| WT + MUT2 | 2 | 1 |
| WT + MUT1 + MUT2 | 1 | 0 |
| WT + MUT1 + MUT3 | 0 | 1 |
| No WT or MUT band | 0 | 1 |
WT, wild-type allele; MUT, mutated allele (see Table 2 for the codon sequences).
The concordance between the results of the GenoType HelicoDR test and the results of genotyping by real-time PCR was 99.5%. The discordant case showed a wild-type band at the rrl locus and showed a mixture of wild-type and mutated alleles by real-time PCR, which was in accordance with the observation of resistance (MIC, 256 μg/ml). The concordance between the results of the GenoType HelicoDR test and clarithromycin susceptibility (phenotype) is presented in Table 3. Of the seven discordant cases, five were due to the mixing of susceptible bacteria with a wild-type rrl genotype and resistant bacteria with a mutated rrl genotype (see Heterogeneity of the H. pylori population below).
TABLE 3.
Concordance for clarithromycin results between GenoType HelicoDR and susceptibility testing for the H. pylori included in the phase 2 study
| Reference resultsa | GenoType HelicoDR result |
||
|---|---|---|---|
| n | Mutation alleleb | Wild-type allelec | |
| Strains (n = 92) | |||
| Cla resistant | 48 | 43 | 5d |
| Cla susceptible | 44 | 1e | 43 |
| Biopsy specimens (n = 105) | |||
| Cla resistant | 49 | 48 | 1f |
| Cla susceptible | 56 | 0 | 56 |
| Total (n = 197) | |||
| Cla resistant | 97 | 91 | 6 |
| Cla susceptible | 100 | 1 | 99 |
Reference phenotype (Etest MIC using a breakpoint of 0.5 μg/ml).
At least one of the following mutation bands: 23S-MUT1, 23S-MUT2, or 23S-MUT3.
Presence of the 23S-WT band and no 23S mutation band.
For three strains and one biopsy specimen, a mixture of wild-type and resistant populations was assessed; for the two remaining strains, the clarithromycin MICs were 6 and 24 μg/ml, respectively.
A MIC of 0.016 μg/ml and a double-genotype population were assessed by real-time PCR.
Clarithromycin MIC of 1 μg/ml.
The sensitivity and specificity for the detection of a clarithromycin-resistant strain were 94% and 99%, respectively, and the concordance score was 0.96 for clarithromycin. According to the global prevalence of H. pylori resistance to clarithromycin observed in the phase 2 study, which included consecutive strains and positive biopsy specimens (46% for biopsy specimens and strains together), the positive and negative predictive values of the GenoType HelicoDR test for detecting clarithromycin resistance were 99% and 94%, respectively.
GenoType HelicoDR results for levofloxacin resistance detection.
There were 197 results given by GenoType HelicoDR for gyrA genotyping, with examples of strips shown in Fig. 2. For each H. pylori strain, the results for the two codons, 91 and 87, have to be taken into account in order to classify it as a wild-type genotype or a mutated genotype. As for rrl mutation detection, several times (see Heterogeneity of the H. pylori population below) more than one genotype was detected per strain or biopsy specimen.
A mutated gyrA genotype was observed in 60 H. pylori strains (30%): 30 with one mutation at codon 87; 25 with one mutation at codon 91; and 4 with a gyrA mutation at both codons 87 and 91, 1 of which had two mutations at codon 91; and the remaining 1 with two mutations at codon 91. Detailed results are presented in Table 4. The most frequent mutation was D91N (MUT1), followed by the T87I mutation. This mutation was detected as “no wild-type band and no MUT band” at codon 87, and the mutation was assessed by PCR sequencing.
In the remaining two cases of no band detection at codon 87, N87Y was observed in one case. For the remaining strain, the modified codon was not codon 87 but codon 88, where the wild-type triplet was GCA instead of GCG. This probably led to weaker hybridization with the oligonucleotides targeting codon 87.
Probing with the four wild-type alleles at codon 87 showed that the preponderant codon was that of the wt1 profile (AAC), followed by wt2 (AAT), wt3 (ACC), and wt4 (ACT) (Table 4). Overall, threonine codon 87 (wt3 or wt4 triplet) was present for 35 H. pylori strains detected, which represented 25% of the H. pylori strains with a wild-type gyrA allele.
