Abstract
Staphylococcus aureus is a major human pathogen responsible for increasing the prevalence of community- and hospital-acquired infections. Protein A (SpA) is a key virulence factor of S. aureus and is highly conserved. Sequencing of the variable-number tandem-repeat region of SpA (spa typing) provides a rapid and reliable method for epidemiological studies. Rarely, non-spa-typeable S. aureus strains are encountered. The reason for this is not known. In this study, we characterized eight non-spa-typeable bacteremia isolates. Sequencing of the entire spa locus was successful for five strains and revealed various mutations of spa, all of which included a deletion of immunoglobulin G binding domain C, in which the upper primer for spa typing is located, while two strains were truly spa negative. This is the first report demonstrating that nontypeability of S. aureus by spa sequencing is due either to mutation or to a true deficiency of spa.
Staphylococcus aureus is an important human pathogen responsible for many community- and hospital-acquired infections (22). S. aureus can cause various diseases, ranging from superficial skin infections to severe and life-threatening diseases, such as pneumonia, osteomyelitis, endocarditis, and sepsis (36). Recently, the prevalence of community- and hospital-acquired methicillin (meticillin)-resistant S. aureus (MRSA) infections has increased (2). MRSA infections, especially, are responsible for enhanced mortality and significant increases in the length of hospitalization and hospital charges (4).
Protein A (SpA) represents an important virulence factor for S. aureus (7, 8, 29) and has recently been shown to be involved in the pathogenesis of S. aureus pneumonia (10, 11), to be expressed in invasive diseases (21), and to play a role in proteinaceous biofilm formation (25). SpA is a protein of 42 kDa and comprises several regions with different functions: The signal sequence (S region) in the N-terminal part is followed by four or five highly homologous immunoglobulin G (IgG)-binding domains in tandem (the E, D, A, B, and C regions) (20). The C-terminal region, or X region, is divided into two domains: (i) the repeat region XR, consisting of variable repeats with mostly octapeptide structures, which are used for spa typing, and (ii) the XC region, consisting of a conserved sequence, which confers anchoring to the cell wall via an LPXTG-binding motif (31, 32). The best-studied function of SpA is the interaction with human IgG by binding to the Fc part, thereby compromising the host immune system (6, 13). Furthermore, SpA can bind to various host structures, such as the von Willebrand factor and the receptor gC1qR/p33 on platelets (15, 27), which promote adhesion to platelets.
In recent years, spa (S. aureus protein A) typing has been used frequently as a typing method. As a single-locus sequence-based typing method, it combines a number of technical benefits, such as rapidity, reproducibility, and portability (33), thereby allowing easy interlaboratory comparability via the Internet and synchronization to a central server (http://www.ridom.de/spaserver/) (1, 14). spa typing uses the sequence of the polymorphic X region, which consists of a variable number of tandem repeats of 24 bp, as a genetic marker (9). This region has been shown to be rather stable and allows distinguishing of strains to a degree comparable to that of pulsed-field gel electrophoresis (PFGE) and whole-genome DNA microarray (19). Interestingly, mutations of the repeat region, including insertions, deletions, and point mutations, have been observed only after long-term persistence of S. aureus in vivo in the airways of cystic fibrosis patients, allowing calculation of the clock speed of this region (18) and to establish an algorithm for the analysis of this region (24).
So far, no spa-deficient clinical strains have been described. Although some studies demonstrate 100% spa typeability (12), non-spa-typeable strains have been detected recently (23, 34). The reason for this is not known. Therefore, this study was performed to investigate the underlying mechanism of the nontypeability of eight invasive S. aureus strains, which were isolated from bacteremic patients with different infections. We sequenced the whole spa locus and investigated the expression of SpA by Western blot analysis and real-time PCR.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Seven of 148 methicillin-susceptible S. aureus (MSSA) strains (4.7%) and 1 particular MRSA strain (9 of 1,300 MRSA strains [0.7%] were nontypeable), which were cultured in the Department of Clinical Microbiology, Hvidovre Hospital, in 2004, were non-spa typeable and were further analyzed. The strains were isolated from blood cultures of patients with invasive infections (Table 1). For cultivation of S. aureus, tryptic soy broth (Difco, now Becton Dickinson GmbH, Heidelberg, Germany) and brain heart broth (Merck, Darmstadt, Germany) were used. For the growth curve analysis, cultures were grown at 37°C on a rotary shaker. For determination of the optical density at 578 nm beyond a value of 2, the optical density value was calculated from readings of diluted cultures.
