Abstract
Tracking novel influenza viruses which have the potential to cause pandemics, such as the pandemic (H1N1) 2009 virus, is a public health priority. Pandemic (H1N1) 2009 virus was first identified in Mexico in April 2009 and spread worldwide over a short period of time. Well-validated diagnostic tools that are rapid, sensitive, and specific for the detection and tracking of this virus are needed. Three real-time reverse transcription PCR (RT-PCR) assays for the amplification and detection of pandemic (H1N1) 2009 virus were developed, and their performance characteristics were compared with those of other published diagnostic assays. Thirty-nine samples confirmed to be positive for pandemic (H1N1) 2009 virus from Alberta, Canada, and six additional samples that were positive for influenza A virus but that were not typeable by using published seasonal influenza H1/H3 virus assays were available for this validation. Amplification and direct sequencing of the products was considered the “gold standard” for case identification. The new assays were sensitive and able to reproducibly detect virus in a 10−6 dilution of 4 × 106 50% tissue culture infective doses/ml when 5 μl was used as the template. They showed 100% specificity and did not cross-react with other respiratory viruses or seasonal influenza A virus subtypes. The coefficient of variation in crossing cycle threshold values for the detection of different template concentrations of pandemic (H1N1) 2009 virus was ≤3.13%, showing good reproducibility. The assays had a wide dynamic range for the detection of pandemic (H1N1) 2009 virus and utilized testing platforms appropriate for high diagnostic throughput with rapid turnaround times. We developed and validated these real-time PCR procedures with the goal that they will be useful for diagnosis and surveillance of pandemic (H1N1) 2009 virus. These findings will contribute to the informed management of this novel virus.
Novel influenza viruses introduced into the human population that are able to spread efficiently from human to human have the potential to cause pandemics with significant morbidity and mortality (5, 9, 16). A novel subtype of influenza A virus called pandemic (H1N1) 2009 virus was identified in Mexico and was reported by the Centers for Disease Control and Prevention (CDC; Atlanta, GA) and WHO in April 2009 (2, 3, 8). Cases of swine influenza virus infection in humans have been reported previously, but these viruses did not show evidence of efficient transmission between human hosts (11). However, pandemic (H1N1) 2009 virus is a novel subtype virus that transmits easily between humans, with 21 countries reporting cases within a month of the initial identification of the virus (4). It is essential that public health laboratories around the world undertake detailed surveillance to monitor the spread and impact of pandemic (H1N1) 2009 virus as well as to try to predict future changes in its virulence (8). Methods for the rapid diagnosis, case identification, and tracking of this novel pathogen in the human population are required to develop appropriate management strategies to mitigate morbidity and mortality.
Novel influenza viruses are first identified by amplification and sequencing of specific genes; these sequences can be compared to those already deposited in databases. Such a methodological approach is important for the identification of new viruses but generally requires high viral loads (or cultured virus) for good-quality sequence data to be obtained. The turnaround time required for this type of analysis negates its use as a frontline diagnostic test, especially with large numbers of clinical samples. Once sequences are available, it is important that public health laboratories develop and validate high-throughput sensitive, specific, and rapid assays for use for diagnostics and surveillance.
As a rapid response to the outbreak of pandemic (H1N1) 2009 virus, the CDC provided a detailed methodology for real-time PCR amplification and detection of this virus (2). Other conventional and real-time reverse transcription (RT)-PCR protocols for the detection of this virus have also recently been published (13). We have developed three real-time RT-PCR assays for the detection of pandemic (H1N1) 2009 virus using primers and hydrolysis probes targeting the hemagglutinin (HA) and matrix (M) genes. We report on the development, validation, and performance of these assays and compare the results with those obtained by using the CDC real-time procedures as well as a multiplex suspension microarray assay (the xTAG respiratory virus panel [RVP] assay) used for the detection of multiple respiratory viruses, including influenza A virus. Patient samples collected between 24 April and 18 May 2009 were used for the validation study.
Improved detection methods will aid with the identification of new cases of pandemic (H1N1) 2009 virus and ensure optimal management to minimize transmission to vulnerable individuals, as well as aid with surveillance activities to provide an understanding of the full impact of this virus in the community.
MATERIALS AND METHODS
Samples tested.
Samples (n = 5,436) submitted to the Provincial Laboratory for Public Health (ProvLab) from Alberta, Northwest Territory, Nunavut, and Yukon from 24 April to 18 May 2009 for testing for respiratory viruses were included in this analysis. Specimens from patients for whom a significant travel history, epidemiological links to pandemic (H1N1) 2009 virus, or severe respiratory infection (SRI) was indicated on the requisition were identified so that the respiratory virus testing algorithm could be adjusted to enhance its ability to detect influenza A virus in these specimens, as described in the next section.
Diagnostic testing algorithm for human seasonal influenza and pandemic (H1N1) 2009 virus.
Specimens from patients with a history of travel or exposure to pandemic (H1N1) 2009 virus or SRI were tested concurrently by the RVP assay on the Luminex platform (Luminex Molecular Diagnostics, Inc., Toronto, Ontario, Canada) and a real-time RT-PCR assay targeting the M gene developed by the CDC (2). The CDC referred to the assay as InfA in the original publication (2), but we use the term CDC-M assay in this report. Other specimens from patients with no history related to pandemic (H1N1) 2009 virus were tested by the RVP assay only. The total numbers of specimens tested by the different assays are provided below. The RVP assay can detect seasonal influenza A viruses and can specifically identify the circulating H1 and H3 subtypes (12); it has not been validated for use for the detection of pandemic (H1N1) 2009 virus. The CDC-M assay detects only influenza A viruses and does not provide additional subtyping information. A direct fluorescent-antigen (DFA) test (Imagen; Dako Diagnostics Ltd., Ely, Cambridgeshire, United Kingdom) for influenza A virus, influenza B virus, parainfluenza virus, and respiratory syncytial virus was also performed with samples for which rapid screening would be useful, such as samples from patients with SRI.