The concordance between the GenoType HelicoDR test and fluoroquinolone susceptibility, defined by either ciprofloxacin or levofloxacin susceptibility, is presented in Table 5. The discordant results were mainly a wild-type allele with a high MIC (nine discordant results). For the two biopsy specimens for which a mutated allele was detected in a fluoroquinolone-susceptible H. pylori strain, there was no band hybridizing at codon 87 in one case (sequencing revealed a silent mutation at codon 88, as described above) and no band at codon 91 in the second case. By repeating the test, we observed weak hybridization to the WT91 probe, and sequencing confirmed a wild-type gyrA allele.
TABLE 5.
Concordance of fluoroquinolone results between GenoType HelicoDR and susceptibility testing for the H. pylori strains included in the phase 2 study
| Phenotypea | GenoType HelicoDR result |
||
|---|---|---|---|
| n | Mutation alleleb | Wild typec | |
| Strains (n = 92) | |||
| FQ resistant | 41 | 34 | 7d |
| FQ susceptible | 51 | 0 | 51 |
| Biopsy specimens (n = 105) | |||
| FQ resistant | 26 | 24 | 2e |
| FQ susceptible | 79 | 2f | 77 |
| Total (n = 197) | |||
| FQ resistant | 67 | 58 | 9 |
| FQ susceptible | 130 | 2 | 128 |
Ciprofloxacin or levofloxacin susceptibility testing using breakpoint of 1 μg/ml. FQ, fluoroquinolone.
One MUT band or no WT band at codon 87 or at codon 91.
Presence of at least one WT band at codons 87 and 91 and no MUT band.
MIC, 32 μg/ml for six strains and 8 μg/ml for one strain.
MIC, 32 μg/ml for the two strains.
MIC < 0.5 μg/ml; silent mutation at codon 88 for one biopsy specimen and absence of wild-type codon 91 for the second.
The sensitivity and specificity for detecting resistance were 87% and 98.5%, respectively, and the concordance score was 0.94.
According to the 25% prevalence of levofloxacin resistance in our study, positive and negative predictive values were 96% and 96%, respectively.
Heterogeneity of the H. pylori population.
In 66 (33%) of the cases (30 strains and 36 biopsy specimens), more than one hybridization band was observed at one codon. This indicated that there were at least two alleles amplified during the PCR. The mixture was shown with the rrl probes or the gyrA probes. It concerned a resistant and a wild-type sequence, as well as a mixture of two wild-type sequences, but with different triplets for the same amino acid. The results are presented in Table 6, and examples are shown in Fig. 2.
TABLE 6.
Heterogeneity of the H. pylori population per patient as assessed by Genotype HelicoDR
| Heterogeneity | No. (%) in strains (n = 92) | No. (%) in gastric biopsy specimens (n = 105) | Total (%) (n = 197) |
|---|---|---|---|
| No heterogeneity | 62 (67) | 69 (66) | 131 (66.5) |
| Heterogeneous genotypesa | 30 (33) | 36 (32) | 66 (32) |
| Wild-type and resistant rrl alleles | 9 (10) | 8 (8) | 17 (9) |
| Several resistant rrl alleles | 5 (5) | 2 (2) | 7 (3.6) |
| Wild-type and resistant gyrA alleles | 11 (12) | 5 (5) | 16 (8) |
| Several wild-type gyrA alleles | 17 (18.5) | 23 (22) | 40 (20) |
At least two genotypes (details of genotypes are listed in Table 4).
Analytical sensitivity and specificity with the GenoType HelicoDR test for the detection of H. pylori DNA.
All biopsy specimens detected as positive for H. pylori by the real-time PCR detection method and by culture were positive with GenoType HelicoDR. Bacterial suspensions of H. pylori strains with a McFarland density of 1.0 to 8.0 (DNA concentration, 3.6 to 21.4 μg/ml, respectively) gave a positive signal with the GenoType HelicoDR test.
The analytical specificity of the GenoType HelicoDR test was measured by testing DNA extracted from 20 Helicobacter species other than H. pylori. The point results of hybridization are shown in Table 7. Although some amplicons hybridized to some probes (rrl probes more often than gyrA), none of the Helicobacter species hybridized to the H. pylori control probe or to all of the WT or MUT probes, except Helicobacter acinonychis, which is known to be the closest to H. pylori.
TABLE 7.