TABLE 1.
Patient and strain information
| Strain | Susceptibility | Phage group | Age (yr)/sexa | Disease | MLST result |
|---|---|---|---|---|---|
| NT935 | MSSA | II (3C/71) | 72/W | Endocarditis (mitral valve) | NTb |
| NT936 | MSSA | II (3A/3C/55/71) | 52/W | Soft tissue infection (neutropenia caused by chemotherapy) | NT |
| NT937 | MSSA | III (53/93) | 43/M | Osteomyelitis | NT |
| NT938 | MSSA | III (6/53/54/75/83A/84/89) | 58/M | Osteomyelitis (Th10 to L5 plus abscessus epiduralis and subduralis) | NT |
| NT939 | MSSA | II (55) | 40/M | Spondylitis (Th12) | NT |
| NT940 | MSSA | NT | 5/M | Osteomyelitis (left femur) | NT |
| NT941 | MSSA | I (80 complex, a subtype of group I) | 12/M | Osteomyelitis (right clavicular-sternal joint) | NT |
| NT942 | MRSA | NAc | 67/M | Urinary tact infection | ST8 |
W, woman; M, male.
NT, nontypeable.
NA, not analyzed.
Molecular analysis.
Amplification and sequencing of the whole spa locus were performed with chromosomal DNA from each strain as a template. Chromosomal DNA was purified with the PrestoSpin D kit (Molzym GmbH & Co., KG, Bremen, Germany) after cell lysis with lysostaphin (WAK Chemie Medical GmbH, Steinbach/Ts, Germany). The primer sites for PCR amplifications were designed according to a consensus sequence of the sequenced strains N315, Mu50, MW2, MSSA 476, MRSA 252, and 8325-4. The oligonucleotide primers used for PCR are listed in Table 2. Sequencing analysis was performed at Eurofins MWG Operon (Martinsried, Germany).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| spa-1Fa | ATATGGATCCGATGACTTTACAAATACATACAGGGGGTb |
| spa-1Rc | ATATCCCGGGTGAGGCGTTTCAGAAGTTGTTTAGAd |
| spa-2Fa | GATGACTTTACAAATACATACAGGGGGT |
| spa-2Re | AAAATGCACTGAGCAACAAAAGATG |
| spa-3Ff | ATAGCGTGATTTTGCGGTT |
| spa-3Rg | CTAAATATAAATAATGTTGTCACTTGGA |
| rt-spa F | TATCTGGTGGCGTAACA |
| rt-spa R | TAGGCATATTTAACACTTGAT |
| gyrA F | CAACATTACGTCCTTTAGGC |
| gyrA R | GCTACATCAAGCCCTACAAC |
| spa-probe F | CAATTCGTAAACTAGGTGTAGG |
| spa-probe R | TTGGAGCTTGAGAGTCATTA |
Located 34 to 7 bp upstream of the S region.
The SmaI restriction site is underlined.
Located 180 to 156 bp downstream of the stop codon of spa.
The BamHI restriction site is underlined.
Located 139 to 115 bp downstream of the stop codon.
Located 178 to 160 bp upstream of the S region.
Located 332 to 305 bp downstream of the stop codon.
Southern blot hybridization analysis.
Southern blot hybridization analysis of genomic DNA digested by SmaI (Roche Diagnostics GmbH, Mannheim, Germany) was performed using standard procedures. The Southern blot was probed with a 252-bp fragment of the spa gene including the S and E regions (Table 2). The probe was labeled with the PCR DIG Probe Synthesis kit (Roche Diagnostics GmbH, Mannheim, Germany).