Specimens that gave a positive result for influenza A virus by the DFA, CDC-M, or RVP assay but were not subtyped as seasonal H1 or H3 viruses by the RVP assay were subjected to real-time RT-PCR assays, as described previously (2), for the typing of seasonal H1 and H3 subtypes. The protocol for these typing assays was distributed in December 2008 through U.S. Public Health Service laboratories and the WHO Global Influenza Surveillance Network; it has been approved for use by the Food and Drug Administration. All influenza A virus-positive samples for which a valid subtype by the RVP assay or the real-time typing methods was not obtained were subjected to conventional RT-PCR (cRT-PCR) assays targeting the HA and M genes for amplification and sequencing, as described below. Table 1 provides a list of the assays used for this validation study and clarifies the source and purpose of each test.
TABLE 1.
Real-time and conventional RT-PCR assays used for the study
| Assay | Source or reference | Purpose |
|---|---|---|
| CDC-Ma | CDC (2) | Detection of all influenza A virus subtypes |
| CDC-H1-swineb | CDC (2) | Real-time RT-PCR for pH1N1_2009c confirmation |
| In-house HA | This study | Real-time RT-PCR for pH1N1_2009 confirmation |
| In-house M1 | This study | Real-time RT-PCR for pH1N1_2009 confirmation |
| In-house M2 | This study | Real-time RT-PCR for pH1N1_2009 confirmation |
| cRT-PCR (HA) | NML | Gel-based assay for pH1N1_2009 confirmation |
| cRT-PCR (M) | 7 | Gel-based assay for detection of all influenza A virus subtypes |
| NASBA (NPd) | 10 | Detection of all influenza A virus subtypes |
cRT-PCR amplification and confirmation of pandemic (H1N1) 2009 virus by sequencing.
cRT-PCR was performed by using a One-Step RT-PCR kit (Qiagen, Mississauga, Ontario, Canada) in a total volume of 50 μl. This included 10 μl of 5× RT-PCR buffer, 10 μl of Q solution, 2 μl of 10 mM deoxynucleoside triphosphates, 0.125 μl of 40 U/μl RNaseOUT (Invitrogen), 2 μl of One-Step RT-PCR enzyme mix, 3.0 μl each of 10 μM primers, and 15 μl of RNase-free water. Primers H1_F_swine and H1_R_swine were provided by the National Microbiology Laboratory (NML; Public Health Agency of Canada, Winnipeg, Manitoba, Canada; unpublished data) for specific amplification of a 517-bp fragment of the HA gene of pandemic (H1N1) 2009 virus. Primers published previously (7) were used for amplification of a 244-bp fragment of the M gene of all influenza A virus subtypes, including pandemic (H1N1) 2009 virus. The thermal cycling conditions comprised a 30-min RT step at 50°C; a 15-min initial PCR activation step at 95°C; and 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s each, followed by a 10-min extension at 72°C. All thermal cycling was performed with a model 2720 thermal cycler (Applied Biosystems [ABI], Foster City, CA). The PCR products were subjected to gel electrophoresis for visualization of the products. For confirmation, the products were purified with QIAquick columns (Qiagen) and sequenced in both directions by using an ABI Prism BigDye Terminator (version 3.1) cycle sequencing kit in an ABI Prism 3130-Avant genetic analyzer on a 50-cm array. The sequencing reaction was performed according to the manufacturer's protocol in a final volume of 10 μl with approximately 2 ng of amplified product, BigDye Terminator (version 3.1) premix, and 0.64 pmol of the primer used for RT-PCR. Unincorporated nucleotides were purified by using a BigDye XTerminator purification kit (ABI). The data were collected by using ABI software (version 2.0). The sequences were analyzed by using Sequencing Analysis software (version 5.3; ABI), and alignments were performed by using the ClustalW program (version 1.4) included in the BioEdit package (version 7.0.0; http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The sequences were compared to the previously published sequences of pandemic (H1N1) 2009 virus and seasonal influenza virus subtypes available from the GenBank database and the Global Initiative on Sharing Avian Influenza Data (GISAID; http://platform.gisaid.org). Sequences with >99% nucleotide identity to database submissions were confirmed to be positive for pandemic (H1N1) 2009 virus.
Comparison of different assays for confirmation of pandemic (H1N1) 2009 virus.
A subset of the samples confirmed to be positive for pandemic (H1N1) 2009 virus by sequencing (n = 39 of 97 samples identified during this time period) and 6 samples that gave a positive result for influenza A virus but could not be subtyped as seasonal H1 or H3 virus by the real-time typing assays or as pandemic (H1N1) 2009 virus by cRT-PCR and sequencing were included in the evaluation and validation of the three in-house assays described here. The results were compared with those of the swH1 assay published previously (2); that assay is referred to as the CDC-H1-swine assay in this report. Assay details are described below. Table 1 provides the list of assays used in this comparison study. Of the 45 specimens included in this assay validation, 30 were nasopharyngeal swab specimens collected in universal transport medium (UTM; Copan Diagnostics Inc., Murrieta, CA), 10 were throat swab specimens in UTM, 4 were formalin-fixed tissue samples, and 1 was a respiratory swab specimen of unknown source that was received in UTM.
