Abstract
The NucliSENS easyMAG automated system was compared to the column-based Qiagen method for Epstein-Barr virus (EBV) or cytomegalovirus (CMV) DNA extraction from whole blood before viral load determination using the corresponding R-gene amplification kits. Both extraction techniques exhibited a total agreement of 81.3% for EBV and 87.2% for CMV.
Epstein-Barr virus (EBV) and cytomegalovirus (CMV) infections represent a significant clinical threat for immunocompromised patients. The frequent determination of EBV and CMV viral load permits the early diagnosis of infection, start of preemptive or curative therapy, and monitoring of treatment efficiency (5, 17, 20). By comparison to serum, plasma, or white-blood-cell fractions, whole-blood samples are now recognized as the most suitable sample for the determination of viral loads for EBV and CMV (3, 4, 7, 9, 10, 12, 13, 18, 19).
Although the methods relying on silica columns are time-consuming and need trained experimenters, these methods are considered the gold standard for the extraction of nucleic acids from whole-blood samples. Due to the large amount of genetic material in such samples, new extraction methods must be carefully evaluated, including those relying on automated devices (1, 7, 10, 14, 15). The fully automated NucliSENS easyMAG instrument (bioMérieux) using magnetic silica particles (2) allows the simultaneous process of up to 24 extractions. The use of magnetic particles eliminates the several centrifugation steps that could be a source of cross-contamination, and manual steps are limited to the loading of samples, reagents, and disposables. The performance of this method in the extraction of DNA from whole-blood samples prior to viral quantification has not been yet evaluated. The present study was undertaken to answer this question in the clinical context of EBV or CMV infection.
The whole-blood specimens selected for this study included 80 samples for EBV analysis and 94 samples for CMV analysis, taken from patients hospitalized at the University Hospital of Saint-Etienne, Saint-Etienne, France, from December 2007 to September 2008. The samples were kept frozen at −20°C. After the samples were thawed, whole-blood aliquots were tested and kept at 4°C for up to 24 h for potential retest. Two hundred microliters of each selected sample was extracted by two different technicians either by the reference manual method, i.e., QIAamp column DNA blood extraction kit according to the manufacturer's recommendations (Qiagen), or by the new specific B protocol on the NucliSENS easyMAG instrument. The latter protocol consists of the treatment of 200 μl of whole blood in 2 ml of lysis buffer and the capture of nucleic acids by 140 μl of magnetic silica. After incubation and washing procedures, nucleic acids were recovered in 50 μl of elution buffer. EBV and CMV loads were quantified by using the respective R-gene amplification kit (Argene Biosoft) according to the manufacturer's recommendations. Both amplification kits have been previously validated for quantification of EBV and CMV load in whole blood (8, 11). DNA extracts from both methods were amplified in the same run using an ABI 7500 instrument (Applied Biosystems).
Qualitative analysis.
As shown in Table 1, the overall agreement between the two extraction techniques was 81.3% (72.7 to 89.8%) for EBV and 87.2% (80.5 to 94.0%) for CMV. All the discrepant results corresponded to low DNA loads: between 20 and 459 copies/ml for the 15 samples not detected after Qiagen extraction for EBV and less than 1,000 copies/ml for the 12 samples found discordant for CMV results.
TABLE 1.
Qualitative analysis of EBV and CMV DNA detection after extraction using columns (Qiagen) or automated method (NucliSENS easyMAG)
| Virus (no. of samples) | Amplification result after DNA extraction by the NucliSENS easyMAG method | No. of specimens showing the indicated resulta for DNA extraction by Qiagen method: |
|
|---|---|---|---|
| Positive | Negative | ||
| EBV (n = 80) | Positive | 52 | 15 |
| Negative | 0 | 13 | |
| CMV (n = 94) | Positive | 60 | 8 |
| Negative | 4 | 22 | |
Results are expressed as number of specimens exhibiting the corresponding pattern (e.g., positive by the NucliSENS easyMAG method and positive by the Qiagen method).
Quantitative analysis.