Molecular typing analysis of non-spa-typeable isolates.
PFGE was performed as described recently (17). Similarity of strains was evaluated using the Dice coefficient and the criterion of a difference of fewer than 6 bands as described by Tenover et al. (35). Multilocus sequence typing (MLST) was performed as described previously (5). Phage typing was performed by the method of Blair and Williams with the contemporary international set of typing phages and two Danish experimental phages, which have been used since 1969 to differentiate among isolates of group III and the 83A complex (16).
RNA extraction and real-time PCR.
For RNA extraction, a combination of the FastRNA Pro Blue kit (RNApro solution and Lysing Matrix B; Qbiogene [part of MP Biomedicals], Heidelberg, Germany), the RNAprotect Bacteria reagent (Qiagen, Hilden, Germany), and the RNeasy minikit (Qiagen) was used. Cells were mechanically disrupted by a Fast Prep FP120 instrument (Qbiogene). The remaining DNA was eliminated by DNase (Qiagen).
For the real-time PCR, cDNA was synthesized from 25 ng RNA by using the Quantitect reverse transcription kit (Qiagen) according to the manufacturer's recommendations. Real-time amplification was conducted using specific primers (Table 2) and was carried out on an iCycler iQ real-time PCR-system (Bio-Rad, Hercules, CA) using the iQ SYBR green Supermix (Bio-Rad). The levels of mRNA expression of spa were normalized against the expression of the internal control gene gyrA (DNA gyrase subunit A). The transcript quantities are expressed as changes (n-fold) relative to the values of the internal control (3).
Protein A extraction.
Cell wall-bound protein A was extracted from 5-h cultures. After centrifugation, the pellet was washed twice with 25 mM Tris-HCl buffer. Cell lysis was done with lysostaphin (WAK Chemie Medical GmbH), in combination with DNA degradation with DNase (Roche Diagnostics GmbH). After centrifugation, the pellet was used for further investigations.
Secreted protein A from the medium was precipitated from sterile-filtered supernatants (0.2-μm-pore-size filter; Sartorius Biotech GmbH, Göttingen, Germany) of 5-h cultures by trichloroacetic acid. The protein pellets were washed with acetone, air dried, and dissolved in 25 mM Tris-HCl buffer.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% [wt/vol] total acrylamide) and were subsequently electrotransferred (Transblot SD; Bio-Rad) to nitrocellulose membranes (Schleicher & Schüll, Dassel, Germany) using established protocols. To detect SpA, affinity-purified immunoglobulins from rabbit serum were used as primary antibodies, and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulins were used as secondary antibodies (Bio-Rad). Protein A P3838 (Sigma-Aldrich Chemie GmbH, Munich, Germany) was used as a positive control, and the PageRuler Plus prestained protein ladder from Fermentas GmbH (St. Leon-Rot, Germany) was used as a marker.
Nucleotide sequence accession numbers.
The sequences of the non-spa-typeable strains NT935, NT937, NT938, NT939, and NT941 have been deposited in GenBank under accession numbers FJ491257 to FJ491261.
RESULTS
Genetic analysis of spa of non-spa-typeable strains.
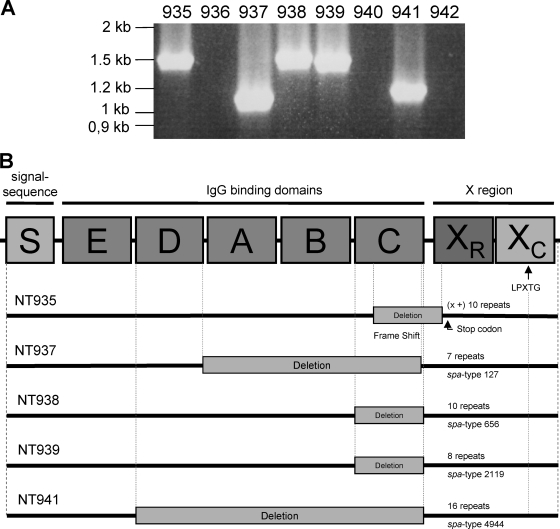
All strains were responsible for invasive S. aureus infections (Table 1). spa was detected in five of eight non-spa-typeable strains by amplifying the entire gene with flanking primers (Fig. 1A).