(i) Sample preparation.
Respiratory samples were pretreated with 25 μl of Qiagen protease (0.01 milli-Anson units/μl) in a thermomixer (Eppendorf, Westbury, NY) at 56°C and 1,000 rpm for 15 min or until the specimen cleared. Nucleic acid was extracted from the treated samples by using an easyMAG automated extractor (bioMérieux, Durham, NC), according to the manufacturer's instructions. The extracted nucleic acid was eluted in 110 μl from a sample input volume of 200 μl. Tissue samples were extracted by using the total nucleic acid extraction protocol for tissue specimens by using the QIAamp kit (Qiagen), according to the manufacturer's instructions.
(ii) Design of primers and probes for in-house real-time RT-PCR assays.
Three sets of primers and probes were designed for this study. All available HA and M gene sequences from GenBank and GISAID (as of 25 April 2009) were aligned, and the three sets of primers and probes were designed to amplify pandemic (H1N1) 2009 virus specifically by using the Primer Express (version 3.0) program (ABI). These primers and probes are listed in Table 2. A review of the sequences submitted to the online databases up to 20 May 2009 confirmed that these in-house-designed primers and probes would detect pandemic (H1N1) 2009 virus sequences available up to that date.
TABLE 2.
Primers and probes designed in-house for detection of pandemic (H1N1) 2009 virus
| Assay and gene target | Primer/probe name | Nucleotide locationa | Primer/probe sequence (5′-3′) |
|---|---|---|---|
| In-house HA assay | |||
| HA | SwineHA-359_For | 359-381 | AGCAATTGAGCTCAGTGTCATCA |
| HA | SwineHA-405_Rev | 405-424 | TGGGCCATGAACTTGTCTTG |
| HA | SwineHA-386_Probe | 386-403 | FAM-AAAGGTTTGAGATATTCC-BHQ1b |
| In-house M1 assay | |||
| M | SwineM-259_For | 259-283 | CGAACAACATGGATAGAGCAGTTAA |
| M | SwineM-307_Rev | 307-326 | GGCCCCATGGAACGTTATTT |
| M | SwineM-286_Probe | 286-305 | FAM-TATACAAGAAGCTCAAAAGA-BHQ1 |
| In-house M2 assay | |||
| M | SwineM-408_For | 408-432 | ACAGAAGCTGCTTTTGGTCTAGTGT |
| M | SwineM-455_Rev | 455-485 | TGAGACCGATGCTGTGAATCA |
| M | SwineM-434_Probe | 434-450 | FAM-TGCCACTTGTGAACAGA-BHQ1 |
The nucleotide positions for the HA and M genes are based on sequences obtained from GISAID corresponding to EPI176470 HA A/California/04/2009 EPI_ISL_295732009712049_seg4H1N1 and EPI176471 MP A/California/04/2009 EPI_ISL_29573 2009712049_seg7 H1N1, respectively.
The minor groove binding hydrolysis probes were labeled with 6-carboxyfluorescein (FAM) at the 5′ end and a black hole quencher (BHQ1) at the 3′ end.
The in-house HA assay targets the 5′ end of the HA gene, and the in-house M1 and M2 assays target the 5′ end of the M gene. These assays are henceforth referred to as the in-house HA, in-house M1, and in-house M2 assays. All assays utilized minor groove binding hydrolysis probes and were labeled with 6-carboxyfluorescein as the reporter dye at the 5′ end and a black hole quencher at the 3′ end. The primers were synthesized at the University Core DNA Services (University of Calgary, Calgary, Alberta, Canada), and the probes were synthesized by ABI.
(iii) Real-time RT-PCR for detection of pandemic (H1N1) 2009 virus.
A one-step RT-PCR method was used for the detection of pandemic (H1N1) 2009 virus by the in-house-developed assays targeting the HA and M genes. The master mixture contained the TaqMan one-step RT-PCR master mix, the MultiScribe enzyme mixture, and the primers and probes at final concentrations of 0.8 μM and 0.2 μM, respectively. RT-PCR was performed in an SDS 7500 system in optical tubes from ABI by using 5 μl of extracted nucleic acid and 20 μl of the master mixture. The cycling conditions followed the TaqMan universal amplification protocol, according to the manufacturer's instructions. The reaction components for the CDC-M assay were the same as those described above. The cycling conditions included a RT reaction at 48°C for 30 min, followed by enzyme inactivation at 95°C for 10 min. The template was denatured at 95°C for 15 s, annealing and data acquisition were performed at 55°C, and extension was performed at 72°C for 45 cycles.
In addition to the three in-house assays, a published real-time RT-PCR assay designed for the subtyping of pandemic (H1N1) 2009 virus (the CDC-H1-swine assay) (2) was also performed with the available positive samples used in this validation study. This assay was performed on the SDS 7500 system by using the one-step RT-PCR master mixture (ABI) and the protocol provided by the CDC. This protocol was optimized by using an Invitrogen SuperscriptIII Platinum one-step quantitative kit that has been shown to have comparable results on thermocycler systems such as ABI real-time PCR systems (systems 7000, 7300, 7500, etc.), Bio-Rad real-time PCR detection systems (the iQ or iQ5 system), or Stratagene quantitative PCR instruments (the MX4000, MX3000, or MX3005 instrument). Our experiments comparing the use of the one-step RT-PCR master mixture from ABI with the Invitrogen SuperscriptIII Platinum one-step quantitative kit on the 7500 SDS system showed that the performance of the assays were similar (data not shown). Table 1 provides a list of the assays used for this validation study and clarifies the source and purpose of each test.