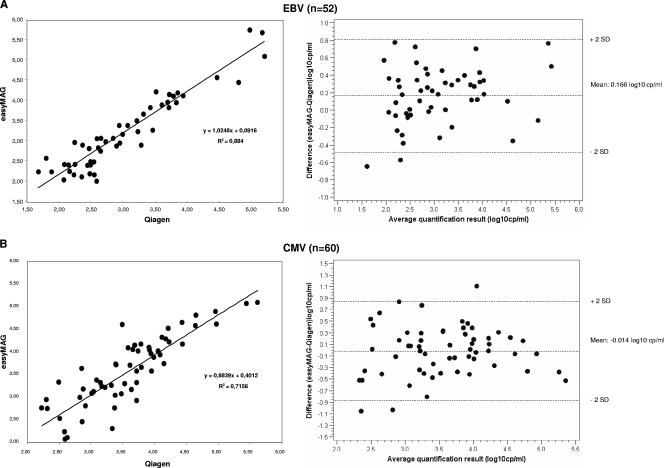
The results of the samples found positive after extraction by both methods were retained for quantitative analysis (Fig. 1). The correlation coefficients between the viral loads were 0.88 for EBV (P < 0.001) and 0.72 for CMV (P < 0.001). The mean between the difference in viral loads was 0.166 log10 copies/ml to the benefit of the automated extraction method for EBV (P < 0.001) and −0.014 log10 copies/ml for CMV (not statistically significant). All the results were included in the interval between the mean ± 2 standard deviations, except for two and three samples for EBV and CMV, respectively. Both methods gave equivalent levels of quantification (difference of less than 0.5 log copies/ml) in 43 out of 52 samples (82.7%) for EBV and in 50 out of 60 samples (83.3%) for CMV. Seven of the nine samples with discrepancies of at least 0.5 log copies/ml for EBV load exhibited a higher quantification after easyMAG extraction, whereas the results of the 10 samples with discrepancies of at least 0.5 log copies/ml for CMV load were equally distributed in both directions.
FIG. 1.
Quantitative analysis of EBV (A) and CMV (B) detection after extraction using Qiagen columns or NucliSENS easyMAG automated method. (Left) Regression curves; (right) Bland-Altman plots (log10cp/ml, log10 copies/ml). Each point represents the difference observed between the results of the two methods against their mean. The standard deviations (SD) are 0.324 for EBV and 0.427 for CMV.
QCMD panel analysis.
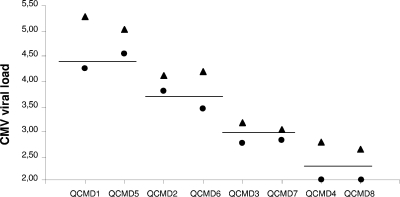
Eight samples of the 2006 Quality Control for Molecular Diagnostics (QCMD, Scotland, United Kingdom) CMV Proficiency Program were also tested. The results depicted in Fig. 2 show that the range of quantification was proportional by both techniques but that, for the four specimens tested in duplicate, the viral load was consistently higher by the automated method. The theoretical load of each sample (illustrated by the horizontal bars in Fig. 2) was close to the results of the Qiagen extraction, which is not surprising, since this technique or a related one was probably used for the extraction step of the experiment performed to define the reference values. Despite the small number of specimens and the limitation that the medium in which the different concentrations of CMV DNA were diluted was not whole blood, these results suggest a better ability of the automated extraction method to recover viral DNA.
FIG. 2.
Quantification results (log10) obtained after an extraction step using either the NucliSENS easyMAG automated method or Qiagen columns for eight lyophilized samples from QCMD containing 200 to 25,000 copies/ml (two samples in duplicate for each of the four tested combinations). Viral loads after Qiagen and NucliSENS easyMAG extraction are indicated by circles and triangles, respectively. The horizontal bars show the theoretical viral loads given by the manufacturer of the proficiency panel.
Detection of PCR inhibitors and gain in operational steps.
Among all of the whole-blood specimens analyzed (n = 174), an invalid result was found for two samples after DNA extraction by the NucliSENS easyMAG method and in two other samples by the Qiagen method; reextraction was successful for all of these samples. Evaluation of user convenience of both extraction methods showed that the turnaround time necessary to treat 24 samples was approximately 60 min by using the off-board protocol for the NucliSENS easyMAG instrument, including 30 min of hands-on time, whereas the extraction of 24 samples by the Qiagen technique was completed in 90 min of technician time starting from the addition of the lysis buffer.