FIG. 1.
(A) Amplification of the entire spa locus. By use of primers flanking the entire spa locus, spa could be amplified in five of eight non-spa-typeable strains: NT935, NT937, NT938, NT939, and NT941. (B) Results of sequencing of the spa locus for five non-spa-typeable S. aureus strains: NT935, NT937, NT938, NT939, and NT941. The deletions are indicated by gray rectangles. Strain NT935 revealed a deletion from the middle part of IgG-binding domain C up to the beginning of the repeat region XR, resulting in a frameshift mutation, presumably leading to a premature stop codon. In strain NT937, a deletion of the three IgG-binding domains A, B, and C occurred; in strains NT938 and NT939, a deletion of IgG-binding domain C was observed; and in strain NT941, a deletion of the four IgG-binding domains D, A, B, and C was demonstrated.
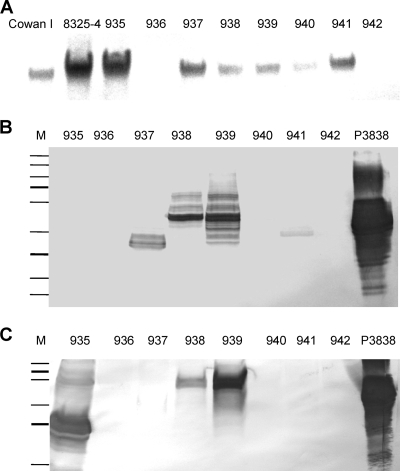
Sequence analysis of the amplicons of the non-spa-typeable strains revealed deletions, upstream of the repeat region XR, of one to four IgG-binding domains, including region C, the region where the upstream primer for spa typing is located. In strain NT935, a deletion from the middle part of IgG-binding domain C up to the beginning of the repeat region XR resulted in a frameshift mutation with a premature stop codon (Fig. 1B). Southern blot analysis revealed the presence of spa in one additional strain, NT940 (Fig. 2A).
FIG. 2.
(A) Southern blot hybridization analysis. SmaI-digested chromosomal DNAs of non-spa-typeable strains were probed with a 252-bp fragment of spa, which detects the S and E regions. As positive controls, DNAs of the S. aureus strains Cowan I and 8325-4 were used. (B and C) Western blot analyses. Cell wall extracts (B) and secreted proteins (C) of the non-spa-typeable strains were incubated with affinity-purified immunoglobulins from rabbit serum as primary antibodies and with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulins as secondary antibodies. As a marker, the PageRuler Plus prestained protein ladder from Fermentas GmbH was used. Protein A P3838 (Sigma-Aldrich Chemie GmbH) was used as a positive control.
Expression of protein A by real-time PCR and Western blot analysis.
spa transcription could be detected by RT-PCR, though with different values, in all spa-positive strains (Table 3). The typical pattern of spa transcription, with the highest transcription in the early- or late-logarithmic growth phase, was observed. Interestingly, the strains with three to four deletions of IgG-binding domains displayed the highest spa transcription levels (strains NT937 and NT941). However, transcription of these mutated genes resulted in detection of only low levels of protein or almost no detectable proteins as assessed by Western blot analysis (Fig. 2B and C). In contrast, low levels of spa transcription yielded large amounts of cell wall-associated protein and secreted protein A, as demonstrated for strain NT938 (Fig. 2B and C; Table 3).
TABLE 3.