Results of the Genotype HelicoDR test for strains of Helicobacter species other than H. pylori
| Non-pylori Helicobacter species | Hybridization results for the probes present on the DNA stripa |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HP | gyrA | gyr87 WT1 | gyr87 WT2 | gyr87 WT3 | gyr87 WT4 | gyr87 MUT | gyr91 WT | gyr91 MUT1 | gyr91 MUT2 | gyr91 MUT3 | 23S | 23S WT | 23S MUT1 | 23S MUT2 | 23S MUT3 | |
| H. winghamensis | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| H. cholecystus | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | + |
| H. mustelae | − | + | − | − | − | − | − | + | ± | − | − | + | + | − | − | − |
| H. cinaedi | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − |
| H. canadensis | − | + | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| H. fennelliae | − | + | − | − | − | − | − | − | − | − | − | + | + | − | − | − |
| H. acinonychis | + | + | + | + | − | − | − | + | − | − | − | + | + | − | − | − |
| H. hepaticus | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| H. canis | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − |
| H. salomonis | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − |
| H. pametensis | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − |
| H. muridarum | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − |
| H. rappini | − | − | − | − | − | − | − | − | − | − | − | ± | + | − | − | − |
| H. bizzozeronii | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − |
| H. ganmani | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| H. pullorum | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| H. felis | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − |
| H. bilis | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| H. typhlonius | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − |
| H. cetorum | − | + | − | − | − | − | − | − | − | − | − | + | + | − | − | − |
+, positive signal; −, negative signal; ±, weakly positive signal.
For the 23 gastric biopsy specimens that were negative for H. pylori by culture, molecular detection (real-time PCR), and histology, GenoType HelicoDR results were negative for the H. pylori control band, except for one biopsy specimen. Other bands were also observed for the gyrA and the rrl control probes and the rrl alleles (WT and MUT1 plus MUT3). The gastric histology results for the patient with the positive biopsy showed atrophy and inflammation in the antrum but no H. pylori. This suggests that the biopsy specimen may have contained H. pylori DNA, even though other reference methods of detection (culture and real-time PCR) were negative. For seven biopsy specimens, although some bands were positive (mostly the rrl control probe), the absence of the H. pylori control band and of most of the control bands led to a negative result.
DISCUSSION
Because the success of H. pylori eradication treatment is closely related to prior recognition of antibiotic resistance and because susceptibility testing is rarely done due to fastidious growth of the bacterium, we developed and evaluated a new molecular test that rapidly detects antibiotic resistance in H. pylori. The test, based on DNA strip technology, was developed to be applicable to strains, as well as to gastric biopsy specimens, and to be affordable and easy to perform in clinical microbiology laboratories.
The detection of clarithromycin and fluoroquinolone resistance was targeted, since (i) clarithromycin and levofloxacin are both effective drugs to treat H. pylori infection (6) and (ii) resistance to these drugs has been clearly shown to be correlated with eradication failure (21). Since H. pylori resistance mechanisms for these drugs are due to point mutations in a small region of one gene, it was possible to design a DNA strip test based on the few oligonucleotides detecting mutations. Detection of metronidazole resistance, although the most frequent, could not be included in the test, since it involves several genes (rdxA, frxA, and fdxB) without clear correlation with the resistance phenotype so far (14, 21).
Clarithromycin resistance is correlated with rrl mutations (A2146G, A2146C, or A2147G) for more than 90% of the clarithromycin-resistant clinical strains (21). Few mutations have been described outside the two positions 2146 and 2147, and their association with clarithromycin resistance is not yet consensual (3, 16, 28, 32). By using the GenoType HelicoDR test, we showed that 94% of the clarithromycin-resistant H. pylori strains harbored one of the three mutations listed above (37 out of the 40 strains in the development study and 91 out of 97 H. pylori strains in the evaluation study).
Since 1995, fluoroquinolone resistance in H. pylori has been related to gyrA gene mutations at positions 87 and 91 (H. pylori numbering system) (17, 23, 30). Fluoroquinolone (levofloxacin, gatifloxacin, and ciprofloxacin) resistance was correlated with gyrA mutations in 80 to 100% of the fluoroquinolone-resistant strains (1, 4, 15, 25). The mutations are generally found in codons 87 and 91 and rarely in codon 86 (4). This is concordant with the present results of GenoType HelicoDR testing in 67 fluoroquinolone-resistant H. pylori strains, 87% of which showed a mutation either at codon 87 (57% of the mutations) or at codon 91 (48%). In some cases, the GenoType HelicoDR results showed a pattern concordant with a resistant allele but with no band corresponding to a precise mutation, and also without any wild-type-positive band. In most of the cases, sequencing showed a missense mutation at codon 86 or 87 that was not specifically present on the DNA strip. Similar observations were made for other GenoType strips, such as the GenoType MTBDRplus strip (13). Thus, the test can detect a broad spectrum of mutations mediating resistance, and not only those present as mutated alleles on the strip. However, control sequencing is recommended when no band is seen, since in one case in our study, the gyrA sequence showed a silent mutation.