Different automated extraction methods have been previously evaluated for EBV or CMV DNA extraction from whole blood (1, 3, 7, 9, 10, 13-16). To our knowledge, the present study is the first one that evaluated the performance of the NucliSENS easyMAG platform for determination of CMV and EBV loads from whole blood. Of note, the specific A protocol recommended by the manufacturer of the automated method for other clinical samples is not accurate for DNA-rich specimens such as whole blood (data not shown); the volume of silica particles and that of the lysis buffer were increased (specific B protocol) in order to recover the large amount of nucleic acids present in whole-blood specimens. This new protocol was shown to be at least as efficient as the manual Qiagen method to extract EBV and CMV DNA from whole-blood samples (Table 1 and Fig. 1), with a slightly better sensitivity of the automated extraction technique for EBV DNA, as has been previously reported for different clinical samples (6). The results of analysis of the specimens from the CMV QCMD panel also suggests a higher extraction rate obtained by the automated technique (Fig. 2).
In addition to a higher sensitivity, notably with regard to low values, the advantages of this automated extraction technique include (i) a significant reduction of technical time, (ii) an absence of the risk of cross-contamination due to centrifugation steps, and (iii) an improvement in standardization, traceability, and quality control assessment.
Acknowledgments
We are grateful to C. Menet, N. Plotton, V. Sauron, and C. Soler for excellent technical assistance. We thank the following individuals and companies whose reagents are involved in this study for helpful discussions and gifts of products: C. Baranger, P. Bourgeois, M. Joannès, and A. Nessali for Argene Biosoft, and Y. Aron, N. Brun, O. Laffrique, O. Paour, and S. Roux for bioMérieux.
Footnotes
Published ahead of print on 26 August 2009.
REFERENCES
- 1.Beuselinck, K., M. van Ranst, and J. van Eldere. 2005. Automated extraction of viral-pathogen RNA and DNA for high-throughput quantitative real-time PCR. J. Clin. Microbiol. 43:5541-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bressollette-Bodin, C., M. Coste-Burel, B. Besse, E. André-Garnier, V. Ferre, and B. M. Imbert-Marcille. 2009. Cellular normalization of viral DNA loads on whole blood improves the clinical management of cytomegalovirus or Epstein Barr virus infections in the setting of pre-emptive therapy. J. Med. Virol. 81:90-98. [DOI] [PubMed] [Google Scholar]
- 4.Deback, C., A. M. Fillet, N. Dhedin, B. Barrou, S. Varnous, F. Najioullah, F. Bricaire, and H. Agut. 2007. Monitoring of human cytomegalovirus infection in immunosuppressed patients using real-time PCR on whole blood. J. Clin. Virol. 40:173-179. [DOI] [PubMed] [Google Scholar]
- 5.Dehee, A., C. Asselot, T. Piolot, C. Jacomet, W. Rozenbaum, M. Vidaud, A. Garbarg-Chenon, and J. C. Nicolas. 2001. Quantification of Epstein-Barr virus load in peripheral blood in human immunodeficiency virus-infected patients using real-time PCR. J. Med. Virol. 65:543-552. [PubMed] [Google Scholar]
- 6.Dundas, N., N. K. Leos, M. Mitui, P. Revell, and B. B. Rogers. 2008. Comparison of automated nucleic acid extraction methods with manual extraction. J. Mol. Diagn. 10:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fafi-Kremer, S., K. Brengel-Pesce, G. Barguès, M. J. Bourgeat, O. Genoulaz, J. M. Seigneurin, and P. Morand. 2004. Assessment of automated DNA extraction coupled with real-time PCR for measuring Epstein-Barr virus load in whole blood, peripheral mononuclear cells and plasma. J. Clin. Virol. 30:157-164. [DOI] [PubMed] [Google Scholar]