Limit of detection, sensitivity, specificity, reproducibility, and efficiency of the real-time RT-PCR assays.
The pandemic (H1N1) 2009 virus was propagated to 4 × 106 50% tissue culture infectious doses (TCID50)/ml at the NML, and nucleic acid extracted from this harvest was kindly provided to the public health laboratories involved in pandemic (H1N1) 2009 virus testing in Canada for use as control material. This extract was used as the template for all sensitivity studies. Tenfold serial dilutions from 10−4 to 10−8 of the extract were made in PCR-grade water to which carrier RNA (Qiagen) was added to a final concentration of 1 μg/μl. All extracts were tested in triplicate using three independent runs by the new in-house RT-PCR assays (the HA, M1, and M2 assays) and the CDC RT-PCR assays (the CDC-M and CDC-H1-swine assays) described above with 5 μl of template per reaction mixture (5 μl of template at a dilution of 10−4 contains 2 × 100 TCID50). The sensitivity is reported in terms of the TCID50, as a cloned plasmid is not yet available; the ratio between the TCID50 values and the genomic copy numbers can vary between strains and harvests; thus, these numbers cannot be considered absolute and provide only a relative comparison between the different assays used. These dilutions were also tested by the RVP assay and an influenza A virus nucleic acid sequence-based amplification (NASBA) assay targeting the nucleoprotein gene (10) to determine the limit of detection for pandemic (H1N1) 2009 virus. Six specimens that were influenza A virus positive by the screening assays but that could not be subtyped as human H1 or H3 virus by the real-time typing assays or confirmed to be the pandemic (H1N1) 2009 virus by cRT-PCR were tested by the new in-house and CDC-H1-swine assays to investigate if these assays could identify additional pandemic (H1N1) 2009 cases.
The specificities of the newly developed assays were determined by testing extracts of cultures from a variety of influenza A virus strains, including A/Beijing/95-like, A/Shangdong/11742/95-like, A/Taiwan/95-like, A/Texas/95-like, A/Johannesburg/96-like, A/Sydney/98-like, A/New Caledonia/00-like, A/Panama/03-like, and A/Brisbane/59/07-like strains. In addition, 22 extracts from harvests of patient specimens that were positive for influenza A virus, including subtype H1N1, H1N2, and H3N2 viruses recovered from 1995 to 2009, were used to determine that the pandemic (H1N1) 2009 virus-specific assays did not detect seasonal influenza A viruses. These specimens included four influenza A viruses of the H3N2 subtype and two of the H1N1 subtype that were cocirculating in the community during the same time period as the pandemic (H1N1) 2009 virus after 24 April 2009. The extract from an influenza A virus (H3N2) of probable swine origin designated A/Canada/1158/2006 isolated from a child living on a farm in Canada (14) was tested to determine the specificities of the assays. Samples with high copy numbers of common respiratory pathogens, including influenza B virus; parainfluenza virus types 1, 2, 3, and 4B; respiratory syncytial viruses A and B; human coronavirus NL63, 229E, and OC43; rhinovirus type 1B; coxsackievirus type A16; human metapneumovirus; adenovirus type 2; bocavirus; Chlamydophila pneumoniae; Legionella pneumophila; and Mycoplasma pneumoniae, were also tested by the in-house HA, M1, and M2 assays and the CDC-H1-swine assay.
The reproducibilities of all the real-time RT-PCR assays described above on the SDS 7500 system were evaluated in two independent runs by using three positive clinical specimens with a range of viral loads tested in five replicates.
Tenfold serial dilutions of the pandemic (H1N1) 2009 virus control nucleic acid diluted in carrier RNA were used to determine the dynamic range of the in-house assays and the CDC-M and CDC-H1-swine assays. The efficiency of the PCR was calculated on the basis of the crossing cycle threshold (CT) values obtained.
Data analysis.
Samples which gave equivocal results by the RVP assay were excluded from the sensitivity and specificity analyses; equivocal results were defined by the manufacturer to be those with a median fluorescent intensity (MFI) of between 150 and 300. The McNemar test was used to test for a significant difference between the detection of influenza A virus by the RVP assay and the CDC-M assay by using any nucleic acid amplification test (NAT) result positive for influenza A virus as a true-positive result. The differences in CDC-M CT values for samples that gave negative, equivocal, or positive results by the RVP assay were compared by the Kruskal-Wallis test. The CT values of the various real-time assays for the 39 samples confirmed to be positive for pandemic (H1N1) 2009 virus were compared by the Friedman test.
RESULTS
Detection of influenza A virus by RVP and CDC-M assays.
During the 25-day study period, 5,436 respiratory specimens from Alberta, Northwest Territories, Yukon, and Nunavut were tested for respiratory viruses at ProvLab by the RVP and/or CDC-M assay. A total of 217 (4.0%) samples were positive for influenza A virus. Of the 1,465 specimens tested by this method, 3 samples were initially identified to be positive for virus by DFA, 156 were positive by both the RVP and the CDC-M assays, 29 were positive by the CDC-M assay (of which 17 were negative and 12 gave an equivocal result by the RVP assay), 1 was positive by the RVP assay but negative by the CDC-M assay, and 28 were positive by the RVP assay but no CDC-M assay was performed with these samples. The H-typing results for the 217 influenza A virus-positive samples were as follows: 97 were confirmed to be pandemic (H1N1) 2009 virus; 80 were confirmed to be seasonal H3 virus; 28 were confirmed to be seasonal H1 virus; and 9 were unresolved, likely because of low viral loads. HA typing of the virus in the remaining three specimens was not completed because they were duplicate samples from patients who had tested positive for pandemic (H1N1) 2009 virus.