The results of the GenoType HelicoDR test were in concordance with those of clarithromycin and fluoroquinolone susceptibility testing. The discordances observed were mainly due to discrepancies between the genotyping and phenotyping methods, especially for fluoroquinolone. There was agreement of the GenoType HelicoDR test with genotyping performed by another method (PCR sequencing or real-time PCR). Susceptibility testing methods, although standardized, were not demonstrated to be fully reliable for bacteria such as H. pylori because of its fastidious growth and difficulties in assessing an adequate inoculum (9).
In our study, we showed that more than 30% of the H. pylori strains present in gastric biopsy specimens or isolated as strains were in fact mixtures of several genotypes. The presence of more than one genotype in a single biopsy specimen has been shown to be the consequence of either different strains coexisting in the gastric mucosa or the presence of mutated and wild-type alleles in the same strain (26, 32). Using culture, a similar proportion of 20 to 25% of the H. pylori cases was found. When a mixture of susceptible and resistant genotypes has been found, it has been related to clinical failure so far (26). In some cases, the results of genotyping by our test showed a mixture of wild-type alleles. Recently, Sheu et al. showed that infections with more than one strain are associated with more severe disease (29).
Other genotyping methods have been developed for the detection of antibiotic resistance in H. pylori based on the LiPA technology (32, 33), real-time PCR (2, 18, 19, 27), fluorescence in situ hybridization (11), microarray (5, 35), or others (34). Most of them are in-house tests that are not commercially available, and they detect only clarithromycin resistance. This may explain why detection of antibiotic resistance is rarely implemented in microbiology laboratories. However, there is a need to diagnose resistance in developed countries, where ulcers and gastric cancers are still frequent, as well as in developing countries, where most of the population is infected and 1/10 suffer from gastroduodenal ulcers.
We developed a test that is easy to perform and that is based on the worldwide experience of M. tuberculosis testing (GenoType MTBDRplus kit) (12). It requires a thermocycler, which nowadays is present in most laboratories, and a specific incubator for hybridization. Although we did not evaluate the quantity of bacteria necessary to obtain a positive GenoType HelicoDR test result, the analytical sensitivity with biopsy specimens was not lower than that with real-time PCR or culture, since we tested biopsy specimens known to be H. pylori positive by these methods, and in all cases the GenoType HelicoDR test led to a positive result. Since the GenoType HelicoDR test has not been tested on feces, we do not recommend using it, especially because the presence of large amounts of other bacteria may hamper the specificity of the multiplex PCRs and of the hybridization.
Despite the recommendation for antibiotic treatment of H. pylori infection since 1995 (6, 20) and the increase in antibiotic resistance, susceptibility testing of H. pylori is rarely done and is limited to a few laboratories with expertise. Moreover, it is recommended only for patients suffering from their second relapse. Although phenotypic susceptibility testing was performed in the 1990s, it was for the most part abandoned because of the fastidious growth of the organism and also because of a small impact at that time on the treatment and outcome. Now, since primary antibiotic resistance has reached over 20% for clarithromycin, it is difficult to avoid performing the detection of antibiotic resistance even in new cases of H. pylori infection. Clarithromycin is still the first-line therapy recommended worldwide. Since the GenoType HelicoDR test accurately predicts clarithromycin resistance, it can be used to decide the first-line treatment without facing the 80% failure observed in clarithromycin-resistant cases (7). This will allow the development of new strategies of antibiotic therapy based on susceptibility tests, and it will improve H. pylori eradication rates, which have decreased to 65 to 70% in the last few years. In relapse cases, where the rate of resistance to fluoroquinolones is increasing (4, 8, 25), the detection of fluoroquinolone resistance will provide new information for the choice of antibiotics to be used in third-line therapies, often a combination of amoxicillin with levofloxacin or gatifloxacin, tetracycline, or rifabutin; resistance to the last two antibiotics is still very rare.
Acknowledgments
We thank Romain Roth Dit Bettoni for technical assistance and Marc Tordjemann for helpful discussion.