- 8.Fafi-Kremer, S., P. Morand, C. Barranger, G. Barguès, S. Magro, J. Bés, P. Bourgeois, M. Joannes, and J. M. Seigneurin. 2008. Evaluation of the Epstein-Barr virus R-gene quantification kit in whole blood with different extraction methods and PCR platforms. J. Mol. Diagn. 10:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrigue, I., S. Boucher, L. Couzi, A. Caumont, C. Dromer, M. Neau-Cransac, R. Tabrizi, M. H. Schrive, H. Fleury, and M. E. Lafon. 2006. Whole blood real-time quantitative PCR for cytomegalovirus infection follow-up in transplant recipients. J. Clin. Virol. 36:72-75. [DOI] [PubMed] [Google Scholar]
- 10.Gouarin, S., A. Vabret, E. Gault, J. Petitjean, A. Regeasse, B. Hurault de Ligny, and F. Freymuth. 2004. Quantitative analysis of HCMV DNA load in whole blood of renal transplant patients using real-time PCR assay. J. Clin. Virol. 29:194-201. [DOI] [PubMed] [Google Scholar]
- 11.Gouarin, S., A. Vabret, C. Scieux, F. Agbalika, J. Cherot, C. Mengelle, C. Deback, J. Petitjean, J. Dina, and F. Freymuth. 2007. Multicentric evaluation of a new commercial cytomegalovirus real-time PCR quantitation assay. J. Virol. Methods 146:147-154. [DOI] [PubMed] [Google Scholar]
- 12.Hakim, H., C. Gibson, J. Pan, K. Srivastava, Z. Gu, M. J. Bankowski, and R. T. Hayden. 2007. Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood. J. Clin. Microbiol. 45:2151-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullberg-Lindh, C., S. Olofsson, M. Brune, and M. Lindh. 2008. Comparison of serum and whole blood levels of cytomegalovirus and Epstein-Barr virus DNA. Transpl. Infect. Dis. 10:308-315. [DOI] [PubMed] [Google Scholar]
- 14.Mengelle, C., F. Legrand-Abravanel, J. M. Mansuy, C. Barthe, I. Da Silva, and J. Izopet. 2008. Comparison of two highly automated DNA extraction systems for quantifying Epstein-Barr virus in whole blood. J. Clin. Virol. 43:272-276. [DOI] [PubMed] [Google Scholar]
- 15.Mengelle, C., K. Sandres-Sauné, C. Pasquier, L. Rostaing, J. M. Mansuy, M. Marty, I. Da Silva, M. Attal, P. Massip, and J. Izopet. 2003. Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay. J. Clin. Microbiol. 41:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michelin, B. D. A., I. Hadžisejdić, M. Bozic, M. Grahovac, M. Hess, B. Grahovac, E. Marth, and H. H. Kessler. 2008. Detection of cytomegalovirus (CMV) DNA in EDTA whole-blood samples: evaluation of the quantitative [i]artus[r] CMV LightCycler PCR kit in conjunction with automated sample preparation. J. Clin. Microbiol. 46:1241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowe, D. T., S. Webber, E. M. Schauer, J. Reyes, and M. Green. 2001. Epstein-Barr virus load monitoring: its role in the prevention and management of post-transplant lymphoproliferative disease. Transpl. Infect. Dis. 3:79-87. [DOI] [PubMed] [Google Scholar]
- 18.Wada, K., N. Kubota, Y. Ito, H. Yagasaki, K. Kato, T. Yoshikawa, Y. Ono, H. Ando, Y. Fujimoto, T. Kiuchi, S. Kojima, Y. Nishiyama, and H. Kimura. 2007. Simultaneous quantification of Epstein-Barr virus, cytomegalovirus, and human herpesvirus 6 DNA in samples from transplant recipients by multiplex real-time PCR assay. J. Clin. Microbiol. 45:1426-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadowsky, R. M., S. Laus, M. Green, S. A. Webber, and D. Rowe. 2003. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by Taqman PCR and in peripheral blood lymphocytes by competitive PCR. J. Clin. Microbiol. 41:5245-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner, H. J., L. Fischer, W. J. Jabs, M. Holbe, K. Pethig, and P. Bucsky. 2002. Longitudinal analysis of Epstein-Barr viral load in plasma and peripheral blood mononuclear cells of transplanted patients by real-time polymerase chain reaction. Transplantation 74:656-664. [DOI] [PubMed] [Google Scholar]