For the 4,128 samples tested by both the CDC-M and the RVP assays, the sensitivity of the RVP assay was 90.2% (157/174 samples; 95% confidence interval [CI], 84.8 to 94.2%). The sensitivity of the CDC-M assay was 99.4% (173/174 samples; 95% CI, 96.8 to 100%). The specificity was 100% (3,954/3,954 samples) for both assays (n = 4,128; P < 0.001, McNemar test).
Of the 17 specimens that gave a negative result for influenza A virus by the RVP assay but a positive result by the CDC-M assay, 2 were typed as pandemic (H1N1) 2009 virus, 5 as seasonal H3 virus, and 1 as seasonal H1 virus; the types in the remaining 9 specimens were unresolved because of low viral loads. Of the 12 specimens with an equivocal result by the RVP assay but a positive result by the CDC-M assay, 7 were typed as pandemic (H1N1) 2009 virus and 5 as seasonal H3 virus. The median CT values obtained by the CDC-M assay for samples that were negative by the RVP assay and equivocal by the RVP assay or that had concordant influenza A virus-positive results by both the RVP and the CDC-M assays were 36.0 (range, 31.6 to 38.0), 34.7 (range, 29.7 to 36.6), and 26.4 (range, 13.5 to 36.2), respectively (P < 0.001, Kruskal-Wallis test). The one sample that was influenza A virus positive by the RVP assay but negative by the CDC-M assay was confirmed to be pandemic (H1N1) 2009 virus.
Assessment of pandemic (H1N1) 2009 virus RT-PCR assay performance.
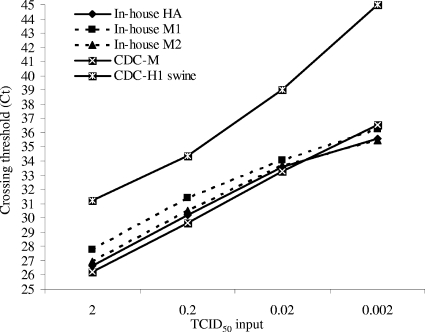
The results for the 10-fold serial dilutions tested in nine replicates are shown in Table 3 and Fig. 1. Table 3 shows the number of replicates that were positive at each template concentration. The NASBA signal and MFI for the RVP assay are also shown in Table 3. The CT values obtained for the real-time RT-PCR assays performed on the SDS 7500 system at each template concentration are given in Fig. 1. All assays except the NASBA assay had comparable end-point sensitivities and were able to detect the control pandemic (H1N1) 2009 virus-positive nucleic acid reproducibly with an input of 2 × 10−2 TCID50; the NASBA assay detected seven of the nine replicates.
TABLE 3.
Limit of detection analysis for the different assays
| Virus concn (no. of TCID50s/5 μla) | No. of positive replicates of nine tested for the following amplification assay: |
||||||
|---|---|---|---|---|---|---|---|
| In-house HA | In-house M1 | In-house M2 | CDC-M | CDC-H1-swine | RVP (MFI) | NASBA (signalb) | |
| 2 × 100 | 9 | 9 | 9 | 9 | 9 | 9 (7,302) | 9 (2.89) |
| 2 × 10−1 | 9 | 9 | 9 | 9 | 9 | 9 (2,680) | 9 (2.73) |
| 2 × 10−2 | 9 | 9 | 9 | 9 | 9 | 9 (433) | 7 (2.46) |
| 2 × 10−3 | 4 | 4 | 5 | 6 | 2 | 0 | 3 (2.28) |
Five microliters of template was used per reaction mixture.
The fluorescence signal above the background.
FIG. 1.
Comparison of CT values for detection of serial dilutions of pandemic (H1N1) 2009 virus. The CT values are shown for the hydrolysis probe-based assays performed on the SDS 7500 system. The mean CT value from the replicates is indicated. The number of replicates with a positive result for each dilution is indicated in Table 3.
The newly designed assays did not amplify representative specimens of seasonal influenza A viruses, including subtypes H1N1, H1N2, and H3N2 recovered from 1995 to 2008; antigenic drift variants; an H3N2 virus of swine origin isolated in 2006; and the other viral and bacterial respiratory pathogens tested. This indicates that the newly designed assays show 100% specificity for the detection of pandemic (H1N1) 2009 virus. The assays did not detect the human influenza A virus H1N1 and H3N2 subtypes, which were cocirculating with pandemic (H1N1) 2009 virus between 24 April and 18 May 2009 in Alberta.
Three clinical specimens were tested by using the SDS 7500 system in five replicates in two independent runs. The coefficient of variation for the CT values ranged from 0.36% to 3.13% for all five assays tested, showing that the assays have good reproducibilities.