This work was supported by Hain Lifescience (expenses for the materials used for microbiological, biochemical, and genetic analyses).
Vera Allerheiligen (Hain Lifescience Research Department) designed the probes and amplification protocol and controlled the development of the prototype kit.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Bogaerts, P., C. Berhin, H. Nizet, and Y. Glupczynski. 2006. Prevalence and mechanisms of resistance to fluoroquinolones in Helicobacter pylori strains from patients living in Belgium. Helicobacter 11:441-445. [DOI] [PubMed] [Google Scholar]
- 2.Burucoa, C., M. Garnier, C. Silvain, and J. L. Fauchere. 2008. Quadruplex real-time PCR assay using allele-specific scorpion primers for detection of mutations conferring clarithromycin resistance to Helicobacter pylori. J. Clin. Microbiol. 46:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burucoa, C., C. Landron, M. Garnier, and J. L. Fauchere. 2005. T2182C mutation is not associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 49:868-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattoir, V., J. Nectoux, C. Lascols, L. Deforges, J. C. Delchier, F. Megraud, C. J. Soussy, and E. Cambau. 2007. Update on fluoroquinolone resistance in Helicobacter pylori: new mutations leading to resistance and first description of a gyrA polymorphism associated with hypersusceptibility. Int. J. Antimicrob. Agents 29:389-396. [DOI] [PubMed] [Google Scholar]
- 5.Chen, S., Y. Li, and C. Yu. 2008. Oligonucleotide microarray: a new rapid method for screening the 23S rRNA gene of Helicobacter pylori for single nucleotide polymorphisms associated with clarithromycin resistance. J. Gastroenterol. Hepatol. 23:126-131. [DOI] [PubMed] [Google Scholar]
- 6.Chey, W. D., and B. C. Wong. 2007. American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am. J. Gastroenterol. 102:1808-1825. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach, L., and E. L. Evans. 2007. Meta-analysis: the effect of antibiotic resistance status on the efficacy of triple and quadruple first-line therapies for Helicobacter pylori. Aliment. Pharmacol. Ther. 26:343-357. [DOI] [PubMed] [Google Scholar]
- 8.Glocker, E., H. P. Stueger, and M. Kist. 2007. Quinolone resistance in Helicobacter pylori isolates in Germany. Antimicrob. Agents Chemother. 51:346-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glupczynski, Y., F. Megraud, M. Lopez-Brea, and L. P. Andersen. 2001. European multicentre survey of in vitro antimicrobial resistance in Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 20:820-823. [DOI] [PubMed] [Google Scholar]
- 10.Graham, D. Y., and A. Shiotani. 2008. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat. Clin. Pract. Gastroenterol. Hepatol. 5:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guimaraes, N., N. F. Azevedo, C. Figueiredo, C. W. Keevil, and M. J. Vieira. 2007. Development and application of a novel peptide nucleic acid probe for the specific detection of Helicobacter pylori in gastric biopsy specimens. J. Clin. Microbiol. 45:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillemann, D., S. Rusch-Gerdes, and E. Richter. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 45:2635-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillemann, D., M. Weizenegger, T. Kubica, E. Richter, and S. Niemann. 2005. Use of the genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 43:3699-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaakoush, N. O., C. Asencio, F. Megraud, and G. L. Mendz. 2009. A redox basis for metronidazole resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 53:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J. M., J. S. Kim, N. Kim, H. C. Jung, and I. S. Song. 2005. Distribution of fluoroquinolone MICs in Helicobacter pylori strains from Korean patients. J. Antimicrob. Chemother. 56:965-967. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J. M., J. S. Kim, N. Kim, Y. J. Kim, I. Y. Kim, Y. J. Chee, C. H. Lee, and H. C. Jung. 2008. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J. Microbiol. Biotechnol. 18:1584-1589. [PubMed] [Google Scholar]
- 17.Kist, M. 2007. Helicobacter pylori: primary antimicrobial resistance and first-line treatment strategies. Euro. Surveill. 12:E1-E2. [DOI] [PubMed] [Google Scholar]