Overview of the results for non-spa-typeable S. aureus strains
| Strain | PCR/sequencing result | Deletion | spa type | Result(s) by: |
||
|---|---|---|---|---|---|---|
| Southern blotting | Real-time PCRa | Western blottingb | ||||
| NT 935 | +/+ | C domain+ frameshift + stop codon | Unknown; CCTGGT + 10 repeats | + | EL, 0.44; LL, 0.47; SP, 0.08 | Ex |
| NT 936 | −/− | − | ND | |||
| NT 937 | +/+ | A, B, and C domains | t127 | + | EL, 59.71; LL, 19.70; SP, 7.73 | CW |
| NT 938 | +/+ | C domain | t656 | + | EL, 0.01; LL, ND; SP, 0.01 | Ex + CW |
| NT 939 | +/+ | C domain | t2119 | + | EL, 0.57; LL, 0.09; SP, 0.08 | Ex + CW |
| NT 940 | −/− | + | EL, 0.02; LL, 0.05; SP, 0.01 | |||
| NT 941 | +/+ | D, A, B, and C domains | A4944 | + | EL, 168.9; LL, 6.28; SP, 0.71 | CW |
| NT 942 | −/− | − | ND | |||
EL, early-log phase; LL, late-log phase; SP, stationary phase; ND, nondetectable.
Ex, extracellular; CW, cell wall associated.
Epidemiologic relationships of non-spa-typeable strains.
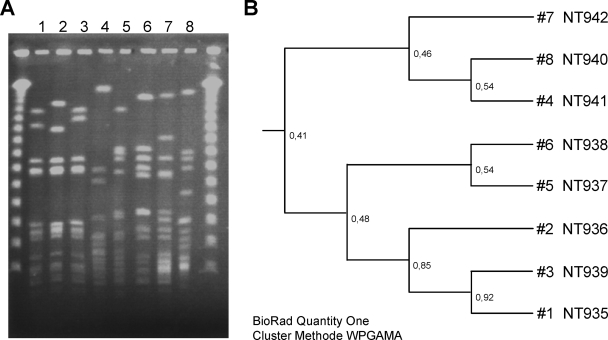
PFGE allowed all non-spa-typeable strains to be distinguished (Fig. 3). The fragment patterns of the strains revealed that three of the eight strains analyzed were closely related (strains NT936, NT935, and NT939), while the other strains were genetically unrelated (Fig. 3).
FIG. 3.
Phylogenetic tree of the non-spa-typeable S. aureus strains. (A) PFGE results. The molecular weight marker used consists of concatemers of lambda phage DNA. (B) Deduced phylogenetic tree after SmaI digestion of chromosomal DNA from the non-spa-typeable strains.
DISCUSSION
This is the first report demonstrating the existence of clinical S. aureus strains in which either protein A is missing or deletions of one or several IgG-binding domains have occurred. SpA is a versatile molecule of ca. 42 kDa that exists mostly in a cell wall-associated form and has the ability to interact with several host components (7). Its polymorphic region is used as a single-locus sequence-typing method due to its discriminatory power (9). Recently, the development of an open Web-based database (www.ridom.de/spaserver/) (14) in which data of more than 83,000 strains from 63 countries are collected (March 2009) has resulted in a rapid increase in spa typing for molecular analyses and epidemiological studies. Most S. aureus strains are typeable by using standard spa primers for PCR and sequencing (12). Rarely, strains that are nontypeable by this method are encountered (23, 34). To find the underlying reasons for this nontypeability, we performed a thorough analysis of eight non-spa-typeable strains, which were collected from blood cultures of patients with invasive S. aureus infections in Denmark.