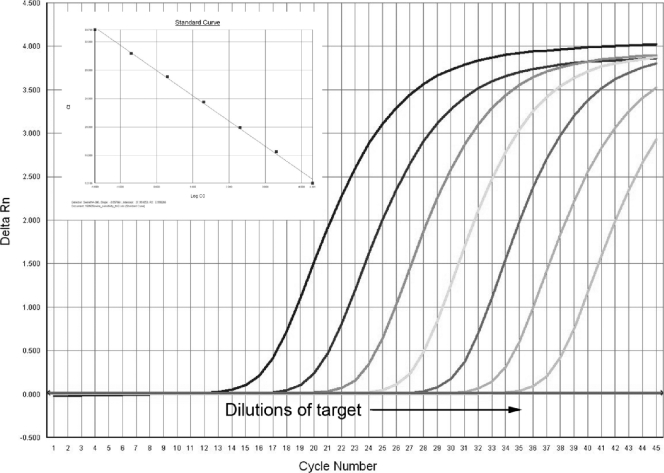
All the assays were able to detect 10-fold serial dilutions of the control material over 7 log units of template dilution from 4 × 106 TCID50/ml to 4 × 100 TCID50/ml when 5 μl of template was used. On the basis of these CT values, the PCR amplification efficiency of the different assays ranged from 82.17% to 91.04%. A representative example of an amplification curve for the in-house HA assay and the standard curve generated by using these dilutions is given in Fig. 2.
FIG. 2.
Representative amplification curves for 10-fold dilutions of template RNA obtained by the in-house HA assay and the standard curve generated by using these dilutions. Tenfold serial dilutions of template starting at 4 × 106 TCID50/ml were tested by using 5 μl of template per reaction mixture. The features of the standard curve were as follows: slope, −3.56; R2, 0.99.
Comparison of RT-PCR results for detection of pandemic (H1N1) 2009 virus-positive specimens.
A subset of all positive samples detected was used for the validation study. Extracts from 39 specimens positive for influenza A virus by the CDC-M screening assay and confirmed to be positive for pandemic (H1N1) 2009 virus by cRT-PCR were retrospectively tested by the CDC-H1-swine assay and the three in-house PCR assays. The CT values obtained by the CDC-M assay ranged from 17.1 to 36.6 (median, 27.1). Of these 39 specimens, 35 were influenza A virus positive, 3 were equivocal, and 1 was negative by the RVP assay. As expected, none of the pandemic (H1N1) 2009 virus-positive samples were typed by the RVP assay (subtyping is based on human influenza virus sequences). The MFI values for the positive samples ranged from 313 to 9,835 (median, 7,151). One specimen containing the pandemic (H1N1) 2009 virus sequence was negative for influenza A virus by the RVP assay but gave a CT of 33.53 by the CDC-M assay, suggesting the presence of a low viral load. These results are summarized in Table 4.
TABLE 4.
Analysis of influenza A virus-positive specimens tested for pandemic (H1N1) 2009 virus
| Assaya | Influenza A_pH1N1_2009b (n = 39) |
No. of influenza A_unresolved samples (n = 6) | % Sensitivityd | 95% CId | ||
|---|---|---|---|---|---|---|
| No. of samples |
CTc |
|||||
| Median | Range | |||||
| RVP | 35e | 0 | ||||
| CDC-M | 39 | 27.1 | 17.1-36.6 | 6 | ||
| CDC-H1-swine | 34 | 32.1 | 21.9-45.0 | 1 | 83.33 | 68.6-93.0 |
| In-house HA | 38 | 25.9 | 17.0-45.0 | 2 | 95.24 | 83.8-99.4 |
| In-house M1 | 36 | 29.1 | 19.5-45.0 | 2 | 90.48 | 77.4-97.3 |
| In-house M2 | 38 | 27.8 | 18.8-45.0 | 3 | 97.62 | 87.4-99.9 |
The CDC-M and RVP assays were performed for the detection of influenza A virus, and gel/sequencing methods (cRT-PCR) were used for the prospective confirmation of pandemic (H1N1) 2009 virus. Detection of the pandemic (H1N1) 2009 virus by the real-time assays was performed retrospectively.
pH1N1_2009, abbreviation for pandemic (H1N1) 2009 virus.
The median and range of CT values is indicated for the different assays (P < 0.001, Friedman test). A CT value of 45 was used to represent a negative result.
The sensitivity and 95% CI calculations were performed by using 42 samples with positive results (39 samples confirmed to be positive by cRT-PCR and 3 samples positive by one or more pandemic [H1N1] 2009 real-time typing assays).
Of the 39 samples tested by the RVP assay, 35 were positive, 3 had a MFI defined as equivocal, and 1 was negative by the RVP assay.
The CT values obtained by the different assays (the CDC-M, CDC-H1-swine, and three in-house assays) for the detection of virus in these 39 samples are shown in Table 4 (P < 0.001, Friedman test). The virus could not be detected in one specimen by any of the real-time assays for pandemic (H1N1) 2009 virus. This specimen had given a weak positive result for influenza A virus by the CDC-M assay (CT value, 36.57) and an equivocal MFI of 153 by the RVP assay. The reason that the virus in this sample was missed by the pandemic (H1N1) 2009 virus-specific assays was likely due to the presence of a low viral load combined with additional freeze-thawing, leading to nucleic acid degradation before the sample was tested by the pandemic (H1N1) 2009 virus-specific real-time assays. The in-house HA and M2 assays gave positive results for 38 specimens, and the CT values were comparable to those from the CDC-M assay (Table 4). The in-house M1 assay gave positive results for 36 of the 39 samples containing pandemic (H1N1) 2009 virus sequences (CT values for the samples in which virus was not detected by the CDC-M assay, 34.80, 34.96, and 36.57). The CT values obtained by the in-house M1 assay were higher than those obtained by the CDC-M and in-house HA and M2 assays. The CDC-H1-swine assay gave positive results for 34 samples containing pandemic (H1N1) 2009 virus sequences (CT values, 34.53, 33.73, 34.65, 34.42, and 36.57 by the CDC-M assay for the 5 samples in which virus was not detected by the CDC-H1-swine assay). These results suggest that the in-house HA and M2 assays have sensitivities comparable to the sensitivity of the CDC-M assay and provide slightly better sensitivities than the in-house M1 and CDC-H1-swine assays for the detection of pandemic (H1N1) 2009 virus in clinical samples, as summarized in Table 4.