- 18.Lascols, C., D. Lamarque, J. M. Costa, C. Copie-Bergman, J. M. Le Glaunec, L. Deforges, C. J. Soussy, J. C. Petit, J. C. Delchier, and J. Tankovic. 2003. Fast and accurate quantitative detection of Helicobacter pylori and identification of clarithromycin resistance mutations in H. pylori isolates from gastric biopsy specimens by real-time PCR. J. Clin. Microbiol. 41:4573-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lottspeich, C., A. Schwarzer, K. Panthel, S. Koletzko, and H. Russmann. 2007. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J. Clin. Microbiol. 45:1718-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malfertheiner, P., F. Megraud, C. O'Morain, F. Bazzoli, E. El-Omar, D. Graham, R. Hunt, T. Rokkas, N. Vakil, and E. J. Kuipers. 2007. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut 56:772-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Megraud, F. 2004. Helicobacter pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut 53:1374-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megraud, F., and P. Lehours. 2007. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin. Microbiol. Rev. 20:280-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore, R. A., B. Beckthold, S. Wong, A. Kureishi, and L. E. Bryan. 1995. Nucleotide sequence of the gyrA gene and characterization of ciprofloxacin-resistant mutants of Helicobacter pylori. Antimicrob. Agents Chemother. 39:107-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishizawa, T., H. Suzuki, K. Kurabayashi, T. Masaoka, H. Muraoka, M. Mori, E. Iwasaki, I. Kobayashi, and T. Hibi. 2006. Gatifloxacin resistance and mutations in gyrA after unsuccessful Helicobacter pylori eradication in Japan. Antimicrob. Agents Chemother. 50:1538-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishizawa, T., H. Suzuki, A. Umezawa, H. Muraoka, E. Iwasaki, T. Masaoka, I. Kobayashi, and T. Hibi. 2007. Rapid detection of point mutations conferring resistance to fluoroquinolone in gyrA of Helicobacter pylori by allele-specific PCR. J. Clin. Microbiol. 45:303-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noguchi, N., E. Rimbara, A. Kato, A. Tanaka, K. Tokunaga, T. Kawai, S. Takahashi, and M. Sasatsu. 2007. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J. Med. Microbiol. 56:1174-1180. [DOI] [PubMed] [Google Scholar]
- 27.Oleastro, M., A. Menard, A. Santos, H. Lamouliatte, L. Monteiro, P. Barthelemy, and F. Megraud. 2003. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J. Clin. Microbiol. 41:397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rimbara, E., N. Noguchi, T. Kawai, and M. Sasatsu. 2008. Novel mutation in 23S rRNA that confers low-level resistance to clarithromycin in Helicobacter pylori. Antimicrob. Agents Chemother. 52:3465-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheu, S. M., B. S. Sheu, C. C. Lu, H. B. Yang, and J. J. Wu. 2009. Mixed infections of Helicobacter pylori: tissue tropism and histological significance. Clin. Microbiol. Infect. 15:253-259. [DOI] [PubMed] [Google Scholar]
- 30.Tankovic, J., C. Lascols, Q. Sculo, J. C. Petit, and C. J. Soussy. 2003. Single and double mutations in gyrA but not in gyrB are associated with low- and high-level fluoroquinolone resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 47:3942-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vakil, N. 2005. Primary and secondary treatment for Helicobacter pylori in the United States. Rev. Gastroenterol. Disord. 5:67-72. [PubMed] [Google Scholar]
- 32.van der Ende, A., L. J. van Doorn, S. Rooijakkers, M. Feller, G. N. Tytgat, and J. Dankert. 2001. Clarithromycin-susceptible and -resistant Helicobacter pylori isolates with identical randomly amplified polymorphic DNA-PCR genotypes cultured from single gastric biopsy specimens prior to antibiotic therapy. J. Clin. Microbiol. 39:2648-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Doorn, L. J., Y. Glupczynski, J. G. Kusters, F. Megraud, P. Midolo, N. Maggi-Solca, D. M. Queiroz, N. Nouhan, E. Stet, and W. G. Quint. 2001. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob. Agents Chemother. 45:1500-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woo, H. Y., D. I. Park, H. Park, M. K. Kim, D. H. Kim, I. S. Kim, and Y. J. Kim. 2009. Dual-priming oligonucleotide-based multiplex PCR for the detection of Helicobacter pylori and determination of clarithromycin resistance with gastric biopsy specimens. Helicobacter 14:22-28. [DOI] [PubMed] [Google Scholar]
- 35.Xing, J. Z., C. Clarke, L. Zhu, and S. Gabos. 2005. Development of a microelectronic chip array for high-throughput genotyping of Helicobacter species and screening for antimicrobial resistance. J. Biomol. Screen. 10:235-245. [DOI] [PubMed] [Google Scholar]