Sequencing of the entire spa locus revealed the presence of various deletions in all strains, all of which included a deletion of the IgG-binding domain C, in which the upstream primer used for spa typing is located. Only two strains actually failed to give a positive result, indicating that these strains were true spa-deficient strains. Although spa has been shown to be an important virulence factor in the pathogenesis of S. aureus infections (29), the two spa-deficient strains were isolated from patients with severe S. aureus-related infections, indicating that the strains were still virulent and invasive in spite of being spa deficient. Peacock et al., in their study of 334 S. aureus carriage and invasive isolates, detected the presence of spa in 90 and 94% of strains, respectively, by PCR analysis (30). However, their analyses also depended on a PCR approach that might have missed spa-positive S. aureus strains with mutations in the primer binding region. Southern blot analysis may have detected a higher number of spa-positive strains.
In addition to strain NT935, which exclusively secreted SpA into the medium due to a loss of the LPXTG region, two more strains (NT938 and NT939) also released SpA into the medium in measurable amounts as detected by Western blot analysis. Usually, most of the protein is bound to the cell wall. However, in some strains, SpA has been shown to be released from the cell wall into the medium, presumably due to autolysis during stationary phase (26).
spa expression is tightly regulated, and a number of different regulators that either repress (agr, sarA) or stimulate (mgrA, sarS, sarT, arl, and rot) the transcription of spa are involved in its regulation (28). The fact that the transcription of spa in all strains followed the pattern usually observed, with the highest levels of transcripts in the early- and late-logarithmic phases, suggests that no major deletions of upstream regulators occurred in the strains investigated.
PFGE analysis revealed that only three of eight strains were genetically related, indicating that (i) there was no outbreak of a specific clone with non-spa typeability in Denmark at the time of strain collection and (ii) mutations within the spa region can occur in different genetic backgrounds.
In summary, in this study we showed that spa DNA and RNA were present in six out of eight non-spa-typeable strains. However, sequencing was successful for only five of these strains. The spa gene revealed deletions between 161 and 705 bp, all of which included the region of the upstream primer binding site usually used for spa typing, thus explaining the impossibility of typing. Expression analysis of SpA revealed that four of five strains covalently bound SpA to the cell wall, while one strain exclusively secreted the protein into the medium due to a premature stop codon and a missing LPXTG region. Although in all non-spa-typeable strains investigated, either spa mutations occurred or spa was absent, these strains were still able to induce invasive disease, demonstrating the redundancy of virulence factors in S. aureus.
Acknowledgments
This project was funded by a grant from the German Research Foundation to B.C.K., KA 2249/1-3 (www.dfg.de).
We thank Muzaffar Hussain, Andreas Fischer, and Matthias Grundmeier for helpful suggestions and Michaela Brück and Brigitte Schuhen for expert technical assistance.
Footnotes
Published ahead of print on 16 September 2009.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher, H. W., and G. R. Corey. 2008. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46(Suppl. 5):S344-S349. [DOI] [PubMed] [Google Scholar]
- 3.Chatterjee, I., M. Herrmann, R. A. Proctor, G. Peters, and B. C. Kahl. 2007. Enhanced post-stationary-phase survival of a clinical thymidine-dependent small-colony variant of Staphylococcus aureus results from lack of a functional tricarboxylic acid cycle. J. Bacteriol. 189:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosgrove, S. E., G. Sakoulas, E. N. Perencevich, M. J. Schwaber, A. W. Karchmer, and Y. Carmeli. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin. Infect. Dis. 36:53-59. [DOI] [PubMed] [Google Scholar]
- 5.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster, T. J. 2005. Immune evasion by staphylococci. Nat. Rev. Microbiol. 3:948-958. [DOI] [PubMed] [Google Scholar]
- 7.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 8.Fournier, B., and D. J. Philpott. 2005. Recognition of Staphylococcus aureus by the innate immune system. Clin. Microbiol. Rev. 18:521-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frénay, H. M., A. E. Bunschoten, L. M. Schouls, W. J. Van Leeuwen, C. M. Vandenbroucke-Grauls, J. Verhoef, and F. R. Mooi. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15:60-64. [DOI] [PubMed] [Google Scholar]