Six specimens that were influenza A virus positive by the CDC-M assay but that could not be subtyped as human H1, H3, or pandemic (H1N1) 2009 virus were tested by the new real-time assays to investigate if they provided enhanced sensitivity. These specimens gave positive results for influenza A virus by the CDC-M assay and had CT values that ranged from 31.64 to 37.64 (median, 36.02), but they were all influenza A virus negative by the RVP assay. Of these six nontypeable specimens, one was positive for pandemic (H1N1) 2009 virus by all four real-time PCR assays, one gave a positive result for pandemic (H1N1) 2009 virus by the three in-house assays, and the in-house M2 assay detected an additional pandemic (H1N1) 2009 virus-positive specimen. The CT values (by the CDC-M assay) of three samples which could not be typed by the new pandemic (H1N1) 2009 virus RT-PCR assays ranged from 36.00 to 37.64, suggesting that they had very low viral loads. Thus, additional testing by our newly developed assays identified three new cases of pandemic (H1N1) 2009 virus infection.
When the 42 positive specimens (39 confirmed to be positive by cRT-PCR and 3 that were positive by one or more pandemic [H1N1] 2009 virus real-time typing assays) are included, the sensitivities for the cRT-PCR, CDC-H1-swine, in-house HA, in-house M1, and in-house M2 assays were 92.86% (95% CI, 80.5 to 98.5%), 83.33% (95% CI, 68.6 to 93.0%), 95.24% (95% CI, 83.8 to 99.4%), 90.48% (95% CI, 77.4 to 97.3%), and 97.62% (95% CI, 87.4 to 99.9%), respectively. This information is also included in Table 4.
DISCUSSION
Influenza A virus pandemics can arise when genetic reassortment of the influenza A virus in bird and swine populations leads to the emergence of a novel virus to which majority of the population is susceptible. Two distinct lineages of swine influenza A viruses of the H1N1 subtype currently circulate within the North American and European pig populations (6). Available sequence data indicate that six segments of the currently circulating pandemic (H1N1) 2009 virus are related to swine viruses from North America and that the NA and M genes are related to swine viruses from Europe/Asia (15). Early reports concerning the pandemic potential of the circulating pandemic (H1N1) 2009 virus have suggested that the transmissibility of this virus is substantially higher than that of seasonal influenza virus (8).
NATs are increasingly being used for the identification of respiratory virus infections, including those caused by influenza viruses. NATs provide a rapid and sensitive means of detection of the etiological agent and are not easily compromised by sample quality or the timing of collection. Compared with traditional methods for respiratory virus detection (culture, antigen detection), NATs have enhanced sensitivities, specificities, and turnaround times. For novel viruses such as pandemic (H1N1) 2009 virus, it is important that a rapid diagnosis be provided without the need for culture to ensure that these tests can be performed even in laboratories without biosafety level 2 facilities.
In our diagnostic algorithm, the CDC-M and RVP assays were used to screen patient specimens for influenza A virus. Positive specimens were subtyped to identify seasonal H1 and H3 viruses and pandemic (H1N1) 2009 virus. The higher sensitivity of the CDC-M assay for the detection of influenza A virus compared with that of the RVP assay did not seem to be related to the H types but seemed to be more directly linked to the amount of virus in the sample, as estimated by the CT values. Sensitive real-time assays for the subtyping of seasonal H1 and H3 viruses but not pandemic (H1N1) 2009 virus were already available at the time of this study. Initial identification and confirmation of pandemic (H1N1) 2009 virus were performed by sequencing, which is labor-intensive, involves multiple steps, and is not readily available in many diagnostic laboratories. In order to facilitate rapid subtyping, we designed three real-time RT-PCR assays based on the available sequences for pandemic (H1N1) 2009 virus recovered at the outset of the outbreak. Comparison of the performance characteristics of those assays to the performance characteristics of assays provided by the CDC for the detection of all influenza virus subtypes (the CDC-M assay) and the specific detection of pandemic (H1N1) 2009 virus (CDC-H1-swine assay) showed that these in-house assays have comparable (and perhaps enhanced) sensitivity for the identification of individuals infected with pandemic (H1N1) 2009 virus. Due to the limited availability of positive specimens, a relatively small number was used to validate the new assays, but these tests will be monitored prospectively for additional validation data. Although we have not compared our newly developed assay with those reported previously, the limit-of-detection studies suggest that the assay has a sensitivity comparable to that of other reported methods for the detection of nucleic acid for pandemic (H1N1) 2009 virus (13). These new assays will be used in conjunction with the influenza A virus screening and seasonal H1/H3 subtyping assays for the diagnosis and surveillance of pandemic (H1N1) 2009 virus. The assays were specific and did not show any cross-reaction with common respiratory viruses or a variety of influenza A viruses circulating from 1995 to 2009, including human H1 and H3 virus subtypes cocirculating with the pandemic (H1N1) 2009 virus.