- 10.Gómez, M. I., A. Lee, B. Beddy, A. Muir, G. Soong, A. Pitt, A. Cheung, and A. Prince. 2004. Staphylococcus aureus protein A induces airway epithelial inflammatory responses by activating TNFR1. Nat. Med. 10:842-848. [DOI] [PubMed] [Google Scholar]
- 11.Gómez, M. I., M. O'Seaghdha, M. Magargee, T. J. Foster, and A. S. Prince. 2006. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J. Biol. Chem. 281:20190-20196. [DOI] [PubMed] [Google Scholar]
- 12.Hallin, M., A. Deplano, O. Denis, R. De Mendonca, R. De Ryck, and M. J. Struelens. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45:127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p. 310. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 14.Harmsen, D., H. Claus, W. Witte, J. Rothgänger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in the university hospital setting using a novel software for spa-repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartleib, J., N. Köhler, R. B. Dickinson, G. S. Chhatwal, J. J. Sixma, O. M. Hartford, T. J. Foster, G. Peters, B. E. Kehrel, and M. Herrmann. 2000. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood 96:2149-2156. [PubMed] [Google Scholar]
- 16.Jessen, O., K. Rosendal, P. Bülow, V. Faber, and K. R. Eriksen. 1969. Changing staphylococci and staphylococcal infections. A ten-year study of bacteria and cases of bacteremia. N. Engl. J. Med. 281:627-635. [DOI] [PubMed] [Google Scholar]
- 17.Kahl, B. C., A. Duebbers, G. Lubritz, J. Haeberle, H. G. Koch, B. Ritzerfeld, M. Reilly, E. Harms, R. A. Proctor, M. Herrmann, and G. Peters. 2003. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 41:4424-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahl, B. C., A. Mellmann, S. Deiwick, G. Peters, and D. Harmsen. 2005. Variation of the polymorphic region X of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J. Clin. Microbiol. 43:502-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discrimination among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löfdahl, S., B. Guss, M. Uhlen, L. Philipson, and M. Lindberg. 1983. Gene for staphylococcal protein A. Proc. Natl. Acad. Sci. USA 80:697-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loughman, J. A., S. A. Fritz, G. A. Storch, and D. A. Hunstad. 2009. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 23.Mellmann, A., A. W. Friedrich, N. Rosenkotter, J. Rothganger, H. Karch, R. Reintjes, and D. Harmsen. 2006. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 3:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellmann, A., T. Weniger, C. Berssenbrugge, J. Rothganger, M. Sammeth, J. Stoye, and D. Harmsen. 2007. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiol. 7:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merino, N., A. Toledo-Arana, M. Vergara-Irigaray, J. Valle, C. Solano, E. Calvo, J. A. Lopez, T. J. Foster, J. R. Penades, and I. Lasa. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191:832-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Movitz, J. 1976. Formation of extracellular protein A by Staphylococcus aureus. Eur. J. Biochem. 68:291-299. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen, T., B. Ghebrehiwet, and E. I. Peerschke. 2000. Staphylococcus aureus protein A recognizes platelet gC1qR/p33: a novel mechanism for staphylococcal interactions with platelets. Infect. Immun. 68:2061-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oscarsson, J., C. Harlos, and S. Arvidson. 2005. Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC 8325-4. Int. J. Med. Microbiol. 295:253-266. [DOI] [PubMed] [Google Scholar]
- 29.Palmqvist, N., T. Foster, A. Tarkowski, and E. Josefsson. 2002. Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathog. 33:239-249. [DOI] [PubMed] [Google Scholar]
- 30.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. J. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70:4987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneewind, O., A. Fowler, and K. F. Faull. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103-106. [DOI] [PubMed] [Google Scholar]
- 32.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 33.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strommenger, B., C. Braulke, D. Heuck, C. Schmidt, B. Pasemann, U. Nubel, and W. Witte. 2008. spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J. Clin. Microbiol. 46:574-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldvogel, F. A. 2000. Staphylococcus aureus (including toxic shock syndrome), p. 2069-2092. In G. L. Mandell, R. G. Douglas, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, Philadelphia, PA.