As the influenza season begins in the Southern Hemisphere, there may be further opportunities for the pandemic (H1N1) 2009 virus to reassort and mutate. A range of good diagnostic tools for tracking cases will be important in our future planning for the pandemic and the management of this (and other) novel influenza viruses. Assays targeting conserved genes (e.g., the M, nucleoprotein, and polymerase genes) are useful for the screening of patient specimens for influenza A virus but will not differentiate between seasonal influenza A viruses and viruses such as the pandemic (H1N1) 2009 virus. However, they are less likely to be prone to problems relating to sequence variations and point mutations. Assays targeting surface genes, such as the HA gene, are important for subtyping and antigenic characterization but may be subject to sequence variation, especially in the region where short hydrolysis probes bind to the target sequence. Single point mutations may disrupt primer or probe binding. Thus, a combination of screening and typing assays provides an optimal diagnostic algorithm for the detection of influenza A viruses and monitoring of pandemic (H1N1) 2009 virus infections.
The future evolution of pandemic (H1N1) 2009 virus and, thus, its transmissibility, antigenicity, virulence, and antiviral resistance are impossible to predict (8, 9). These factors can vary depending on whether the antigenic changes in the virus are incremental or if the mutations in the viral genome go from point to point in a confined space (1, 17). The tracking of sequence changes will be vital to anticipating the pandemic potential of this novel subtype of influenza A virus.
Acknowledgments
We thank all staff at ProvLab for their efforts with accessioning, data entry, testing, and reporting of results for all the samples during the time of enhanced laboratory testing and surveillance. We thank the technologists in the Molecular Diagnostics Division for excellent technical support. Special thanks go to Kara Tokaryk and Sandy Shokoples for their technical support.
Footnotes
Published ahead of print on 2 September 2009.
REFERENCES
- 1.Allen, J. E., S. N. Gardner, E. A. Vitalis, and T. R. Slezak. 2009. Conserved amino acid markers from past influenza pandemic strains. BMC Microbiol. 9:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2009. Outbreak of swine-origin influenza A (H1N1) virus infection—Mexico, March-April 2009. MMWR Morb. Mortal. Wkly. Rep. 58:467-470. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2009. Update: novel influenza A (H1N1) virus infections—worldwide, May 6, 2009. MMWR Morb. Mortal. Wkly. Rep. 58:453-458. [PubMed] [Google Scholar]
- 5.de Wit, E., and R. A. M. Fouchier. 2008. Emerging influenza. J. Clin. Virol. 41:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunham, E. J., V. G. Dugan, E. K. Kaser, S. E. Perkins, I. H. Brown, E. C. Holmes, and J. K. Taubenberger. 2009. Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J. Virol. 83:5485-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., T. M. Bestebroer, S. Herfst, K. L. Van Der, G. F. Rimmelzwaan, and A. D. Osterhaus. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J. Clin. Microbiol. 38:4096-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser, C., C. A. Donnelly, S. Cauchemez, W. P. Hanage, K. Van, T. D. Hollingsworth, J. Griffin, R. F. Baggaley, H. E. Jenkins, E. J. Lyons, T. Jombart, W. R. Hinsley, N. C. Grassly, F. Balloux, A. C. Ghani, N. M. Ferguson, A. Rambaut, O. G. Pybus, H. Lopez-Gatell, C. M. Pluche-Aranda, I. B. Chapela, E. P. Zavala, D. M. Guevara, F. Checchi, E. Garcia, S. Hugonnet, and C. Roth. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallaher, W. R. 2009. Towards a sane and rational approach to management of influenza H1N1 2009. Virol. J. 6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore, C., S. Hibbitts, N. Owen, S. A. Corden, G. Harrison, J. Fox, C. Gelder, and D. Westmoreland. 2004. Development and evaluation of a real-time nucleic acid sequence based amplification assay for rapid detection of influenza A. J. Med. Virol. 74:619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers, K. P., C. W. Olsen, and G. C. Gray. 2007. Cases of swine influenza in humans: a review of the literature. Clin. Infect. Dis. 44:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pabbaraju, K., K. L. Tokaryk, S. Wong, and J. D. Fox. 2008. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46:3056-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poon, L. L., K. H. Chan, G. J. Smith, C. S. Leung, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2009. Molecular detection of a novel human influenza (H1N1) of pandemic potential by conventional and real-time quantitative RT-PCR assays. Clin. Chem. 55:1555-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson, J. L., B. E. Lee, J. Patel, N. Bastien, K. Grimsrud, R. F. Seal, R. King, F. Marshall, and Y. Li. 2007. Swine influenza (H3N2) infection in a child and possible community transmission, Canada. Emerg. Infect. Dis. 12:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trifonov, V., H. Khiabanian, B. Greenbaum, and R. Rabadan. 2009. The origin of the recent swine influenza A(H1N1) virus infecting humans. Euro. Surveill. 14:19193.. [PubMed] [Google Scholar]
- 16.Tumpey, T. M., and J. A. Belser. 2009. Resurrected pandemic influenza viruses. Annu. Rev. Microbiol. 63:79-98. [DOI] [PubMed] [Google Scholar]
- 17.Wikramaratna, P. S., and S. Gupta. 2009. Influenza outbreaks. Cell. Microbiol. 11:1016-1024. [DOI] [PubMed] [Google Scholar]